Abstract
Structure-function relationships for inhibition of human cytochrome P450s (P450s) 1A1, 1A2, 1B1, 2C9, and 3A4 by 33 flavonoid derivatives were studied. Thirty-two of the 33 flavonoids tested produced Reverse Type I binding spectra with P450 1B1, and the potencies of binding were correlated with the abilities to inhibit 7-ethoxyresorufin O-deethylation activity. The presence of a hydroxyl group in flavones, e.g. 3-, 5-, and 7-monohydroxy- and 5,7-dihydroxyflavone, decreased the 50% inhibition concentration (IC50) of P450 1B1 from 0.6 µM to 0.09, 0.21, 0.25, and 0.27 µM, respectively, and 3,5,7-trihydroxyflavone (galangin) was the most potent, with an IC50 of 0.003 µM. The introduction of a 4’-methoxy- or 3’,4’-dimethoxy group into 5,7-dihydroxyflavone yielded other active inhibitors of P450 1B1 with IC50 values of 0.014 and 0.019 µM, respectively. The above hydroxyl- and/or methoxy-groups in flavone molecules also increased the inhibition activity with P450 1A1 but not always towards P450 1A2, where 3-, 5-, or 7-hydroxyflavone, and 4’-methoxy-5,7-dihydroxyflavone were less inhibitory than flavone itself. P450 2C9 was more inhibited by 7-hydroxy-,5,7-dihydroxy-, and 3,5,7-trihydroxyflavones than by flavone but was weakly inhibited by 3-and 5-hydroxyflavone. Flavone and several other flavonoids produced Type I binding spectra with P450 3A4, but such binding was not always related to the inhibitiory activities towards P450 3A4. These results indicate that there are different mechanisms of inhibition for P450s 1A1, 1A2, 1B1, 2C9, and 3A4 by various flavonoid derivatives and that the number and position of hydroxyl and/or methoxy groups highly influence the inhibitory actions of flavonoids towards these enzymes. Molecular docking studies suggest that there are different mechanisms involved in the interaction of various flavonoids with the active site of P450s, thus causing differences in inhibition of these P450 catalytic activities by flavonoids.
Introduction
Many plant flavonoids are found in nature and it has been reported that these natural products have various biological properties, e.g. anti-oxidative and anti-mutagenic activities, thus preventing cancer, heart disease, bone loss, and a number of diseases (1–3). Recent studies have shown that these biological activities vary with the number and substitution positions of hydroxyl and/or methoxy groups in the flavonoid molecules (4–6). Inhibition of P450 enzymes by structurally diverse flavonoids has been extensively studied in several laboratories (4–10). Human P450s 1A1, 1A2, 1B1, 2C9, and 3A4 are reported to be inhibited by various flavonoids (11–17), and these phenomena have been suggested to influence human health, since these P450 enzymes play important roles in the activation and detoxification of endogenous and xenobiotic chemicals (18–22).
We have previously shown that a variety of chemical inhibitors—e.g. structural derivatives of pyrene, naphthalene, phenanthrene, biphenyl, and flavone—interact with human P450 1B1 (P450 1B1) to produce reverse type I binding spectra and that the spectral dissociation constants (Ks) and the magnitudes of the binding (ΔA/Ks) of these compounds are well correlated with potencies of these chemicals to inhibit P450 1B1, as determined by 7-ethoxyresorufin O-deethylation (EROD) as a model reaction (23). A structure-function relationship study has revealed that the substitution of pyrene with acetylenic groups decreases the affinity to P450 1B1 while the substitution of naphthalene, phenanthrene, and biphenyl with acetylenic groups or propargyl ethers causes increases in their affinities to P450 1B1. In the cases of flavonoids, substitutions of 3’-monomethoxy-, 4’-monomethoxy-, or 3’,4’-dimethoxy groups with 5,7-dihydroxyflavone cause strong interactions with P450 1B1 while 7,8-dihydroxylation markedly decreases the affinity towards P450 1B1 (23).
In this study, we further examined and compared the structure-function relationships of the inhibition of human P450s 1A1, 1A2, 1B1, 2C9 and 3A4 with a total of 33 flavonoids by measuring EROD for the former three enzymes and flurbiprofen 1’-hydroxylation and midazolam 4-hydroxylation, respectively, for the latter two enzymes. The flavonoids used were flavone, nine hydroxylated flavones, four methoxylated flavones, eight methoxylated hydroxyflavones, flavanone, 4’,5,7-trihydroxylflavanone and its glycoside, 4’,5,7-trihydroxyisoflavone, and 4’-methoxy-5,6-dihydroxyisoflavone. We also determined the effects of addition of 2’- and 4’-propargyl ethers on the inhibition of P450s 1A1, 1A2, 1B1, 2C9 and 3A4 caused by ANF and BNF. Spectral interaction of these flavonoids with five P450s were examined and compared with their inhibitory potencies towards P450s 1A1, 1A2, 1B1, 2C9, and 3A4. Molecular docking studies of the interaction of these flavonoids with active sites of individual P450 enzymes are also reported and considered.
Experimental Procedures
Chemicals
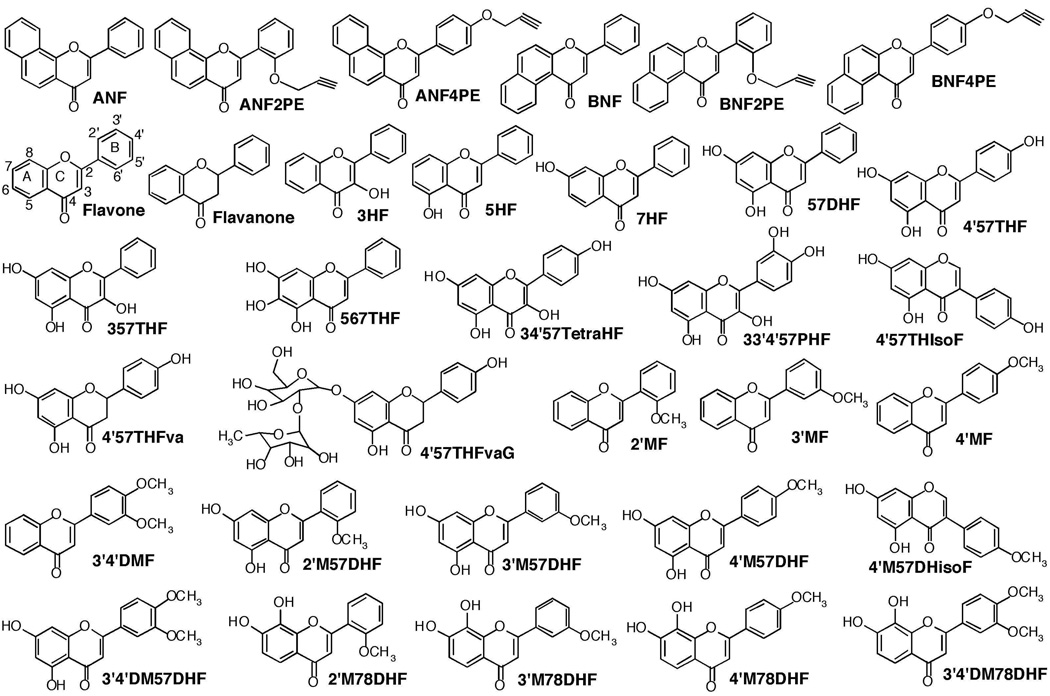
A total of 33 flavonoid derivatives was used for the inhibition studies of P450s 1A1, 1A2, 1B1, 2C9, and 3A4 (Figure 1). 7-Ethoxyresorufin, resorufin, α-naphthoflavone (ANF), β–naphthoflavone (BNF), flavone, flavanone, 3-hydroxyflavone (flavonol) (3HF), 5-hydroxyflavone (5HF), 7-hydroxyflavone (7HF), 5,7-dihydroxyflavone (chrysin) (57DHF), 3,5,7-trihydroxyflavone (galangin) (357THF), 4’5,7-trihydroxyflavone (apigenin) (4’57THF), 4’,5,7-trihydroxyisoflavone (genistein) (4’57THIsoF), 4’,5,7-trihydroxyflavanone (naringenin) (4’57THFva), 4’,5,7-trihydroxyflavanone glycoside (naringin) (4’57THFvaG), 5,6,7-trihydroxyflavone (baicalein) (567THF), 3,4’,5,7-tetrahydroxyflavone (kaempferol) (34’57TetraHF), 3,3’,4’,5,7-pentahydroxyflavone (quercetin) (33’4’57PHF), 4’-methoxy-5,7-dihydroxyflavone (acacetin) (4’M57DHF), and 4’-methoxy-5,7-dihydroxyisoflavone (biochain A) (4’M57DHIsoF) were obtained from Sigma Chemical Co. (St. Louis, MO) or Wako Pure Chemical (Osaka) or Kanto Kagaku Co. (Tokyo). 2’-Methoxyflavone (2’MF), 3’-methoxyflavone (3’MF), 4’-methoxyflavone (4’MF), 3’,4’-dimethoxyflavone (3’4’DMF), 2’-methoxy-5,7-dihydroxyflavone (2’M57DHF), 3’-methoxy-5,7-dihydroxyflavone (3’M57DHF), 3’4’-dimethoxy-5,7-dihydroxyflavone (3’4’DM57DHF), 2’-methoxy-7,8-dihydroxyflavone (2’M78DHF), 3’-methoxy-7,8-dihydroxyflavone (3’M78DHF), 4’-methoxy-7,8-dihydroxyflavone (4’M78DHF), and 3’,4’-dimethoxy-7,8-dihydroxyflavone (3’4’DM78DHF) were synthesized as described previously (23,24). Methods for the synthesis of α-naphthoflavone 2’-propargyl ether (ANF2’PE), α-naphthoflavone 4’-propargyl ether (ANF4’PE), β–naphthoflavone 2’-propargyl ether (BNF2’PE), and β–naphthoflavone 4’-propargyl ether (BNF4’PE) were described elsewhere (25). All of the flavonoids and substrates for P450 assay were dissolved in (CH3)2SO and added directly to the incubation mixtures; the final concentration of organic solvent in the assay was <0.4%. Other chemicals and reagents used in this study were obtained from the sources described previously or were of the highest quality commercially available (23,26–28).
Figure 1.
Structures of various flavonoid derivatives used in this study.
Enzymes
Bacterial "bicistronic" P450 1B1, 1A1, 1A2, 1B1, 2C9, and 3A4 systems were prepared as described (23,26–30). To facilitate expression and purification of P450s, six histidine residues were introduced at the position before the termination codon (23,31,32). The plasmids for the expression of P450s 1A1, 1A2, 1B1, 2C9, or 3A4 plus human NADPH-P450 reductase were introduced into Escherichia coli DH5α cells, and the bacterial membranes were prepared and suspended in 10 mM Tris-HCl buffer (pH 7.4) containing 1.0 mM EDTA and 20% glycerol (v/v) as described (23).
For purification of the P450 1B1, 1A1, and 1A2 enzymes, the bacterial membranes were solubilized in 0.10 M potassium phosphate buffer (pH 7.4) containing 20% glycerol (v/v), 0.5 M NaCl, 10 mM β-mercaptoethanol, 0.5% sodium cholate (w/v), 1% Triton N-101 (w/v), and 30 µM ANF. The solubilized membranes were centrifuged, the supernatant was applied to a nickel-nitrilotriacetic acid column (Qiagen), and the P450 proteins were purified by the methods described previously (23,31,32). Methods for purification of P450 2C9 and 3A4 from the bacterial membranes have been described elsewhere (29,30).
Enzyme Assays
The 50% inhibition concentration (IC50) of EROD activities of P450 1A1, 1A2, and 1B1 was determined in a standard incubation mixture (0.5 mL) consisting of P450 1A1 (0.03 µM), P450 1A2 (0.05 µM), or P450 1B1 (0.04 µM) in bacterial membranes co-expressing human NADPH-P450 reductase, chemical inhibitors, 100 mM potassium phosphate buffer (pH 7.4), and an NADPH-generating system consisting of 0.5 mM NADP+, 5 mM glucose 6-phosphate, and 0.5 unit of yeast glucose 6-phosphate dehydrogenase/mL (23). 7-Ethoxyresorufin (2.5 µM) was added to start the reaction and formation of resorufin was determined in a Hitachi F-4500 spectrofluorometer using an excitation wavelength of 571 nm and an emission wavelength of 585 nm. Control EROD activities (without flavonoids) by P450 1A1, 1A2, and 1B1 were 44 ± 3.8, 3.2 ± 0.47, and 18 ± 1.9 nmol product formed/min/nmol P450, respectively. The minimum rate of detection for EROD assay was <0.1 nmol resurufin fromed/min/nmol P450.
