Abstract
The oncoproteins of small DNA tumor viruses promote tumorigenesis by complexing with cellular factors intimately involved in the control of cell proliferation. The major oncogenic determinants for human adenovirus type 9 (Ad9) and high-risk human papillomaviruses (HPV) are the E4-ORF1 and E6 proteins, respectively. These seemingly unrelated viral oncoproteins are similar in that their transforming activities in cells depend, in part, on a carboxyl-terminal PDZ domain-binding motif which mediates interactions with the cellular PDZ-protein DLG. Here we demonstrated that both Ad9 E4-ORF1 and high-risk HPV E6 proteins also bind to the DLG-related PDZ-protein MAGI-1. These interactions resulted in MAGI-1 being aberrantly sequestered in the cytoplasm by the Ad9 E4-ORF1 protein or being targeted for degradation by high-risk HPV E6 proteins. Transformation-defective mutant viral proteins, however, were deficient for these activities. Our findings indicate that MAGI-1 is a member of a select group of cellular PDZ proteins targeted by both adenovirus E4-ORF1 and high-risk HPV E6 proteins and, in addition, suggest that the tumorigenic potentials of these viral oncoproteins depend, in part, on an ability to inhibit the function of MAGI-1 in cells.
Keywords: adenovirus E4-ORF1, MAGI-1, papillomavirus E6, PDZ
Introduction
Human adenoviruses are associated primarily with respiratory, gastrointestinal, and eye infections in people but, in rodents, some of these viruses have the capacity to induce tumors (Shenk, 1996). Based on the types of tumors elicited and the oncoproteins that determine their tumorigenicity, two different classes of oncogenic human adenoviruses can be distinguished. Human adenoviruses from subgroups A and B induce primarily undifferentiated sarcomas at the site of injection, and the tumorigenic potential of these viruses depends solely on their nuclear E1A and E1B transforming proteins (Shenk, 1996). In contrast, subgroup D human adenovirus type 9 (Ad9) generates exclusively estrogen-dependent mammary tumors (Javier et al., 1991), and the tumorigenic potential of this virus relies on its cytoplasmic Ad9 E4-ORF1 (9ORF1) transforming protein (Javier, 1994; Thomas et al., 1999).
Human papillomaviruses (HPV) are the etiological agents of warts in people. With regard to HPVs that infect the genital tract, high-risk HPVs (types 16, 18, 31, and 45) are strongly associated with cervical cancer whereas low-risk HPVs (types 6 and 11) are weakly or not associated with this disease (Howley, 1996). In addition, the major oncogenic determinants of high-risk HPVs are their E7 and E6 gene products. Interestingly, the tumorigenic potentials of high-risk HPV E7 and E6 and adenovirus E1A and E1B, as well as SV40 large T-antigen, similarly depend in part on their capacity to complex with and inactivate the tumor suppressor proteins pRb and p53 (Nevins and Vogt, 1996). Such findings have revealed that seemingly unrelated oncoproteins from DNA tumor viruses often target common cellular factors having critical roles in the control of cellular proliferation. We and others recently showed that the seemingly unrelated adenovirus E4-ORF1 and high-risk HPV E6 proteins, as well as the human T-cell leukemia virus type I (HTLV-1) Tax protein, likewise target a common cellular factor (Lee et al., 1997). It was found that these viral oncoproteins similarly bind to the cellular PDZ domain-containing protein DLG (Kiyono et al., 1997; Lee et al., 1997), which is a mammalian homolog of the Drosophila tumor suppressor protein dlg (Lue et al., 1994; Muller et al., 1995).
PDZ domains are 80–90 amino-acid protein-protein interaction modules most often found within cellular factors that function in signal transduction (Fanning and Anderson, 1999). These domains typically bind specific sequence motifs located at the extreme carboxyl-terminus of target proteins, although they also participate in other types of protein interactions. Three different types of carboxyl-terminal PDZ domain-binding motifs are recognized and, at their extreme carboxyl-termini, adenovirus E4-ORF1, high-risk HPV E6, and HTLV-1 Tax proteins possess a type I motif having the consensus sequence -(T/S)-X-(V/I)-COOH (X, any amino-acid [aa] residue) (Lee et al., 1997). These PDZ domain-binding motifs mediate interactions with one or more PDZ domains of DLG and, for the 9ORF1 and high-risk HPV-16 E6 proteins, disruption of this motif abolishes their transforming activity (Kiyono et al., 1997; Lee et al., 1997). These findings suggest that transformation by these viral oncoproteins depends in part on their ability to block the function of DLG.
Drosophila dlg has been designated as a tumor suppressor protein because, for larvae carrying homozygous dlg mutations, imaginal disc epithelial cells exhibit loss of polarity and neoplastic outgrowth and, in addition, certain neuronal cells of the brain undergo hyperplastic growth (Woods and Bryant, 1991). The fact that DLG rescues the phenotypic defects of Drosophila unable to express functional dlg indicates that these two proteins are functionally homologous (Thomas et al., 1997). These two closely-related proteins are members of the membrane-associated guanylate kinase (MAGUK) family of proteins, which typically have a domain structure consisting of one or more amino-terminal PDZ domains, an internal SH3 domain, and a carboxyl-terminal guanylate kinase-homology domain (Craven and Bredt, 1998). In general, this family of polypeptides functions to properly localize membrane and cytosolic proteins to the plasma membrane at specialized regions of cell-cell contact, as well as to organize these targets into large signaling complexes (Fanning and Anderson, 1999). These results, together with our recent finding that high-risk HPV E6 proteins target DLG for degradation in cells (Gardiol et al., 1999), suggest a model whereby the proposed cell signaling regulatory activities of DLG function to suppress inappropriate proliferation of cells.
Our previous findings with the 9ORF1 oncoprotein suggest that its oncogenic potential depends not only on interactions with DLG (Lee et al., 1997), but also with other unidentified cellular PDZ proteins (p220, p180, p160, p155) (Weiss et al., 1997a). In this paper, we screened a panel of large cellular PDZ proteins for an ability to bind the 9ORF1 protein in order to identify additional cellular factors that contribute to 9ORF1-induced transformation. We found that 9ORF1, as well as high-risk HPV E6 oncoproteins, selectively complex with the widely-expressed cellular PDZ-protein MAGI-1, a MAGUK protein related to DLG. Additional results showed that MAGI-1 is aberrantly sequestered in the cytoplasm of cells by the 9ORF1 protein and is targeted for degradation in cells by high-risk HPV E6 proteins, suggesting that the transforming potentials of two unrelated viral oncoproteins depend in part on an ability to inactivate this cellular PDZ protein.
Results
9ORF1 complexes with the PDZ-protein MAGI-1 in cells
The transforming activity of the 9ORF1 oncoprotein depends on its carboxyl-terminal PDZ domain-binding motif (Table 1) (Lee et al., 1997), which mediates direct interactions with multiple large cellular polypeptides (p220, p180, p160, p155, and p140/p130) (Weiss et al., 1997a). Whereas 9ORF1-associated protein p140/p130 was previously identified as the PDZ-protein DLG (Lee et al., 1997), the identities of the remaining 9ORF1-associated proteins have not been determined. Reasoning that, like DLG, these unidentified 9ORF1-associated polypeptides contain PDZ domains and also considering their predicted sizes (155–220 kD), we examined a group of cellular PDZ-proteins that included FAP-1 (273 kD) (Sato et al., 1995), ZO-1 (220 kD) (Willott et al., 1993), AF-6 (182 kD) (Prasad et al., 1993), hINADL (167 kD) (Philipp and Flockerzi, 1997), MAGI-1 (152 kD) (Dobrosotskaya et al., 1997), and ZO-3 (130 kD) (Haskins et al., 1998) for binding to 9ORF1. Using a variety of assays, we demonstrated that 9ORF1 bound to the widely-expressed PDZ-protein MAGI-1 (see below), but not to the other PDZ-proteins indicated above (data not shown).
Table 1.
