Abstract
Adeno-associated virus vectors have been shown to mediate persistent transduction in animal models of gene therapy. However, clinical trials with AAV vectors have shown that an immune response to AAV capsid protein can result in clearance of transduced cells. One source of capsid antigen is from the delivered vector virions, but expression of cap DNA impurities in AAV vector preparations might provide an alternative and persistent source of capsid antigen. Here we show that DNA without any AAV sequences can be packaged in AAV virions, and that both cap and rep DNA are packaged into AAV vectors produced by standard methods. Using a sensitive complementation assay, we also observed significant expression of capsid in cultured cells transduced with AAV vectors. In an attempt to solve this problem, we inserted a large intron into the cap gene to generate a capsid expression cassette (captron) that is too large for packaging into AAV virions. Both complementation assays and quantitative reverse-transcription PCR analysis showed that cultured cells infected with AAV vectors made with the captron plasmid expressed no detectible capsid. Elimination of transfer of capsid-expressing DNA may reduce immune responses to AAV vector-transduced cells and promote long-term expression of therapeutic proteins.
Keywords: AAV vectors, vector immune responses, capsid gene transfer
Introduction
Vectors based on adeno-associated viruses (AAV) have been used in many applications in vivo because they promote persistent gene expression in multiple somatic tissues of animals.1–4 However, recent studies have shown that the lack of an immune response seen in many mouse and some large animal studies has not been duplicated in human trials. In a clinical trial for hemophilia B, two of seven subjects given an AAV vector expressing clotting factor IX (FIX) developed a transient self-limiting increase in liver transaminases, followed by clearance of the FIX-expressing cells at 4 to 8 weeks after delivery.5 A cytotoxic lymphocyte (CTL) response to AAV capsid but not to FIX was detected in peripheral blood mononuclear cells. In another clinical trial involving lipoprotein lipase, clearance of AAV-transduced cells coincided with a CTL response towards the capsid, but not the transgene.6
It is important to know how this immune response is generated in order to prevent it. One hypothesis suggests that persistence of Cap proteins from the vector inoculum results in clearance of the transduced cells. An alternative suggests that Cap expression from cap DNA packaged in AAV vectors is responsible for the immune response. A clinical protocol that uses transient immunosuppression may be successful if the former hypothesis is true, but if the later is true, an immune response is likely to occur when immunosuppression is lifted. It is also possible that both mechanisms occur, and in this case, successful long-term transduction will require both transient immunosuppression and methods to prevent contamination with capsid-expressing DNA.
The observation of a capsid-directed immune response even after 16 weeks of immunosuppression in a canine model of muscle gene transfer7 strongly suggests a stable source of antigen. We also have observed an immune response to AAV6 capsid that significantly decreased transgene expression within 3 weeks in a canine model of lung gene transfer.8 Immune suppression promoted long term gene expression (4 months) but that was lost after immune suppression was lifted.8
The development of AAV vectors for clinical use is marked by continuing efforts to improve efficiency and to remove impurities. Early methods of AAV vector production resulted in contamination with replication-competent AAV. While several strategies were used to prevent such contamination,9–11 packaging of other DNA sequences can still occur. For example, bacterial ampicillin resistance gene DNA from plasmids used to make AAV vectors has been found in AAV vector preparations at 0.5–7% of the level of vector genomes (vg).12 The ampicillin resistance gene DNA was also found in tissues of mice, dogs, and non-human primates up to 5 months after vector delivery, demonstrating that DNA impurities in AAV vector preparations can persist in vivo.12 Furthermore, the presence of cap DNA sequences in clinical lots of AAV vectors at a level of 0.00018 cap copies per vg has been reported by Hauck et al.13 However, they did not detect capsid expression in recipient mice or cultured cells, measured by quantitative PCR (qPCR) of reverse-transcribed mRNA.
Here we confirm that AAV capsids can package DNA that does not contain AAV sequences, and show that the packaged DNA can express protein in vector-transduced cells. Furthermore, we were able to detect cap and rep DNA in several AAV vectors made using standard techniques, and found expression of Cap proteins in cells transduced with AAV vectors by using a sensitive complementation assay. Finally, we show that introduction of a large intron in the cap gene (captron) resulted in undetectable Cap expression in cells exposed to vectors made with the captron plasmid.
Results
Transduction of a gene without flanking AAV sequences in AAV virions
We compared the rate of AAV-mediated transduction of a gene that encodes human placental alkaline phosphatase (AP), either flanked by AAV ITRs in the standard AAV vector configuration (pARAP4), or without any flanking AAV sequences (pRAP). AAV virions were produced by cotransfection of each of these plasmids with a plasmid that expresses AAV Rep and Cap and adenovirus helper proteins (pDGM6), and the virions were purified by using heparin columns. HTX cells were then exposed to the purified AAV preparations and foci of AP-positive cells were quantitated. AAV made with the plasmid that does not contain AAV sequences (pRAP) did indeed induce AP+ foci in HTX cells (Table 1, row 2). However, the number of AP+ foci produced by virus made with the pRAP plasmid was 4 to 5 orders of magnitude lower than that of the standard vector plasmid pARAP4 (Table 1, row 1). Virus preparations made with a control plasmid (pUC18) instead of the plasmid encoding AAV Rep and Cap proteins (pDGM6) exhibited low levels of background AP+ foci (Table 1, row 3).
Table 1.
Transfer of genes with and without flanking AAV terminal repeatsa
| AP+ foci produced per transfected 10-cm dish: | ||
|---|---|---|
| Transfected plasmids | before benzonase treatment | after benzonase treatment |
| pARAP4 + pDGM6 | 1.5 × 107 | 1.3 × 107 |
| pRAP + pDGM6 | 7.5 × 102 | 3.6 × 102 |
| pRAP + pUC18 | 0.25 | 0.25 |
| pRAP + pUC18, pDGM6 + pUC18 b | Not done | 0.30 |
| pRAP + pACZn + pDGM6 | Not done | 1.8 × 103 |
293 cells were transfected with the indicated plasmids one day after seeding the cells in 10-cm dishes. Three days after transfection, AAV virions were harvested and purified as described in Materials and Methods. Purified virions were left untreated or were treated with benzonase to remove DNA that might be bound to the outside of virions. Virions capable of expressing AP were measured using HTX cells as targets for infection. Results are means of duplicate determinations in a representative experiment.