Flurbiprofen 4’-hydroxylation and midazolam 1’- and 4-hydroxylation were determined in a standard incubation mixtures (0.2 mL or 0.25 mL, respectively) containing P450 2C9 (0.03 µM) or P450 3A4 0.04 µM) in bacterial membranes co-expressing human NADPH-P450 reductase, 100 mM potassium phosphate buffer (pH 7.4), and an NADPH-generating system described above. Flurbiprofen (0.1 mM) or midazolam (0.1 mM) was added to start the reaction and product formation was determined in a spectrofluorimeter using an excitation wavelength of 260 nm and an emission wavelength of 320 nm (33) in the former case and in a UV detector at 220 nm (34) in the latter case. Control flurbiprofen 4’-hydroxylation activity (without flavonoids) by P450 2C9 was 3.6 ± 0.44 nmol/min/nmol P450, and control midazolam 1’- and 4-hydroxylation activities by P450 3A4 were 3.2 ± 0.33 and 6.3 ± 0.55 nmol product formed/min/nmol P450, respectively. The minimum rate of detection for flurbiprofen 4’hydroxylation activity was <0.1 nmol product fromed/min/nmol P450 and that for midazolam 1’- and 4-hydroxylation was <0.2 nmol/min/nmol P450.
Spectral Binding Titrations
Purified P450 enzymes were diluted to 1–3 µM in 0.10 M potassium phosphate buffer (pH 7.4) containing 20% glycerol (v/v) and the binding spectra were recorded with subsequent additions of chemical inhibitors in a Jasco V-550 spectrophotometer as described previously (23). The chemical inhibitors were added in a buffer solution and the spectra were recorded between 350 and 700 nm. The P450 spectra with or without inhibitors were obtained by subtracting the blank spectra (in the absence of P450) from the P450 spectra, and the difference spectra of the interaction of chemicals with P450 1B1 were obtained. Spectral dissociation constants (Ks) were estimated using GraphPad Prism software (GraphPad Software, San Diego, CA).
Other Assays
P450 and protein concentrations were estimated by the methods described previously (35,36).
Kinetic analysis
Kinetic parameters for inhibition of P450-dependent substrate oxidation activities by flavonoids were estimated by nonlinear regression analysis using the program KaleidaGraph (Synergy Softwaare, Reading, PA) or Graphpad Prism (Graphpad, San Diego, CA). Statistical correlation coefficients were obtained by power equation analysis (New Cricket III).
Docking Simulation of flavonoids into human P450 enzymes
The crystal structures of P450 1A2, 2C9, and 3A4 have recently been reported (37–39). The human P450 1A1 and 1B1 primary sequences were aligned with human P450 1A2 (Protein Data Bank code 2HI4) in the MOE software (ver. 2009.10, Chemical Computing Group, Montreal, Canada) for modeling of a three-dimensional structure. Prior to docking, the energy of the P450 structures was minimized using the CHARMM22 force field. Docking simulation was carried out for flavonoid binding to P450 enzymes using the MMFF94x force field distributed in the MOE Dock software. Twenty solutions were generated for each docking experiment and ranked according to total interaction energy (U value).
Results
Inhibition of P450 1B1 by Flavonoid Derivatives
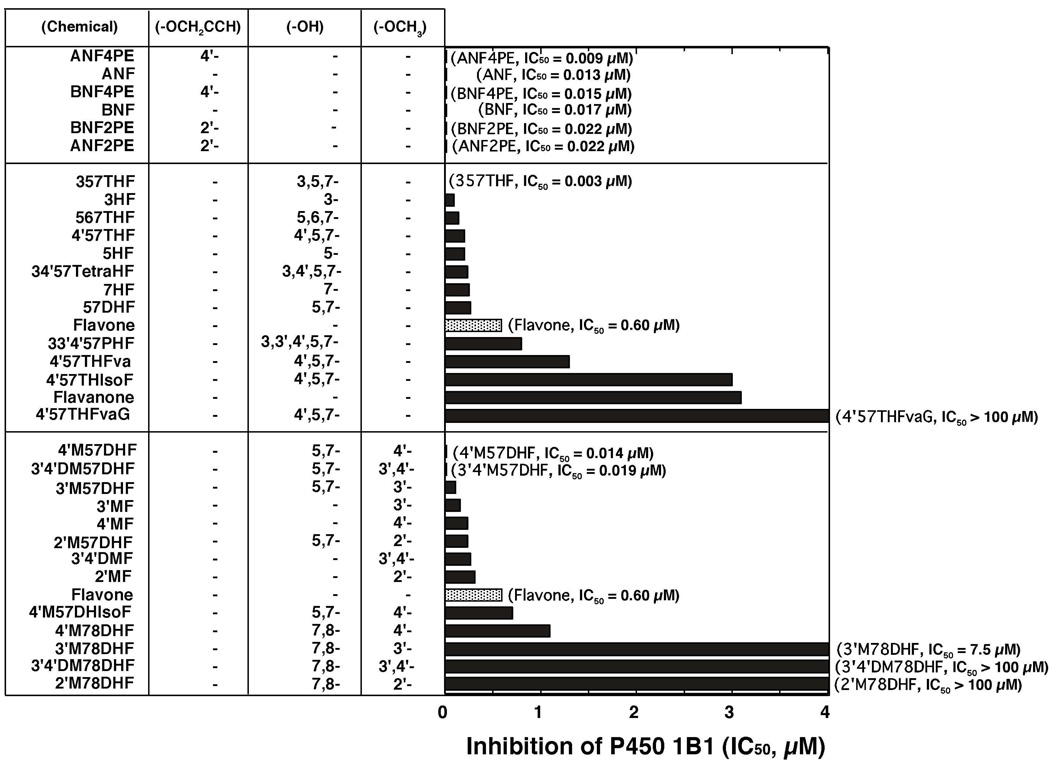
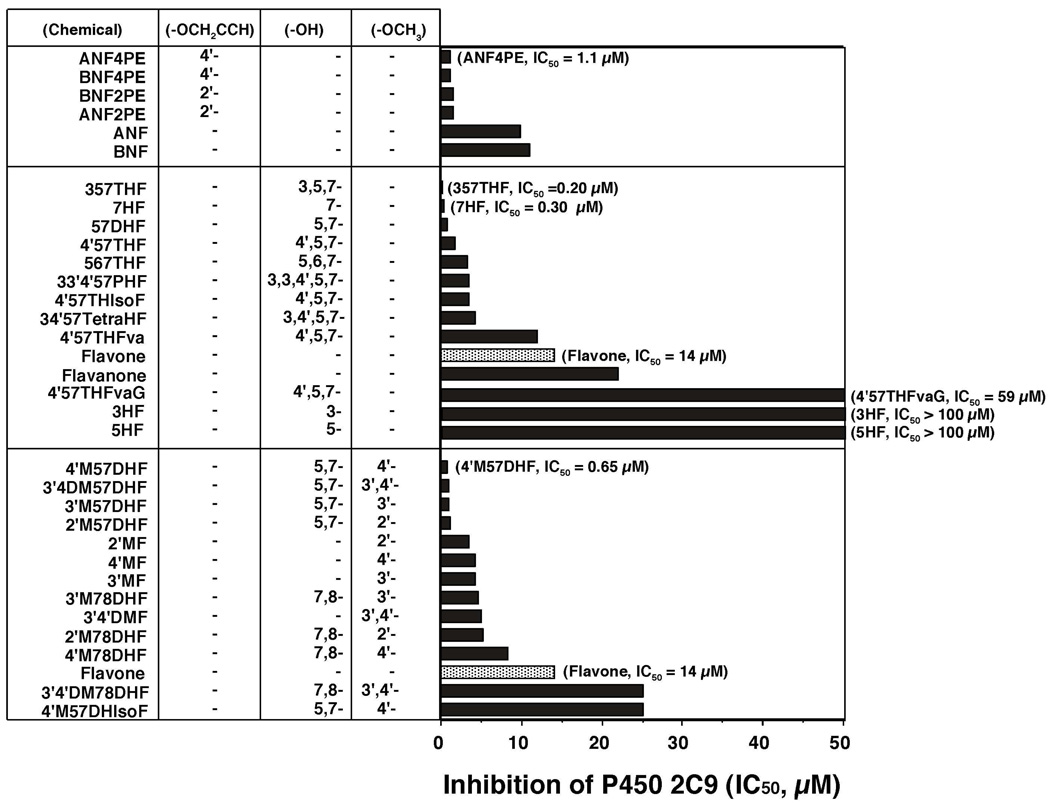
Inhibition of P450 1B1 by 33 flavonoid derivatives was determined in systems containing E. coli membranes expressing both P450 1B1 and NADPH-P450 reductase by measuring EROD activity as a model reaction (Figure 2). In that figure (and also in Figures 5, 6, 8, and 9), the flavonoids were divided into three groups: a) ANF and BNF and their 2’- and 4’-propargyl ethers, b) flavonoids and their hydroxylated derivatives, and c) methoxylated flavonoids (with or without hydroxyl groups). In each group, the chemicals are listed in order from lower to higher IC50 values.
Figure 2.
Inhibition of P450 1B1-dependent EROD activities by flavonoids. Flavonoids are grouped into three sets, i.e. i) ANF and BNF and their propagyl ethers, ii) flavonoids and hydroxylated derivatives, and iii) flavonoids and methoxylated derivatives. The substituted positions of propargyl ether, hydroxyl-, and methoxy- groups are indicated. The IC50 values were obtained from experiments using different concentrations of flavonoids (n = 8–15).
Figure 5.
Inhibition of P450 1A1-dependent EROD activities by various flavonoid derivatives. The IC50 values were obtained from the experiments using different concentrations of flavonoids (n=8~15). Other details are as in Figure 2.
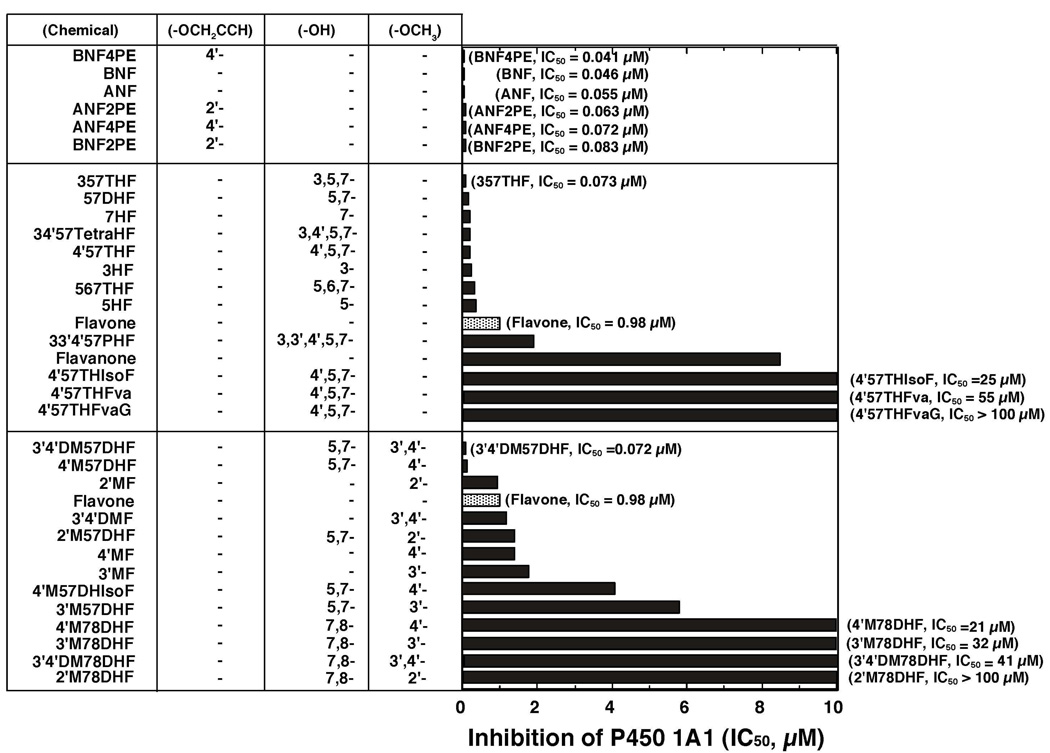
Figure 6.
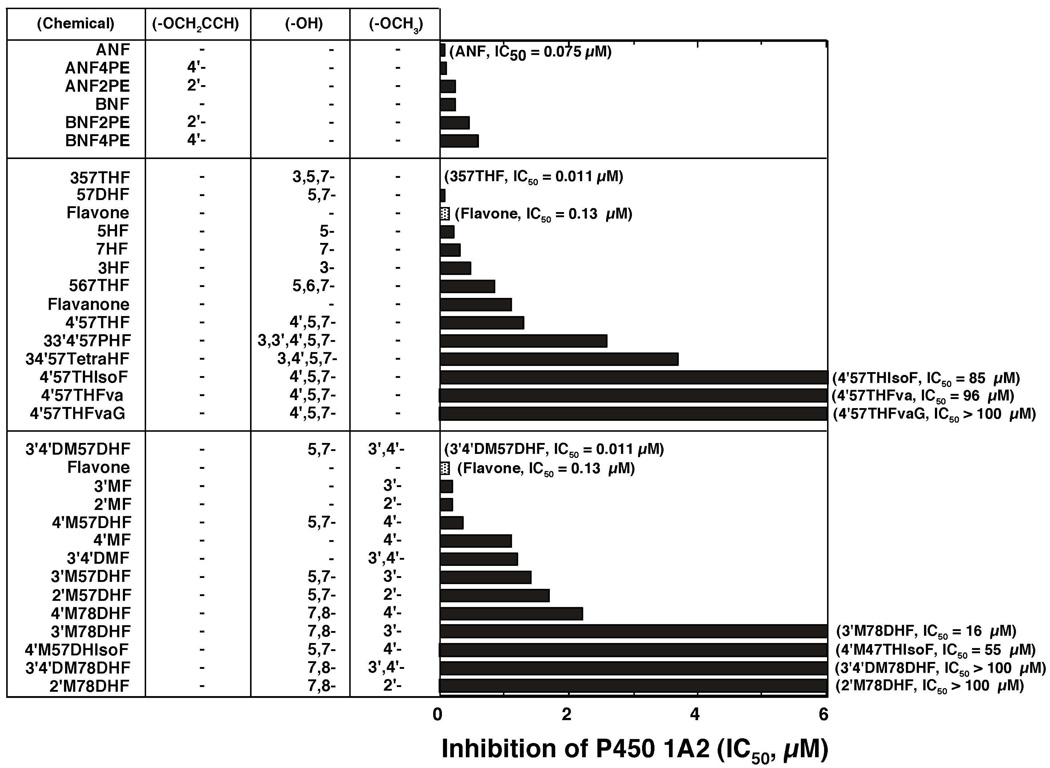
Inhibition of P450 1A2-dependent EROD activities by various flavonoid derivatives. The IC50 values were obtained from the experiments using different concentrations of flavonoids (n=8~15). Other details are as in Figure 2.
