Summary
Sphingolipids are a structurally diverse group of molecules based on long-chain sphingoid bases found in animal, fungal and plant cells. In contrast to the situation in animals and yeast, we know much less about the spectrum of sphingolipid species in plants and the roles they play in mediating cellular processes. Here, we report the cloning and characterization of a plant ceramidase from rice (Oryza sativa spp. Japonica cv. Nipponbare). Sequence analysis suggests that the rice ceramidase (OsCDase) is similar to mammalian neutral ceramidases. We demonstrate that OsCDase is a bona fide ceramidase following heterologous expression in the yeast double knockout mutant, Δypc1Δydc1, which lacks the yeast ceramidases, YPC1p and YDC1p. Biochemical characterization of OsCDase showed that it exhibited classical Michaelis-Menten kinetics with an optimum activity ranging from pH 5.7 to 6.0. OsCDase activity was enhanced in the presence of Ca2+, Mg2+, Mn2+, Zn2+ but inhibited in the presence of Fe2+. OsCDase appears to use ceramide instead of phytoceramide as a substrate. Sub-cellular localization showed that OsCDase is localized to the endoplasmic reticulum and Golgi, suggesting that these organelles are sites of ceramide metabolism in plants..
Keywords: Sphingolipid, ceramide, ceramidase, reverse ceramidase activity
Introduction
Sphingolipids are ubiquitous components of eukaryotic cells where they form key structural components of membranes, and function as important mediators of physiological responses from apoptosis to cell survival and proliferation (Pyne and Phye. 2000; Dickson and Lester, 2002; Hannun and Obeid, 2002, 2008; Spiegel and Milstien, 2003; Futerman and Hannun, 2004). Sphingolipid metabolism has been intensely investigated in animal cells and yeast, and much is known about the different metabolic intermediates and the enzymes responsible for catalyzing their formation (Pyne and Phye. 2000; Dickson and Lester, 2002; Hannun and Obeid, 2002, 2008; Spiegel and Milstien, 2003; Futerman and Hannun, 2004). In yeast, sphingolipid metabolites like dihydrosphingosine and phytosphingosine have been shown to increase the ability of yeast to tolerate and grow under heat-stress conditions (Dickson et al., 1997; Jenkins et al., 1997; Mao et al., 1999). Sphingosine-1-phosphate has been shown in animal cells to be important in mediating cellular survival and proliferation, whereas ceramide has been implicated in mediating programmed cell death (Hannun and Obeid, 2002, 2008; Spiegel and Milstien, 2003; Futerman and Hannun, 2004). This has led to the suggestion that the dynamic balance between levels of sphingolipid metabolites within cells function to regulate cell fate although the situation is likely to be highly complex due to the inter-convertibility of sphingolipid metabolites (Futerman and Hannun, 2004; Hannun and Obeid, 2008). This is particularly true for ceramide and sphingosine/sphingosine-1-phosphate because sphingosine is formed by the action of ceramidases, placing ceramide as a central mediator of sphingolipid-based signalling systems in cells (Hannun and Obeid, 2002, 2008).
In contrast to the situation in animal cells and in yeast, we know much less about the roles of sphingolipid metabolites in regulating cellular processes. Plant membranes are made up of significant amounts of sphingolipids, and it has been estimated that 7 to 26% of plant membrane lipids are sphingolipids (Lynch, 1993, Schmid nad Ohlrogge, 1996). The first information on plant sphingolipids was mainly obtained from analyzing the lipid composition of seeds (Carter et al., 1958; Ito et al., 1985) although there are indications that sphingolipids may be important cellular mediators in plants. It was reported that glucosylceramides (cerebrosides A and C) from wheat grain could induce fruiting in Schizophyllum commune, a fungus that causes wood decay (Kawai et al., 1986; Kawai, 1989), and the same glucosylceramides from the pathogenic rice fungus, Magnaporthe grisea are elicitors of hypersensitive cell death and phytolexin accumulation in rice plants (Koga et al., 1998). More recently, it was reported that sphingosine-1-phosphate is a calcium-mobilizing messenger active in the responses of stomatal guard cells to the drought hormone, abscisic acid (Ng et al., 2001) and that the effects of sphingosine-1-phosphate is dependent on heterotrimeric G-proteins (Coursol et al., 2003). Progress in the understanding of sphingolipid metabolism in plants has been aided by the characterization of Arabidopsis mutants affected in sphingolipid metabolism (Chen et al., 2006; Tsegaye et al., 2007; Shi et al., 2007; Dietrich et al., 2008) and from the development of mass spectrometric methods for the analysis of plant sphingolipids (Markham et al., 2006; Teng et al., 2008). Mutant analyses in Arabidopsis have revealed important roles for sphingolipid metabolites in mediating sensitivity towards the mycotoxin, fumonisin B1 (Tsegaye et al., 2007), and sphingolipid metabolites have been shown to be important regulators of gametophytic and sporophytic development (Imamura et al., 2007; Dietrich et al., 2008; Teng et al., 2008), thereby affecting fertility.
Ceramide has been shown to induce programmed cell death in Arabidopsis cells (Liang et al., 2003; Townley et al., 2005), suggesting that ceramide may be an important regulator of plant development. In order to further elucidate the role of ceramide in plants, it is important to understand the way in which ceramide is metabolized in plant cells. Ceramidases are enzymes responsible for metabolizing ceramide to sphingosine by cleaving the N-acyl linkage between the sphingoid base and the fatty acid. There are three different types of ceramidases, and they have been classified as acid, neutral and alkaline according to their pH optima for activity (Mao et al., 2002).
Here we report the cloning of a plant ceramidase from rice. We demonstrate that OsCDase is a bona fide ceramidase that appears to use ceramide instead of phytoceramide as a substrate. We also used a lipidomic approach to show that OsCDase exhibits reverse ceramidase activity, resulting in the formation of phytoceramides with very long chain fatty acids. Sub-cellular localization showed that OsCDase-DsRed2 protein fusion is targeted to the endoplasmic reticulum and Golgi, suggesting that these organelles are sites of sphingolipid metabolism in plants.
