Abstract
We used 204 nm excitation UV Resonance Raman (UVRR) spectroscopy to examine the role of side chain electrostatic interactions in determining the conformation of poly-L-lysine (PLL). We examined the pH and ionic strength dependence of the UVRR. The pH dependence of PLL UVRR spectra between pH 7.1 and 11.7 cannot be described by a two-state model, but requires at least one additional state. The AmIII3 region fitting with pH 7.1 and 11.7 basis spectra reveals a small pH induced decrease in the relative fraction of the 2.51-helix conformation compared to the PPII conformation. We performed a 2D general correlation analysis on the PLL pH dependence UVRR spectra. The asynchronous spectrum shows enhanced spectral resolution. The 2D asynchronous spectrum reveals multiple components in the Cα-H b band and the AmII band whose origins are unclear. The cross peaks in the 2D asynchronous spectrum between the AmIII band and the other bands reveals that increasing pH induces three new structures: π-helix, α-helix and some turn structure. We find that 2.5 M NaCl does not change the equilibrium between the PPII and 2.51-helix conformations by screening sidechain electrostatic repulsion. The result indicates that NaCl does not penetrate the region between the sidechain and the peptide backbone. We also compared PLL conformations induced by high pH to that induced by 0.8 M ClO4−. Both conditions induce α-helix-like conformations. 0.8 M ClO4− induces 6% more α-helix-like conformations than at pH 12.4. Higher pH gives rise to longer α-helices and less turn structures.
INTRODUCTION
Developing an understanding of the mechanism of protein folding is one of the most important unsolved problems in structural biology.1–5 The major underlying assumption is that the protein native structure is the thermodynamically most stable structure.6–8 Further, this, native structure is thought to be defined by the primary amino acid sequence and the protein solution environment. Thus, the protein primary sequence is expected to contain all of the information necessary to specify the protein native structure and its folding mechanisms.
Determining the mechanism of protein folding involves determining the energy landscape along the folding coordinates and determining the folding process intermediates.9 Computational modeling has made important contributions to the understanding of protein folding mechanisms.10–12 The conformational space accessible to a peptide can now be searched by molecular dynamics. The mechanisms of protein folding have been probed experimentally by techniques such as CD, NMR, IR, Raman and a variety of temperature jump spectroscopies.
In the work here we are studying the conformational transitions of poly-L-lysine (PLL) induced by solution pH and salt concentration changes. At neutral and low pH values, the lysine side chains are positively charged. We previously demonstrated that under these conditions PLL exists in an unfolded state in an equilibrium between a PPII and 2.51-helix conformation.13 The PPII conformation is mainly stabilized by peptide-water hydrogen bonding,14 whereas the 2.51-helix conformation is stabilized by electrostatic repulsion between lysine side chains.13,15 Raising the pH neutralizes the side chains, and PLL folds into compact α-helix-like conformations.
Charged PLL also forms α-helix-like conformations in the presence of ClO4−; Ma et al. observed that in the presence of ClO4−, PLL adopts multiple α-helix-like conformations including pure α-helix and π-bulge/helix conformations.16 High temperature converts the high pH folded PLL to a β-sheet conformation.17 Recently, Jiji et al’s temperature-jump UVRR spectroscopy study of PLL indicated that the PPII and extended β-strand conformations may be involved as intermediates in the α-helix to β-sheet conformational transition.18
In our UVRR studies of the salt concentration dependence of PLL conformations we surprisingly find that high concentrations of NaCl negligible impact the low pH PLL conformations. This indicates that NaCl does not effectively screen PLL sidechain repulsions. We discuss the impact of this result on the partitioning of in the region between the sidechains and the peptide backbone.
EXPERIMENTS AND METHODS
Materials
Poly-L-Lysine HCl (MWvis = 20900, DPvis= 127, MWMALLS = 11400, DPMALLS = 69, where DPvis and DPMALLS refer to the degree of polymerization measured by viscosity and multi-angle laser light scattering, respectively) was obtained from Sigma and used without further purification. NaClO4 and NaOD (40% wt. solution in D2O, 99 + atom %D), purchased from Sigma, were used without further purification. The Ac-KKKKKKKKKK-NH2 peptide (K10) was obtained from the Pittsburgh Peptide Synthesis Facility (>98% pure). Small aliquots of concentrated fresh NaOH and NaOD solutions were used to adjust the solution pH and pD values.
UVRR Instrument
We used a Coherent Infinity Nd: YAG laser (Coherent, Inc.) to produce 355 nm light pulses (3rd harmonic) at a 100 Hz repetition rate with a pulse width of 3 ns. This beam was Raman shifted to 204 nm (5th anti-Stokes) by using a 1-m tube filled with hydrogen (60 psi), giving 2 mW average power.19,20 A Pellin Broca prism was used to select the 204 nm excitation. The sample was circulated in a free surface, temperature controlled stream to avoid heating or accumulation of photochemical degradation products formed by the high peak power laser pulses. The Raman scattered light was imaged into a subtractive double spectrometer.19 The dispersed UV light was detected by the liquid nitrogen cooled, Lumogen coated back-thinned CCD with a reported >30% quantum efficiency in the deep UV (Princeton Instruments Spec-10:400B).