Carboxyl-terminal amino-acid sequences of wild-type and mutant adenovirus E4-ORF1 and human papillomavirus (HPV) E6 proteins*
| Proteins | Carboxyl-terminal amino-acid sequence Consensus type I PDZ domain-binding motif | |||
|---|---|---|---|---|
| X | (S/T) | X | (V/I/L)-COOH | |
| Adenovirus E4-ORF1 | ||||
| wt 9ORF1 | A | T | L | V |
| mutant IIIA | A | P | ||
| mutant IIIC | D | T | L | V |
| mutant IIID | A | T | P | V |
| wt 5ORF1 | A | S | N | V |
| wt 12ORF1 | A | S | L | I |
| HPV E6 | ||||
| wt 18E6 | E | T | Q | V |
| mutant 18E6- | E | T | Q | A |
| V158A | ||||
| wt 16E6 | E | T | Q | L |
| mutant 16E6- | E | D | Q | A |
| T149D/L151A | ||||
| wt 11E6 | D | L | L | P |
The carboxyl-terminal sequences of Ad9 E4 ORF1 (9ORF1), HPV-18 E6 (18E6), and HPV-16 E6 (16E6) define a type I PDZ domain-binding motif, which is not present at the carboxyl-terminus of HPV-11 E6 (11E6). Substitution mutations are indicated by bold amino-acid residues
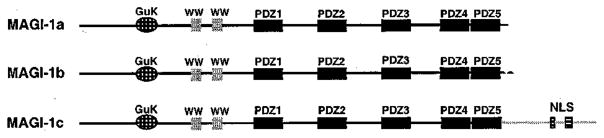
Although its function is not known, MAGI-1 is a MAGUK protein related to DLG (Dobrosotskaya et al., 1997). MAGI-1 is structurally inverted relative to DLG, however, as MAGI-1 has a guanylate kinase-homology domain at its amino-terminus and five PDZ domains at its carboxyl-terminus (Figure 1). In addition, MAGI-1 possesses two WW domains rather than the SH3 domain of DLG. Three MAGI-1 isoforms (a, b and c), identical except for sequences carboxyl-terminal to PDZ5, have been identified (Figure 1). The presence of a consensus bipartite nuclear localization signal within the unique carboxyl-terminal sequences of MAGI-1c hints that this particular isoform may under certain conditions have functions in the nucleus.
Figure 1.
Three isoforms of MAGI-1. MAGI-1 has an inverted MAGUK domain structure with a guanylate kinase-homology domain (GuK) at its amino-terminus and PDZ domains at its carboxyl-terminus. MAGI-1a, -1b, and -1c isoforms are identical except that their sequences diverge carboxyl-terminal to PDZ5. WW, WW domain; NLS, putative bipartite nuclear localization signal
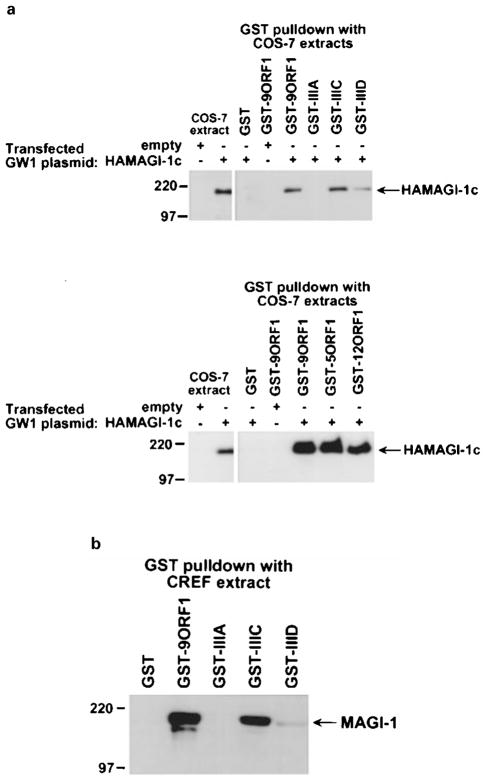
To show binding of MAGI-1 to the 9ORF1 protein, we subjected extracts of COS-7 cells expressing HA epitope-tagged mouse MAGI-1b (HAMAGI-1b) or MAGI-1c (HAMAGI-1c) to GST-pulldown assays with a wild-type 9ORF1 fusion protein. We found that wild-type 9ORF1 complexed similarly with both MAGI-1b (data not shown) and MAGI-1c (Figure 2a; upper panel). Like 9ORF1, the related E4-ORF1 transforming proteins of adenovirus types 5 (5ORF1) and 12 (12ORF1) also possess carboxyl-terminal PDZ domain-binding motifs (Table 1) and likewise bound to both MAGI-1b (data not shown) and MAGI-1c in these assays (Figure 2a; lower panel). With the use of MAGI-1-specific antibodies (Dobrosotskaya et al., 1997), we also showed that 9ORF1 is similarly able to associate with endogenous MAGI-1 protein derived from extracts of CREF rat embryo fibroblasts (Figure 2b). Which MAGI-1 isoform(s) is expressed in CREF cells was not determined, but the size of the detected polypeptide is most consistent with that of MAGI-1c. Also notable was that MAGI-1 and 9ORF1-associated protein p180 co-migrated in protein gels, suggesting that these proteins are the same (Figure 3).
Figure 2.
Binding of 9ORF1 to MAGI-1 in vitro. (a) 9ORF1 protein binding to mouse MAGI-1c detected in GST-pulldown assays. Extracts of RIPA buffer-lysed COS-7 cells transfected with 5 μg of empty GW1 or 5 μg of GWI-HAMAGI-1c plasmid were used in GST-pulldown reactions with the indicated GST fusion protein, and recovered proteins were immunoblotted with HA antibodies. Upper panel, MAGI-1 binding to wild-type and mutant 9ORF1 proteins (see Table 1). Lower panel, MAGI-1 binding to the wild-type E4-ORF1 proteins of Ad9, Ad5 (5ORF1), and Ad12 (12ORF1). COS-7 extracts representing one-half the amount used in GST pulldown reactions were also directly immunoblotted with HA antibodies as a control. (b) 9ORF1 binding to endogenous rat MAGI-1 of CREF cells using GST-pulldown assays. CREF cell extracts in RIPA buffer were subjected to GST-pulldown assays and then immunoblotted with MAGI-1 antibodies
Figure 3.
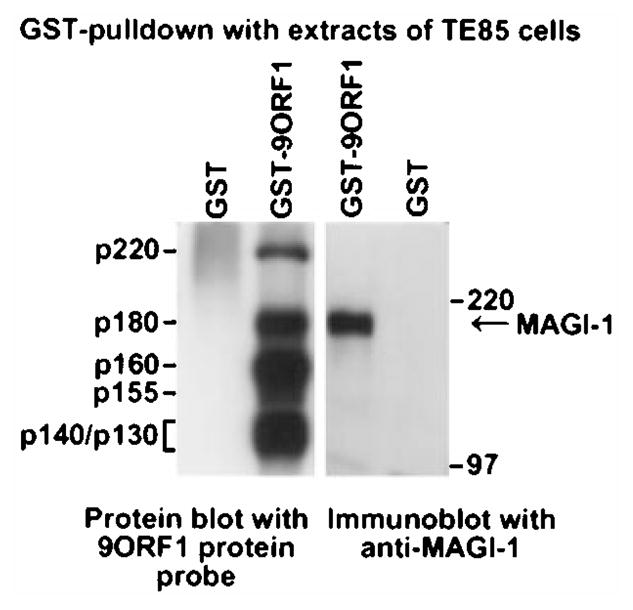
Co-migration of MAGI-1 and 9ORF1-associated protein p180 in a protein gel. GST-pulldown reactions using GST or GST-9ORF1 protein were performed with extracts of human TE85 cells in RIPA buffer. Recovered proteins from duplicate GST-pulldown reactions were separated in parallel by SDS–PAGE and transferred to a membrane, and membranes were either blotted with a radiolabeled 9ORF1 protein probe (left) or with MAGI-1 antibodies (right)
The specificity of the binding results described above in Figure 2 was demonstrated by inclusion of 9ORF1 mutant proteins in the same experiments. The PDZ domain-binding motif of severely transformation-defective 9ORF1 mutant IIIA is disrupted by deletion (Table 1), which renders this mutant unable to complex with any 9ORF1-associated proteins (Weiss et al., 1997a; Weiss and Javier, 1997). In contrast, the PDZ domain-binding motifs of the weak-transforming 9ORF1 mutants IIIC and IIID have less disruptive missense mutations (Table 1), which permit these mutants to bind a subset of 9ORF1-associated proteins, albeit at substantially reduced levels in most cases (Weiss et al., 1997a; Weiss and Javier, 1997). In GST-pulldown assays, we found that MAGI-1 failed to complex with mutant IIIA, yet complexed with mutant IIIC at approximately wild-type levels or with mutant IIID at substantially reduced levels (Figures 2a, b). The fact that this binding profile of MAGI-1 to wild-type and mutant 9ORF1 proteins was identical to that previously observed for 9ORF1-associated protein p180 (Weiss and Javier, 1997) provided further support for the idea that these proteins are the same. More important, these findings indicated that 9ORF1 binding to MAGI-1 is specific and depends on a functional 9ORF1 PDZ domain-binding motif.