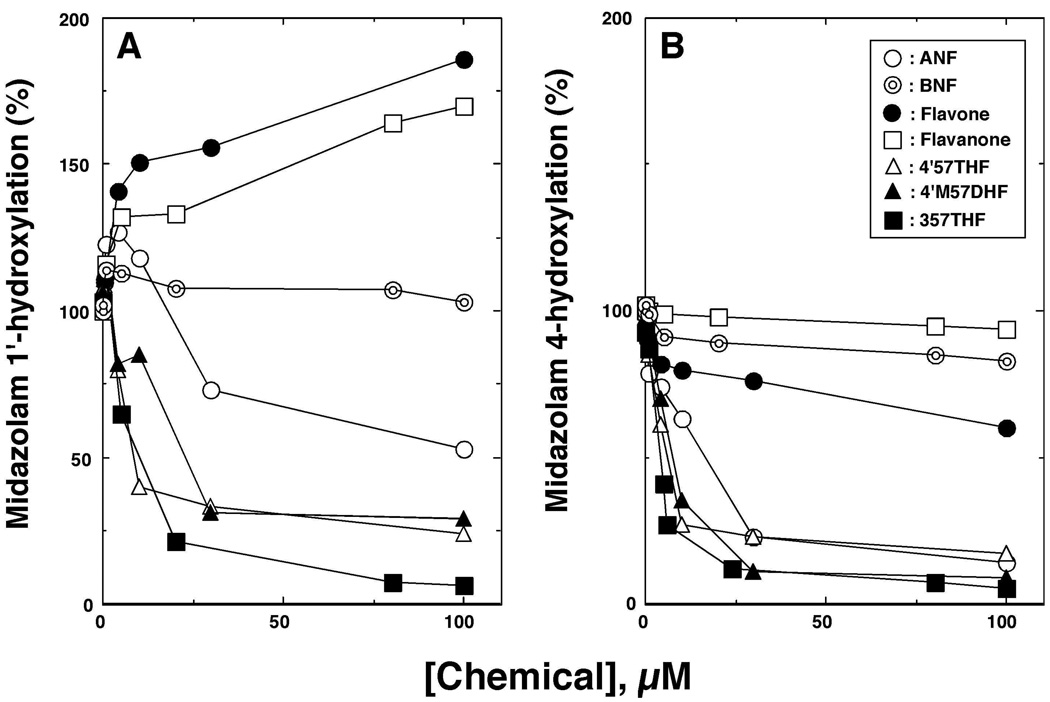
Figure 8.
Effects of several flavonoids on midazolam 1’- (A) and 4-hydroxylation (B) activities catalyzed by P450 3A4.
Figure 9.
Inhibition of P450 3A4-dependent midazolam 4-hydroxylation activities by flavonoid derivatives. The IC50 values were obtained from the experiments using different concentrations of flavonoids (n=6~8). Other details are as in Figure 2.
Both ANF and BNF were very potent inhibitors with IC50 values of 0.013 and 0.017 µM, respectively, and our results showed that four propargyl ether metabolites of ANF and BNF were also potent inhibitors of P450 1B1 (upper panel of Figure 2).
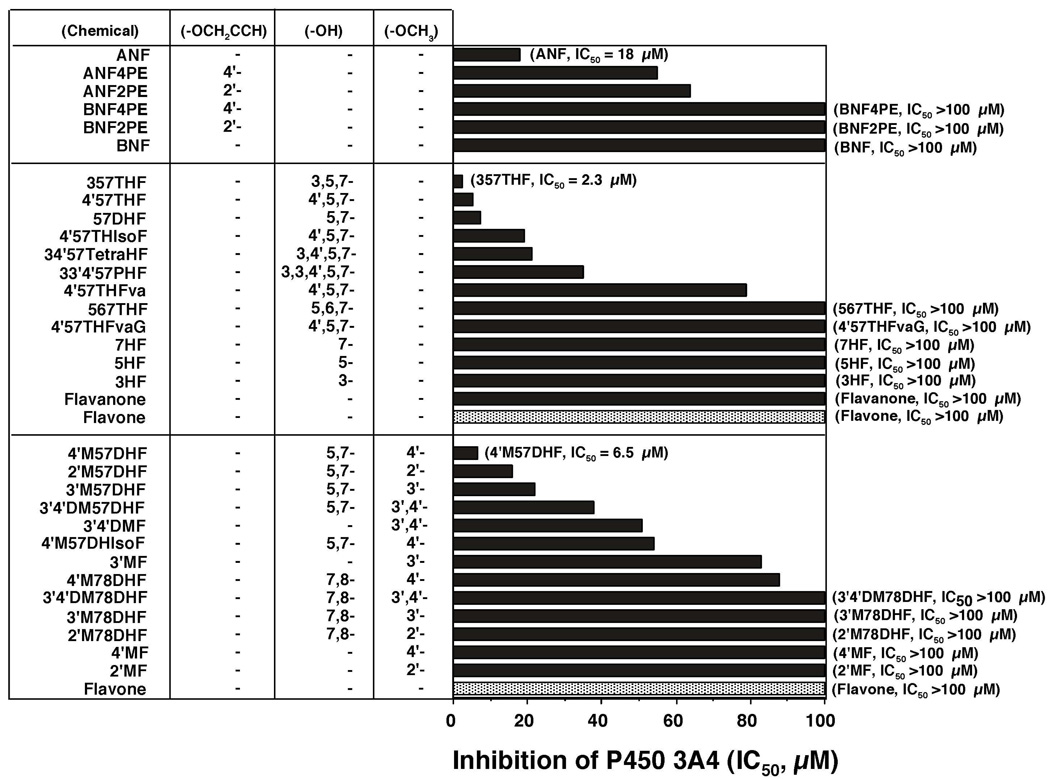
The potencies (IC50 values) for inhibiting P450 1B1-dependent EROD activities by flavone were compared with those of flavonoid derivatives that were substituted with either hydroxyl and/or methoxy groups in the molecules (Figure 2, middle and lower panels, respectively). Hydroxylation of flavone at positions 3-, 5-, 7-, and 5,7- decreased the IC50 values to 15, 35, 42, and 45%, respectively, of that of flavone itself (0.6 µM). 3,5,7THF (galangin) was found to be the most active compound inhibiting P450 1B1, with an IC50 value of 0.003 µM (Figure 2, middle panel). 567THF, 4’57THF, and 34’57tetraHF had IC50 values 23, 33, and 38% of that of flavone. Flavanone, 4’57THF, and 4’57THIsoF were not as potent in inhibiting P450 1B1 as flavone and 4’,5,7-trihydroxyflavone. 4’57THFvaG (naringin) was a weak inhibitor of P450 1B1.
Substitution with 3’-, 4’-, and 2’-methoxy and 3’,4’-dimethoxyl groups on flavone caused decreases in IC50 values from 0.60 µM to 0.15, 0.23, 0.32, and 0.26 µM, respectively (Figure 2, lower part). Interestingly, the addition of 4’-methoxy (acacetin) and 3’,4’-dimethoxy moieties to 57DHF (chrysin) significantly decreased the IC50 values from 0.27 µM to 0.014 and 0.019 µM, respectively. The IC50 values of 3’M57DHF and 2’M57DHF were 0.11 and 0.24 µM, respectively, and 4’M57DHIsoF (biochain A) was not as potent in inhibiting P450 1B1 (IC50 = 0.70 µM) as 4’M57DHF. Our results also showed that 2’M78DHF, 3’M78DHF, 4’M78DHF and 3’4’DM78DHF had decreased potencies towards inhibition of P450 1B1 as compared with 2’MF, 3’MF, 4’MF, and 3’4’DMF.
Spectral Interaction of Flavonoids with P450 1B1
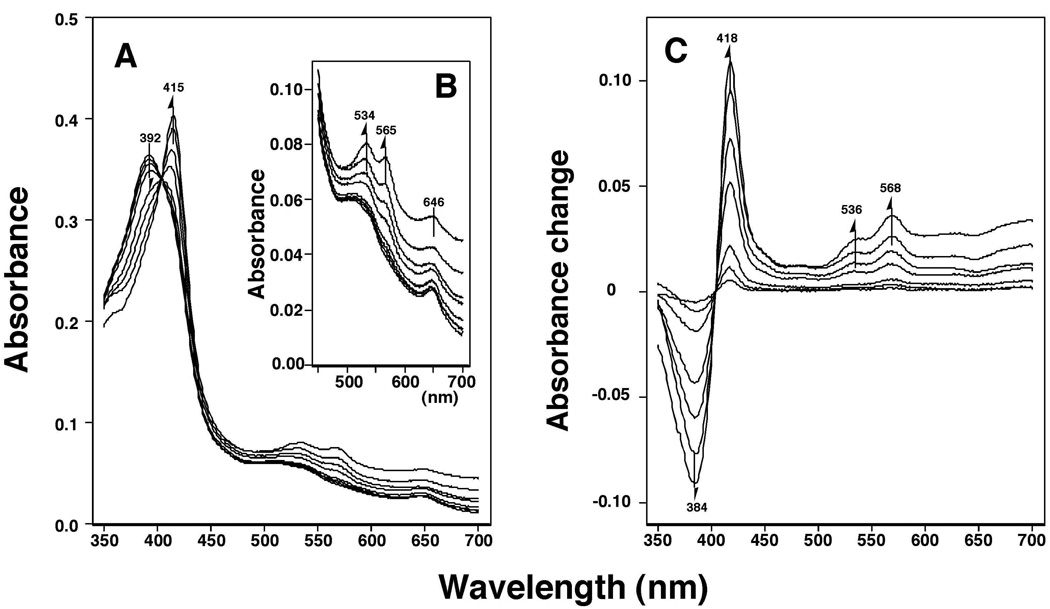
Previously we showed that a variety of chemical inhibitors, including 14 flavonoids and ANF and BNF, induce reverse Type I binding spectra and that the spectral dissociation constants (Ks) and the efficiencies of binding (ΔA/Ks) correlate with the abilities of inhibition (IC50) of P450 1B1 by these chemicals (23). We further studied the spectral interaction and inhibition of P450 1B1 using 33 flavonoids and six other molecules, including ANF and BNF and their derivatives (Figure 3). Flavone produced Reverse Type I binding spectra; the Soret band (peak wavelength) was shifted from 392 to 415 nm and α- and β-bands appeared at 534 and 565 nm, respectively (Figure 3A and 3B). The difference spectra indicated a typical Reverse Type I binding spectra (Figure 3C).
Figure 3.
Flavone-induced Reverse Type I binding spectra of P450 1B1. Purified P450 1B1 was diluted to a final concentration of 3 µM in 0.10 M potassium phosphate buffer (pH 7.4) containing 20% glycerol (v/v) and 10 mM EDTA and the spectra were recorded after addition of various concentrations of flavone. A) Absolute spectra, B) changes in the α- and β-bands, and C) difference spectra of P450 1B1 after addition of flavone.