The UVRR spectra of 1 mg/ml PLL and K10 H2O solution were measured at various pH values at 10°C. ClO4− could not be used as an internal standard, since it induces α-helical conformations.16 Instead, all Raman spectra were normalized to the integrated intensities of the AmI bands, because they show the least sensitivity to secondary structural changes.21
The D2O PLL solutions utilized 2 mg/ml concentrations. The UVRR spectra of PLL in D2O were measured at pD 6.5 and 11.5 at 10 °C. Spectra were normalized to the integrated intensity of the AmI’ band.
2D Correlation Analysis
The measured UVRR spectra at different pH values were fit by using the peak fitting routine in GRAMS software (Thermo Galactic, Grams version 8). The fit spectra were used to construct the data matrices for the 2D correlation analysis. Synchronous and asynchronous correlation intensities were computed from the fitted spectra at different pH values by using a Matlab program that we wrote that utilized Noda’s generalized 2D correlation algorithm.22,23
RESULTS AND DISCUSSION
pH dependence of PLL UVRR spectra in H2O
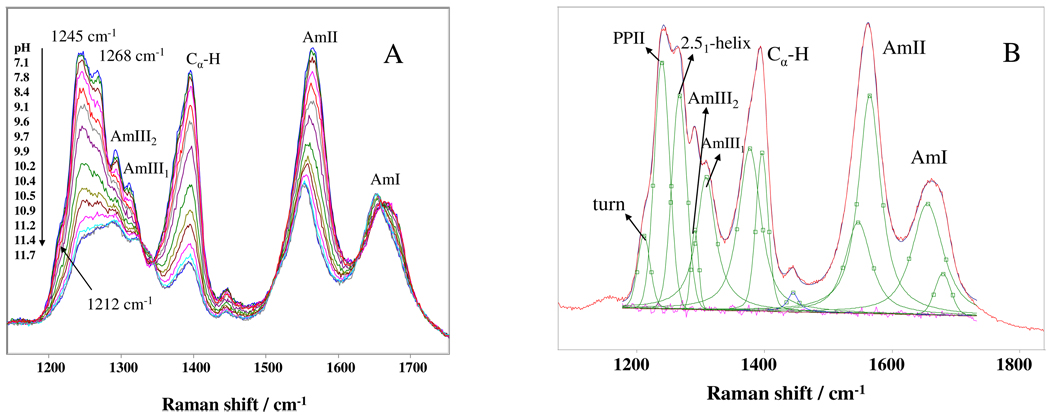
Fig. 1A shows the pH dependence of the 204 nm UVRR spectrum of PLL. The spectra show the four characteristic UVRR bands of the peptide bond: the AmI band located at ~ 1660 cm−1(mainly C=O s), the AmII band at ~ 1567 cm−1 (mainly out of phase combination of C-N s and N-H b), the ~1400 cm−1 Cα-H b band (complex vibration involving Cα-H b and N-H b motions), and the AmIII bands occurring in the range of 1200 to 1350 cm−1 (mainly involving N-H b and C-N s).
Figure 1.
A) pH dependence of the 204 nm UVRR spectra of PLL at 10 °C. The spectra were normalized to the AmI band integrated intensities.16 B) Spectral deconvolution of the 10 °C 204 nm UVRR PLL spectrum at pH 7.1. The excellence of the fit is evident from the flat residuals shown below.
As the pH increases, the AmII, the Cα-H b and the AmIII band intensities decrease, which indicates formation of α-helical-like conformations. For 204 nm excitation, the α-helix UVRR cross sections are smaller than for extended conformations, due to the hypochromism that results from α-helix excitonic interactions between the peptide bond π-π* electronic transitions.24,25 The Cα-H b band shows an additional intensity decrease relative to the AmII and AmIII bands because the Cα-H b bands are only resonantly enhanced for PPII-like, β-sheet, and β-strand-like conformations, but not for α-helix-like conformations.
As the pH increases, the AmI band downshifts by 14 cm−1 and its bandshape narrows and it becomes more symmetric. The AmI band shows a clear isosbestic point, presumably indicating a transition between only two different carbonyl hydrogen bonding states. The smaller AmI high pH α-helix bandwidths result from the better defined α-helix conformation hydrogen bonding, while the AmI frequency decrease results from stronger α-helix carbonyl hydrogen bonding.