Results
Cloning, phylogenetic, and expression analysis of OsCDase
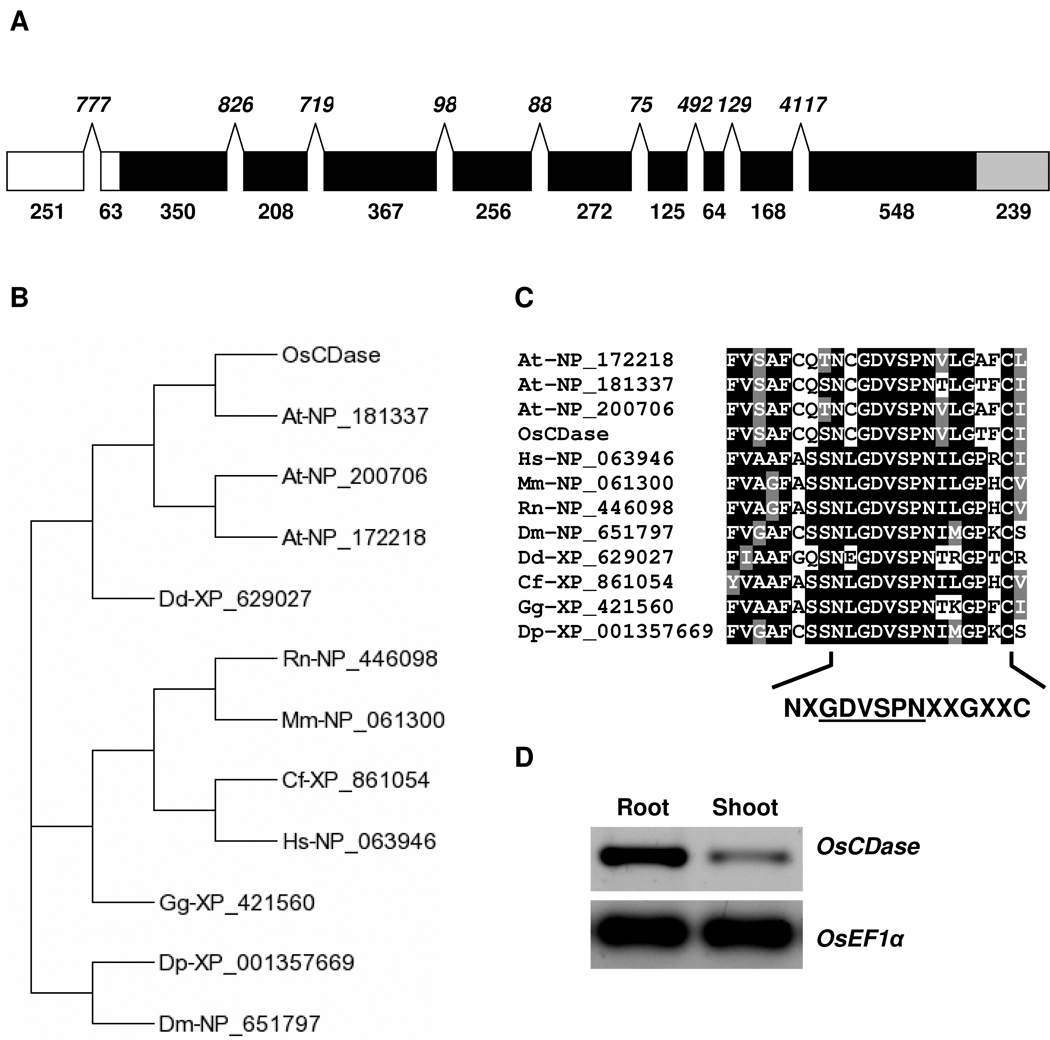
We used the Blast program to search the rice genome database for sequences bearing similarities to the human neutral ceramidase gene, ASAH2 (GenBank Accession No.: NP_063946) and identified a single copy gene on Chromosome 1 designated Os01g43520. The predicted amino acid sequence of the putative rice ceramidase (OsCDase) has a 42% sequence identity and 59% sequence similarity to ASAH2, suggesting that OsCDase may be a neutral ceramidase. We then used RT-PCR and 5’- and 3’-RACE PCR to obtain the full-length transcript for OsCDase. OsCDase is made up of 10 exons (Figure 1A). Interestingly the 5’-UTR of OsCDase contains a 777 bp intron although the Ensembl Exon report on Gramene (http://www.gramene.org) predicted an intron of 255 bp. In silico analysis of the 5’-UTR of OsCDase using Regulatory RNA Motifs and Elements Finder (RegRNA; http://regrna.mbc.nctu.edu.tw/index.html) showed the presence of a 90 bp (position -32 to -122) internal ribosomal entry site (IRES) element. Phylogenetic analysis of neutral ceramidases from various organisms indicates that OsCDase is clustered with the putative ceramidases from Arabidopsis thaliana and the neutral ceramidase homologue of Dictyostelium discoideum (Figure 1B). ClustalW alignment of the amino acid sequence of OsCDase with ceramidases from various organisms (Supplemental Figure 1) showed the presence of the highly conserved hexapeptide sequence, GDVSPN within the larger conserved amidase domain, NXGDVSPNXXC (Figure 1C), important for ceramidase activity (Galadari et al., 2006). Expression analysis indicated that steady-state levels of OsCDase transcripts are expressed throughout the seedling with a higher level of expression in roots compared to shoots (Figure 1D).
Figure 1. Cloning, sequence analysis and expression of rice ceramidase (OsCDase) in planta.
(A) Schematic representation of the sequence organization of OsCDase. The sizes of the 5’-UTR (white), exons (black), and 3’UTR (grey) are indicated by numbers below the boxes. Sizes of introns are shown in italics. (B) Phylogenetic tree of the neutral ceramidases from various organisms (Os: Oryza sativa; At: Arabidopsis thaliana; Dd: Dictyostelium discoideum; Rn: Rattus norvegicus; Mm: Mus musculus; Cf: Canis familiaris; Hs: Homo sapiens; Gg: Gallus gallus; Dp: Drosophila pseudoobscura; Dm: Drosophila melanogaster) were generated using T-Rex (http://www.trex.uqam.ca/). (C) Clustal alignment of the neutral ceramidases from various organisms showing the highly conserved hexapeptide sequence, GDVSPN within the larger conserved amidase domain, NXGDVSPNXXC, important for ceramidase activity. (D) RT-PCR expression analysis of OsCDase in shoot and roots of 2-week old seedlings. Housekeeping control gene: OsEF1α.
Biochemical characterization of OsCDase
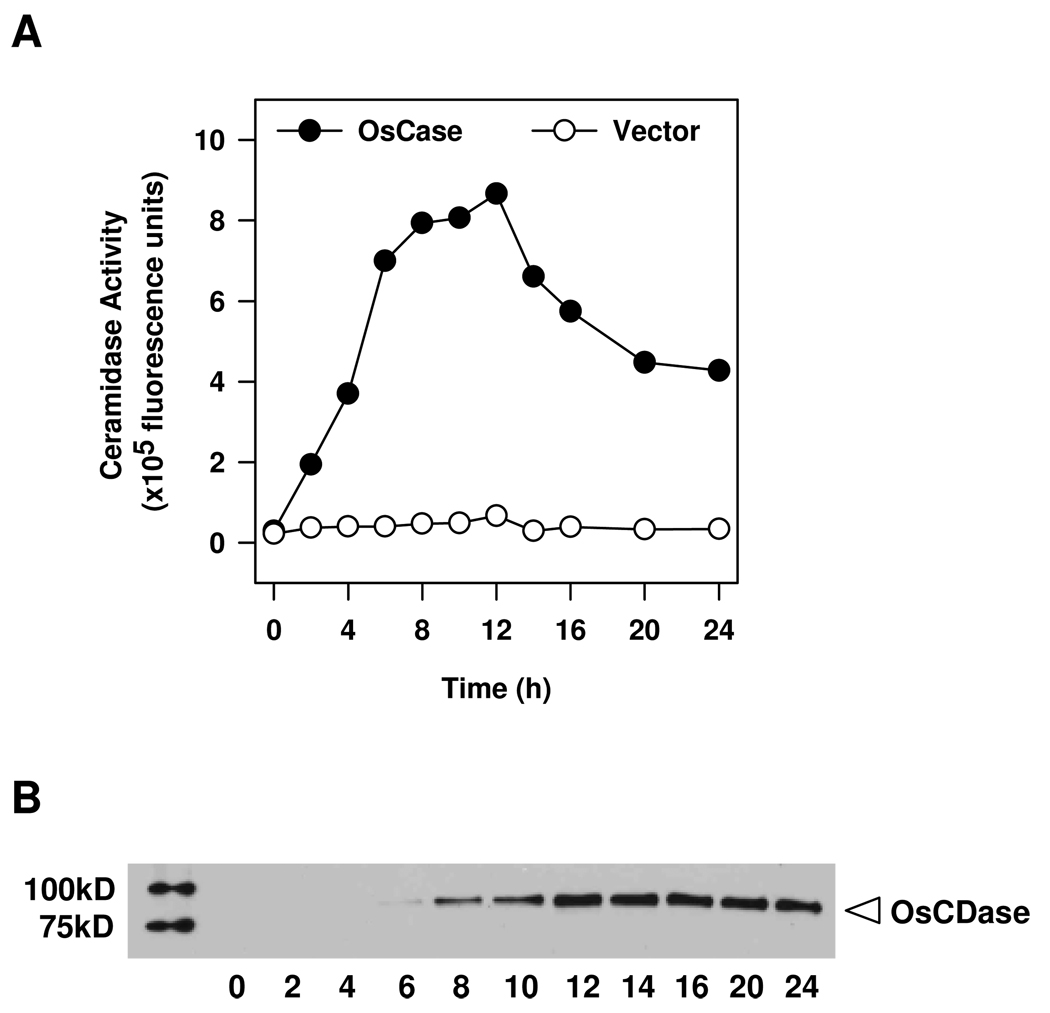
We cloned the full-length OsCDase CDS into the yeast expression vector, pYES2/CT under the control of the Gal1 promoter, and the resulting plasmid was transformed into the yeast double knockout mutant, Δypc1Δydc1, which lacks the yeast CDases, YPC1p and YDC1p (Mao et al., 2000). This is important as it allowed us to express recombinant OsCDase and minimize contamination from endogenous yeast CDase activity that may interfere with subsequent activity characterization of OsCDase. Expression of recombinant OsCDase was induced by culturing the transformed yeast in medium containing galactose, and cells were harvested at different time points to determine the optimal time for induction of OsCDase expression. Homogenates were obtained from harvested cells and assayed for CDase activity using NBD-C12-ceramide as a substrate (Figure 2A). CDase activity was detectable 2 h following induction with galactose, peaking at 10–12 h, and decreasing thereafter. We could not detect any CDase activity after induction with galactose in yeast containing the vector alone (Figure 2A). The amount of recombinant OsCDase was determined in western blot using Anti-His antibody, and the amount of recombinant OsCDase peaked at about 12 h (Figure 2B), in agreement with optimal activity at 12 h post-induction (Figure 2A). On the basis of these data, we selected 12 h has the duration of induction for expression of OsCDase for subsequent biochemical characterization. Importantly, these results demonstrate that OsCDase exhibits robust ceramidase activity.