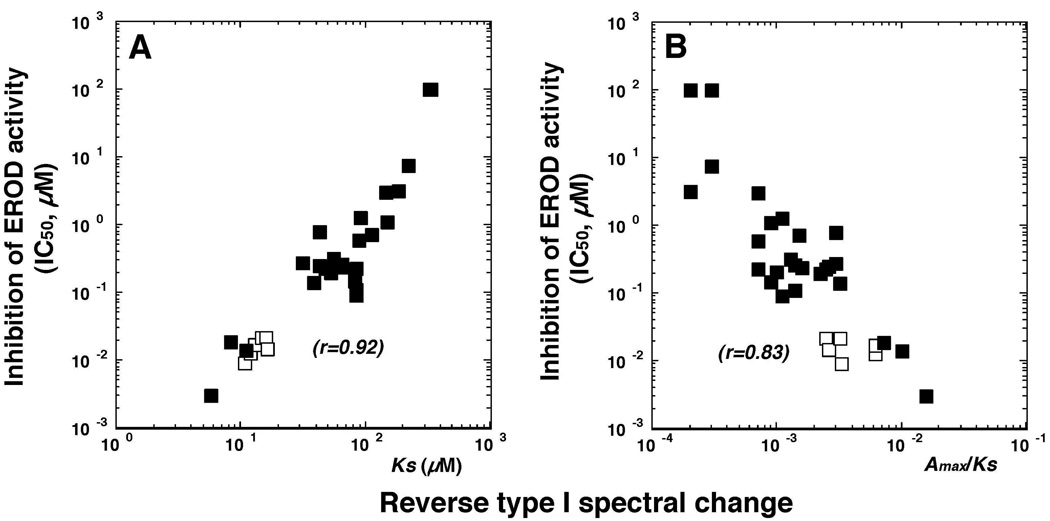
The Ks and ΔA/Ks for interaction of these flavonoids (except 2M78DHF, which did not show spectra) with P450 1B1 were found to be well correlated with the inhibition of EROD activities catalyzed by P450 1B1 (r = 0.92 and 0.83, respectively) (Figure 4). It was also noted that ANF and BNF and their 2’- and 4’-propynyl ether derivatives had high affinities to produce spectral interaction with and to inhibit P450 1B1 (Figure 4, open squares).
Figure 4.
Relationship between efficiency of reverse Type I binding spectra and inhibition of EROD activity of P450 1B1 by various flavonoid derivatives. Since there were no binding spectra with 2’M78DHF, the points indicated were with 32 chemicals. Closed squares (■) are the results from ANF, BNF, and their propargyl ethers and open squares (ρ) are the remaining 26 flavonoids.
Interaction and Inhibition of P450 1A1 and 1A2 with Flavonoids
As reported previously (23), neither P450 1A1 nor 1A2 induced spectral changes in interacting with several of the flavonoid derivatives examined at concentrations up to 100 µM (results not shown).
P450 1A1 EROD activity was strongly inhibited by ANF and BNF and their propargyl ethers; there was little difference in the IC50 values among free and propargyl ether derivatives of ANF and BNF (Figure 5, upper panel).
Among the free and hydroxylated flavone, flavanone, and isoflavone derivatives examined, 357THF (galangin) was the most active in inhibiting P450 1A1, followed by 5,7DHF, 7HF, 34’57TetraF, 4’57THF, 3HF, 567THF, and 5HF; these chemicals were more active than flavone (Figure 5, middle panel). However, 33’4’57PentaHF, flavanone, 4’57THFva, and 4’57THisoF were less potent than flavone. We also found that both 3’4’DM57DHF (IC50 0.072 µM) and 4’M57DHF (IC50 0.10 µM), but not 2’M57DHF (IC50 1.4 µM) or 3’M57DHF (IC50 5.8 µM), were slightly more active than 57DHF (IC50 0.17 µM) in inhibiting P450 1A1 (Figure 5, lower panel). Other methoxylated flavones, e.g. 2’MF, 3’MF, 4’MF, and 3’4’DMF, showed similar abilities to flavone in inhibiting P450 1A1. All of the methoxylated derivatives of 7,8-dihydroxyflavone were less potent than those of the respective 5,7-dihydroxyflavone.
P450 1A2 EROD activity was inhibited by ANF (IC50 0.075 µM), ANF2’PE (IC50 0.24 µM), and ANF4’PE (IC50 0.10 µM) but only weakly by BNF, BNF2’PE, and BNF4’PE (Figure 6, upper panel). As in the cases of P450 1B1 and 1A1, 357THF and 3’4’DM57DHF were potent inhibitors of P450 1A2 with IC50 values of 0.011 µM (in both cases) (Figure 6). Although 57DHF was more potent in inhibiting P450 1A2 than flavone, other hydroxylated flavonoids were not as active as flavone itself. Methoxylated flavones (except for 3’4’DM57DHF) and other derivatives of flavanone and isoflavone were also weak inhibitors as compared with flavone (Figure 6, lower panel).
Comparative Analysis of Inhibition of P450 1A1-, 1A2-, and 1B1-Dependent EROD Activity by Several Flavonoids
Kinetic analysis of the inhibition of P450 1B1 by 357THF, 4’57THFva, and 4’M57DHisoF at concentrations of 0.02, 8.0, and 8.0 µM, respectively, showed that these chemicals inhibited the activities in a mixed-type pattern (results not shown). Similar inhibition mechanisms were also noted for P450 1A1 with 0.08 µM 357THF, 4.0 µM 4’M57DHIsoF, and 20 µM 4’57THFvaG and for P450 1A2 with 0.01 µM 357THF, 40 µM 4’M57DHIsoF, and 40 µM 4’57THFva (results not shown).
Interaction and Inhibition of P450 2C9 with Flavonoids
We examined the spectral interaction of flavonoids with P450 2C9 and found that flavone, 5HF, 7HF, 57DHF, 357THF, 4’57THF, 34’57TetraHF, 4’M57DHF, 4’M57DHIsoF, ANF, and BNF and their propargyl ether derivatives did not induce spectral changes with P450 2C9 at concentrations up to 100 µM (results not shown). On the other hand, P450 2C9 was found to interact with a typical substrate flurbiprofen to produce Type I spectra with a spectral dissociation constant (Ks) of 5.5 µM (results not shown).
Inhibition of P450 2C9 by flavonoids was determined by measuring flurbiprofen 4’-hydroxylation activity (Figure 7). Interestingly, four propargyl ether derivatives of ANF and BNF were found to be more potent in inhibiting P450 2C9 than the parent compounds (Figure 7, upper panel). Several hydroxylated flavonoids, e.g. 357THF, 7HF, 57DHF, 4’57THF, 567THF, 33’4’57PHF, 4’57THIsoF, and 34’57TetraHF were more potent in inhibiting P450 2C9 than flavone, but flavanone, 3HF, 5HF, and 4’57THFvaG were less active (Figure 7, middle panel). The addition of 2’-, 3’, 4’, and 3’,4’-methoxy groups to 57DHF did not produce more active inhibitors of P450 2C9 (Figure 7, lower panel). 2’MF, 3’MF, 4’MF, and 3’4’DMF were slightly more active than flavone in inhibiting P450 2C9.
Figure 7.
Inhibition of P450 2C9-dependent flurbiprofen 4’-hydroxylation activities by flavonoid derivatives. The IC50 values were obtained from the experiments using different concentrations of flavonoids (n=6~8). Other details are as in Figure 2.
Interaction of P450 3A4 with Flavonoids
P450 3A4 oxidized midazolam to form 1’- and 4-hydroxylated metabolites; we found that some of the flavonoids, such as flavone and flavanone, enhanced the formation of 1’-hydroxymidazolam but inhibited the formation of 4-hydroxymidazolam (Figure 8). It was also found that 4’57THF, 4’M57DHF, and 357THF significantly inhibited P450 3A4-catalyzed midazolam 4-hydroxylation but slightly activated the formation of 1’-hydroxylation at lower concentration (Figure 8).
Because none of the 33 flavonoids used here activated—but mostly inhibited—the midazolam 4-hydroxylation activity, the inhibitory potencies of these flavonoids towards P450 3A4 were compared (Figure 9). ANF inhibited midazolam 4-hydroxylation with an IC50 value of 18 µM, and ANF2’PE and ANF4’PE were found to be less active in inhibiting P450 3A4 (IC50 values of 64 and 55 µM, respectively). BNF and its derivatives were not active inhibitors of P450 3A4. Both flavone and flavanone were weak in inhibiting P450 3A4 with IC50 values >100 µM (Figure 9). As in the cases of other P450s, 357THF was the most potent inhibitor of P450 3A4 with an IC50 of 2.3 µM. 4’57THF, 57DHF, 4’57THIsoF, 3,4’57TetraHF, and 3,3’4’57PHF inhibited P450 3A4 with IC50 values between 5.2 and 35 µM. 3HF, 5HF, 7HF, 4’57THF, and 567THF were rather inactive in inhibiting P450 3A4, with IC50 values >100 µM. 57DHF inhibited midazolam 4-hydroxylation (IC50 7.4 µM) and 2’M57DHF, 3’M57DHF, and 3’4’DM57DHF were found to inhibit P450 3A4, with IC50 values of 2, 16, 6.5, and 38 µM, respectively. 2’MF, 3’MF, 4’MF, and 3’4’DMF were not very effective inhibitors of P450 3A4.
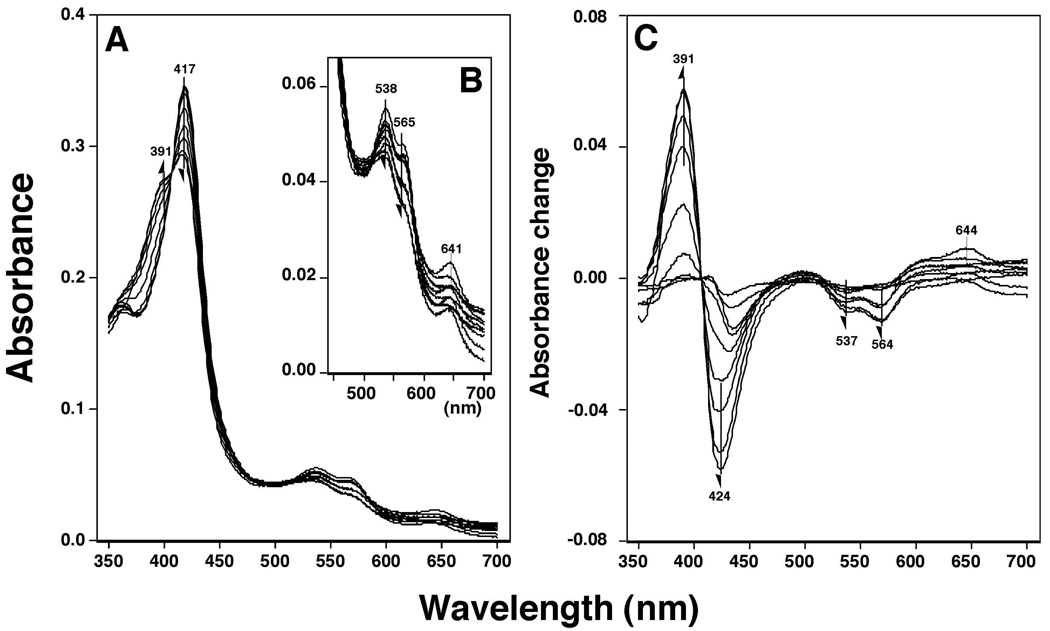
Flavone produced a Type I spectral change with P450 3A4 showing a spectral shift of the Soret peak from 417 to 391 nm and decreases in both the α and β peaks at 538 and 565 nm, respectively (Figures 10A and 10B). The Ks (µM) and ΔAmax values obtained from difference spectra (Soret band) were determined to be 380 ± 81 µM and 0.12 ± 0.02, respectively (Figure 10C). Because most of other flavonoids had spectral bands near the Soret peak and interfered with the spectral measurements at higher concentrations, we monitored the decreases in the α and β bands for interaction of several of the flavonoid derivatives with P450 3A4 (Figure 10B). Our results showed that 357THF, 2’MF, 3’MF, 4’MF, 3’57DHF, and 4’M57DHF induced spectral changes, but flavanone, 57DHF, 4’57THF, 3’4’DM57DHF, and 3’4’DM78DHF did not even at 100 µM concentrations.
Figure 10.
Flavone-induced Type I binding spectra with P450 3A4. Purified P450 3A4 was diluted to a final concentration of 3 µM in 0.10 M potassium phosphate buffer (pH 7.4) containing 20% glycerol (v/v) and 10 mM EDTA, and spectra were recorded after addition of various concentrations of flavone. A) Absolute spectra, B) changes in α- and β-bands, and C) difference spectra of P450 3A4 with flavone.
ANF, ANF2’PE, ANF4’PE, and BNF also interacted with P450 3A4 and yielded Type I binding spectra with Ks values of 6.5, 3.7, 6.8, and 6.3 µM respectively; the values much lower compared with those of flavone and testosterone (260 µM). BNF2PE and BNF4PE did not produce Type I binding spectra with P450 3A4 (results not shown).