293 cells in 10-cm dishes were independently transfected with pUC18 and pRAP, or pUC18 and pDGM6. After harvest of cells and medium, cell lysates were combined, treated with benzonase, co-purified on a heparin column, and then assayed for AP+ FFU.
To test the possibility that the pRAP DNA was transferred to cells by binding to the surface of the AAV virions, vector preparations were treated with benzonase to destroy DNA outside of virions and were repurified on heparin columns. Benzonase treatment did not destroy the ability of virions made with pRAP to transduce target cells (Table 1, last column), indicating that the AP gene was packaged within the AAV virions.
To further rule out transfer of pRAP by interactions with the surface of the capsid, two separate cell lysates were made, one from cells transfected with only the non-ITR pRAP plasmid and one from cells transfected with only the helper plasmid expressing the AAV capsid proteins. The two preparations were combined, treated with benzonase and purified. Infection of HTX cells with the combined preparation yielded only low background levels of AP, showing that the transfer of AP that we observed could not be explained by interactions between DNA and fully formed particles (Table 1, row 4).
We next tested whether transfer of the AP gene in pRAP would still occur in competition with a genuine AAV vector, to address the possibility of transfer of genes without AAV sequences during routine AAV vector production. For this experiment we used the AAV vector ACZn that expresses a nuclear-localized β-galactosidase (βgal) to allow independent quantitation of gene transfer by pRAP and the AAV vector. Indeed, the AP gene was transferred by virus produced by cotransfection of pRAP, pACZn and pDGM6 (Table 1, row 5). The rate of AP gene transfer was 5-fold higher in the presence of the ACZn AAV vector than in its absence, while the titer of the ACZn vector produced in the presence of pRAP was similar to that of other preparations made without it (2.5 × 105 βgal+ focus-forming units (FFU) per 10-cm dish, data not shown). In summary, our experiments confirm that DNA sequences without AAV ITRs can be packaged into AAV virions, and in this case, are transferred and expressed at a rate that is 4 to 5 orders of magnitude lower than that of a typical AAV vector.
AAV vector virions contain cap and rep DNA
The above results suggested that other genes without flanking AAV ITRs, such as those used for expression of Cap and Rep during AAV vector production, could be transferred and expressed in AAV vectors. To address this issue, we first tested for the presence of cap and rep DNA in several AAV vectors by qPCR. The primers used to detect cap and rep amplify products of 539 and 416 base pairs, respectively. The qPCR results showed that 7 to 170 copies of cap DNA and 9 to 150 copies of rep DNA were packaged for every million vector genomes (Table 2). The rate of both cap and rep packaging was similar and correlated inversely with the AAV vector yield; AAV vectors that routinely had higher production yields (ACF3’B) showed less cap and rep packaging, while those with lower yields (ACWRZN) had more. This is consistent with a model in which the rate of cap and rep packaging is constant and independent of AAV vector packaging, and that normalization of cap and rep copy number to the vector copy number generates this inverse relationship. AAV preparations tested were made using the single plasmid pDGM6 or using plasmids pMTrep2, pCMVcap6, and pLadeno5, and we found cap and rep in virions made with either plasmid combination. One vector (ARAP4) was made with both methods, and we did not observe a difference in packaging of cap or rep related to the helper plasmids used.
Table 2.
Quantitation of rep and cap DNA in AAV vector virions by qPCR a
| Helper | rep and cap | Copies of cap | Copies of rep | |
|---|---|---|---|---|
| AAV vector | plasmid | plasmids | per 106 vg | per 106 vg |
| ACWRZn | pLadeno5 | pMTrep2, pCMVcap6 | 170 | 150 |
| ARAP4 prep 1 | pLadeno5 | pMTrep2, pCMVcap6 | 58 | 70 |
| ARAP4 prep 2 | pLadeno5 | pMTrep2, pCMVcap6 | 14 | 16 |
| ARAP4 prep 3 | pLadeno5 | pMTrep2, pCMVcap6 | 36 | 32 |
| ARAP4 prep 4 | pDGM6 | pDGM6 | 32 | 38 |
| ACAGhAAT prep 1 | pDGM6 | pDGM6 | 26 | 27 |
| ACAGhAAT prep 2 | pDGM6 | pDGM6 | 49 | 63 |
| ACF3’B | pDGM6 | pDGM6 | 7.0 | 9.1 |
Virus was prepared for qPCR by treating 1010 to 1011 genome-containing particles (determined by Southern analysis) with benzonase. Next virion DNA was extracted as described in Materials and Methods and was subjected to qPCR. Results are expressed as copies of DNA per 106 vector genomes as determined by Southern analysis. For the ACWRZn vector, the genome number was checked by qPCR after benzonase treatment and purification of virions, and was 63% of the value determined by Southern analysis. Results are means of two experiments done in triplicate.