Figure 2.
Time course of induction of rice ceramidase (OsCDase) expression in the yeast double-knockout strain, Δypc1Δydc1. OsCDase was cloned into the vector, pYES2/CT and the construct was transformed into the yeast double-knockout strain, Δypc1Δydc1, which lacks the yeast CDases. (A) Expression of OsCDase was induced by growth in galactose medium, and at different time points the culture was terminated and whole cell lysates were assayed for CDase activity using D-erythro-C12-NBD-ceramide as substrate. Values represent means from two independent experiments. (B) Western blot of whole cell lysates (10 µg protein per lane) using anti-His antibody at different time points following growth in galactose medium.
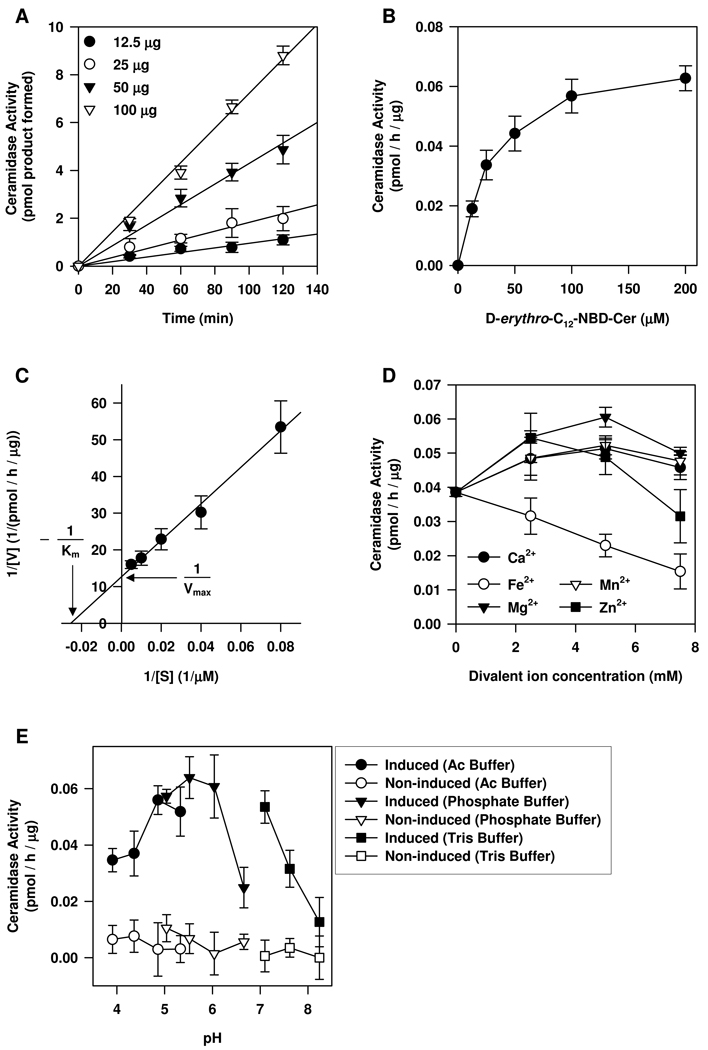
We next examined the biochemical properties of OsCDase and determined the effects of different amounts of protein on CDase activity at different times (Figure 3A). The results indicated that the reaction is linear over a period of 120 min (Figure 3A). The reaction catalyzed by OsCDase exhibited classical Michaelis-Menten kinetics (Figure 3B) and showed an apparent Km of 39.34 µM (1.23 mol%) and an apparent Vmax of 0.079 pmol/h/µg protein as determined using the Lineweaver-Burk plot (Figure 3C). OsCDase activity was enhanced in the presence of Mg2+, Ca2+, Mn2+ (from 2.5 to 7.5 mM) and Zn2+ (2.5 to 5 mM) but inhibited in the presence of Fe2+ (Figure 3D). Because ceramide has been shown to induce micromolar increases in cytosolic free calcium in plants (Townley et al., 2005), we examined the effects of micromolar concentrations of Ca2+ on OsCDase activity in vitro. We showed that OsCDase activity was only weakly activated by free calcium ions in the micromolar range and we observed a significant peak activation of OsCDase activity in the presence of 5 mM Ca2+ (t-test, p < 0.01) (Supplemental Figure 2). We observed OsCDase activity over a broad pH range, with an optimum pH from pH 5.7 to 6.0 (Figure 3E).
Figure 3.
Biochemical characterization of the rice CDase (OsCDase) in the yeast double-knockout strain, Δypc1Δydc1. OsCDase was cloned into the vector, pYES2/CT and the construct was transformed into the yeast double-knockout strain, Δypc1Δydc1, which lacks the yeast CDases. Expression of OsCDase was induced by growth in galactose medium for 12 h. (A) The effect of different amounts of yeast protein on CDase activity at different assay times. For (B) to (E), 50 µg of total yeast protein was used for CDase activity assay and the reaction time was 1 h. (B) Michaelis-Menten representation for OsCDase activity towards increasing concentrations of D-erythro-C12-NBD-ceramide. (C) Lineweaver-Burk plot of OsCDase activity. (D) Effects of increasing concentrations of cations on OsCDase activity. (E) pH optimum determination and the buffers used are described in the ‘Experimental’ section. Values are means ± S.D. of four replicates from two independent experiments.