We next performed reciprocal co-immunoprecipitation assays with extracts of COS-7 cells co-expressing HAMAGI-1c and either wild-type or mutant 9ORF1 protein. We found that MAGI-1 co-precipitated with wild-type 9ORF1 but failed to co-precipitate with mutant IIIA (Figure 4). MAGI-1 also co-precipitated with mutants IIIC and IIID, but at levels slightly below or substantially below, respectively, that of the wild-type 9ORF1 protein (Figure 4). These results were concordant with those of the GST-pulldown assays (see Figure 2) and also indicated that 9ORF1 and MAGI-1 form specific complexes in cells.
Figure 4.

Binding of 9ORF1 to MAGI-1 in vivo. Co-immuno-precipitation assays were performed with extracts of COS-7 cells transfected with 4 μg of GW1-HAMAGI-1 plasmid and either 4 μg empty GW1 or 4 μg of GW1 plasmid expressing wild-type or the indicated mutant 9ORF1 protein. COS-7 extracts in RIPA buffer were subjected to immunoprecipitation with either HA antibodies (upper panel) or 9ORF1 antibodies (lower panel), and recovered proteins were separately immunoblotted with the same two antibodies. In the lower panel, immunoprecipitation of the COS-7 extract with pre-immune serum (pre) was included as a negative control
9ORF1 interacts primarily with MAGI-1 PDZ1 and PDZ3
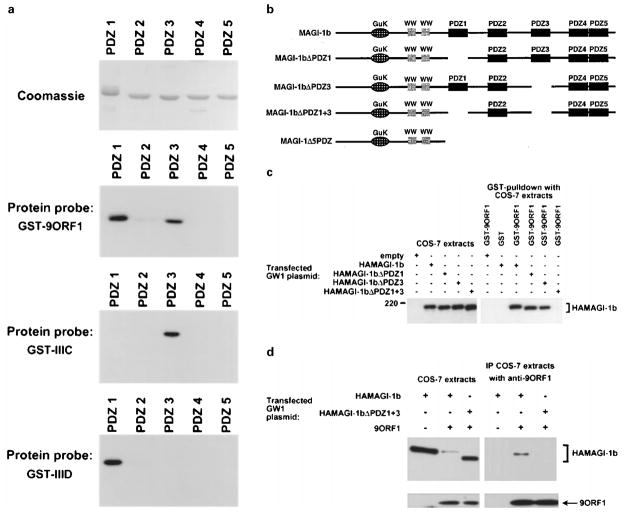
The fact that disrupting the PDZ domain-binding motif of 9ORF1 impairs its binding to MAGI-1 implies that this viral protein binds one or more MAGI-1 PDZ domains. To verify this prediction, we performed protein blotting assays by incubating membrane-immobilized fusion proteins of individual MAGI-1 PDZ domains with a radiolabeled 9ORF1 protein probe. In these assays, the wild-type 9ORF1 protein probe bound strongly with PDZ1 and PDZ3, weakly with PDZ2, but failed to bind either PDZ4 or PDZ5 (Figure 5a). Additionally, none of these MAGI-1 PDZ domains reacted with a mutant IIIA protein probe (data not shown), indicating that the detected binding was specific and dependent on a functional 9ORF1 PDZ domain-binding motif.
Figure 5.
Binding of 9ORF1 to MAGI-1 PDZ1 and PDZ3. (a) Strong binding of 9ORF1 to two of five MAGI-1 PDZ domains. GST proteins fused to individual MAGI-1 PDZ domains were separated by SDS– PAGE, immobilized on duplicate membranes, and either stained with coomassie or protein blotted with the indicated wild-type or mutant 9ORF1 fusion protein probe. (b) Illustration of MAGI-1 deletion mutants. (c) A MAGI-1 mutant missing PDZ1 and PDZ3 fails to interact with 9ORF1 in GST pulldown assays. Extracts of RIPA buffer-lysed COS-7 cells transfected with 5 μg of GW1 plasmid expressing either HA-tagged wild-type or the indicated mutant MAGI-1 protein were subjected to GST-pulldown reactions with either GST or GST-9ORF1 fusion protein, and recovered proteins were immunoblotted with HA antibodies. COS-7 extracts representing one-tenth the amount used in GST pulldown reactions were also directly immunoblotted with HA antibodies as a control. (d) A MAGI-1 mutant missing PDZ1 and PDZ3 fails to interact with 9ORF1 in co-immunoprecipitation assays. Co-immunoprecipitation assays were performed with extracts of COS-7 cells transfected with 4 μg of wild-type or mutant GW1-HAMAGI-1 plasmid and either 4 μg empty GW1 or 4 μg of GW1-9ORF1 plasmid. COS-7 extracts in RIPA buffer were subjected to immunoprecipitation with 9ORF1 antibodies, and recovered proteins were separately immunoblotted with either HA antibodies (upper panels) or 9ORF1 antibodies (lower panels)
As 9ORF1 mutants IIIC and IIID displayed nearly wild-type or reduced binding, respectively, to MAGI-1 (see Figures 2 and 4), these mutant proteins were also used as probes in protein blotting assays. In these experiments, mutants IIIC and IIID displayed reciprocal defects in binding to MAGI-1 PDZ1 and PDZ3. Specifically, mutant IIIC reacted with PDZ3 but not with PDZ1 and mutant IIID reacted with PDZ1 but not with PDZ3 (Figure 5a). Also, mutant IIID bound to MAGI-1 PDZ2, but mutant IIIC did not interact detectably with this domain. These results revealed that, although they are able to bind MAGI-1, mutants IIIC and IIID both have impaired domain interactions with this PDZ protein.
To confirm that MAGI-1 PDZ1 and PDZ3 primarily determine binding of 9ORF1 to the full-length MAGI-1 polypeptide, we constructed a MAGI-1 double-deletion mutant missing both PDZ1 and PDZ3 (HAMAGI-1ΔPDZ1+3) (Figure 5b). In agreement with the results of protein blotting assays (see Figure 5a), 9ORF1 failed to bind HAMAGI-1ΔPDZ1+3 both in GST-pulldown assays (Figure 5c) and in co-immunoprecipitation assays (Figure 5d). Although 9ORF1 can bind to MAGI-1 PDZ2 (see Figure 5a), this weak interaction was presumably too low to detect in these experiments. These findings showed that MAGI-1 PDZ1 and PDZ3, and no other region of MAGI-1, largely mediate binding to 9ORF1.
Contrary to results obtained with the HAMAGI-1ΔPDZ1+3 double-deletion mutant, MAGI-1 single-deletion mutants missing either only PDZ1 (HAMA-GI-1ΔPDZ1) or only PDZ3 (HAMAGI-1ΔPDZ3) (Figure 5b) associated with 9ORF1 at approximately wild-type levels (Figure 5c), demonstrating that either PDZ1 alone or PDZ3 alone is sufficient to confer upon MAGI-1 wild-type binding to 9ORF1. This finding also suggested that the strong binding of mutant IIIC to MAGI-1 (see Figures 2 and 4) is due to this mutant retaining approximately wild-type affinity for PDZ3 and that the weak binding of mutant IIID to MAGI-1 (see Figures 2 and 4) is due to this mutant having reduced affinity for PDZ1. The reason that the predicted reduced affinity of mutant IIID for PDZ1 was not revealed in protein blotting assays (see Figure 5a) is not clear, but it may be due to differences in the specific activity of each protein probe.