Correlation of inhibition of P450s 1B1, 1A1, 1A2, 2C9, and 3A4 with 33 flavonoids
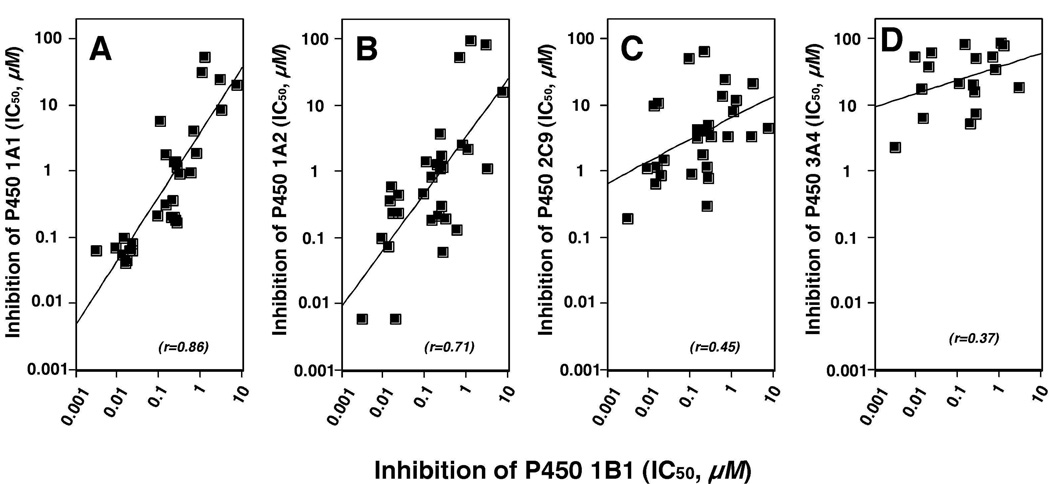
All of the above results suggested that these flavonoids inhibit substrate oxidations catalyzed by the five P450s examined, depending on the flavonoid and the P450 used. We compared IC50 values for inhibition of P450 1B1 with those for inhibition of P450s 1A1, 1A2, 2C9, and 3A4 by 33 flavonoids, except for the compounds with IC50 values >100 µM (Figure 11). The correlation between IC50 values of P450 1B1 and 1A1 with these flavonoids was very high (r = 0.86), followed by P450 1A2 (r = 0.71), P450 2C9 (r = 0.45), and P450 3A4 (r = 0.37). The r value was 0.75 for comparison between P450 1A1 and 1A2.
Figure 11.
Correlation of inhibition of P450 1B1 with inhibition of P450 1A1 (A), 1A2 (B), 2C9 (C), and 3A4 (D) by various flavonoids. Some of the 33 flavonoids inhibited very weakly (IC50 values >100 µM) and were deleted from the analysis.
Docking Simulation of the Interaction of Flavonoids with Human P450 Enzymes
Molecular docking studies were performed on six selected flavonoids with human P450 enzymes using the reported crystal structures of P450 1A2 (37), P450 2C9 (38), and P450 3A4 (39). The structures of P450s 1A1 and 1B1 were constructed by homology modeling based on the crystal structure of P450 1A2 (37). The six flavonoids examined were five active inhibitors—flavone, 57DHF, 357THF, 4’M57DHF, 3’4’DM57DHF—and one inactive inhibitor 3’4’DM78DHF. In the figures, the amino acid residues in the substrate recognition site (SRS) are indicated based on the results of interaction of ANF with P450 1A2 (37), flurbiprofen with P450 2C9 (38), and erythromycin with P450 3A4 (39) (Figures 12–14). The aligned amino acid residues of P450 1A1 and 1B1—based on the amino acid sequence of P450 1A2 (37)—are also indicated in Figures 15 and 16, respectively.
Figure 12.
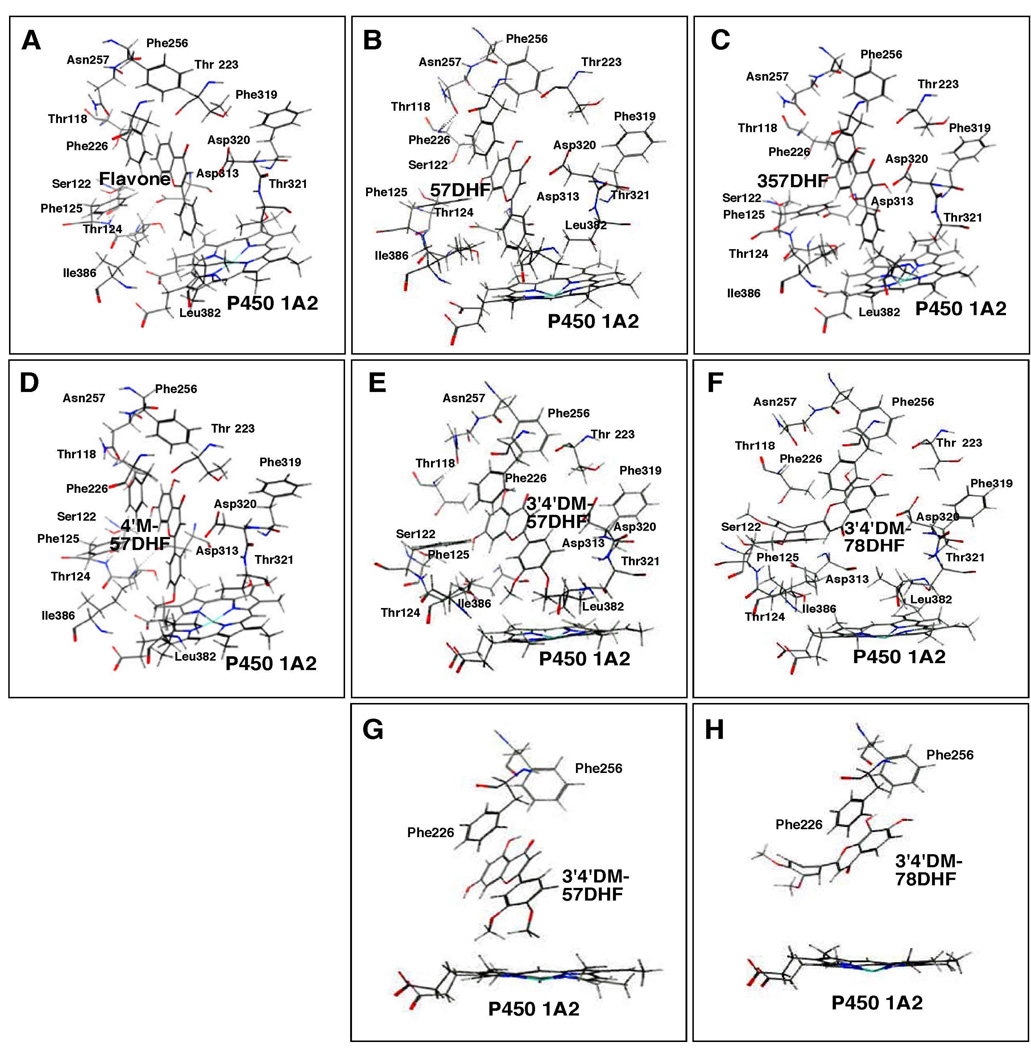
Docking simulation of flavone (A), 57DHF (B), 357THF (C), 4’M57DHF (D), 3’4’DM57DHF (E), and 3’4’DM78DHF (F) into P450 1A2. Different views of interaction of 3’4’DM57DHF (G) and 3’4’DM78DHF (H) with the active site of P450 1A2 are also shown in the figure. The heme group of the P450 is shown at the lower part of each of the figure and the amino acid residues that may interact with these flavonoids are presented. In the figure, oxygen, nitrogen, sulfur, and iron atoms are colored with red, blue, yellow, and light blue, respectively.
Figure 14.
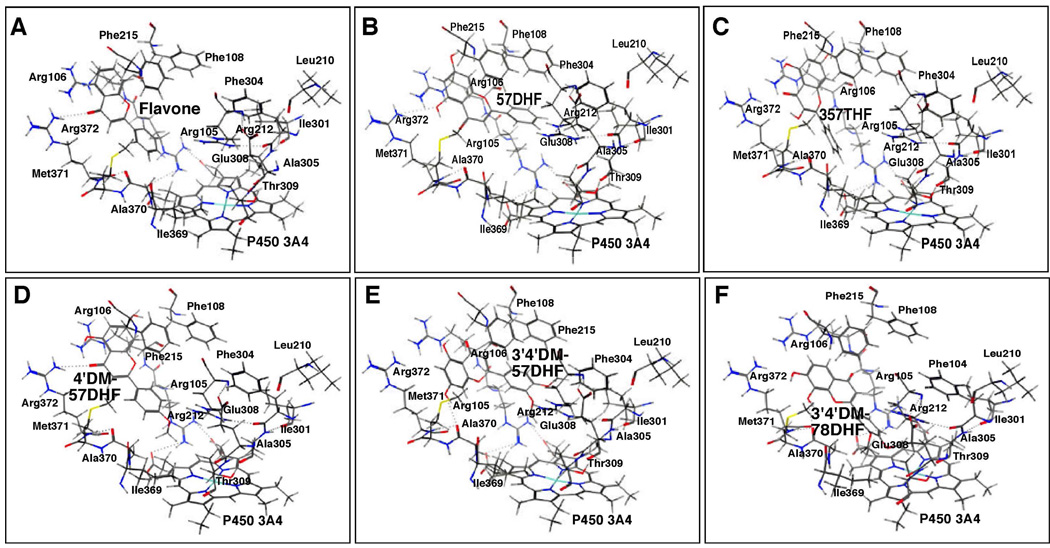
Docking simulation of flavone (A) and 57DHF (B), 357THF (C), 4’M57DHF (D), 3’4’DM57DHF (E), and 3’4’DM78DHF (F) into P450 3A4. Other details are the same as in the legend to Figure 12.
Figure 15.
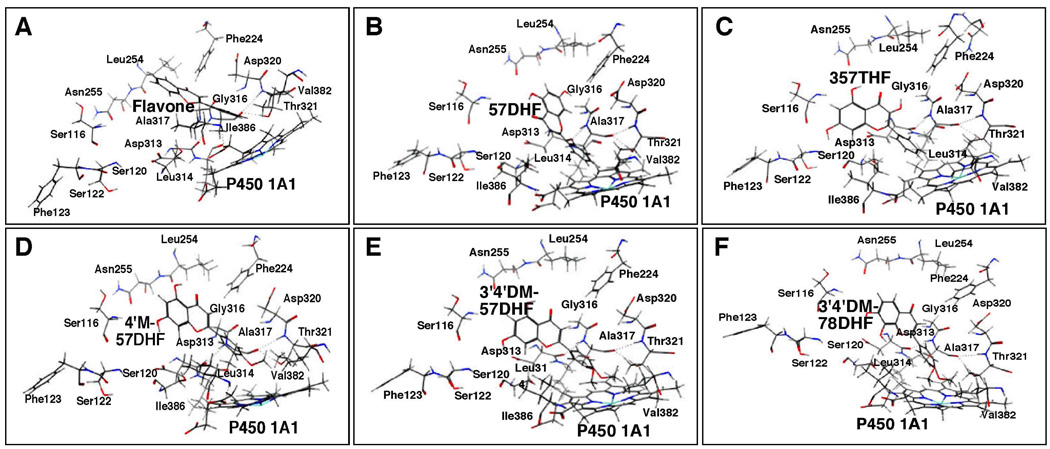
Docking simulation of flavone (A) and 57DHF (B), 357THF (C), 4’M57DHF (D), 3’4’DM57DHF (E), and 3’4’DM78DHF (F) into P450 1A1. Other details are the same as in the legend to Figure 12.
Figure 16.
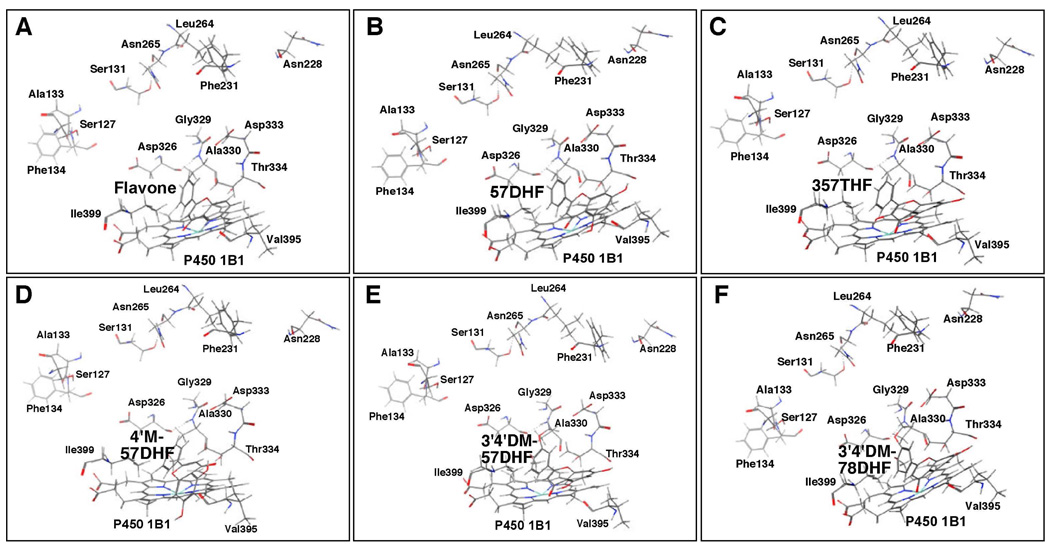
Docking simulation of flavone (A) and 57DHF (B), 357THF (C), 4’M57DHF (D), 3’4’DM57DHF (E), and 3’4’DM78DHF (F) into P450 1B1. Other details are the same as in the legend to Figure 12.