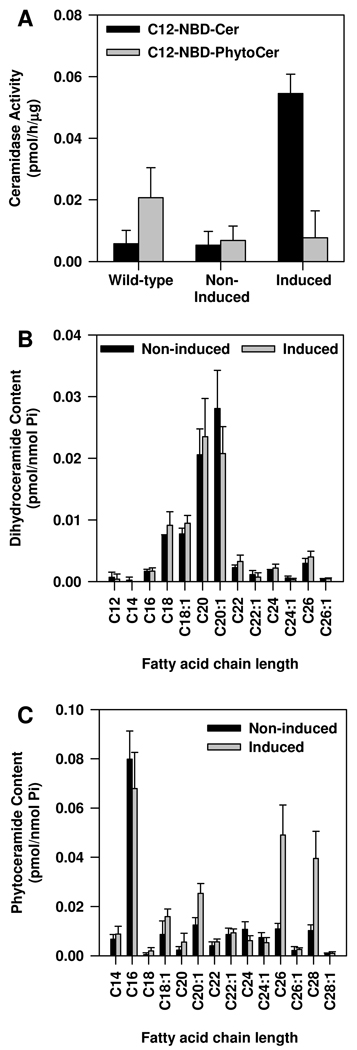
We also examined the substrate specificity of OsCDase by substituting D-erythro-C12-NBD-ceramide with NBD-C12-phytoceramide. Phytoceramides are the predominant ceramide species in plants, including rice (Ohnishi et al., 1985; Sperling and Heinz, 2003; Dunn et al., 2004; Lynch and Dunn, 2004). Interestingly, our results suggest that phytoceramide is not a substrate for OsCDase (Figure 4A). We also measured endogenous levels of dihydroceramide (Figure 4B) and phytoceramide (Figure 4C) in the yeast double knockout mutant Δypc1Δydc1 expressing OsCDase. Our results suggest that dihydroceramide is unlikely to be a substrate for OsCDase (Figure 4B). Additionally, measurements of endogenous levels of phytoceramide suggests that OsCDase does not use phytoceramide as a substrate (Figure 4C), an observation consistent with the lack of activity when NBD-C12-phytoceramide was presented as a substrate in vitro (Figure 4A). We also measured changes in the endogenous levels of α-OH-phytoceramide and showed that OsCDase does not use α-OH-phytoceramide as a substrate (data not shown). Interestingly, we observed that endogenous levels of phytoceramide with fatty acid chain lengths of C26 and C28 were elevated in Δypc1Δydc1 when expression of OsCDase was induced by growth in galactose. This suggests that OsCDase may exhibit reverse ceramidase activity and catalyze the formation of phytoceramide with very long chain fatty acids (Figure 4C).
Figure 4.
Substrate utilization by rice CDase (OsCDase). OsCDase was cloned into the vector, pYES2/CT and the construct was transformed into the yeast double-knockout strain, Δypc1Δydc1, which lacks the yeast CDases. (A) Expression of OsCDase in the yeast double-knockout strain, Δypc1Δydc1 was induced by growth in galactose medium for 12 h and whole cell lysates were assayed for CDase activity using D-erythro-NBD-C12-ceramide (C12-NBD-Cer) or NBD-C12-Phytoceramide (C12-NBD-PhytoCer) as substrates. Wild-type yeast expressing YPC1p and YDC1p was used as a control to show that the yeast CDases uses NBD-C12-Phytoceramide as a substrate. Endogenous levels of (B) dihydroceramide and (C) phytoceramide following induction of OsCDase expression in the yeast double-knockout strain, Δypc1Δydc1. Values are means ± S.D. of four replicates from two independent experiments.
Sub-cellular localization of OsCDase
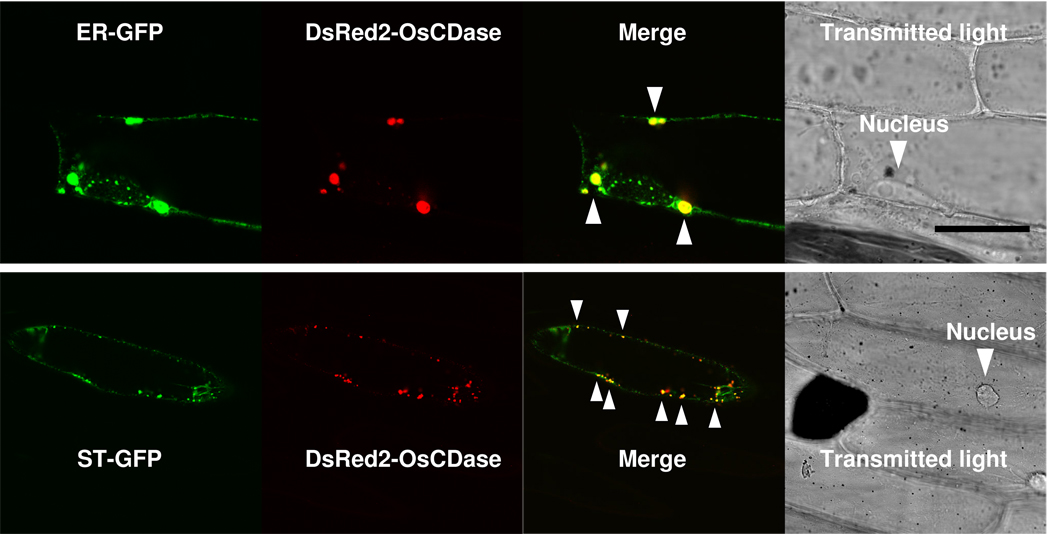
In silico analysis of the amino acid sequence of OsCDase using TMHMM (http://www.cbs.dtu.dk/services/TMHMM-2.0/) indicated that OsCDase is a transmembrane protein. Additionally, we used SLP-Local (Sub-cellular Location Predictor based on Local Features of Amino Acid Sequence; http://sunflower.kuicr.kyoto-u.ac.jp/~smatsuda/slplocal.html) to analyse the OsCDase sequence, and OsCDase is predicted to be localized to the secretory pathway. In order to determine the sub-cellular localization of OsCDase in planta, we cloned the full-length OsCDase CDS into the expression vector, pGDR (Goodin et al, 2000) containing the red fluorescent protein, DsRed2 as the reporter driven by the constitutive CaMV35S promoter from cauliflower mosaic virus for transient expression in onion epidermal cells following biolistic transformation. To determine the sub-cellular localization of DsRed2-OsCDase, we co-transformed onion epidermal cells with green fluorescent protein (GFP) targeted to the endoplasmic reticulum (ER-GFP) or Golgi (ST-GFP). We observed co-localization of DsRed2-OsCDase with ER-GFP (Figure 5A) and ST-GFP (Figure 5B), indicating that OsCDase is localized to the ER and Golgi, and that these organelles are sites of ceramide metabolism in planta.
Figure 5.
Sub-cellular localization of OsCDase fused to the red fluorescent protein (DsRed2) following biolistic transformation of onion epidermal cells. OsCDase was cloned into the vector, pGDR and the plasmid was used for biolistic transformation of onion epidermal cells. ER-GFP (ER-targeted/retained Green Fluorescent Protein) and ST-GFP (Golgi-targeted Green Fluorescent Protein) were used for co-localization studies. Co-localization is visualized in yellow (and indicated with arrows) in the ‘Merge’ panels. Localisation of the nucleus is shown in the transmitted light micrograph. Images were acquired using confocal laser scanning microscopy and are representative of 30 to 40 transformed cells. Scale bar = 50 µm.