9ORF1 aberrantly sequesters MAGI-1 in the cytoplasm of cells
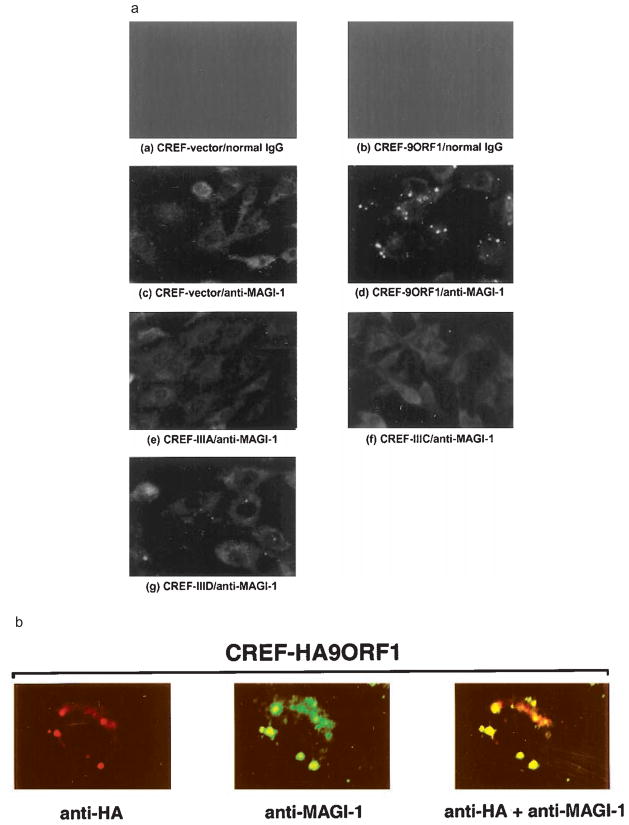
Indirect immunofluorescence (IF) microscopy assays were used to ascertain the subcellular distribution of MAGI-1 in cells. We found that, in normal CREF fibroblasts, MAGI-1 was primarily distributed diffusely within the cytoplasm (Figure 6a). Other PDZ proteins also localize in the cytoplasm (Wu et al., 1998; Yang et al., 1998), although these types of polypeptides more often associate with the plasma membrane at sites of cell-cell contact in epithelial cells (Fanning and Anderson, 1999). Because 9ORF1 is present within punctate bodies in the cytoplasm of CREF cells (Weiss et al., 1996), we hypothesized that the staining pattern of MAGI-1 in 9ORF1-expressing CREF cells would be dramatically altered from that of normal CREF cells, resembling that of 9ORF1. As expected, MAGI-1 was redistributed within punctate bodies in the cytoplasm of CREF cells expressing wild-type 9ORF1 (CREF-9ORF1) (Figure 6a). This aberrant localization of MAGI-1 was due to association of 9ORF1 with MAGI-1, as these proteins co-localized in these cells (Figure 6b).
Figure 6.
Aberrant sequestration of MAGI-1 in the cytoplasm of 9ORF1-expressing CREF cells. (a) Localization of MAGI-1 in normal CREF cells or CREF cells expressing wild-type or mutant 9ORF1 proteins. Indirect immunofluorescence assays were performed with normal CREF cells (panels a and c) or CREF cells stably expressing wild-type 9ORF1 (panels b and d), mutant IIIA (panel e), mutant IIIC (panel f), or mutant IIID (panel g). Cells were reacted with either normal rabbit IgG (panels a–b) or MAGI-1 antibodies (panels c–g) and visualized by fluorescence microscopy. (b) Co-localization of 9ORF1 and MAGI-1 proteins in CREF cells. Double-label indirect immunofluorescence assays were performed by reacting CREF cells stably expressing HA epitope-tagged 9ORF1 (CREF-HA9ORF1) with both HA and MAGI-1 antibodies. Each of the three panels represents the same field containing a single representative cell stained for 9ORF1 (left panel), MAGI-1 (center panel), or the merged images (right panel)
We also examined the subcellular distribution of MAGI-1 in CREF cell lines stably expressing mutant 9ORF1 proteins (see Table 1) which, similar to wild-type 9ORF1, exhibit punctate cytoplasmic staining in CREF cells (Weiss et al., 1997a). Despite the fact that each of the different CREF cell lines expressed 9ORF1 protein at comparable levels (Figure 7, upper panel), the staining pattern of MAGI-1 in the CREF cell line expressing mutant IIIA (CREF-IIIA; Figure 6a), which fails to bind MAGI-1 (see Figure 2), was similar to that of normal CREF cells, whereas substantially less aberrant punctate staining for MAGI-1 was detected in the cytoplasm of the CREF cell line expressing mutant IIID (CREF-IIID; Figure 6a), which binds weakly to MAGI-1 (see Figure 2). Interestingly, the CREF cell line expressing mutant IIIC (CREF-IIIC), which binds MAGI-1 at nearly wild-type levels (see Figure 2), showed a MAGI-1 staining pattern similar to that of normal CREF cells (Figure 6a). Therefore, in addition to having defective PDZ-domain interactions with MAGI-1 (see Figure 5a), mutants IIIC and IIID also either failed or showed a substantially reduced capacity, respectively, to sequester MAGI-1 within punctate bodies in the cytoplasm of CREF cells.
Figure 7.
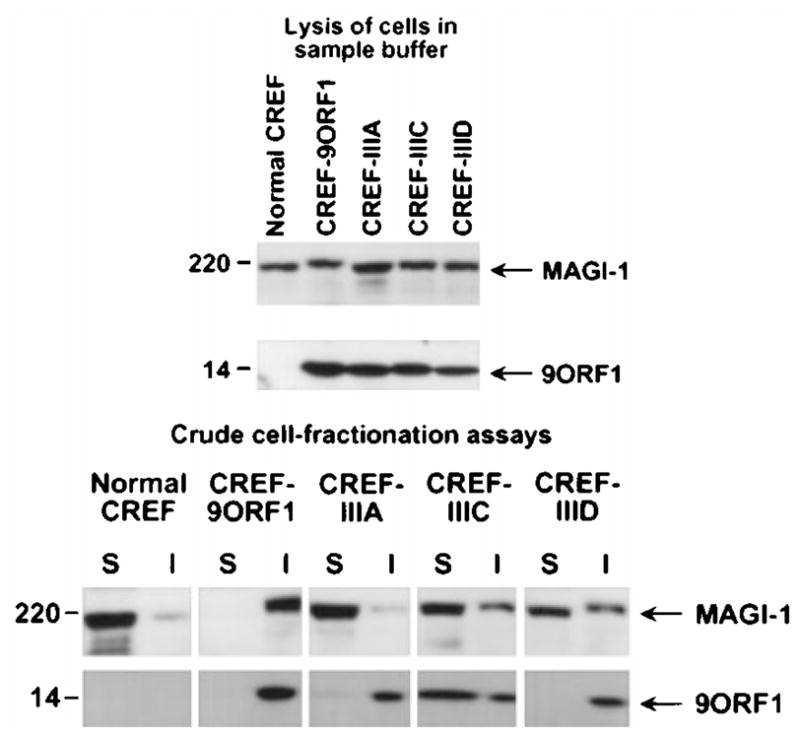
Aberrant sequestration of MAGI-1 within RIPA buffer-insoluble complexes in 9ORF1-expressing CREF cells. Normal CREF cells or CREF cells stably expressing wild-type or mutant 9ORF1 protein were lysed either in sample buffer (upper panel) or in RIPA buffer and subsequently centrifuged to yield RIPA buffer-soluble supernatant (S) and RIPA buffer-insoluble pellet (I) fractions (lower panel). Extracts of sample buffer-lysed cells or equal volumes of the S and I fractions from RIPA buffer-lysed cells were separately immunoblotted with MAGI-1 antibodies or 9ORF1 antiserum
To confirm the aberrant sequestration of MAGI-1 by 9ORF1 detected in IF assays, we performed crude cell-fractionation assays with the same CREF cell lines. In these experiments, cells lysed in RIPA buffer were separated by centrifugation into RIPA buffer-soluble supernatant and RIPA buffer-insoluble pellet fractions, each of which was immunoblotted for the presence of MAGI-1 protein. In normal CREF cells, the majority of MAGI-1 protein was detected in the RIPA buffer-soluble fraction but, in CREF-9ORF1 cells, MAGI-1 was exclusively present within the RIPA buffer-insoluble fraction (Figure 7, lower panel). It was this inability to recover soluble MAGI-1 protein from extracts of CREF-9ORF1 cells that prevented us from showing co-immunoprecipitation of 9ORF1 and MAGI-1 with this particular cell line. This observation also explained the substantially lower levels of MAGI-1 protein recovered in lysates of COS-7 cells co-expressing MAGI-1 and 9ORF1 compared to those recovered in COS-7 cells expressing MAGI-1 alone (see Figure 5d). Additionally, the results of crude cell-fractionation assays with CREF cells expressing mutant 9ORF1 proteins were in accordance with the IF findings because MAGI-1 protein was recovered primarily in the RIPA buffer-soluble fraction of the CREF-IIIA, CREF-IIIC, and CREF-IIID cell lines (Figure 7, lower panel).