Flavone, 57DHF, 357THF, 4’M57DHF, 3’4’DM57DHF, and 3’4’DM78DHF were docked into P450 1A2 and the results showed that the former five flavonoids (except for 3’4’DM78DHF) gave similar orientations in interacting with the iron center of the heme in P450 1A2 (Figures 12A–12E). In these former five cases, the B-ring of the flavonoid molecules was placed near the active site of P450 1A2 and the flavonoid molecules were surrounded by amino acid residues Thr-118, Ser-122, Thr-124, and Phe-125 in SRS1, Thr-223 and Phe-226 in SRS2, Phe-256 and Asn-257 in SRS3, Asp-313, Phe-319, Asp-320, and Thr-321 in SRS 4, and Leu-382 and Ile-386 in SRS5 which have been reported to be key amino acids in the interaction of ANF with P450 1A2 (37). The ligand-P450 interaction energies (U values) were −25.2, −27.7, −21.7, −19.4, and −9.5 for flavone, 57DHF, 357DHF, 4’M57DHF, and 3’4’DM57DHF, respectively (Table 1). These U values were not significantly correlated with the IC50 values for the inhibition of P450 1A2 (Table 1). 3’4’DM78DHF, which did not inhibit P450 1A2 at a 100 µM concentration, was docked into P450 1A2 with a different orientation (Figures 12F and 12H) than seen in the former five flavonoids including 3’4’DM57DHF (Figures 12E and 12G); the 3’4’DM78DHF molecule seemed to be blocked by various amino acids in SRS1~SRS4 from entering into the active site cavity. Thus the U-value was very high (256) as compared with other flavonoids (Table 1).
Table 1.
Relationship between inhibition of substrate oxidation by P450s (IC50) and flavonoid-P450 interaction energy (U)
| P450 | flavonoid | inhibition of substrate oxidation by P450s (IC50, µM) | flavonoid-P450 interaction energy (U, kcal mol−1) |
|---|---|---|---|
| P450 1A1 | flavone | 0.98 ± 0.21 | −26.4 |
| 57DHF | 0.17 ± 0.03c) | −32.6 | |
| 357DHF | 0.073 ± 0.02c) | −32.6 | |
| 4’M57DHF | 0.10 ± 0.02c) | −33.5 | |
| 3’4’DM57DHF | 0.072 ± 0.011c) | −22.2 | |
| 3’4’DM78DHF | 41 ± 10c) | −20.4 | |
| P450 1A2 | flavone | 0.13 ± 0.03 | −25.2 |
| 57DHF | 0.060 ± 0.021 | −27.7 | |
| 357DHF | 0.011 ± 0.003c) | −21.7 | |
| 4’M57DHF | 0.36 ± 0.04 | −19.4 | |
| 3’4’DM57DHF | 0.011 ± 0.003c) | −9.5 | |
| 3’4’DM78DHF | >100* | 255 | |
| P450 1B1 | flavone | 0.60 ± 0.021 | 2.9 |
| 57DHF | 0.27 ± 0.041a) | 6.5 | |
| 357DHF | 0.003 ± 0.001c) | 24.4 | |
| 4’M57DHF | 0.014 ± 0.003c) | 40.3 | |
| 3’4’DM57DHF | 0.019 ± 0.003c) | 39.2 | |
| 3’4’DM78DHF | >100* | 40.7 | |
| P450 2C9 | flavone | 14 ± 1.2 | −25.3 |
| 57DHF | 0.80 ± 0.14c) | −31.9 | |
| 357DHF | 0.20 ± 0.03c) | −32.6 | |
| 4’M57DHF | 0.65 ± 0.13c) | −20.8 | |
| 3’4’DM57DHF | 0.88 ± 0.21b) | −25.8 | |
| 3’4’DM78DHF | 25 ± 4.2a) | −30.0 | |
| P450 3A4 | flavone | >100* | −30.7 |
| 57DHF | 7.4 ± 1.1 | −31.9 | |
| 357DHF | 2.3 ± 0.3 | −32.4 | |
| 4’M57DHF | 6.5 ± 1.1 | −36.9 | |
| 3’4’DM57DHF | 38 ± 5.9 | −38.5 | |
| 3’4’DM78DHF | >100* | −33.7 |
IC50 values were obtained by measuring 7-ethoxyresorufin O-deethylation activities for P450s 1A1, 1A2, and 1B1, flurbiprofen 4’-hydroxylation for P450 2C9, and midazolam 4-hydroxylation for P450 3A4. Data for IC50 values represent means ± SD.
indicates that IC50 > 100 µM.
P<0.05,
P<0.01, and
P<0.001, significantly different as compared with the IC50 values of flavone.
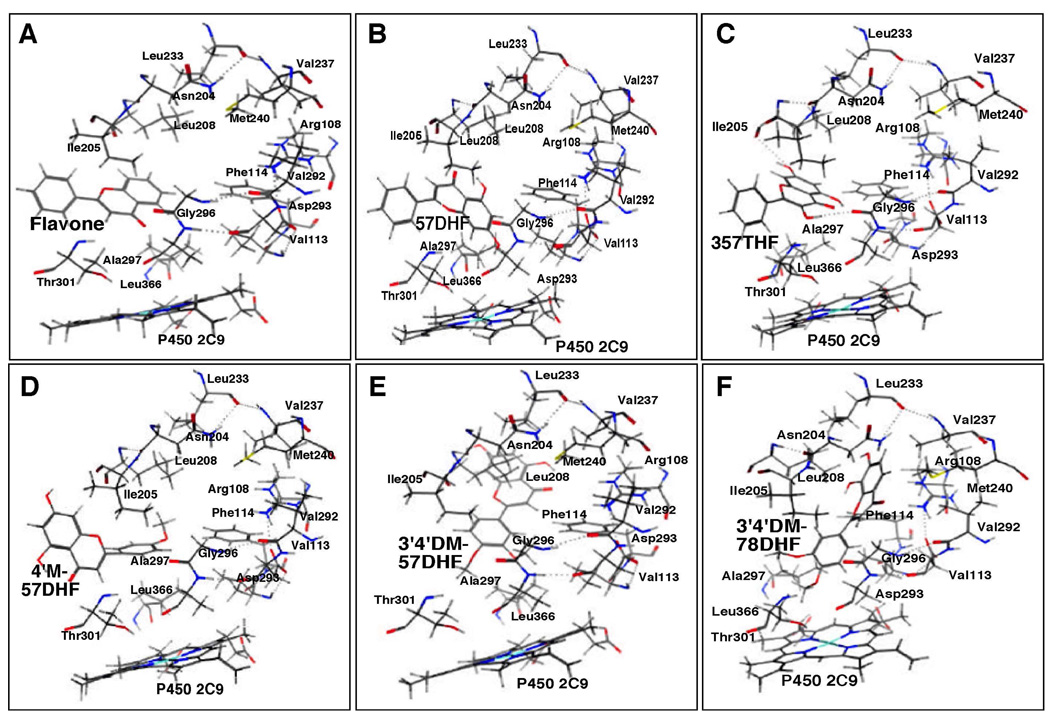
The A- and/or C-rings of flavone, 57DHF, and 357THF were docked into the active site of P450 2C9 with U- values of −25.3, −31.9, and −32.6, respectively (Figures 13A–13C and Table 1). Substitution of 4’-methoxy and 3’,4’-dimethoxy groups into 57DHF and 78DHF caused an interaction of the B-ring of these flavonoids with the active site of P450 2C9 (Figures 13D–13F). All of six flavonoids were surrounded by Arg-108, Val-113, and Phe-114 in SRS1, Asp-204, Ile-205, and Leu-208 in SRS2, Leu-233, Val-237, and Met-240 in SRS3, Val-292, Asp-293, Gly-296, Ala-297, and Thr-301 in SRS4, and Leu-366 in SRS5; these amino acids have been identified in the interaction of flurbiprofen with P450 2C9 (38).
Figure 13.
Docking simulation of flavone (A) and 57DHF (B), 357THF (C), 4’M57DHF (D), 3’4’DM57DHF (E), and 3’4’DM78DHF (F) into P450 2C9. Other details are the same as in the legend to Figure 12.
Our results also showed that the B-rings of flavone, 57DHF, 357THF, 4’M57DHF, and 3’4’DM57DHF were docked into the active site of P450 3A4 (Figure 14A–14E). 3’4’DM78DHF, which was not inhibitory for 3A4 at a 100 µM concentration, was also docked into P450 3A4 in a similar fashion to the hydroxyl and/or methoxy flavones (Figure 14F). These six flavonoids were found to be surrounded by Arg-105, Arg-106, and Phe-108 in SRS1, Leu-210 and Arg-212 in SRS2, Ile-301, Phe-304, Ala-305, Glu-308, and Thr-309 in SRS4, and Ile-369, Ala-370, Met-371, and Arg-372 in SRS5; these amino acids have been identified in interaction of erythromycin with P450 3A4 (39).
The results of molecular docking with P450 1A1 showed that the B-rings of flavone, 57DHF, 357THF, 4’M57DHF, 3’4’DM57DHF, and 3’4’DM78DHF were docked into the active site of P450 (Figure 15). The U values obtained were between −20.4 and −33.5 in these cases, and we did not find any correlation between these U values and IC50 values for the inhibition of P450 1A1 by six flavonoids (Table 1). These flavonoid molecules were surrounded by Ser-116, Ser-120, Ser-122, and Phe-123 in SRS1, Phe-224 in SRS2, Leu-254 and Asn 255 in SRS3, Asp 313, Leu-314, Gly-316, Ala-317, Asp-320, and Thr-321 in SRS4, and Val 382 and Ile-386 in SRS5 (Figure 15).
Molecular docking was also examined in the interaction of six flavonoids with P450 1B1 and we found that the A- and/or C-rings of flavone, 57DHF, 357THF, 4’M57DHF, 3’4’DM57DHF, and 3’4’DM78DHF were placed near at the active site of P450 1B1 and that the interaction energy (U value) was very high compared with those seen in other P450 enzymes (Figure 16 and Table 1). The flavonoid molecules were surrounded by several amino acids in SRS4 and SRS5 but were placed relatively far from the amino acids in SRS1, SRS2, and SRS3 (Figure 16). Our results also showed that P450 1B1 may have a relatively widen pocket for flavonoids, which were associated with several intermolecular hydrogen bonds to amino acid residues around the heme, even with high interaction energy.
Discussion
We first characterized structure-function relationships of inhibition of P450 1B1-dependent EROD activities by flavonoid derivatives, because 32 of 33 flavonoid derivatives interacted with this enzyme to produce reverse Type I binding spectra and the spectral dissociation constant (Ks) and the efficiency of binding (ΔAmax/Ks) correlated well with their inhibitory potencies toward P450 1B1. Comparisons between structures of flavonoids, efficiencies of the spectral binding with P450 1B1, and inhibition potencies of P450 1B1 EROD activities indicated that i) the introduction of 5,7-dihydroxyl groups into flavone increased the potency of inhibition towards P450 1B1, ii) further addition of a 3-hydroxyl group into 57DHF (chrysin) yielded a highly inhibitory flavonoid, 357THF (galangin) (the inhibition potency with 357THF was compatible to those of potent inhibitors, such as ANF, BNF, and their 2- and 4-propargyl ethers), and iii) the addition of 4’-methoxy-and 3’,4-dimethoxy groups to 57DHF produced potent inhibitors of P450 1B1 (i.e., 4’M57DHF and 3’4’DM57DHF). The higher inhibition potencies produced by these 4’-mono- and 3’,4’-dimethoxyl groups in the B-ring of 57DHF suggest the possible roles of substituted hydroxyl and/or methoxy groups in interacting with the active site of P450 1B1. The decreased affinities of 2’M78DHF, 3’M78DHF, 4’M78DHF, and 3’4’DM78DHF (as compared with the respective 57DHF analogues) with P450 1B1 further support the importance of hydroxylation position on the A-ring of flavone.
Our current studies also showed that very potent P450 1B1 inhibitors (e.g., 57DHF, 357THF, and 3’4’DM57DHF) are active inhibitors of P450s 1A1 and 1A2 and that several weak P450 1B1 inhibitors (e.g., 2’M78DHF, 3’M78DHF, 4’M78DHF, and 3’4’DM78DHF) inhibited P450 1A1 and 1A2 at slower rates than the respective analogues (e.g., 2’M57DHF, 3’M57DHF, 4’M57DHF, and 3’4’DM57DHF), indicating that the chemical structures of flavonoids affect their inhibitory actions towards these P450 enzymes. The inhibition patterns of P450 1B1 with 33 flavonoid derivatives (except for flavonoids whose IC50 values >100 µM) were compared with those of P450s 1A1 and 1A2 and the correlation coefficients (r) for inhibition potencies with these P450 enzymes were 0.88 and 0.71, respectively; the values were higher than those seen with P450 2C9 (r = 0.45) and P450 3A4 (r = 0.37) (Figure 11), indicating that similar mechanisms may exist by which these flavonoids inhibit P450s 1A1, 1A2, and 1B1 (18–20). These lines of evidence support the view that the presence of a 5,7-dihydroxyl group in the A-ring of the flavone molecule increases the potency to inhibit P450 1-dependent EROD activity and that the further introduction of a 3-hydroxyl group into the C-ring of 57DHF produces a highly inhibitory flavonoid, 357THF. Our results also showed that introduction of a 3’,4’-dimethoxy group into the B-ring of 57DHF (but not 78DHF) produced another highly inhibitory flavonoid, 3’4’DM57DHF (active towards P450 1A1, 1A2, and 1B1) but 3’4’DM78DHF was not very inhibitory of P450 1 catalytic activities.