Packaged cap DNA is expressed in infected cells
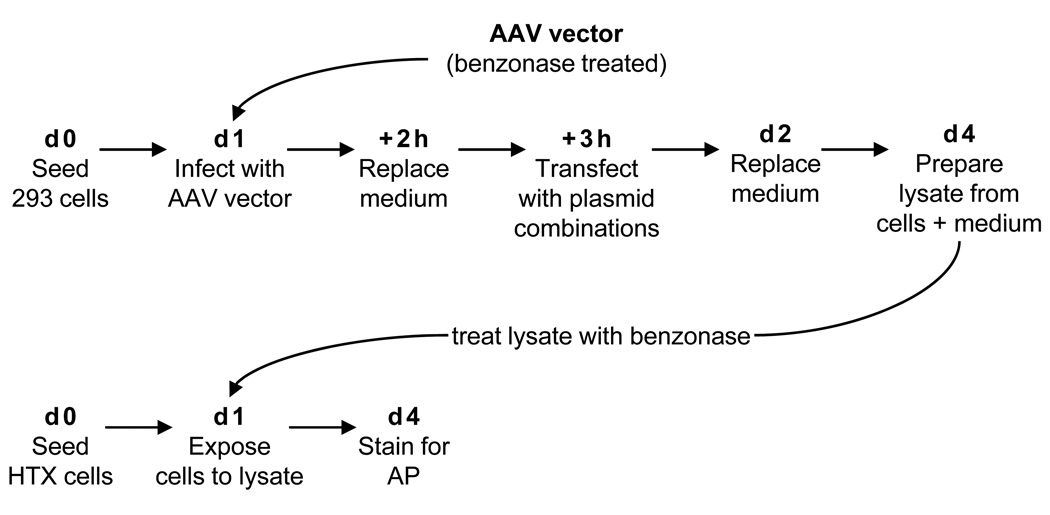
To determine whether the packaged cap and rep DNAs are expressed in vector infected cells, we developed a Cap and Rep complementation assay (Figure 1). In this assay, purified benzonase-treated AAV vector preparations were used to infect 293 cells. Soon afterwards, cells were transfected with plasmid combinations that included various combinations of an AP-expressing AAV vector plasmid (pARAP4) and the plasmids used for AAV vector production. As a positive control, the AAV vector and all 3 AAV production plasmids were used, while remaining combinations lacked one or more plasmids. Cells and medium were collected and lysates were prepared, and HTX cells were used to titer ARAP4 vector present in the lysates. Production of AP+ HTX cells using plasmid combinations lacking cap and/or rep DNA would indicate provision of these functions by the AAV vector stock used initially to infect the 293 cells.
Figure 1. Cap and Rep complementation assay.
Several AAV vectors were examined using the complementation assay (Table 3). Infection with ACF3’B followed by transfection with plasmid combination 2 that contained vector, replication and helper functions but lacked a capsid-expressing plasmid, yielded 104 AP+ FFU per ml of lysate, indicating transfer of functional cap genes by the ACF3'B vector. Only background levels of AP+ foci were observed with plasmid combinations 4 or 5.
Table 3.
Transfer of cap and rep function following infection with AAV vectors a
| AP+ FFU per ml for plasmid combination: | |||||||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | |||
| pLadeno5 | + | + | + | + | |||
| pMTrep6 | + | − | − | − | |||
| AAV | Amount | pCMVcap6 | − | + | − | − | |
| Expt | vector | (vg) | pARAP4 | + | + | + | − |
| 1 | ACF3’B (AAV6) | 2 × 1012 | ND | 1.0 × 104 | ND | 7 | 3 |
| None | ND | 36 | ND | ND | ND | ||
| 2 | ACAGhAAT (AAV6) | 7 × 1010 | ND | 4.5 × 103 | ND | 4 | 6 |
| None | 1.2 × 106 | 66 | 17 | ND | ND | ||
| 3 | ACAGhAAT (AAV6) | 7 × 1010 | ND | 7.1 × 103 | 8 | 4 | 6 |
| None | ND | 89 | ND | ND | ND | ||
| 4 | ACWRZn (AAV6) | 7 × 1010 | 3.6 × 105 | 3.1 ×104 | 3.9 × 103 | ND | ND |
| ACWRZn (AAV2) | 7 × 1010 | 4.4 × 105 | 1.4 ×104 | 8 | 5 | ND | |
| None | 5.5 × 105 | <1 | 2 | ND | ND | ||
Cap and Rep complementation assays were done as shown in Fig. 1 and described in Materials and Methods, except that the crude cell lysate was filtered (0.2 µm-pore-size) to reduce background AP in experiment 4. Results are means of duplicate determinations. ND, not done.
We also detected transfer of cap gene function by the second vector, ACAGhAAT. In two experiments, using 7 × 1010 vg for each infection, an average vector titer of 5.8 × 103 AP+ FFU per ml was observed after transfection of plasmid combination 2 that lacks the cap expression plasmid. No production of AP vector was seen from transfection combination 3 that lacked rep, showing that rep function was not transferred by the ACAGhAAT vector.
Note that when no AAV vector was added in the above experiments, the plasmid combination that did not contain the cap construct produced 36, 66 and 89 AP+ FFU per ml (Table 3, Expts. 1–3, plasmid combination 2). This was higher than the background AP values observed in combinations 3, 4 and 5, and we hypothesize that this is due to AP protein transfer (pseudotransduction). The Rep protein in this combination likely caused replication of the ARAP4 vector DNA resulting in higher levels of AP protein in the lysate added to the target cells.
To test whether cap function could be transferred by an AAV vector with a different AAV serotype, we tested for transfer of cap function by an AAV vector packaged with AAV6 or AAV2 capsid proteins, and found vectors with both serotypes transferred cap gene function (Table 3, Expt. 4). In one case we also detected transfer of rep gene function, indicating that both the cap and rep genes can be transferred and expressed, as might have been expected based on the presence of both cap and rep gene sequences in AAV virions (Table 2). Note that in this experiment, we filtered the virus produced by the transfected 293 cells once through a 0.2 µm-pore-size filter in an attempt to reduce transfer of AP protein (pseudotransduction). Indeed, this substantially reduced the background AP+ foci obtained for plasmid combination 2 when no AAV vector was used (Table 3, last row) compared to that found for experiments 1 to 3 (Table 3).
To estimate how many cap-transducing particles are in AAV vector preparations, we performed the cap complementation assay with different amounts of the ACAGhAAT (AAV6) vector. Two independent assays showed cap complementation by 1010 vg and higher amounts of the vector, with no cap complementation at 4 × 109 vg or lower amounts (data not shown). This provides a crude estimate of about one cap-transducing virion in 1010 vg of the ACAGhAAT (AAV6) vector preparation.