High-risk human papillomavirus E6 oncoproteins also complex with MAGI-1
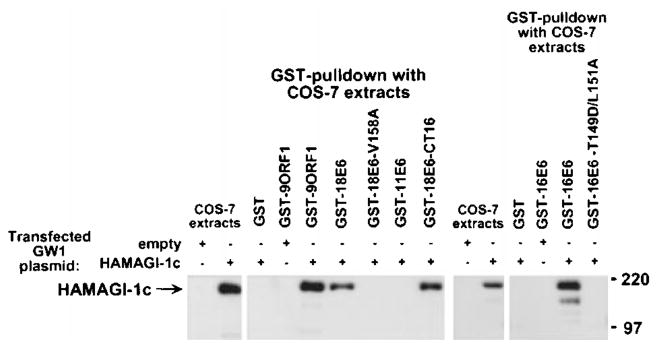
We (Lee et al., 1997) and others (Kiyono et al., 1997) previously showed that, like 9ORF1, high-risk but not low-risk HPV E6 oncoproteins contain functional PDZ domain-binding motifs at their carboxyl-termini (Table 1) and bind DLG. To determine whether high-risk HPV E6 oncoproteins also bind MAGI-1, we performed GST-pulldown assays with lysates of COS-7 cells expressing the HAMAGI-1c protein. The results indicated that wild-type high-risk 16E6 and 18E6, but not low-risk HPV-11 E6 (11E6), bind MAGI-1 in these assays (Figure 8). Moreover, the mutants 16E6-T149D/L151A and 18E6-V158A, which have disrupted PDZ domain-binding motifs (Table 1), failed to bind MAGI-1, indicating that the interactions between wild-type high-risk E6 oncoproteins and MAGI-1 were specific and required a functional PDZ domain-binding motif. We further demonstrated that a 16 aa-residue carboxyl-terminal 18E6 peptide (18E6-CT16) containing the PDZ domain-binding motif was sufficient to mediate binding to MAGI-1 (Figure 8).
Figure 8.
Binding of high-risk HPV E6 oncoproteins to MAGI-1 in vitro. Extracts of COS-7 cells transfected with 5 μg of empty GW1 or 5 μg of GWI-HAMAGI-1c plasmid were subjected to GST-pulldown reactions with the indicated GST fusion protein, and recovered proteins were immunoblotted with HA antibodies. GST pulldown assays with the indicated GST fusion protein were performed with COS-7 extracts in RIPA buffer or NETN buffer. COS-7 extracts representing one-tenth the amount used in GST pulldown reactions were also directly immunoblotted with HA antibodies as a control
High-risk HPV E6 oncoproteins target MAGI-1 for degradation
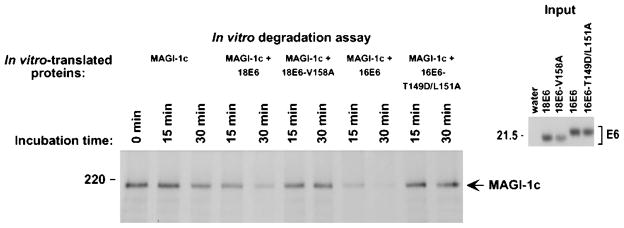
High-risk HPV E6 oncoproteins promote degradation of the p53 tumor suppressor protein in cells by ubiquitin-dependent proteolysis (Scheffner et al., 1990). As we recently showed that DLG is also targeted for degradation by high-risk HPV E6 proteins (Gardiol et al., 1999), it was of interest to determine whether MAGI-1 is similarly affected by these viral proteins. This possibility was initially examined by mixing and incubating in vitro-translated FLAG epitope-tagged MAGI-1 with wild-type or mutant 18E6 and 16E6 proteins. We found that, following incubation with either wild-type 18E6 or 16E6, MAGI-1 protein levels were substantially more reduced than following incubation with mutant 18E6-V158A or 16E6-T149D/L151A, or with a water-primed in vitro-translation reaction (Figure 9).
Figure 9.
HPV-18 E6-induced degradation of MAGI-1 in vitro. In vitro-translated FLAG epitope-tagged MAGI-1c (MAGI-1c) was mixed with in vitro-translated wild-type 18E6, mutant 18E6-V158A, wild-type 16E6, mutant 16E6-T149D/L151A, or control water-primed lysates and incubated at 30°C for the indicated times. At each time point, reactions were immunoprecipitated with FLAG antibodies, and recovered proteins were separated by SDS–PAGE and visualized by autoradiography (left panel). The amount of in vitro-translated E6 protein used in each assay is shown (right panel)
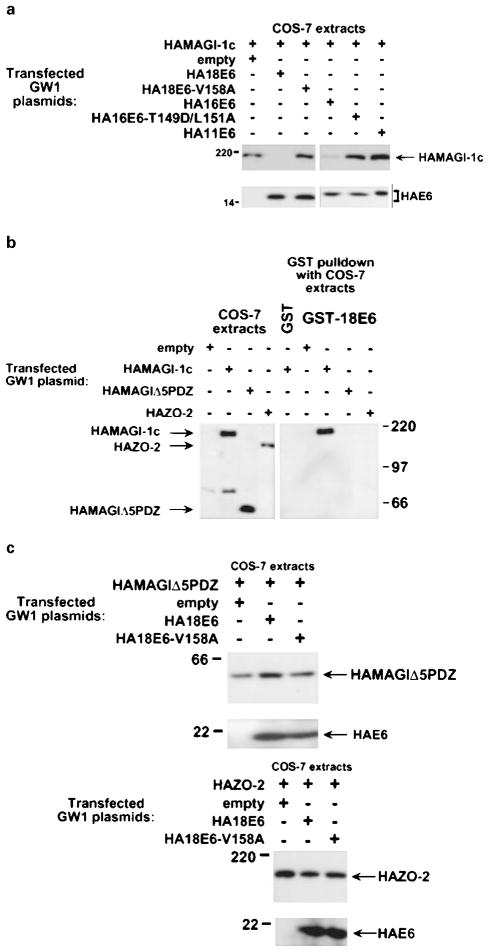
To determine whether high-risk HPV E6 proteins also specifically target MAGI-1 for degradation in cells, we compared the steady-state protein levels of MAGI-1 in COS-7 cells either expressing MAGI-1 alone or co-expressing MAGI-1 and wild-type or mutant E6 proteins. Consistent with our in vitro results, MAGI-1 protein levels were substantially reduced in cells expressing either wild-type 18E6 or 16E6 (Figure 10a). The results of additional experiments indicated that the reduction of MAGI-1 protein levels in cells expressing these wild-type E6 proteins was specific and due to proteolysis. First, no decrease in MAGI-1 protein levels was detected in cells expressing either the 18E6-V158A or 16E6-T149D/L151A mutant protein, or the low-risk 11E6 protein (Figure 10a). Second, consistent with our inability to detect binding of 18E6 to a truncated MAGI-1 protein lacking all five PDZ domains (MAGI-1Δ5PDZ) (see Figure 5b) or to the wild-type MAGUK-family PDZ-protein ZO-2 (Jesaitis and Goodenough, 1994) (Figure 10b), 18E6 failed to reduce the protein levels of either polypeptide in cells (Figure 10c). Finally, the results of pulse-chase experiments in COS-7 cells expressing MAGI-1 alone or co-expressing MAGI-1 and 18E6 demonstrated that the half-life of the MAGI-1 protein is drastically decreased from approximately 24 h in normal cells to approximately 1 h in 18E6-expressing cells (Figure 11). From these results, we conclude that high-risk HPV E6 oncoproteins specifically target the PDZ-protein MAGI-1 for degradation in cells.
Figure 10.
Selective reduction in MAGI-1 protein levels in vivo by high-risk HPV E6 proteins. (a) Decrease in the steady-state protein levels of MAGI-1 induced by high-risk HPV E6 oncoproteins. Extracts of RIPA buffer-lysed COS-7 cells transfected with 0.5 μg of GW1-HAMAGI-1c plasmid alone or in combination with 4 μg of a GW1 plasmid expressing the indicated wild-type or mutant HPV E6 protein were immunoblotted with HA antibodies. (b) Failure of 18E6 to bind a MAGI-1 deletion mutant missing all five PDZ domains (MAGI-1Δ5PDZ) or the wild-type ZO-2 protein. Extracts of RIPA buffer-lysed COS-7 cells transfected with 5 μg of the indicated GW1 expression plasmid were subjected to GST-pulldown reactions with either GST or GST-18E6 protein. Recovered proteins were immunoblotted with HA antibodies. COS-7 extracts representing one-tenth the amount used in GST pulldown reactions were also directly immunoblotted with HA antibodies as a control. (c) Inability of 18E6 to reduce the steady-state protein levels of MAGI-1Δ5PDZ or ZO-2 in cells. Extracts of RIPA buffer-lysed COS-7 cells transfected with 0.1 μg of GWI-HAMAGIΔ5PDZ plasmid (upper panel) or with 0.01 μg of GW1-HAZO-2 plasmid (lower panel), alone or in combination with 4 μg of GW1 plasmid expressing wild-type or mutant HA18E6, were immunoblotted with HA antibodies
Figure 11.