In order to understand the basis of structural requirements for flavonoid derivatives to inhibit P450 1 enzymes, we examined the molecular docking of selected six flavonoids (flavone, 57DHF, 357DHF, 4’M57DHF, and 3’4’DM57DHF) as relatively strong inhibitors and 3’4’DM78DHF (as a weak inhibitor) with the active sites of P450 1 enzymes using the reported crystal structure of P450 1A2 and structures of P450 1A1 and 1B1 constructed by homology modeling based on the P450 1A2 structure (37). Our molecular docking studies indicate that there are different mechanisms in the interaction of inhibitory flavonoid derivatives with P450 1A1, 1A2, and 1B1. In the case of P450 1A2, the B-rings of flavone, 57DHF, 357THF, 4’M57DHF, and 3’4’DM57DHF could be docked into the iron center of the P450 heme and were surrounded by amino acids in SRS1–SRS4, but the weak inhibitor 3’4’DM78DHF was docked into P450 1A2 with a different orientation than seen in the former five flavonoids; the 3’4’DM78DHF molecule seemed to be blocked by various amino acids in SRS1–SRS4 from entering into the active site cavity and thus the U-value was found to be very high (256), compared with the other five flavonoids. It can be speculated that the phenyl ring of Phe-226 on helix F in P450 1A2 produces a parallel substrate binding surface as it is in contact with the substrate binding cavity at the active site (37). A possible reasons is that the apparent increased affinity of 57DHF toward P450 1A2 may be due to the presence of phenyl ring in Phe-226 for the observed π-π stacking with the meta-hydroxyl substitution of the 57DHF derivatives (Fig. 12B). This good interaction could not be achieved by ortho-substitutions of 78DHF derivatives.
In the case of P450 1A1, although B-rings of flavone, 57DHF, 357DHF, 4’M57DHF, and 3’4’DM57DHF were docked into the active heme moiety (as in a case of P450 1A2), the weak inhibitor 3’4’DM78DHF was also oriented in a similar fashion as in cases of the other five flavonoids and the U value was not very different with those of other flavonoids. Our molecular docking studies also showed that there are different mechanisms in the interaction of six flavonoids with P450 1B1 in comparison with those of P450 1A1 and 1A2, based on the following lines of evidence. First, the A-rings of all of the six flavonoids docked into the active site of P450 1B1 with essentially similar orientations, despite the fact that there were large differences in IC50 values in inhibiting P450 1B1 catalytic activity. Second, the interaction energy (U value) was very high as compared with those in other P450 enzymes, indicating that interaction between these flavonoids and the active site of P450 1B1 is not so tight as to inhibit the catalytic activity. In addition, the U values were not correlated with the IC50 values for inhibition of P450 1B1 in these six flavonoids. Finally, molecular docking results showed that these six flavonoids were located near the SRS4 and SRS5 regions but were far from the amino acids of SRS1, SRS2, and SRS3, indicating that they do not enter the active site of P450 1B1 with a favorable orientation and that inhibition may occur at different sites rather than the active site of P450 1B1. The presence of two or more binding sites in P450 1B1 for these flavonoids may be considered to explain how these flavonoids inhibit P450 1B1 catalytic activities. The molecular docking studies also indicated that P450 1B1 may have a relatively widen pocket for flavonoids which are associated with several intermolecular hydrogen bonds to amino acid residues around the heme, even with high interaction energy. The above results collectively indicate that there are different mechanisms in the interaction of flavonoids with P450 1A1, 1A2, and 1B1 despite the fact that relatively similar inhibition patterns for flavonoids exist with these P450 enzymes. One of the mechanisms of inhibition of P450 1A2 is suggested to be due to the direct interaction of flavonoids with the active site of P450 1A2, although such mechanisms must be ruled out for P450 1A1 and 1B1 because of the observation of possible interaction of 3’4’DM78DHF, a weak inhibitor, with these P450s.
357THF, 7HF, 57DHF, 4’M57DHF, 3’4’DM57DHF, 3’M57DHF, and 2’M57DHF were also found to be strong inhibitors of P450 2C9; however 3HF and 5HF were very weak in inhibiting P450 2C9 even at 100 µM concentration. Molecular docking studies using the reported crystal structure of P450 2C9 (38) revealed that the A- and/or C-rings of flavone, 57DHF, and 357THF could be dockedinto active site of P450 2C9, while the substitution of 4’-dimethoxy- and 3’,4’-dimethoxy groups into 57DHF and 78DHF caused an interaction at the B-rings with P450 2C9. Si et al. (13) recently reported that 4’57THF, 567THF, and 33’4’57PenHF could interact with P450 2C9 with a similar binding site as that of the typical substrate flurbiprofen, but the non-competitive inhibitor 6-hydroxyflavone has different inhibition characteristics with P450 2C9.
Of the five P450s examined thus far, P450 3A4 was found to be the most resistant to inhibition by these flavonoids, e.g. the IC50 values of P450 3A4 with 357THF, 3’4’DM57DHF, 4’M57DHF, and 57DHF were 11-, 43-, 10-, and 9-fold higher, respectively, than those of P450 2C9 and were 760-, 2000-, 4600-, and 27-fold higher, respectively, than those for P450 1B1. Interestingly, several of these flavonoids enhanced P450 3A4-catalyzed midazolam 1’-hydroxylation at low concentrations. P450 3A4 has been shown to be activated in vitro by various endogenous and xenobiotic chemicals through homo- and heterotropic cooperativity (40,41). Mechanisms underlying the enhancement of P450 3A4-supported catalytic activities have been extensively studied and several studies support the view that two or more of these chemicals fit into the binding site of P450 3A4, thus causing activation as well as inhibition of enzyme activity (42,43). Molecular docking studies with the reported P450 3A4 structure (39) revealed that the B-rings of all of the six flavonoids could be docked into the active site of P450 3A4 with U-values between −30.7 and −38.5. A very weak inhibitor, 3’4’DM78DHF, was also docked into the active site of P450 3A4 in a similar fashion with five other flavonoids, indicating that inhibition of P450 3A4 by flavonoids occurs at different sites of interaction rather than the active site of P450 3A4. The presence of two or more binding sites in P450 3A4 for substrates and inhibitors has been reported (44–46).
ANF has long been known to be a potent inhibitor of family 1 P450 enzymes (18,19,47), and it was found in this study that ANF and BNF and their 2’- and 4’-propargyl ether metabolites strongly inhibit P450s 1B1, 1A1, and 1A2. Among these three enzymes, P450 1B1 was more susceptible to these compounds and the inhibitory potential was not changed by substitution of propargyl ether groups on the ANF and BNF molecules. This was also the case for P450 1A1, but ANF was found to be slightly more active than BNF in inhibiting P450 1A2. An interesting finding is the observation that the 2’- and 4’-propargyl ether derivatives of ANF and BNF were more potent in inhibiting P450 2C9 than the parent structures and that BNF and its derivatives—but not ANF and its derivatives—were very weak inhibitors of P450 3A4. The latter finding was somewhat unexpected because BNF produced Type I binding spectra with P450 3A4 with a Ks of 6.3 µM, a value comparable to ANF and its derivatives. These results suggest that the mechanisms of inhibition of the P450 1 family of enzymes differ from those of P450 2C9 and 3A4 with regard to ANF and BNF and their derivatives; P450 1B1 interacts with these naphthoflavones (as analyzed by spectral binding) and its catalytic activity was strongly inhibited; P450 1A1 and 1A2 were also strongly inhibited by all six naphthoflavone derivatives. The 2’- and 4’-propargyl ether derivatives inhibited more strongly than the parent compounds, and these findings support different mechanisms of inhibition compared with the P450 Family 1 enzymes. In addition, inhibition of P450 3A4 by ANF and BNF and the derivatives was not so potent; ANF inhibited P450 3A4 with an IC50 of 18 µM, although these chemicals (except for BNF2’PE and BNF4’PE) interacted spectrally with P450 3A4 with Ks values <7 µM.
In conclusion, our present studies show that structurally diverse flavonoid derivatives inhibit human P450 1A1, 1A2, 1B1, 2C9, and 3A4 to different degrees, depending on the enzymes and inhibitors, and that there are different mechanisms of inhibition of these P450s by flavonoids. The presence of a 5,7-dihydroxyl group in the A-ring of flavone was found to increase the inhibition potency towards these P450 enzymes and the presence of a 3-hydroxyl group in the C-ring of 57DHF produced a highly potent inhibitor of these P450 enzymes, 357THF (galangin). Our results also show that the presence of a 3’4’-dimethoxy group in the B-ring of 57DHF resulted in another potent inhibitor of P450 enzymes. Molecular docking studies suggest that there are different orientations in the interaction of the six flavonoids with the five P450 enzymes examined and that two or more mechanisms are possible to explain how various flavonoids inhibit individual P450 enzymes differently.
Acknowledgments
This work was supported in part by Grants from the Ministry of Education, Science, and Culture of Japan, the Ministry of Health and Welfare of Japan (T.S., K.T., S.T., H.Y., M.K.), NIH grant S06 GM08008 and DOE grant DE-FC26-00NT40843 (M.K.F.), and NIH grants R37 CA090426 and P30 ES000267 (F.P.G.).
Abbreviations
- EROD
7-ethoxyresorufin O-deethylation
- ANF
α-naphthoflavone
- ANF2’PE
α-naphthoflavone 2’-propargyl ether
- ANF4’PE
α-naphthoflavone 4’-propargyl ether
- BNF
β–naphthoflavone
- BNF2’PE
β–naphthoflavone 2’-propargyl ether
- BNF4’PE
β–naphthoflavone 4’-propargyl ether
- 3HF
3-hydroxyflavone (flavonol)
- 5HF
5-hydroxyflavone
- 7HF
7-hydroxyflavone
- 57DHF
5,7-dihydroxyflavone (chrysin)
- 357THF
3,5,7-trihydroxyflavone (galangin)
- 4’57THF
4’5,7-trihydroxyflavone (apigenin)
- 4’57THIF
4’,5,7-trihydroxyisoflavone (genistein)
- 4’57THFva
4’,5,7-trihydroxyflavanone (naringenin)
- 4’57THFvaG
4’,5,7-trihydroxyflavanone glycoside (naringin)
- 567THF
5,6,7-trihydroxyflavone (baicalein)
- 34’57TetraHF
3,4’,5,7-tetrahydroxyflavone (kaempferol)
- 33’4’57PHF
3,3’,4’,5,7-pentahydroxyflavone (quercetin)
- 4’M57DHF
4’-methoxy-5,7-dihydroxyflavone (acacetin)
- 4’M57DHIsoF
4’-methoxy-5,7-dihydroxyisoflavone (biochain A)
- 2’MF
2’-methoxyflavone
- 3’MF
3’-methoxyflavone
- 4’MF
4’-methoxyflavone
- 3’4’DMF
3’,4’-dimethoxyflavone
- 2’M57DHF
2’-methoxy-5,7-dihydroxyflavone
- 3’M57DHF
3’-methoxy-5,7-dihydroxyflavone
- 3’4’DM57DHF
3’4’-dimethoxy-5,7-dihydroxyflavone
- 2’M78DHF
2’-methoxy-7,8-dihydroxyflavone
- 3’M78DHF
3’-methoxy-7,8-dihydroxyflavone
- 4’M78DHF
4’-methoxy-7,8-dihydroxyflavone
- 3’4’DM78DHF
3’,4’-dimethoxy-7,8-dihydroxyflavone
References
- 1.Arct J, Pytkowska K. Flavonoids as components of biologically active cosmeceuticals. Clin. Dermatol. 2008;26:347–357. doi: 10.1016/j.clindermatol.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 2.Kale A, Gawande S, Kotwal S. Cancer phytotherapeutics: role for flavonoids at the cellular level. Phytother. Res. 2008;22:567–577. doi: 10.1002/ptr.2283. [DOI] [PubMed] [Google Scholar]
- 3.Zhang S, Yang X, Coburn RA, Morris ME. Structure activity relationships and quantitative structure activity relationships for the flavonoid-mediated inhibition of breast cancer resistance protein. Biochem. Pharmacol. 2005;70:627–639. doi: 10.1016/j.bcp.2005.05.017. [DOI] [PubMed] [Google Scholar]