Discussion
We have cloned a ceramidase from rice (OsCDase) and sequence analysis showed that it bears similarities to neutral ceramidases from a variety of organisms (Figure 1 and Supplemental Figure 1). Interestingly, we observed the presence of an intron of 777 bp within the 5’-UTR of OsCDase. This is in contrast to the predicted intron within the 5’-UTR of 255 bp. Introns in plant genes are characterized by a strong nucleotide bias towards T proximal to the AG intron acceptor site and that there is an A/T bias throughout the intron relative to the adjacent exon. It has been proposed that such nucleotide biases are required for efficient intron recognition and splicing by the spliceosome (Lorkovic et al., 2000). However, the situation is less clear for introns within the 5’-UTR of plant genes as there appears to be no nucleotide bias to distinguish introns from exon sequences (Chung et al., 2006). The lack of a nucleotide bias for introns in 5’-UTRs appears to hold true for human genes as well (Eden and Brunak, 2004). This may account for the discrepancy between the predicted size of the intron with the results we have obtained from using 5’-RACE PCR. The significance of the long intron within the 5’-UTR of OsCDase is unclear although evidence to date suggests that the presence of 5’-UTR introns may enhance gene expression and that the length of the intron may also influence the level of gene expression in plants (Rose, 2000, 2004; Chung et al., 2006).
In silico analysis of the 5’-UTR of OsCDase showed the presence of a 90 bp (position -32 to -122) internal ribosomal entry site (IRES) element. This is interesting as IRES elements have been found in the 5’-UTRs of mRNAs of proteins involved in cellular proliferation and apoptosis (Holcik et al., 2000). IRES elements were first discovered in piconavirus mRNAs where they function to initiate translation of uncapped viral mRNAs (Pelletier and Sonenberg, 1998). It has been proposed that IRES-dependent (and m7G cap-independent) translation of mRNA may be an important regulatory point for eukaryotic cells to fine-tune their responses to stress through IRES-dependent translation of survival or pro-apoptotic factors (Holcik et al., 2000). Given the recent observations that ceramide can initiate programmed cell death in plant cells (Liang et al., 2003; Townley et al., 2005), the identification of an IRES element in the 5’-UTR of OsCDase is interesting, and suggests that OsCDase may be subject to translational regulation as part of the programmed cell death machinery in plant cells.
In order to determine the biochemical characteristics of OsCDase, we expressed recombinant OsCDase in the yeast double knockout mutant, Δypc1Δydc1, which lacks the yeast CDases, YPC1p and YDC1p (Mao et al., 2000). Our results showed that OsCDase is indeed a bona fide ceramidase and it exhibits typical Michealis-Menten kinetics. OsCDase activity was enhanced in the presence of Ca2+, Mg2+, Mn2+ and Zn2+ but inhibited in the presence of Fe2+ (Figure 3D). We showed that OsCDase activity was significantly activated in the presence of 5 mM Ca2+ (Supplemental Figure 2) whereas Lynch (2000) reported a 100% increase in CDase activity in the presence of 1mM Ca2+. We also tested the effects of micromolar concentrations of Ca2+ on OsCDase activity and showed that OsCDase activity was only weakly activated by micromolar concentrations of Ca2+ (Supplemental Figure 2). It is unlikely that the activity of the ER/Golgi-localised OsCDase is influenced by changes in cytosolic-free calcium concentrations and our observation of significant activation of OsCDase activity in the presence of 5 mM Ca2+ is consistent with the ER being an important intracellular calcium store. The divalent ion-dependent enhancement of CDase activity is in agreement with studies showing that Ca2+, Mg2+, Mn2+ also enhanced the activity of the human neutral CDase (Galadari et al., 2006). Interestingly, while Zn2+ (2.5 to 5 mM) appeared to enhance the activity of OsCDase, the same concentrations inhibited the activity of the human neutral CDase (Galadari et al., 2006). Additionally, the activity of the Dictyostelium discoideum neutral CDase homologue does not appear to be affected by the presence of the following divalent ions, Ca2+, Mg2+, Mn2+, and Zn2+ (Monjusho et al., 2003). The observation that OsCDase exhibits a pH optimum of 5.7 to 6.0 is interesting as it bears sequence similarities to neutral ceramidases. The observation of an acidic pH-dependency of enzymes of the neutral ceramidase family has been reported in Dictyostelium discoideum where it was demonstrated that the neutral CDase homologue exhibited a pH optimum of 3 (Monjusho et al., 2003). Lynch (2000) also reported that CDase activity from plant membrane fractions exhibited an acidic pH dependency of between 5.2 to 5.6. This appears to be in agreement with our observation that OsCDase exhibited an acidic pH optimum of between pH 5.7 to 6.0. The slight acidic pH dependency of OsCDase may explain the observation that it is phylogenetically clustered with the CDase from Dictyostelium discoideum (Figure 1B). Further work on putative members of the neutral ceramidase family from other plants, for example, the 3 candidates from Arabidopsis, is likely to provide greater insights into the acidic pH-dependency of the neutral ceramidase family.
Analysis of substrate utilization by OsCDase revealed a substrate preference for ceramide and not phytoceramide (Figure 4A). This is interesting as phytoceramide is a major ceramide species in fungi and plants (Ohnishi et al., 1985; Sperling and Heinz, 2003; Dunn et al., 2004; Lynch and Dunn, 2004). The substrate preference of sphingolipid metabolizing enzymes from plants for metabolites of low abundance is not without precedence. For example, Coursol and co-workers showed that sphingosine kinase from Arabidopsis exhibited a substrate preference for sphingosine, which is of low abundance compared to dihydrosphingosine and phytosphingosine, both of which are predominant sphingolipid metabolites in plants (Coursol et al., 2003, 2005). Analysis of yeast ceramide species following expression of OsCDase in the yeast double knockout mutant, Δypc1Δydc1 also showed that both dihydroceramide and phytoceramide are unlikely to be substrates for OsCDase (Figs. 4B & 4C). Reverse ceramidase activity has been reported in members of the neutral ceramidase family (Okino et al., 1998; Kita et al., 2000; El Bawab et al., 2001; Wu et al., 2007). Analyses of yeast sphingolipids following induction of OsCDase expression in the double knockout mutant, Δypc1Δydc1 showed elevated levels of phytoceramide with fatty acid chain lengths of C26 and C28 (Figure 4C). This observation points to the possibility that OsCDase may exhibit reverse ceramidase activity, leading to elevated levels of these 2 phytoceramide species.
Neutral ceramidases have been shown to be localized to the plasma membrane, endosome-like organelle, mitochondria and endoplasmic reticulum/Golgi compartments (El Bawab et al., 2000; Mitsutake et al., 2001; Yoshimura et al., 2004). Additionally, neutral ceramidases may also be secreted (Romiti et al., 2000). We showed that OsCDase is localized to the endoplasmic reticulum and Golgi in planta, suggesting that the ER/Golgi compartments are sites of ceramide metabolism in plants. This observation is consistent with in silico prediction of OsCDase as a transmembrane protein localized to the secretory pathway.
In conclusion, we have cloned a bona fide plant ceramidase from rice and showed that it exhibited classical Michaelis-Menten kinetics with an optimum activity ranging from pH 5.7 to 6.0. OsCDase activity was enhanced in the presence of Ca2+, Mg2+, Mn2+ and Zn2+ but inhibited in the presence of Fe2+. Expression analysis indicated that steady-state levels of OsCDase transcripts are expressed throughout the seedling with a higher level of expression in roots compared to shoots. Sub-cellular localization showed that OsCDase is localized to the ER/Golgi compartments, suggesting that these organelles are sites of sphingolipid metabolism in planta. Given the observations that ceramide is an important sphingolipid metabolite involved in the regulation of programmed cell death in plants (Liang et al., 2003; Townley et al., 2005), our characterization of the rice ceramidase is an important first step in understanding how ceramide is metabolized and provide the basis for further investigations into how sphingolipid metabolism is co-ordinated with developmental processes in plants.