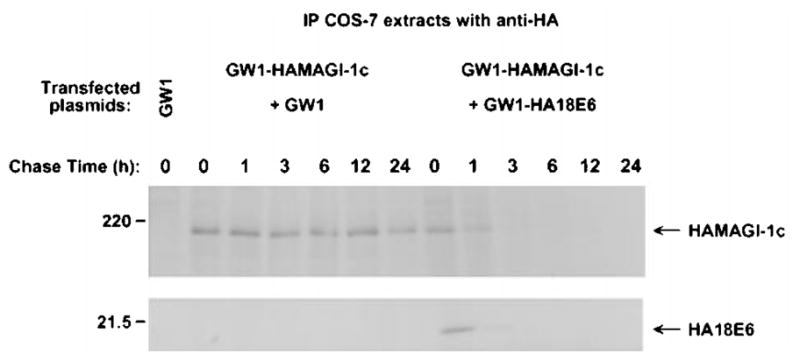
Decrease in MAGI-1 protein half-life induced by the high-risk HPV-18 E6 protein. COS-7 cells transfected with 0.5 μg of GW1-HAMAGI-1c plasmid in combination with 4 μg of empty GW1 or GW1-HA18E6 plasmid were pulse-labeled for 15 min with [35S] EXPRESS protein label and subsequently chased with unlabeled culture medium for the indicated times. At each time point, cell extracts were immunoprecipitated with HA antibodies. Recovered MAGI-1 protein was visualized by autoradiography and quantified by phosphorimager analysis
Discussion
Interactions of the 9ORF1 oncoprotein with the MAGUK-protein DLG and with several other unidentified cellular factors (p220, p180, p160, p155) correlate with the ability of this viral protein to transform cells (Weiss and Javier, 1997). Thus, identification and characterization of the unidentified cellular proteins is expected to aid in fully revealing the mechanisms of 9ORF1-induced transformation. In this study, we showed that, in addition to DLG, 9ORF1 also binds to the related MAGUK-protein MAGI-1 and that this cellular PDZ protein likely represents the previously unidentified 9ORF1-associated protein p180. Particularly noteworthy is that all transformation-defective 9ORF1 mutants having an altered PDZ domain-binding motif displayed impaired interactions with MAGI-1. For example, the severely transformation-defective mutant IIIA and the weak-transforming mutant IIID failed or showed a substantially reduced capacity, respectively, to bind MAGI-1. In addition, although the weak-transforming mutant IIIC bound to MAGI-1 at nearly wild-type levels in cells, this mutant protein was impaired both for interacting with certain MAGI-1 PDZ domains and for aberrantly sequestering MAGI-1 within RIPA buffer-insoluble complexes in the cytoplasm of cells. With respect to the latter observation, we noted that, in contrast to wild-type 9ORF1, substantial amounts of mutant IIIC protein exist in the RIPA buffer-soluble fraction of cells, suggesting that this mutant may be inherently unable to sequester PDZ proteins in the cytoplasm of cells. Taken together, results with transformation-defective 9ORF1 mutants argue that the abilities of 9ORF1 to bind and aberrantly sequester MAGI-1 in cells contribute to 9ORF1-mediated cellular transformation. The fact that transformation-defective 9ORF1 mutants also fail to bind other 9ORF1-associated PDZ-proteins (Lee et al., 1997), however, suggests that interactions of 9ORF1 with additional PDZ proteins are likewise important.
It is also noteworthy that, among six different PDZ proteins examined, only MAGI-1 was found to interact with 9ORF1. This finding indicates that 9ORF1 targets only select PDZ proteins in cells. This idea is further evidenced by the select interaction of 9ORF1 with two of the five PDZ domains of MAGI-1 and two of the three PDZ domains of DLG (Lee et al., 1997). These observations, coupled with the fact that 9ORF1 mutants IIIC and IIID bind only one MAGI-1 PDZ domain, argue that precise sequence requirements both within the PDZ domain-binding motif of 9ORF1 and within each PDZ domain of the 9ORF1-associated targets determine these highly specific protein-protein interactions.
The fact that MAGI-1 is a MAGUK-family protein suggests that this PDZ protein functions to assemble numerous cellular targets into large signaling complexes in cells. In contrast to many PDZ proteins, however, MAGI-1 was found to localize predominantly in the cytoplasm of CREF fibroblasts. Although we cannot discount the possibility that a minor fraction of MAGI-1 is present at the membrane of CREF cells, this finding may indicate that MAGI-1 functions primarily in the cytoplasm. Alternatively, it is feasible that MAGI-1 does function at the membrane, but its translocation to this site occurs only following specific cellular stimuli. Besides possible cytoplasmic and membrane activities, the MAGI-1c isoform, which contains a consensus bipartite nuclear localization signal (Dobrosotskaya et al., 1997), may additionally function in the nucleus, perhaps to regulate the transcription of certain cellular genes. Again, we did not detect MAGI-1 in the nucleus of cells, but nuclear localization may occur only under specific conditions, as has been reported for the related MAGUK-family protein ZO-1 (Gottardi et al., 1996). Regardless of where in the cell MAGI-1 functions, however, this cellular factor would likely be inactivated in 9ORF1-expressing cells, as we found that 9ORF1 aberrantly sequesters MAGI-1 in the cytoplasm of cells. Also considering that 9ORF1 binds strongly to MAGI-1 PDZ1 and PDZ3, 9ORF1 would be expected to block MAGI-1 from complexing with the normal cellular targets of these PDZ domains. Therefore, aberrant sequestration and disruption of protein complexes, as well as perturbation of associated protein activities, may all be envisioned as possible mechanisms by which 9ORF1 could inhibit the normal functions of MAGI-1 in cells.
Like 9ORF1, all known high-risk HPV E6 oncoproteins also possess a carboxyl-terminal PDZ domain-binding motif (Lee et al., 1997). Significantly, disruption of the PDZ domain-binding motif of 16E6 renders this viral protein transformation-defective in rat 3Y1 fibroblasts (Kiyono et al., 1997). As infections with high-risk HPV-16 and HPV-18 are associated with approximately 75% of cervical carcinomas (Bosch et al., 1995), our finding that both the 16E6 and 18E6 oncoproteins, but not the low-risk 11E6 protein, utilize their carboxyl-terminal PDZ domain-binding motif to bind the PDZ-protein MAGI-1 is also likely to be important. This idea is further underscored by the facts that 16E6 and 18E6 mutant proteins having disrupted PDZ domain-binding motifs fail to bind MAGI-1 and that the wild-type viral proteins do not complex with the related MAGUK proteins ZO-1 (unpublished results) and ZO-2. Such specific and selective interactions suggest that the ability of high-risk HPV E6 oncoproteins to associate with MAGI-1 in cells may contribute to the development of HPV-associated cancers in people.
A common mechanism by which the oncoproteins of DNA tumor viruses promote tumorigenesis is the inactivation of cellular tumor suppressor proteins. For example, both the adenovirus E1B and high-risk HPV E6 oncoproteins functionally inactivate the tumor suppressor protein p53, yet by distinct mechanisms. In this regard, E1B sequesters p53 in an inactive state (Shenk, 1996), whereas high-risk HPV E6 targets p53 for ubiquitin-mediated proteolysis (Howley, 1996). Likewise, we found that the 9ORF1 oncoprotein sequesters MAGI-1 in the cytoplasm of cells whereas the high-risk HPV E6 proteins target MAGI-1 for degradation in cells. These findings argue that adenovirus E4-ORF1 and high-risk HPV E6 oncoproteins similarly inactivate MAGI-1 by distinct mechanisms. As MAGI-1 is related to DLG and both proteins are selectively targeted by two otherwise unrelated viral oncoproteins, it seems plausible that these PDZ proteins have related functions in cells. Therefore, we hypothesize that MAGI-1 similarly functions to suppress inappropriate cellular proliferation and, consequently, represents a new candidate tumor suppressor protein.