Insertion of a large intron prevents cap gene transduction by AAV vectors
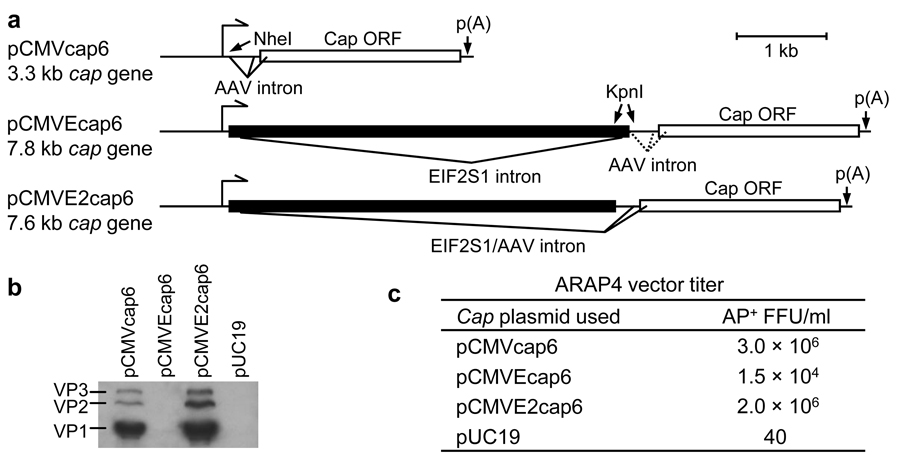
One feature of AAV vectors, often considered to be a limitation, is their small packaging capacity. In contrast, this property can be exploited in a strategy to minimize the packaging of functional cap genes. Although packaging of cap DNA would still occur, expression of capsid would be limited by introducing an intron between the cap gene and the promoter such that the expression cassette would be too large to package into AAV virions. To test this possibility, a large intron (4.3 kb) from the 5’ untranslated region of the human EIF2S1 gene was inserted between the CMV promoter and the alternatively spliced intron in the AAV cap gene to make the plasmid pCMVEcap6 (Figure 2a). Unfortunately, this plasmid did not express functional capsid protein (Figure 2b), and further investigation using northern analysis and RT-PCR revealed that the EIF2S1 intron prevented proper excision of the native AAV intron (data not shown). A new construct with a hybrid EIF2S1/AAV intron was generated by removing a 200 bp fragment containing the EIF2S1 splice acceptor and the AAV splice donor to make the plasmid pCMVE2cap6. The size of this gene from promoter to the end of the cap ORF is about 7.5 kb, much larger than the 4.7 kb AAV packaging limit. This construct expressed all three capsid forms, verifying proper alternative splicing at the AAV splice acceptors (Figure 2b). We measured the titer of crude AAV preparations made by transient transfections of the different cap plasmids, and we observed only a minor change in yield between pCMVE2cap6 and pCMVcap6, even though the 2 µg of pCMVE2cap6 transfected contains about half as many copies as that of the 2 µg of pCMVcap6 (Figure 2c).
Figure 2. Intron-containing cap constructs.
(a) Schematic of intron-containing cap constructs. (b) Western blot of cell lysate of 293T cells transfected with 10 µg DNA using anti-AAV VP1+VP2+VP3 mouse monoclonal antibody (American Research Products). 10 µl of lysate was loaded per lane. (c) Crude AAV lysates were collected three days after transfection of 293T cells with 4 µg pARAP4, 4 µg pDGM6Δcap and 2 µg of a cap plasmid. Cells and medium were freeze/thawed three times, centrifuged at 1,000 × g for 10 min to remove cells and debris, and filtered (0.2 µm-pore-size). ARAP4 titers from one experiment were determined by infecting HTX cells and staining for AP+ foci after three days.
We used the Cap and Rep complementation assay to test the effect of the intron on capsid expression by comparing parallel AAV productions made with pCMVcap6 and the captron plasmid, pCMVE2cap6 (Table 4). Transfection of cells with all 4 plasmids produced lysates with similar titers regardless of whether a standard vector, a captron vector, or no AAV vector was used to infect the cells (Table 4, combination 1), showing that prior infection of 293 cells with an AAV vector did not inhibit production of AAV virions. No transfer of AP was seen without rep (Table 4, combination 3), or without rep and cap (Table 4, combination 4). Cells infected with ACZn and ACMVcFIX AAV preparations made using the original pCMVcap6 did demonstrate expression of Cap protein by production of ARAP4 AAV vector (Table 4, combination 2). In contrast, cells infected with AAV preparations made using the captron plasmid did not produce Cap protein, as only a very low, background level of AP+ cells was observed (Table 4, combination 2). The third vector, ACF3’B, routinely had at least a 10-fold higher vector yield than other vectors, and therefore a lower cap to vg ratio. When 7 × 1010 vg was used for infection, no complementation was observed (data not shown). When 5-fold more vector was used, ACF3’B made with pCMVcap6 showed a detectible level of cap complementation while the AAV preparation made with the captron construct still exhibited no detectible transfer of cap function (Table 4). In summary, inclusion of a large intron sequence in the cap construct reduced Cap expression from all AAV vectors tested to undetectable levels as measured by this highly sensitive complementation assay.
Table 4.
AAV captron vectors do not transfer cap or rep function a
| AP+ FFU per ml for plasmid combination: | ||||||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | |||
| pLadeno5 | + | + | + | |||
| pMTrep6 | + | − | − | |||
| Amount | pCMVcap6 | − | + | − | ||
| AAV Vector | cap plasmid | (vg) | pARAP4 | + | + | + |
| ACZn | CMVcap6 | 7 × 1010 | 9 × 104 | 5.3 × 102 | 0 | 0 |
| Captron | 7 × 1010 | 1 × 105 | 2 | 1 | 1 | |
| None | None | 6 × 104 | 0 | 0 | 0 | |
| ACMVcFIX | CMVcap6 | 7 × 1010 | 1.6 × 105 | 2.8 × 103 | 3 | 3 |
| Captron | 7 × 1010 | 1.9 × 105 | 3 | 1 | 3 | |
| None | None | 2.2 × 105 | 2 | 3 | 1 | |
| ACF3’B | CMVcap6 | 3.8 × 1011 | ND | 41 | 0 | 0 |
| Captron | 3.8 × 1011 | ND | 0 | 3 | 3 | |
The cap and rep complementation assay was performed as shown in Figure 1 and described in Materials and Methods, except that the crude cell lysate was filtered (0.2 µm-pore-size) to reduce background AP. Results are means from one experiment done in duplicate (ACZn and ACF3’B) and means from two, five or four experiments done in duplicate (ACMVcFIX made with the CMVcap6 plasmid, the captron plasmid or none, respectively). ND, not done.