Experimental procedures
Plant growth
Rice (Oryza sativa ssp japonica cv. Nipponbare) seeds were germinated in full-strength Murashige and Skoog media (Sigma, UK) at pH 5.8 in a plant growth cabinet (Snijders Microclima 1750, Snijders, Netherlands) under the following conditions: PPFD: 1000 µmol/m2/s, 12h light/12h darkness with a day temperature of 28°C and a night temperature of 24°C under 85% relative humidity.
cDNA cloning and determination of 5’- and 3’-untranslated regions (UTRs)
Total RNA was isolated from 2-week old seedlings using the RNeasy® Mini Kit (Qiagen, UK) according to manufacturer’s instructions. 500 ng of DNAse-treated (Turbo DNA-free™ Kit, Ambion, USA) total RNA was used for cDNA synthesis using Accuscript™ High-fidelity RT-PCR system (Stratagene, UK). Three µl of the product was used for PCR amplification of the full-length with PfuUltra HF DNA polymerase (Stratagene, UK) using the following primers:
forward primer: 5’-ATGGAGGCTTCATCTTGGTTGTG-3’
reverse primer: 5’-ACGCACCGCGAAAGCACGAGA-3’
Cloning of the 5’- and 3’-UTRs was achieved using the SMART-RACE cDNA amplification kit (Clontech) according to manufacturer’s instructions and using the following gene specific primers.
Gene-specific primer for 5’-RACE: 5’-GAAGCTCCCCATACCGACCAG-3’
Gene-specific primer for 3’-RACE: 5’-CAAGGCCCGCAAGATTGAGTTC-3’
Full-length coding sequence (CDS) and 5’- and 3’-UTRs were cloned into pGEM T-Easy vector (Promega, UK) before sequencing (Eurofins MWG GmbH, Germany).
Expression of OsCDase in yeast
Full-length OsCDase CDS was sub-cloned into the yeast expression vector, pYES2/CT (Invitrogen, UK) and transformed into the yeast double knockout mutant, Δypc1Δydc1, which lacks the yeast ceramidases YPC1p and YDC1p (Mao et al., 2000) using the lithium acetate method as previously described (31). Transformed yeast cultures were grown in synthetic minimal media enriched in dextrose and complemented by a selective uracil-deficient dropout supplement (Clontech, USA), and cell lysates were isolated as previously described (Galadari et al., 2006).
OsCDase enzyme assays and biochemical characterization
Enzyme activity was measured using D-erythro-C12-NBD-ceramide (Avanti Polar Lipids, USA) or NBD-C12-phytoceramide (Lipidomics Core Facility, MUSC, USA) at a final concentration of 100 µM (3.13 mol%) in a 100 mM Phosphate buffer, pH 5.7, containing 0.2% Triton-X-100 and 5 mM MgCl2 final in a total volume of 100 µl. For determining pH optima, the substrate was dissolved in the following buffers: pH 3.91–5.33, 100 mM acetate buffer; pH 5.04–6.66, 100 mM Phosphate buffer; pH 7.1–8.24, 100 mM Tris buffer. The enzymatic reaction was continued for 1 hr at 37°C, and the reaction was terminated by addition of chloroform/methanol (1:1, v/v). The mixture was left at room temperature for 5 min, after which it was centrifuged for 5 min at 5500 rpm (Accuspin microR, Fisher, USA), and the organic phase was taken and dried using a SpeedVac (Savant, USA) evaporator. Lipids were dissolved in 15 µl of chloroform/methanol (1:1, v/v), and spotted on to TLC plates (Partisil LK6D, Whatman, USA). The NBD-C12-fatty acid products were separated from the substrate by developing the plate in chloroform/methanol/25% ammonium hydroxide (60:20:0.5, by vol.) solvent system. The TLC plate was scanned using a PhosphorImager (Storm 860, USA) system set at blue fluorescence mode. The NBD-fatty acid products were identified by comparison with NBD-C12-fatty acid standards and were quantified using ImageQuant™ software. The specific activity was determined using standard curves with known concentrations of D-erythro-C12-NBD-ceramide.
Analysis of ceramides by mass spectrometry
Accumulated ceramides were analyzed by MS using reverse-phase, high-performance liquid chromatography (HPLC) coupled to an electrospray-triple quadrupole mass spectrometer, operating at positive ionization, multiple reaction monitoring (MRM) mode. Mass separations were performed using a ThermoFinnigan TSQ 7000 mass spectrometer (Foster City, CA) according to published methodology (Bielawski et al., 2006).
Sub-cellular localization of OsCDase
Full-length OsCDase CDS was cloned downstream of the red fluorescent protein, DsRed2 under the control of the constitutive 35S CaMV promoter in the vector, pGDR (kindly provided by Dr. Michael Goodin, University of Kentucky, USA) (Goodin et al., 2000). The vector was sequenced to ensure that OsCDase has been cloned in-frame to the DsRed2 reporter gene. Biolistic transformation of onion (Allium cepa) epidermal cells was done using 1 µg of plasmid DNA precipitated onto 100 µg of 1 µm gold particles using CaCl2 and spermidine. DNA-coated gold particles were washed with ethanol and resuspended in ethanol for transformation of onion epidermal cells using a biolistic particle delivery system (Bio-Rad PDS-1000/He, Bio-Rad Laboratories, USA) with a pressure set at 1100 psi. After an overnight incubation, the localization was observed using confocal laser scanning microscopy using a Leica TCS-SP2 confocal microscope (Leica Systems, UK). Transmitted light micrographs were obtained with a transmitted light detector. Co-localization was performed with green fluorescent protein targeted to the ER (plasmid, pGFP:C-domain; kindly provided by Dr. Wendy Boss, North Carolina State University, USA) (Persson et al., 2002) or Golgi (plasmid, pVKH18-EN6::ST-GFP; kindly provided by Dr. Chris Hawes, Oxford Brookes University, UK) (Saint-Jore et al., 2002).
Supplementary Material
Acknowledgements
This work is funded by a Science Foundation Ireland grant BR/04/B0581 (to C.K.-Y.N.), a National Institute of Health (NIH) grant CA87584 (to Y.A.H.), a Irish Research Council for Science, Engineering and Technology Embark Postdoctoral Fellowship (to T.C.X.), and parts of the research was conducted in a facility constructed with support from the NIH Grant C06 RR018823 from the Extramural Research Facilities Program of the National Center for Research Resources.
The nucleotide sequence of OsCDase has been submitted to GenBank with the accession number EU422991.