Materials and methods
Cells and cell extracts
CREF (Fisher et al., 1982), TE85 (McAllister et al., 1971), and COS-7 (Gluzman, 1981) cell lines were maintained in culture medium (Dulbecco’s Modified Eagle Medium supplemented with gentamicin (20 μg/ml) and 6 or 10% fetal bovine serum (FBS)). CREF cell pools (group 16) stably expressing wild-type or mutant 9ORF1 protein (Weiss et al., 1997a), as well as a CREF cell pool stably expressing an influenza hemagglutinin (HA) epitope-tagged 9ORF1 protein (Weiss et al., 1997b), were maintained in culture medium supplemented with G418.
Cell extracts were prepared in either RIPA buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 1% (vol/vol) Nonidet P-40, 0.5% (wt/vol) sodium deoxycholate, 0.1% (wt/vol) sodium dodecyl sulfate (SDS)) or NETN buffer (20 mM Tris, pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40 (vol/vol)) as described previously (Lee et al., 1997). Alternatively, cells were lysed directly in sample buffer (0.15 M Tris-HCl pH 6.8, 2% (wt/vol) SDS, 10% (vol/vol) glycerol, 1% (vol/vol) β-mercaptoethanol, 0.0015% (wt/vol) bromophenol blue). For crude cell-fractionation assays, the pellet obtained after centrifugation of RIPA buffer-lysed cells was solublized in sample buffer using the same volume originally used to lyse the cells in RIPA buffer. Protein concentrations of cell extracts were determined by the Bradford assay (Bradford, 1976).
Plasmids
pCDNA3 (Invitrogen) plasmids coding for amino-terminal FLAG epitope-tagged mouse MAGI-1 isoform b (MAGI-1b), MAGI-1 isoform c (MAGI-1c), and MAGI-1b missing either PDZ1 (aa 453–549; MAGI-1bΔPDZ1), PDZ2 (aa 625–702; MAGI-1bΔPDZ2), or PDZ3 (aa 796–875; MAGI-1bΔPDZ3), as well as pGEX-KG (Pharmacia) plasmids coding for MAGI-1 PDZ1 (aa 431–545), PDZ2 (aa 601–702), PDZ4 (aa 925–1034), or PDZ5 (aa 1013–1116) were generously provided by Guy James. cDNA sequences coding for MAGI-1 PDZ3 (aa 763–880) were amplified by PCR and introduced in-frame with the glutathione S-transferase (GST) gene of pGEX-2T to make pGEX-MAGI-1PDZ3. pGEX-2T and pGEX-2TK plasmids coding for wild-type Ad5 and Ad12 E4-ORF1 proteins (5ORF1 and 12ORF1, respectively), as well as for wild-type or mutant 9ORF1 proteins were described previously (Weiss and Javier, 1997). The cDNAs of HPV-18 E6 (18E6) mutant 18E6-V158A, wild-type HPV-16 E6 (16E6), and mutant 16E6-T149D/L151A were introduced into the HindIII and EcoRI sites of pSP64 (Promega) to generate pSP64-18E6-V158A, pSP64-16E6, and pSP64-16E6-T149D/L151A, respectively. An HA-epitope tag was introduced at the amino-terminus of MAGI-1b, MAGI-1c, MAGI-1Δ5PDZ (aa 1–424), 16E6, mutant 16E6-T149D/L151A, and canine ZO-2 by PCR methods. cDNAs coding for the HA-epitope-tagged MAGI-1 and 16E6 proteins were introduced between the HindIII and EcoRI sites of CMV expression plasmid GW1 (British Biotechnology) to generate GW1-HAMAGI-1b, GW1-HAMAGI-1c, GW1-HAMAGI-1Δ5PDZ, GW1-HA16E6, and GW1-HA16E6-T149D/L151A. The HA-epitope-tagged ZO-2 cDNA was introduced into the SmaI site of GW1 to generate GW1-HAZO-2. The deletions of pCDNA3-FLAGMAGI-1bΔPDZ1 and pCDNA3-FLAGMAGI-1bΔPDZ3 were subcloned individually or in combination into GW1-HAMAGI-1b to generate GW1-HAMAGI-1bΔPDZ1, GW1-HAMAGI-1bΔPDZ3, and GW1-HAMAGI-1bΔPDZ1+3. PCR reactions were performed with Pfu polymerase (Stratagene), and plasmids were verified by restriction enzyme and limited sequence analyses. Plasmids GW1-HA18E6, GW1-HA18E6-V158A, GW1-HA11E6, GWI-9ORF1wt, GW1-9ORF1IIIA, GW1-9OR-F1IIIC, GW1-9ORF1IIID, and pSP64-18E6 were described elsewhere (Gardiol et al., 1999).
Antisera and antibodies
Rabbit polyclonal antiserum raised to the unique amino-terminal region of MAGI-1 (aa 2–140) was generously provided by Guy James (Dobrosotskaya et al., 1997). 9ORF1 antiserum was described previously (Javier, 1994). Commercially-available FLAG antibodies (Santa Cruz Biotechnology), 12CA5 HA monoclonal antibodies (BABCO; Boehringer Mannheim), normal rabbit IgG, peroxidase-conjugated goat anti-rabbit or goat anti-mouse IgG (Southern Biotechnology Associates), FITC-conjugated goat anti-rabbit or goat anti-mouse IgG (Gibco BRL), or Texas red-conjugated goat anti-mouse IgG (Molecular Probe) were used.
GST-pulldown, immunoprecipitation, and immunoblot assays
GST-pulldown and immunoprecipitation assays were performed with cell extracts in RIPA buffer as described previously (Lee et al., 1997). Immunoblot assays were carried out as described previously (Weiss et al., 1996) using either 9ORF1 (1 : 5000), HA (0.2 μg/ml), or MAGI-1 (1 μg/ml) primary antibodies and either horseradish peroxidase-conjugated goat anti-rabbit IgG or goat anti-mouse IgG (1 : 5000) secondary antibodies. Immunoblotted assays were developed by enhanced chemiluminescence (Pierce).
Protein blotting assays
Individual MAGI-1 PDZ domains were expressed as GST-fusion proteins in bacteria, purified on glutathione sepharose beads (Pharmacia) (Smith and Corcoran, 1994), separated by SDS-polyacrylamide gel electrophoresis (PAGE), and transferred to a nitrocellulose membrane. Methods for preparing radiolabeled GST fusion probes and for performing protein blotting assays with such probes have been described (Lee et al., 1997).
Immunofluorescence microscopy assays
For indirect immunofluorescence (IF) microscopy assays (Harlow and Lane, 1988), cells were grown on coverslips, fixed in methanol for 20 min at −20°C, blocked in IF buffer (TBS (50 mM Tris-HCl pH 7.5, 200 mM NaCl) containing 10% goat serum (Sigma)) for 1 h at RT and, then, incubated with either MAGI-1 antibodies (5 μg/ml), HA antibodies (0.24 mg/ml) or normal rabbit IgG (5 μg/ml) for 3 h at 37°C. Cells were subsequently washed with TBS, blocked as described above, incubated with either FITC-conjugated goat anti-rabbit or goat anti-mouse IgG antibodies (1 : 250) or Texas red-conjugated goat anti-mouse IgG antibodies (1 : 250) for 1 h at 37°C, and washed with TBS. All antibodies were diluted in IF buffer. Cells were visualized by fluorescence microscopy using a Zeiss Axiophot microscope, and images were processed using Adobe PhotoShop software.
In vitro-degradation assays
pCDNA3-FLAGMAGI-1c, pSP64-18E6, pSP64-18E6-V158A, pSP64-16E6, and pSP64-16E6-T149D/L151A plasmids were transcribed and translated in vitro using the TnT coupled rabbit reticulocyte lysate system (Promega) and 50 μCi [35S]-cysteine (1200 Ci mmol) (Amersham), according to the manufacturer’s instructions. The amount of radio-activity introduced into in vitro-translated proteins per microliter of reaction was determined by resolving an aliquot of each reaction by SDS–PAGE and quantifying CPM within relevant protein bands using a Storm Molecular Dynamics phosphorimager. For in vitro-degradation assays, a reaction volume equivalent to 50 c.p.m. of in vitro-translated FLAGMAGI-1c protein was mixed with a reaction volume equivalent to 250 c.p.m. of in vitro-translated 18E6, 18E6-V158A, 16E6, or 16E6-T149D/L151A protein. All assay volumes were equalized with a water-primed in vitro-translation reaction and, at selected time points, an aliquot from each assay mixture was removed and subjected to immunoprecipitation analysis. Recovered proteins were resolved by SDS–PAGE and visualized by audioradiography.