We next tested whether cap DNA from the captron plasmid is packaged into AAV virions. Analysis of DNA from AAV vector virions by qPCR showed that both cap and rep sequences were still packaged (Table 5).
Table 5.
Quantitation of AAV virions containing rep and cap sequences in AAV vector preparations a
| Copies of cap | Copies of rep | ||
|---|---|---|---|
| AAV Vector | cap plasmid | per 106 vg | per 106 vg |
| ACZn | CMVcap6 | 240 | 240 |
| Captron | 54 | 110 | |
| ACF3’B | CMVcap6 | 4.1 | 9.5 |
| Captron | 9.8 | 15 | |
| ACMVcFIX | CMVcap6 | 140 | 150 |
| Captron | 55 | 110 |
Virion DNA was prepared for qPCR as described for Table 2 and in Materials and Methods. Vector genomes were quantitated by Southern analysis for the ACF3’B vector and by qPCR for the ACZn and ACMVcFIX vectors by using primers for βgal or cFIX, respectively. Results are means of two experiments done in triplicate. Results for ACMVcFIX are means of experiments using two different vector preparations.
We used a qRT-PCR assay to detect Cap mRNA in virus-exposed cells and to confirm the Cap complementation assay results by using a second method. To maximize sensitivity, we infected HTX cells at a multiplicity of infection (MOI) of 4 × 106 vg of ACMVcFIX vector per cell and used 300 ng of total RNA from virus-exposed cells per qRT-PCR reaction. No Cap mRNA could be detected from cells infected with the vector made with the captron construct in any of the triplicate reactions. However, in qRT-PCR reactions done on two mRNA samples from independent infections with AAV made using pCMVcap6, approximately one copy per reaction was detected in one of the triplicate reactions (2/6 positive reactions, with the correct size amplicon confirmed on an agarose gel). Even after 45 cycles, no amplification products were observed in negative controls (300 ng of mRNA from uninfected HTX cells), or in controls performed without RT (to confirm the absence of DNA contamination). These data show that Cap mRNA transcribed from cap impurities in AAV vectors can be detected by qRT-PCR, and indicate that use of the captron plasmid for vector production eliminated expression of Cap mRNA. The level of cFIX mRNA detected in these samples was 4.9 × 105 copies per reaction, suggesting that Cap mRNA is made at a level 106-fold lower than that of the vector transgene. This was at the limit of detection of qRT-PCR, and therefore the result cannot be accurately quantified. However, the result was reproducible and agrees with results from the complementation assay. Thus, data from both the complementation assay and qRT-PCR assays confirm that use of this intron-containing construct eliminated capsid expression from AAV vectors.
Discussion
Our study confirms previous reports of contamination of AAV vector preparations with DNA from plasmids lacking AAV sequences, as well as DNA from cap and rep plasmids used to make AAV vectors. However, in contrast to previous results,13 we show that cap DNA present in AAV vector preparations can express Cap mRNA and protein in vector-transduced cells. To detect Cap protein expression, we used a sensitive complementation assay, and for RNA detection we exposed cells to more vector (4 × 106 vg per cell) and analyzed more cell RNA (300 ng) than previously used (105 vg per cell and 200 ng RNA13). The level of Cap mRNA was at the limit of detection for the qRT-PCR assay we used, while the Cap protein complementation assay showed consistent and significant levels of Cap expression from cells independently infected with multiple AAV vectors.
For the standard AAV6 vectors tested here, we estimate that cap gene transduction occurs at a rate of about 1 in 1010 vector genome-containing virions. However, the vector transduction rate is also low, about 1 in 104 to 105 vector genome-containing virions,14, 15 providing an estimate of the ratio of the cap transduction to vector transduction rates of 1 in 105 to 106. Even though cap transduction is orders of magnitude lower than vector transduction by standard AAV vector preparations, there may be significant levels of capsid expression in animals and humans treated with such vectors because of the high number of virions required for therapeutic effect. In the FIX clinical trial in which a capsid-specific response was observed, a dose of up to 1.4 × 1014 vg for a 70 kg human was used,5 corresponding to about 104 cap-transducing virions. Furthermore, although AAV6 vectors exhibit very poor transduction rates in cultured cells, their transduction rates can be remarkably high in animals,14–16 suggesting that cap transduction in culture may be an underestimate of that in animals or humans. Thus, our results argue that capsid protein made from cap DNA in AAV preparations could be at least partially responsible for the capsid-directed immune response observed in canine gene therapy experiments and in human clinical trials.
Based on data from canine studies showing a capsid-directed immune response following transient immunosuppression, a significant amount of capsid particles would need to persist for greater than 16 weeks in vivo for the response to be entirely due to administered capsid protein. Indeed, intact AAV virions have been detected up to 6 years after AAV gene transfer in the retina of dogs and primates.17 The authors hypothesized that the virions were from the initial inoculum, but our data suggest that virions also can be made from newly synthesized capsid proteins, which we have shown can be made in the absence of adenovirus helper functions.18
While investigating strategies to prevent packaging of cap DNA, we noted that traditional methods of AAV vector preparation used capsid expression cassettes small enough to fit within the AAV virion. Many copies of the cap gene are present within the cell following transfection, increasing the possibility for packaging. Alternative methods using cell lines with integrated copies of rep and cap would most likely not circumvent this problem, as it has been shown that efficient expression of AAV from cell lines required amplification of the rep-cap genes out of their integrated form. Contamination of AAV produced from a stable cell line containing ampicillin resistance gene DNA has already demonstrated that packaging of amplified DNA occurs.12, 19, 20 Much work with AAV has also focused on purification methods, such as CsCl centrifugation protocols.21 These processes can remove DNA that is not packaged in virions and can separate empty particles from DNA-containing particles, which does reduce the amount of injected capsid protein, but it is important to note that particles containing cap DNA are not empty, and are not likely to be completely excluded by any such process.