References
- Bielawski J, Szulc ZM, Hannun YA, Bielawska A. Simultaneous quantitative analysis of bioactive sphingolipids by high-performance liquid chromatography-tandem mass spectrometry. Methods. 2006;39:82–91. doi: 10.1016/j.ymeth.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Carter HE, Celmer WD, Galanos DS, Gigg RH, Lands WEM, Law JH, Mueller KL, Nakayama T, Tomizawa HH, Weber E. Biochemistry of sphingolipides. X. Phytoglyceride, a complex phytosphingosine-containing lipide from plant seeds. J. Am. Oil Chem. Soc. 1958;35:335–343. [Google Scholar]
- Chen M, Han G, Dietrich CR, Dunn TM, Cahoon EB. The essential nature of sphingolipids in plants as revealed by the functional identification and characterization of the Arabidopsis LCB1 subunit of serine palmitoyltransferaase. Plant Cell. 2006;18:3576–3593. doi: 10.1105/tpc.105.040774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung BYW, Simons C, Firth AE, Brown CM, Hellens RP. Effect of 5’-UTR introns on gene expression in Arabidopsis thaliana. BMC Genomics. 2006;7:120. doi: 10.1186/1471-2164-7-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coursol S, Fan LM, Le Stunff H, Spiegel S, Gilroy S, Spiegel S, Assmann SM. Sphingolipid signalling in Arabidopsis guard cells involves heterotrimeric G proteins. Nature. 2003;423:651–654. doi: 10.1038/nature01643. [DOI] [PubMed] [Google Scholar]
- Coursol S, Le Stunff H, Lynch DV, Gilroy S, Assmann SM, Spiegel S. Arabidopsis sphingosine kinase and the effects of phytosphingosine-1- phosphate on stomatal aperture. Plant Physiol. 2005;137:724–737. doi: 10.1104/pp.104.055806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson RC, Lester RL. Sphingolipid functions in Saccharomyces cerevisiae. Biochim. Biophys. Acta. 2002;1583:13–25. doi: 10.1016/s1388-1981(02)00210-x. [DOI] [PubMed] [Google Scholar]
- Dickson RC, Nagiec EE, Skrzypek M, Tillman P, Wells GB, Lester RL. Sphingolipids Are Potential Heat Stress Signals in Saccharomyces. J. Biol. Chem. 1997;272:30196–30200. doi: 10.1074/jbc.272.48.30196. [DOI] [PubMed] [Google Scholar]
- Dietrich CR, Han G, Chen M, Berg RH, Dunn TM, Cahoon EB. Loss-of-function mutations and inducible RNAi suppression of Arabidopsis LCB2 genes reveal the critical role of sphingolipids in gametophytic and sporophytic cell viability. Plant J. 2008;54:284–298. doi: 10.1111/j.1365-313X.2008.03420.x. [DOI] [PubMed] [Google Scholar]
- Dunn TM, Lynch DV, Michaelson LV, Napier JA. A post-genomic approach to understanding sphingolipid metabolism in Arabidopsis thaliana. Ann. Bot. 2004;93:483–497. doi: 10.1093/aob/mch071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden E, Brunak S. Analysis and recognition of 5’ UTR intron splice sites in human pre-mRNA. Nucleic Acids Res. 2004;32:1131–1142. doi: 10.1093/nar/gkh273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Bawab S, Birbes H, Roddy P, Szulc ZM, Bielawska A, Hannun YA. Biochemical characterization of the reverse activity of rat brain ceramidase. A CoA-independent and fumonisin B1-insensitive ceramide synthase. J. Biol. Chem. 2001;276:16758–16766. doi: 10.1074/jbc.M009331200. [DOI] [PubMed] [Google Scholar]
- El Bawab S, Roddy P, Qian T, Bielawska A, Lemasters JJ, Hannun YA. Molecular cloning and characterization of a human mitochondrial ceramidase. J. Biol. Chem. 2000;275:21508–21513. doi: 10.1074/jbc.M002522200. [DOI] [PubMed] [Google Scholar]
- Futerman AH, Hannun YA. The complex life of simple sphingolipids. EMBO Rep.t. 2004;5:777–782. doi: 10.1038/sj.embor.7400208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galadari S, Wu BX, Mao C, Roddy P, El Bawab S, Hannun YA. Identification of a novel amidase motif in neutral ceramidase. Biochem. J. 2006;393:687–695. doi: 10.1042/BJ20050682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodin MM, Dietzgen RG, Schichnes D, Ruzin S, Jackson AO. pGD vectors: versatile tooles for the expression of green and red fluorescent protein fusions in agroinfiltrated plant leaves. Plant J. 2000;31:375–383. doi: 10.1046/j.1365-313x.2002.01360.x. [DOI] [PubMed] [Google Scholar]
- Hannun YA, Obeid LM. The ceramide-centric universe of lipid-mediated cell regulation: stress encounters of the lipid kind. J. Biol. Chem. 2002;277:25847–25850. doi: 10.1074/jbc.R200008200. [DOI] [PubMed] [Google Scholar]
- Hannun YA, Obeid LM. Principles of bioactive lipid signalling: lessons from sphingolipids. Nat. Rev. Mol. Cell Biol. 2008;9:139–150. doi: 10.1038/nrm2329. [DOI] [PubMed] [Google Scholar]
- Holcik M, Sonnenberg N, Korneluk RG. Internal ribosome initiation of translation and the control of cell death. Trends Genet. 2000;16:469–473. doi: 10.1016/s0168-9525(00)02106-5. [DOI] [PubMed] [Google Scholar]
- Imamura T, Kusano H, Kajigaya Y, Ichikawa M, Shimada H. A rice dihydrosphingosine C4 hydroxylase (DSH1) gene, which is abundantly expressed in the stigmas, vascular cells and apical meristem, may be involved in fertility. Plant Cell Physiol. 2007;48:1108–1120. doi: 10.1093/pcp/pcm084. [DOI] [PubMed] [Google Scholar]
- Ito S, Ohnishi M, Fujino Y. Investigation of sphingolipids in pea seeds. Agr. Biol. Chem. Tokyo. 1985;49:539–540. [Google Scholar]
- Jenkins GM, Richards A, Wahl T, Mao C, Obeid L, Hannun YA. Involvement of yeast sphingolipids in the heat stress response of Saccharomyces cerevisiae. J. Biol. Chem. 1997;272:32566–32572. doi: 10.1074/jbc.272.51.32566. [DOI] [PubMed] [Google Scholar]
- Kawai G. Molecular species of cerebrosides in fruiting bodies of Lentinus edodes and their biological activity. Biochim. Biophys. Acta. 1989;1001:185–190. doi: 10.1016/0005-2760(89)90146-x. [DOI] [PubMed] [Google Scholar]
- Kawai G, Ohnishi M, Fujino Y, Ikeda Y. Stimulatory effect of certain plant sphingolipids on fruiting of Schizophyllum commune. J. Biol. Chem. 1986;261:779–784. [PubMed] [Google Scholar]
- Kita K, Okino N, Ito M. Reverse hydrolysis reaction of a recombinant alkaline ceramidase of Pseudomonas aeruginosa. Biochim. Biophys. Acta. 2000;1485:111–120. doi: 10.1016/s1388-1981(00)00029-9. [DOI] [PubMed] [Google Scholar]
- Koga J, Yamauchi T, Shimura M, Ogawa N, Oshima K, Umemura K, Kikuchi M, Osagawara N. Cerebrosides A and C, sphingolipid elicitors of hypersensitive cell death and phytoalexin accumulation in rice plants. J. Biol. Chem. 1998;48:31985–31991. doi: 10.1074/jbc.273.48.31985. [DOI] [PubMed] [Google Scholar]
- Liang H, Yao N, Song JT, Luo S, Lu H, Greenberg JT. Ceramides modulate programmed cell death in plants. Genes Dev. 2003;17:2636–2641. doi: 10.1101/gad.1140503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorkovic ZJ, Wieczorek KDA, Lambermon MH, Filipowicz W. Pre-mRNA splicing in higher plants. Trends Plant Sci. 2000;5:160–167. doi: 10.1016/s1360-1385(00)01595-8. [DOI] [PubMed] [Google Scholar]
- Lynch DV. Sphingolipids. In: Moore TS, editor. Lipid Metabolism in Plants. USA: CRC Press; 1993. pp. 279–302. [Google Scholar]