Pulse-chase labeling of proteins in cells
At 48 h post-transfection, COS-7 cells were incubated with methionine- and cysteine-free DMEM containing 5% dialyzed FBS (5% FBS-DMEM-MC) for 30 min and, then, pulse labeled for 15 min in the same medium containing 0.2 mCi ml [35S] EXPRESS protein label (Dupont). Following several washes with 5% FBS-DMEM-MC, pulse radiolabeled cells were chased by incubation with culture medium containing fivefold excess methionine (15 mg/l) for various times, harvested, and lysed in RIPA buffer. Cell extracts were subjected to immunoprecipitation with HA antibodies, and recovered proteins were resolved by SDS–PAGE and visualized by autoradiography. Amounts of protein immunoprecipitated were quantified using a phosphorimager.
Acknowledgments
We are indebted to Guy James (University of Texas Health Sciences Center, San Antonio) for generously providing MAGI-1 reagents. We also thank Bruce Stevenson for providing the canine ZO-2 expression plasmid. BA Glaunsinger and SS Lee were recipients of a Molecular Virology Training Grant (T32 AI07471) and the US Army Breast Cancer Training Grant (DAMD17-94-J4204), respectively. This work was supported by grants from the National Institutes of Health (RO1 CA58541), the American Cancer Society (RPG-97-668-01-VM), and the US Army (DAMD17-97-1-7082) to RT Javier and an Associazione Italiana per la Ricerca sul Cancro grant to L Banks.
References
- Bosch FX, Manos MM, Munoz N, Sherman M, Jansen AM, Peto J, Schiffman MH, Moreno V, Kurman R, Shah KV. J Natl Cancer Inst. 1995;87:796–802. doi: 10.1093/jnci/87.11.796. [DOI] [PubMed] [Google Scholar]
- Bradford MM. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Craven SE, Bredt DS. Cell. 1998;93:495–498. doi: 10.1016/s0092-8674(00)81179-4. [DOI] [PubMed] [Google Scholar]
- Dobrosotskaya I, Guy RK, James GL. J Biol Chem. 1997;272:31589–31597. doi: 10.1074/jbc.272.50.31589. [DOI] [PubMed] [Google Scholar]
- Fanning AS, Anderson JM. J Clin Invest. 1999;103:767–772. doi: 10.1172/JCI6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher PB, Babiss LE, Weinstein IB, Ginsberg HS. Proc Natl Acad Sci USA. 1982;79:3527–3531. doi: 10.1073/pnas.79.11.3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiol D, Kuhne C, Glaunsinger B, Lee S, Javier R, Banks L. Oncogene. 1999;18:5487–5496. doi: 10.1038/sj.onc.1202920. [DOI] [PubMed] [Google Scholar]
- Gluzman Y. Cell. 1981;23:175–182. doi: 10.1016/0092-8674(81)90282-8. [DOI] [PubMed] [Google Scholar]
- Gottardi CJ, Arpin M, Fanning AS, Louvard D. Proc Natl Acad Sci USA. 1996;93:10779–10784. doi: 10.1073/pnas.93.20.10779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow E, Lane D. In: Antibodies: a laboratory manual. Harlow E, Lane D, editors. Cold Spring Harbor Laboratory; Cold Spring Harbor, N.Y: 1988. pp. 359–420. [Google Scholar]
- Haskins J, Gu L, Wittchen ES, Hibbard J, Stevenson BR. J Cell Biol. 1998;141:199–208. doi: 10.1083/jcb.141.1.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howley PM. In: Fields Virology. Fields BN, Knipe DM, Howley PM, editors. Vol. 2. Lippincott-Raven; Philadelphia: 1996. pp. 2045–2076. [Google Scholar]
- Javier R, Raska K, Jr, Macdonald GJ, Shenk T. J Virol. 1991;65:3192–3202. doi: 10.1128/jvi.65.6.3192-3202.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javier RT. J Virol. 1994;68:3917–3924. doi: 10.1128/jvi.68.6.3917-3924.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jesaitis LA, Goodenough DA. J Cell Biol. 1994;124:949–961. doi: 10.1083/jcb.124.6.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyono T, Hiraiwa A, Fujita M, Hayashi Y, Akiyama T, Ishibashi M. Proc Natl Acad Sci USA. 1997;94:11612–11616. doi: 10.1073/pnas.94.21.11612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SS, Weiss RS, Javier RT. Proc Natl Acad Sci USA. 1997;94:6670–6675. doi: 10.1073/pnas.94.13.6670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lue RA, Marfatia SM, Branton D, Chishti AH. Proc Natl Acad Sci USA. 1994;91:9818–9822. doi: 10.1073/pnas.91.21.9818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister RM, Filbert JE, Nicolson MO, Rongey RW, Gardner MB, Gilden RV, Huebner RJ. Nat New Biol. 1971;230:279–282. doi: 10.1038/newbio230279a0. [DOI] [PubMed] [Google Scholar]
- Muller BM, Kistner U, Veh RW, Cases-Langhoff C, Becker B, Gundelfinger ED, Garner CC. J Neurosci. 1995;15:2354–2366. doi: 10.1523/JNEUROSCI.15-03-02354.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevins JR, Vogt PK. In: Fields Virology. Fields BN, Knipe DM, Howley PM, editors. Vol. 1. Lippincott-Raven Publishers; Philadelphia: 1996. pp. 301–343. [Google Scholar]
- Philipp S, Flockerzi V. FEBS Lett. 1997;413:243–248. doi: 10.1016/s0014-5793(97)00877-6. [DOI] [PubMed] [Google Scholar]
- Prasad R, Gu Y, Alder H, Nakamura T, Canaani O, Saito H, Huebner K, Gale RP, Nowell PC, Kuriyama K, Miyazaki Y, Croce CM, Cacaani E. Cancer Res. 1993;53:5624–5628. [PubMed] [Google Scholar]
- Sato T, Irie S, Kitada S, Reed JC. Science. 1995;268:411–415. doi: 10.1126/science.7536343. [DOI] [PubMed] [Google Scholar]
- Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- Shenk T. In: Fields Virology. Fields BN, Knipe DM, Howley PM, editors. Vol. 2. Lippincott-Raven Publishers; Philadelphia: 1996. pp. 2111–2148. [Google Scholar]
- Smith DB, Corcoran LM. In: Current Protocols in Molecular Biology. Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Vol. 2. John Wiley and Sons, Inc; New York: 1994. pp. 16.7.1–16.7.7. [Google Scholar]
- Thomas DL, Shin S, Jiang BH, Vogel H, Ross MA, Kaplitt M, Shenk TE, Javier RT. J Virol. 1999;73:3071–3079. doi: 10.1128/jvi.73.4.3071-3079.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas U, Phannavong B, Muller B, Garner CC, Gundelfinger ED. Mech Dev. 1997;62:161–174. doi: 10.1016/s0925-4773(97)00658-8. [DOI] [PubMed] [Google Scholar]
- Weiss RS, Gold MO, Vogel H, Javier RT. J Virol. 1997a;71:4385–4394. doi: 10.1128/jvi.71.6.4385-4394.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss RS, Javier RT. J Virol. 1997;71:7873–7880. doi: 10.1128/jvi.71.10.7873-7880.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss RS, Lee SS, Prasad BVV, Javier RT. J Virol. 1997b;71:1857–1870. doi: 10.1128/jvi.71.3.1857-1870.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss RS, McArthur MJ, Javier RT. J Virol. 1996;70:862–872. doi: 10.1128/jvi.70.2.862-872.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willott E, Balda MS, Fanning AS, Jameson B, Van Itallie C, Anderson JM. Proc Natl Acad Sci USA. 1993;90:7834–7838. doi: 10.1073/pnas.90.16.7834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods DF, Bryant PJ. Cell. 1991;66:451–464. doi: 10.1016/0092-8674(81)90009-x. [DOI] [PubMed] [Google Scholar]
- Wu H, Reuver SM, Kuhlendahl S, Chung WJ, Garner CC. J Cell Sci. 1998;111:2365–2376. doi: 10.1242/jcs.111.16.2365. [DOI] [PubMed] [Google Scholar]
- Yang N, Higuchi O, Mizuno K. Exp Cell Res. 1998;241:242–252. doi: 10.1006/excr.1998.4053. [DOI] [PubMed] [Google Scholar]