It has been reported that AAV vectors are able to express large genes by recombination of multiple gene fragments, but this process is relatively inefficient for oversized constructs.22–26 It is also unknown whether the cap DNA fragments recombine at the same rate, or whether vector recombination is due in part to the presence of the AAV ITRs. The requirement of at least two fragments for recombination and the greatly reduced packaging rate of cap gene fragments in comparison to that of AAV vectors argues that recombination events leading to Cap expression should be exceedingly rare. Indeed, we observe no evidence of any capsid expression in cultured cells when using the captron construct. We conclude that AAV captron vectors should be less immunogenic and may promote longer-term transgene expression in animals and humans compared to previous vectors.
Materials and methods
Cell culture
Human embryonic kidney 293 cells (ATCC CRL 1573) and HTX cells, an approximately diploid subclone of human HT-1080 fibrosarcoma cells (ATCC CCL 121), were maintained in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% Cosmic Calf serum (Thermo Scientific) or 10% fetal bovine serum (FBS), respectively.
AAV vector plasmids
The AAV2-based vector ARAP418 expresses AP and contains the RSV promoter/enhancers, the AP cDNA, and SV40 polyadenylation sequences. The plasmid pRAP was generated by deleting the AAV2 ITRs from pARAP4. The ACWRZn15 and ACZn15 vectors express a nuclear-localized βgal protein, and the ACAGhAAT8 vector expresses human α1-antitrypsin. The ACF3’B vector was derived from ARAP4 and contains the 3’ portion of the human CFTR cDNA (2.1 kb region downstream of the HpaI site). ACMVcFIX expresses canine clotting factor FIX and was kindly provided by Katherine High (Children’s Hospital of Philadelphia). The plasmids pDGM6,27 pLadeno5 (contains adenovirus E2A, E4, and VA RNA regions to provide helper functions), pMTrep2,18 pMTrep6,14 pCMVcap2,18 and pCMVcap614 were used to supply helper, replication and packaging functions. The pDGM6Δcap plasmid was made by removing the SwaI/BstZ171 cap fragment of pDGM6. The AAV vector plasmids were propagated in the bacterial strains Sure (Stratagene) or JC811128, and the packaging and helper plasmids were propagated in the DH5α strain of E. coli.
Generation of intron-containing cap expression construct
The 4,460 bp region including the 5’ untranslated region intron from the human EIF2S1 gene was cloned by PCR amplification of DNA from the HTX cell line using primers EIF2S1inFN (GGCTAGCgcggtggagtgagcgaag) and EIF2S1inRN (GGCTAGCttctgcaatttaaacaaaag), and inserted into the NheI site in pCMVcap6 to generate pCMVEcap6 (Figure 2A). The pCMVE2cap6 construct was generated by removal of a 200 bp KpnI fragment of pCMVEcap6 containing the splice acceptor from the EIF2S1 intron and the splice donor of the AAV cap gene (Figure 2A).
AAV vector production and characterization
AAV vectors were generated by cotransfection of vector and helper plasmids into 293 cells seeded at 4 × 106 cells per 10-cm dish the prior day. For the 2-plasmid protocol, the plasmid pDGM6 was co-transfected with the vector plasmids for generating the AAV vectors (20 µg and 10 µg per plate of 293 cells, respectively). For the 4-plasmid protocol, plasmids pLadeno5, pCMVcap6, pMTrep2 (or pMTrep6), were used to supply helper and packaging functions (5 µg of each plasmid per plate of cells). Concentration and purification of AAV vectors was done as previously described except that the centrifugation step in sucrose prior to loading on to a heparin column was omitted.29 Southern analysis was done to determine the number of vector genomes (vg) in the vector preparations that was generally between 1011 and 1012 vg per ml. Procedures for Southern analysis have been described previously.30 AAV6 vectors used in experiments had a vg to FFU ratio in HTX target cells of 2 × 104 to 5 × 104 for the AP-expressing vectors, and 105 for the βgal-expressing vector.
Quantitative PCR
Purified AAV vector stocks containing 1011 to 1.5 × 1012 vg were treated with benzonase (100 units per reaction, 5 mM Tris pH 8.0, 10 µg/ml bovine serum albumin, 0.1 mM MgCl2) in 440 µl volume for 1 h at 37°C. AAV Cap proteins were digested and virion DNA was released by addition of 10 µl proteinase K (20 mg/ml) and 50 µl of 10X SET buffer (10% SDS, 1 M Tris pH 7.5, 0.05 M EDTA pH 8.0) followed by incubation for 1 h at 50°C. Virion DNA was obtained by two phenol/chloroform extractions, and precipitated with 5 µg tRNA and 0.1 M NaCl in ethanol. Virion DNA pellets were resuspended in 100 µl distilled water.
Maxima SYBR Green/ROX kit (Fermentas, Burlington, Canada) was used for the detection of rep and cap genes using the ABI 7900HT Real Time PCR System. Each reaction was done in triplicates with 25 µl volumes containing 12.5 µl of 2X master mix, 150 nM of each primer, and 1 µl template (resuspended vector DNA). The primers RepF (CGGGGTTTTACGAGATTGTG, nucleotide positions 326–345) and RepR1 (CGCCATTTCTGGTCTTTGTG, nucleotides 742-721) were used for amplification of rep yielding a 416 bp PCR product and primers CapF (CCACAAGAGCCAGACTCCTC, nucleotides 2655–2674) and CapR540 (GCCATCATTCGTCGTGACC, nucleotides 3193-3175) were used for cap gene amplification giving a 539 bp product. Cycling conditions were 95°C for 10 min, followed by 40 cycles of 95°C for 30 s, 55°C for 1 min, and 72°C for 40 s. Plasmid pDGM6 representing 102 to 106 copies was used to generate the standard curve to calculate the copy number in the samples for both rep and cap (linearized with NotI for 1 of 2 cap reactions and both rep reactions). Primers for canine FIX and for βgal were used to determine the amount of vector genomes in reaction. For cFIX, primers cFIXF1 (CCACTGTATTGAGCCTGATGTTAAA) and cFIXR2 (GCTCTGTATGTTCCCTCTTCTCGGT) were used, and pCMVcFIX was used for standardization. For βgal, primers lacZqF1 (GCGTTAACTCGGCGTTTCAT) and lacZqR1 (GCGCTCAGGTCAAATTCAGAC) were used, and pACZn was used for standardization. For all other AAV vectors, vg values were determined by Southern analysis. When comparisons were available, vector genome values derived by Southern analysis and by qPCR varied by less than 2-fold.