- 4.Kim H-J, Lee SB, Park S-K, Kim HM, Park YI, Dong M-S. Effects of hydroxy group numbers on the B-ring of 5,7-dihydroxyflavones on the differential inhibition of human CYP 1A and CYP1B1 enzymes. Arch. Pharm. Res. 2005;28:1114–1121. doi: 10.1007/BF02972971. [DOI] [PubMed] [Google Scholar]
- 5.Tsujimoto M, Horie M, Honda H, Takara K, Nishiguchi K. The structure-activity correlation on the inhibitory effects of flavonoids on cytochrome P450 3A activity. Biol. Pharm. Bull. 2009;32:671–676. doi: 10.1248/bpb.32.671. [DOI] [PubMed] [Google Scholar]
- 6.Walle T. Methoxylated flavones, a superior cancer chemopreventive flavonoid subclass? Semin. Cancer Biol. 2007;17:354–362. doi: 10.1016/j.semcancer.2007.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walle UK, Walle T. Bioavailable flavonoids: cytochrome P450-mediated metabolism of methoxyflavones. Drug Metab. Dispos. 2007;35:1985–1989. doi: 10.1124/dmd.107.016782. [DOI] [PubMed] [Google Scholar]
- 8.Moon YJ, Wang X, Morris ME. Dietary flavonoids: effects on xenobiotic and carcinogen metabolism. Toxicol. In Vitro. 2006;20:187–210. doi: 10.1016/j.tiv.2005.06.048. [DOI] [PubMed] [Google Scholar]
- 9.Breinholt VM, Offord EA, Brouwer C, Nielsen SE, Brøsen K, Friedberg T. In vitro investigation of cytochrome P450-mediated metabolism of dietary flavonoids. Food Chem. Toxicol. 2002;40:609–616. doi: 10.1016/s0278-6915(01)00125-9. [DOI] [PubMed] [Google Scholar]
- 10.Hodek P, Trefil P, Stiborová M. Flavonoids-potent and versatile biologically active compounds interacting with cytochromes P450. Chem.-Biol. Interact. 2002;139:1–21. doi: 10.1016/s0009-2797(01)00285-x. [DOI] [PubMed] [Google Scholar]
- 11.Doostdar H, Burke MD, Mayer RT. Bioflavonoids: selective substrates and inhibitors for cytochrome P450 CYP1A and CYP1B1. Toxicology. 2000;144:31–38. doi: 10.1016/s0300-483x(99)00215-2. [DOI] [PubMed] [Google Scholar]
- 12.Otake Y, Walle T. Oxidation of the flavonoids galangin and kaempferide by human liver microsomes and CYP1A1, CYP1A2, and CYP2C9. Drug Metab. Dispos. 2002;30:103–105. doi: 10.1124/dmd.30.2.103. [DOI] [PubMed] [Google Scholar]
- 13.Si D, Wang Y, Zhou YH, Guo Y, Wang J, Zhou H, Li ZS, Fawcett JP. Mechanism of CYP2C9 inhibition by flavones and flavonols. Drug Metab. Dispos. 2008;37:629–634. doi: 10.1124/dmd.108.023416. [DOI] [PubMed] [Google Scholar]
- 14.Zhai S, Dai R, Friedman FK, Vestal RE. Comparative inhibition of human cytochromes P450 1A1 and 1A2 by flavonoids. Drug Metab. Dispos. 1998;26:989–992. [PubMed] [Google Scholar]
- 15.Kimura Y, Ito H, Ohnishi R, Hatano T. Inhibitory effects of polyphenols on human cytochrome P450 3A4 and 2C9 activity. Food Chem Toxicol. 2010;48:429–435. doi: 10.1016/j.fct.2009.10.041. [DOI] [PubMed] [Google Scholar]
- 16.Quintieri L, Palatini P, Nassi A, Ruzza P, Floreani M. Flavonoids diosmetin and luteolin inhibit midazolam metabolism by human liver microsomes and recombinant CYP 3A4 and CYP3A5 enzymes. Biochem. Pharmacol. 2007;75:1426–1437. doi: 10.1016/j.bcp.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 17.Roberts DW, Doerge DR, Churchwell MI, Gamboa da Costa G, Marques MM, Tolleson WH. Inhibition of extrahepatic human cytochromes P450 1A1 and 1B1 by metabolism of isoflavones found in Trifolium pratense (red clover) J. Agric. Food Chem. 2004;52:6623–6632. doi: 10.1021/jf049418x. [DOI] [PubMed] [Google Scholar]
- 18.Guengerich FP. Human cytochrome P450 enzymes. In: Ortiz de Montellano PR, editor. Cytochrome P450: Structure, Mechanism, and Biochemistry. 3rd ed. New York: Kluwer Academic/Plenum Press; 2005. pp. 377–530. [Google Scholar]
- 19.Shimada T. Xenobiotic-metabolizing enzymes involved in activation and inactivation of carcinogenic polycyclic aromatic hydrocarbons. Drug Metab. Pharmacokinet. 2006;21:257–276. doi: 10.2133/dmpk.21.257. [DOI] [PubMed] [Google Scholar]
- 20.Guengerich FP, Chun Y-J, Kim D, Gillam EMJ, Shimada T. Cytochrome P450 1B1: A target for inhibition in anticarcinogenesis strategies. Mut. Res. 2003;523–524:173–182. doi: 10.1016/s0027-5107(02)00333-0. [DOI] [PubMed] [Google Scholar]
- 21.Walle T, Ta N, Kawamori T, Wen X, Tsuji PA, Walle UK. Cancer chemopreventive properties of orally bioavailable flavonoids—methylated versus unmethylated flavones. Biochem. Pharmacol. 2007;73:1288–1296. doi: 10.1016/j.bcp.2006.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Courter LA, Musafia-Jeknic T, Fischer K, Bildfell R, Giovanini J, Pereira C, Baird WM. Urban dust particulate matter alters PAH-induced carcinogenesis by inhibition of CYP1A1 and CYP1B1. Toxicol. Sci. 2007;95:63–73. doi: 10.1093/toxsci/kfl137. [DOI] [PubMed] [Google Scholar]
- 23.Shimada T, Tanaka K, Takenaka S, Foroozesh MK, Murayama N, Yamazaki H, Guengerich FP, Komori M. Reverse type I binding spectra of human cytochrome P450 1B1 induced by flavonoid, stilbene, pyrene, naphthalene, phenanthrene, and biphenyl derivatives that inhibit catalytic activity: a structure-function relationship study. Chem. Res. Toxicol. 2009;22:1325–1333. doi: 10.1021/tx900127s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McKendall ME, Smith TP, Anh K, Ellis J, McGee T, Foroozesh M, Zhu N, Klein Stevens C. Methoxyflavone inhibitors of cytochrome P450. J. Chem. Crystallogr. 2008;38:231–237. [Google Scholar]
- 25.Zhu N, Lightsey K, Foroozesh M, Alworth W, Chaudhary A, Willett KL, Klein Stevens CLK. Naphthoflavone propargyl ether inhibitors of cytochrome P450. J. Chem. Crystallogr. 2006;36:289–296. [Google Scholar]
- 26.Shimada T, Guengerich FP. Inhibition of human cytochrome P450 1A1-, 1A2-, and 1B1-mediated activation of procarcinogens to genotoxic metabolites by polycyclic aromatic hydrocarbons. Chem. Res. Toxicol. 2006;19:288–294. doi: 10.1021/tx050291v. [DOI] [PubMed] [Google Scholar]
- 27.Shimada T, Murayama N, Okada K, Funae Y, Yamazaki H, Guengerich FP. Different mechanisms of inhibition for human cytochrome P450 1A1, 1A2, and 1B1 by polycyclic aromatic inhibitors. Chem. Res. Toxicol. 2007;20:489–496. doi: 10.1021/tx600299p. [DOI] [PubMed] [Google Scholar]
- 28.Shimada T, Murayama N, Tanaka K, Takenaka S, Imai Y, Hopkins NE, Foroozesh MK, Alworth WL, Yamazaki H, Guengerich FP, Komori M. Interaction of polycyclic aromatic hydrocarbons with human cytochrome P450 1B1 in inhibiting catalytic activity. Chem. Res. Toxicol. 2008;21:2313–2323. doi: 10.1021/tx8002998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamazaki H, Nakamura M, Komatsu T, Ohyama K, Hatanaka N, Asahi S, Shimada N, Guengerich FP, Shimada T, Nakajima M, Yokoi T. Roles of NADPH-P450 reductase and apo- and holo-cytochrome b5 on xenobiotic oxidations catalyzed by 12 recombinant human cytochrome P450s expressed in membranes of Escherichia coli. Protein Expr. Purif. 2002;24:329–337. doi: 10.1006/prep.2001.1578. [DOI] [PubMed] [Google Scholar]
- 30.Isin EM, Guengerich FP. Kinetics and thermodynamics of ligand binding by cytochrome P450 3A4. J. Biol. Chem. 2006;281:9127–9136. doi: 10.1074/jbc.M511375200. [DOI] [PubMed] [Google Scholar]
- 31.Krauser JA, Guengerich FP. Cytochrome P450 3A4-catalyzed testosterone 6beta-hydroxylation stereochemistry, kinetic deuterium isotope effects, and rate-limiting steps. J. Biol. Chem. 2005;280:19496–19506. doi: 10.1074/jbc.M501854200. [DOI] [PubMed] [Google Scholar]
- 32.Wu ZL, Sohl CD, Shimada T, Guengerich FP. Recombinant enzymes overexpressed in bacteria show broad catalytic specificity of human cytochrome P450 2W1 and limited activity of human cytochrome P450 2S1. Mol. Pharmacol. 2006;69:2007–2014. doi: 10.1124/mol.106.023648. [DOI] [PubMed] [Google Scholar]
- 33.Yamazaki H, Inoue K, Chiba K, Ozawa N, Kawai T, Suzuki Y, Goldstein JA, Guengerich FP, Shimada T. Comparative studies on the catalytic roles of cytochrome P450 2C9 and its Cys- and Leu-variants in the oxidation of warfarin, flurbiprofen, and diclofenac by human liver microsomes. Biochem. Pharmacol. 1998;56:243–251. doi: 10.1016/s0006-2952(98)00133-6. [DOI] [PubMed] [Google Scholar]
- 34.Emoto C, Murayama N, Yamazaki H. Effects of enzyme sources on midazolam 1'-hydroxylation activity catalyzed by recombinant cytochrome P450 3A4 in combination with NADPH-cytochrome P450 reductase. Drug Metab. Lett. 2008;2:190–192. doi: 10.2174/187231208785425737. [DOI] [PubMed] [Google Scholar]
- 35.Omura T, Sato R. The carbon monoxide-binding pigment of liver microsomes. I. Evidence for its hemoprotein nature. J. Biol. Chem. 1964;239:2370–2378. [PubMed] [Google Scholar]
- 36.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 37.Sansen S, Yano JK, Reynald RL, Schoch GA, Griffin KJ, Stout CD, Johnson EF. Adaptations for the oxidation of polycyclic aromatic hydrocarbons exhibited by the structure of human P450 1A2. J. Biol. Chem. 2007;282:14348–14355. doi: 10.1074/jbc.M611692200. [DOI] [PubMed] [Google Scholar]
- 38.Wester MR, Yano JK, Schoch GA, Yang C, Griffin KJ, Stout CD, Johnson EF. The structure of human cytochrome P450 2C9 complexed with flurbiprofen at 2.0-A resolution. J. Biol. Chem. 2004;279:35630–35637. doi: 10.1074/jbc.M405427200. [DOI] [PubMed] [Google Scholar]
- 39.Yano JK, Wester MR, Schoch GA, Griffin KJ, Stout CD, Johnson EF. The structure of human microsomal cytochrome P450 3A4 determined by X-ray crystallography to 2.05-Å resolution. J. Biol. Chem. 2004;279:38091–38094. doi: 10.1074/jbc.C400293200. [DOI] [PubMed] [Google Scholar]
- 40.Ueng Y-F, Kuwabara T, Chun YJ, Guengerich FP. Cooperativity in oxidations catalyzed by cytochrome P450 3A4. Biochemistry. 1997;36:370–381. doi: 10.1021/bi962359z. [DOI] [PubMed] [Google Scholar]
- 41.Niwa T, Murayama N, Yamazaki H. Heterotropic cooperativity in oxidation mediated by cytochrome P450. Curr Drug Metab. 2008;5:453–462. doi: 10.2174/138920008784746364. [DOI] [PubMed] [Google Scholar]
- 42.Marechal JD, Yu J, Brown S, Kapelioukh I, Rankin EM, Wolf CR, Roberts GC, Paine MJ, Sutcliffe MJ. In silico and in vitro screening for inhibition of cytochrome P450 CYP3A4 by comedications commonly used by patients with cancer. Drug Metab. Dispos. 2006;34:534–538. doi: 10.1124/dmd.105.007625. [DOI] [PubMed] [Google Scholar]
- 43.Fishelovitch D, Shaik S, Wolfson HJ, Nussinov R. Theoretical characterization of substrate access/exit channels in the human cytochrome P450 3A4 enzyme: involvement of phenylalanine residues in the gating mechanism. J. Phys. Chem. B. 2009;113:13018–13025. doi: 10.1021/jp810386z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Isin EM, Guengerich FP. Kinetics and thermodynamics of ligand binding by cytochrome P450 3A4. J. Biol. Chem. 2006;281:9127–9136. doi: 10.1074/jbc.M511375200. [DOI] [PubMed] [Google Scholar]
- 45.Ekroos M, Sjögren T. Structural basis for ligand promiscuity in cytochrome P450 3A4. Proc. Natl. Acad. Sci., U.S.A. 2006;103:13682–13687. doi: 10.1073/pnas.0603236103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Torimoto N, Ishii I, Hata M, Nakamura H, Imada H, Ariyoshi N, Ohmori S, Igarashi T, Kitada M. Direct interaction between substrates and endogenous steroids in the active site may change the activity of cytochrome P450 3A4. Biochemistry. 2003;42:15068–15077. doi: 10.1021/bi034409n. [DOI] [PubMed] [Google Scholar]
- 47.Shimada T, Iwasaki M, Martin MV, Guengerich FP. Human liver microsomal cytochrome P-450 enzymes involved in the bioactivation of procarcinogens detected by umu gene response in Salmonella typhimurium TA1535/pSK1002. Cancer Res. 1989;49:3218–3228. [PubMed] [Google Scholar]