- Lynch DV. Enzymes of sphingolipid metabolism in plants. Methods Enzymol. 2000;311:130–149. doi: 10.1016/s0076-6879(00)11074-2. [DOI] [PubMed] [Google Scholar]
- Lynch DV, Dunn TM. An introduction to plant sphingolipids and a review of recent advances in understanding their metabolism and function. New Phytol. 2004;161:677–702. doi: 10.1111/j.1469-8137.2004.00992.x. [DOI] [PubMed] [Google Scholar]
- Mao C, Obeid LM. Ceramidases: regulators of turnover of ceramide and ceramide-mediated responses. In: Futerman AH, editor. Ceramide Signaling. USA: Landes Bioscience; 2002. pp. 29–40. [Google Scholar]
- Mao C, Saba J, Obeid LM. The dihydrosphingosine-1-phosphate phosphatases of Saccharomyces cerevisiae are important regulators of cell proliferation and heat stress responses. Biochem. J. 1999;342:667–675. [PMC free article] [PubMed] [Google Scholar]
- Mao C, Xu R, Bielawska A, Szulc ZM, Obeid LM. Cloning and characterization of a Saccharomyces cerevisiae alkaline ceramidase with specificity for dihydroceramide. J. Biol. Chem. 2000;275:31369–31378. doi: 10.1074/jbc.M003683200. [DOI] [PubMed] [Google Scholar]
- Markham JE, Jaworski JG. Rapid measurement of sphingolipids from Arabidopsis thaliana by reverse-phase high-performane liquid chromatography coupled to electrospray ionization tandem mass spectrometry. Rapid Commun. Mass Sp. 2007;21:1304–1314. doi: 10.1002/rcm.2962. [DOI] [PubMed] [Google Scholar]
- Markham JE, Li J, Cahoon EB, Jaworski JG. Separation and identification of major plant sphingolipid classes from leaves. J. Biol. Chem. 2006;281:22684–22694. doi: 10.1074/jbc.M604050200. [DOI] [PubMed] [Google Scholar]
- Mitsutake S, Tani M, Okino N, Mori K, Ichinose S, Omori A, Iida H, Nakamura T, Ito M. Purification, characterization, molecular cloning and subcellular distribution of neutral ceramidase from rat kidney. J. Biol. Chem. 2001;276:26249–26259. doi: 10.1074/jbc.M102233200. [DOI] [PubMed] [Google Scholar]
- Monjusho H, Okino N, Tani M, Maeda M, Yoshida M, Ito M. A neutral ceramidase homologue from Dictyostelium discoideum exhibits an acidic pH optimum. Biochem. J. 2003;376:473–479. doi: 10.1042/BJ20030652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng CK-Y, Carr K, McAinsh MR, Powell B, Hetherington AM. Drought-induced guard cell signal transduction involves sphingosine-1-phosphate. Nature. 2001;410:596–599. doi: 10.1038/35069092. [DOI] [PubMed] [Google Scholar]
- Ohnishi M, Ito S, Fujino Y. Structural characterization of sphingolipids in leafy stems of rice. Agr. Biol. Chem. Tokyo. 1985;49:3327–3329. [Google Scholar]
- Okino N, Tani M, Imayama S, Ito M. Purification and characterization of a novel ceramidase from Pseudomonas aeruginosa. J. Biol. Chem. 1998;273:14368–14373. doi: 10.1074/jbc.273.23.14368. [DOI] [PubMed] [Google Scholar]
- Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1998;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- Persson S, Love J, Tsou P-L, Robertson D, Thompson WF, Boss WF. When a day makes a differerence. Interpreting data from endoplasmic reticulum-targeted green fluorescent protein fusions in cells grown in suspension culture. Plant Physiol. 2002;128:341–344. doi: 10.1104/pp.010840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyne S, Pyne NJ. Sphingosine-1-phosphate signalling in mammalian cells. Biochem. J. 2000;349:385–402. doi: 10.1042/0264-6021:3490385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romiti E, Meacci E, Tani M, Nuti F, Farnararo M, Ito M, Bruni P. Neutral/alkaline and acid ceramidase activities are actively released by murine endothelial cells. Biochem. Biophys. Res. Commun. 2000;275:746–751. doi: 10.1006/bbrc.2000.3370. [DOI] [PubMed] [Google Scholar]
- Rose AB. The effect of intron location on intron-mediated enhancement of gene expression in Arabidopsis. Plant J. 2004;40:744–751. doi: 10.1111/j.1365-313X.2004.02247.x. [DOI] [PubMed] [Google Scholar]
- Rose AB, Beliakoff JA. Intron-mediated enhancement of gene expression independent of unique intron sequences and splicing. Plant Physiol. 2000;122:535–542. doi: 10.1104/pp.122.2.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Jore CM, Evins J, Batoko H, Brandizzi F, Moore I, Hawes C. Redistribution of membrane proteins between the Golgi apparatus and endoplasmic reticulum in plants is reversible and not dependent on cytoskeletal networks. Plant J. 2002;29:661–678. doi: 10.1046/j.0960-7412.2002.01252.x. [DOI] [PubMed] [Google Scholar]
- Schmid KM, Ohlrogge JB. Lipid metabolism in palnts. In: Vance DE, Vance JE, editors. Biochemistry of Lipids, Lipoproteins and Membranes. The Netherlands: Elsevier Science; 1996. pp. 363–389. [Google Scholar]
- Shi L, Bielawski J, Mu J, Dong H, Teng C, Zhang J, Yang X, Tomishige N, Hanada K, Hannun YA, Zuo J. Involvement of sphingoid bases in mediating reactive oxygen intermediate production and programmed cell death in Arabidopsis. Cell Res. 2007;17:1030–1040. doi: 10.1038/cr.2007.100. [DOI] [PubMed] [Google Scholar]
- Sperling P, Heinz E. Plant sphingolipids: structural diversity, biosynthesis, first genes and functions. Biochim. Biophys. Acta. 2003;1632:1–15. doi: 10.1016/s1388-1981(03)00033-7. [DOI] [PubMed] [Google Scholar]
- Spiegel S, Milstien S. Sphingosine-1-phosphate: an enigmatic signalling lipid. Nat. Rev. Mol. Cell Biol. 2003;4:397–407. doi: 10.1038/nrm1103. [DOI] [PubMed] [Google Scholar]
- Teng C, Dong H, Shi L, Deng Y, Mu J, Zhang J, Yang X, Zuo J. Serine palmitoyltransferase, a key enzyme for de novo synthesis of sphingolipids, is essential for male gametophyte development in Arabidopsis. Plant Physiol. 2008 doi: 10.1104/pp.107.113506. doi: 10.1104/pp.107.113506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townley HE, McDonald K, Jenkins GI, Knight MR, Leaver CJ. Ceramides induce programmed cell death in Arabidopsis cells in a calcium-dependent manner. Biol. Chem. 2005;386:161–166. doi: 10.1515/BC.2005.020. [DOI] [PubMed] [Google Scholar]
- Tsegaye Y, Richardson CG, Bravo JE, Mulcahy BJ, Lynch DV, Markham JE, Jaworski JG, Chen M, Cahoon EB, Dunn TM. Arabidopsis mutants lacking long chain base phosphate lyase are fumonisin-sensitive and accumulation trihydroxy-18 :1 long chain base phosphate. J. Biol. Chem. 2007;282:28195–28206. doi: 10.1074/jbc.M705074200. [DOI] [PubMed] [Google Scholar]
- Wu BX, Snook CF, Tani M, Büllesbach EE, Hannun YA. Large-scale purification and characterization of recombinant Pseudomonas ceramidase: regulation by calcium. J. Lipid Res. 2007;48:600–608. doi: 10.1194/jlr.M600423-JLR200. [DOI] [PubMed] [Google Scholar]
- Yoshimura Y, Tani M, Okino N, Iida H, Ito M. Molecular cloning and functional analysis of zebrafish neutral ceramidase. J. Biol. Chem. 2004;279:44012–44022. doi: 10.1074/jbc.M405598200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.