qRT-PCR
HTX cells were infected with an AAV6 vector (CMVcFIX) made with either pCMVcap6 plasmid or the captron plasmid pCMVE2cap6 at an MOI of 4 × 106 vg per cell. Total RNA was extracted using TRIZOL (Invitrogen) after three days. The RNA was treated with DNase and cDNA was synthesized using an oligo (dT20) primer using SuperScript III Reverse Transcriptase (Invitrogen). qRT-PCR was conducted in triplicate using Maxima SYBR Green qPCR master mix (Fermentas) with cDNA from 300 ng of RNA. The primers and standards for cap and cFIX were the same as those used for the above qPCR analysis of virion DNA, except that both pDGM6 and pCMVcFIX standards were linearized for all reactions (with NotI and HindIII, respectively). All standard curve reactions included cDNA made from uninfected HTX cells in addition to the known amounts of linearized plasmid. All primers were used at 0.2 µM and cycle conditions for both reactions were 95°C for 10 min, with 45 cycles of 95°C for 15 s and 60°C for 1 min, followed by a dissociation curve.
Cap and Rep complementation assay
293 cells were plated at 5 × 106 cells per 10-cm dish (day 0). The following day, the AAV vector was treated with benzonase (100 units per ml) at 37°C for 1 h in a total volume of 0.5 ml (7 × 1010 to 2 × 1012 vg) per plate of 293 cells. Cells were then infected with purified benzonase-treated AAV vector preparations for 2 h and then the medium was replaced to remove benzonase and residual vector. Cells were transfected 1–2 h later with plasmid combinations. On day 2, the transfection/infection medium was aspirated and replaced with medium with 2 mM added L-glutamine. On day 4, cells and medium were harvested, freeze/thawed 3 times, centrifuged to remove cell debris, and was frozen in 1 ml aliquots. Prior to infection, 1 ml of cell lysate was thawed and treated for 2 h with benzonase. Then the transducing titer of cell lysate was determined by quantitating AP+ FFU on HTX cells. HTX cells were plated at 5 × 104 cells per well in 6-well dishes on day 0. The following day, medium was aspirated, cells rinsed once with PBS containing calcium and magnesium, then exposed to 500 µl of cell lysate with 500 µl of DMEM for 2 h, then 1 ml of DMEM containing 20% FBS was added. The cells were fixed and stained for AP expression 3 days later.
Acknowledgments
We thank Katherine High for providing the ACMVcFIX AAV vector that expresses canine FIX. This work was supported by grants P30 DK47754, UL1 DE19582 and T32 CA09229 from the National Institutes of Health.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
REFERENCES
- 1.Herzog RW, Yang EY, Couto LB, Hagstrom JN, Elwell D, Fields PA, et al. Long-term correction of canine hemophilia B by gene transfer of blood coagulation factor IX mediated by adeno-associated viral vector. Nat Med. 1999;5:56–63. doi: 10.1038/4743. [DOI] [PubMed] [Google Scholar]
- 2.Passini MA, Watson DJ, Vite CH, Landsburg DJ, Feigenbaum AL, Wolfe JH. Intraventricular brain injection of adeno-associated virus type 1 (AAV1) in neonatal mice results in complementary patterns of neuronal transduction to AAV2 and total long-term correction of storage lesions in the brains of β-glucuronidase-deficient mice. J Virol. 2003;77:7034–7040. doi: 10.1128/JVI.77.12.7034-7040.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weber M, Rabinowitz J, Provost N, Conrath H, Folliot S, Briot D, et al. Recombinant adeno-associated virus serotype 4 mediates unique and exclusive long-term transduction of retinal pigmented epithelium in rat, dog, and nonhuman primate after subretinal delivery. Mol Ther. 2003;7:774–781. doi: 10.1016/s1525-0016(03)00098-4. [DOI] [PubMed] [Google Scholar]
- 4.Mochizuki S, Mizukami H, Ogura T, Kure S, Ichinohe A, Kojima K, et al. Long-term correction of hyperphenylalaninemia by AAV-mediated gene transfer leads to behavioral recovery in phenylketonuria mice. Gene Ther. 2004;11:1081–1086. doi: 10.1038/sj.gt.3302262. [DOI] [PubMed] [Google Scholar]
- 5.Manno CS, Arruda VR, Pierce GF, Glader B, Ragni M, Rasko J, et al. Successful transduction of liver in hemophilia by AAV-Factor IX and limitations imposed by the host immune response. Nat Med. 2006;12:342–347. doi: 10.1038/nm1358. [DOI] [PubMed] [Google Scholar]
- 6.Mingozzi F, Meulenberg JJ, Hui DJ, Basner-Tschakarjan E, Hasbrouck NC, Edmonson SA, et al. AAV-1-mediated gene transfer to skeletal muscle in humans results in dose-dependent activation of capsid-specific T cells. Blood. 2009;114:2077–2086. doi: 10.1182/blood-2008-07-167510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Z, Kuhr CS, Allen JM, Blankinship M, Gregorevic P, Chamberlain JS, et al. Sustained AAV-mediated dystrophin expression in a canine model of Duchenne muscular dystrophy with a brief course of immunosuppression. Mol Ther. 2007;15:1160–1166. doi: 10.1038/sj.mt.6300161. [DOI] [PubMed] [Google Scholar]
- 8.Halbert CL, Madtes DK, Vaughan AE, Wang Z, Storb R, Tapscott SJ, et al. Expression of human α1-antitrypsin in mice and dogs following AAV6 vector-mediated gene transfer to the lungs. Mol Ther. 2010;18:1165–1172. doi: 10.1038/mt.2010.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allen JM, Debelak DJ, Reynolds TC, Miller AD. Identification and elimination of replication-competent adeno-associated virus (AAV) that can arise by nonhomologous recombination during AAV vector production. J Virol. 1997;71:6816–6822. doi: 10.1128/jvi.71.9.6816-6822.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao L, Liu Y, During MJ, Xiao W. High-titer, wild-type free recombinant adeno-associated virus vector production using intron-containing helper plasmids. J Virol. 2000;74:11456–11463. doi: 10.1128/jvi.74.24.11456-11463.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nony P, Chadeuf G, Tessier J, Moullier P, Salvetti A. Evidence for packaging of rep-cap sequences into adeno-associated virus (AAV) type 2 capsids in the absence of inverted terminal repeats: a model for generation of rep-positive AAV particles. J Virol. 2003;77:776–781. doi: 10.1128/JVI.77.1.776-781.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chadeuf G, Ciron C, Moullier P, Salvetti A. Evidence for encapsidation of prokaryotic sequences during recombinant adeno-associated virus production and their in vivo persistence after vector delivery. Mol Ther. 2005;12:744–753. doi: 10.1016/j.ymthe.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 13.Hauck B, Murphy SL, Smith PH, Qu G, Liu X, Zelenaia O, et al. Undetectable transcription of cap in a clinical AAV vector: implications for preformed capsid in immune responses. Mol Ther. 2009;17:144–152. doi: 10.1038/mt.2008.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Halbert CL, Allen JM, Miller AD. Adeno-associated virus type 6 (AAV6) vectors mediate efficient transduction of airway epithelial cells in mouse lungs compared to that of AAV2 vectors. J Virol. 2001;75:6615–6624. doi: 10.1128/JVI.75.14.6615-6624.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Halbert CL, Lam SL, Miller AD. High-efficiency promoter-dependent transduction by adeno-associated virus type 6 vectors in mouse lung. Hum Gene Ther. 2007;18:344–354. doi: 10.1089/hum.2006.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blankinship MJ, Gregorevic P, Allen JM, Harper SQ, Harper H, Halbert CL, et al. Efficient transduction of skeletal muscle using vectors based on adeno-associated virus serotype 6. Mol Ther. 2004;10:671–678. doi: 10.1016/j.ymthe.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 17.Stieger K, Schroeder J, Provost N, Mendes-Madeira A, Belbellaa B, Le Meur G, et al. Detection of intact rAAV particles up to 6 years after successful gene transfer in the retina of dogs and primates. Mol Ther. 2009;17:516–523. doi: 10.1038/mt.2008.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allen JM, Halbert CL, Miller AD. Improved adeno-associated virus vector production with transfection of a single helper adenovirus gene E4orf6. Mol Ther. 2000;1:88–95. doi: 10.1006/mthe.1999.0010. [DOI] [PubMed] [Google Scholar]
- 19.Chadeuf G, Favre D, Tessier J, Provost N, Nony P, Kleinschmidt J, et al. Efficient recombinant adeno-associated virus production by a stable rep-cap HeLa cell line correlates with adenovirus-induced amplification of the integrated rep-cap genome. J Gene Med. 2000;2:260–268. doi: 10.1002/1521-2254(200007/08)2:4<260::AID-JGM111>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 20.Liu X, Voulgaropoulou F, Chen R, Johnson PR, Clark KR. Selective Rep-Cap gene amplification as a mechanism for high-titer recombinant AAV production from stable cell lines. Mol Ther. 2000;2:394–403. doi: 10.1006/mthe.2000.0132. [DOI] [PubMed] [Google Scholar]
- 21.Ayuso E, Mingozzi F, Montane J, Leon X, Anguela XM, Haurigot V, et al. High AAV vector purity results in serotype- and tissue-independent enhancement of transduction efficiency. Gene Ther. 2010;17:503–510. doi: 10.1038/gt.2009.157. [DOI] [PubMed] [Google Scholar]
- 22.Duan D, Yue Y, Engelhardt JF. Expanding AAV packaging capacity with trans-splicing or overlapping vectors: a quantitative comparison. Mol Ther. 2001;4:383–391. doi: 10.1006/mthe.2001.0456. [DOI] [PubMed] [Google Scholar]
- 23.Halbert CL, Allen JM, Miller AD. Efficient mouse airway transduction following recombination between AAV vectors carrying parts of a larger gene. Nat Biotechnol. 2002;20:697–701. doi: 10.1038/nbt0702-697. [DOI] [PubMed] [Google Scholar]
- 24.Lai Y, Yue Y, Duan D. Evidence for the failure of adeno-associated virus serotype 5 to package a viral genome ≥8.2 kb. Mol Ther. 2010;18:75–79. doi: 10.1038/mt.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu Z, Yang H, Colosi P. Effect of genome size on AAV vector packaging. Mol Ther. 2010;18:80–86. doi: 10.1038/mt.2009.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dong B, Nakai H, Xiao W. Characterization of genome integrity for oversized recombinant AAV vector. Mol Ther. 2010;18:87–92. doi: 10.1038/mt.2009.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gregorevic P, Blankinship MJ, Allen JM, Crawford RW, Meuse L, Miller DG, et al. Systemic delivery of genes to striated muscles using adeno-associated viral vectors. Nat Med. 2004;10:828–834. doi: 10.1038/nm1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boissy R, Astell CR. An Escherichia coli recBCsbcBrecF host permits the deletion-resistant propagation of plasmid clones containing the 5'-terminal palindrome of minute virus of mice. Gene. 1985;35:179–185. doi: 10.1016/0378-1119(85)90170-2. [DOI] [PubMed] [Google Scholar]
- 29.Halbert CL, Miller AD. AAV-mediated gene transfer to mouse lungs. Methods Mol Biol. 2004;246:201–212. doi: 10.1385/1-59259-650-9:201. [DOI] [PubMed] [Google Scholar]
- 30.Halbert CL, Standaert TA, Aitken ML, Alexander IE, Russell DW, Miller AD. Transduction by adeno-associated virus vectors in the rabbit airway: efficiency, persistence, and readministration. J Virol. 1997;71:5932–5941. doi: 10.1128/jvi.71.8.5932-5941.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]