The AmII band downshifts 12 cm−1 as the pH increases and the α-helix conformation becomes dominant. Similar AmII band frequency shifts have previously been observed in peptides and proteins upon the conformational shift from PPII-like to α-helix conformations.20,21 Although there have been numerous studies of the conformation frequency dependence of the AmII band, there is as yet no simple explanation of the origin of these AmII band downshifts.26–29
The AmIII region is the most sensitive to peptide backbone conformational changes.30 Fig. 1B shows the spectral deconvolution of the UVRR spectrum of PLL at pH 7.1. The AmIII1 and AmIII2 bands are located at ~1313 cm−1 and 1293 cm−1. The AmIII3 band region between 1190 cm−1 and 1285 cm−1 resolves into two peaks at ~1245 cm−1, ~1268 cm−1 and a shoulder at 1212 cm−1. The AmIII3 band frequencies can differentiate α-helix-like structures from extended structures, as well as differentiate between similar α-helix-like structures and extended structures, e.g., between the α-, 310-, π-helix structures and the PPII and 2.51-helix structures.13
We used the method of Mikhonin et al. to determine the peptide bone Ramachandran Ψ angles from the AmIII3 band frequencies.30 The ~1245 cm−1 and ~1268 cm−1 AmIII3 bands derive from extended PPII and 2.51-helix conformations, respectively.13,16 The shoulder probably derives from a turn structure, whose type we cannot as yet specify.
As the pH increases, the AmIII bandshape changes significantly. This occurs because the concentrations of the PPII and 2.51-helix conformations that have large Raman cross sections decrease, while the concentration of the α-helix-like conformation with a low Raman cross section increases. Little spectral changes occur between pH 11.4 and 11.7, indicating the lack of additional pH induced PLL conformational changes. There are two isosbestic points between the AmIII1 and Cα-H b bands.
pD dependence of PLL UVRR spectra
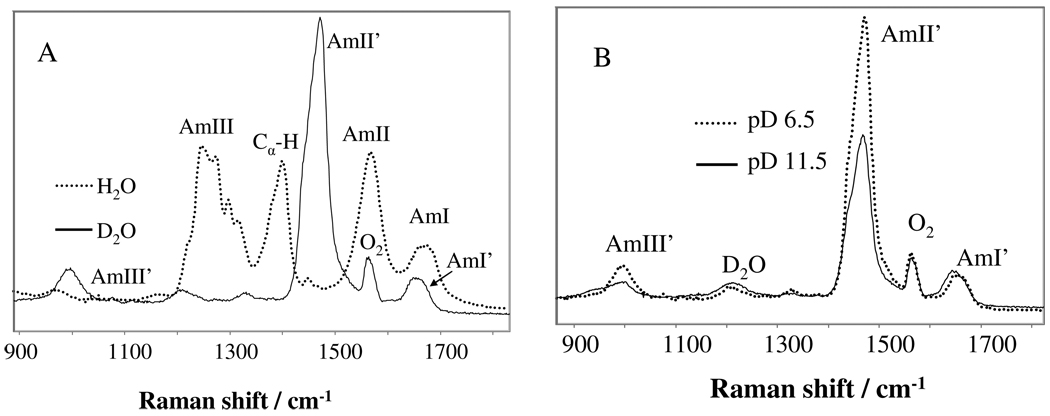
Fig. 2A shows that the pH 7.1 UVRR spectrum of PLL changes dramatically in D2O. The AmIII, AmII and Cα-H b bands disappear and are replaced with a very strong AmII’ band at 1470 cm−1, and a much weaker AmIII’ band at 994 cm−1. These spectral changes result from the deuteration of the peptide bond N-H. The AmII, AmIII and Cα-H b bands involve significant N-H bending; deuteration removes the N-H in plane bending component leaving an almost pure C-N s AmII’ vibration and an AmIII’ vibration which has a large N-D b component.
Figure 2.
(A) 204 nm UVRR spectra of PLL at pH 7.1 in H2O and D2O. (B) 204 nm UVRR spectra of PLL in D2O at pD 6.5 and 11.5. The spectra were normalized to the integrated AmI’ band intensity.
Fig. 2B shows that as the pD increases from 6.5 to 11.5, the AmI’ band downshifts ~ 8 cm−1, which is less than the observed 14 cm−1 AmI band downshift. This indicates that part of the AmI band downshift results from non C=O stretching components.
The AmII’ band of PLL shows only a ~2 cm−1 downshift, much less than the AmII band 12 cm−1 downshift. This smaller AmII’ band shift indicates that the larger AmII band frequency shift mainly derives from changes in N-H hydrogen bonding. The loss of the N-H b component induced by deuteration results in a decreased AmII’ band downshift. This is consistent with the theoretical study of N-methylacetamide by Myshakina et al,28 who proposed that hydrogen bonding to the N-H causes an electron redistribution that dominates the AmII band upshift. The intensities of the AmII’ and AmIII’ bands decrease significantly as the pH increases due to the α-helix hypochromism.
Non-two-state conformational transition of PLL
The pH induced PLL conformational transition previously studied by CD showed an isodichroic point.31 All the CD spectra could be fit by two basis CD spectra measured at the extreme pH values. These results argued for a two-state conformational transition, from an unfolded conformation to an α-helix conformation.
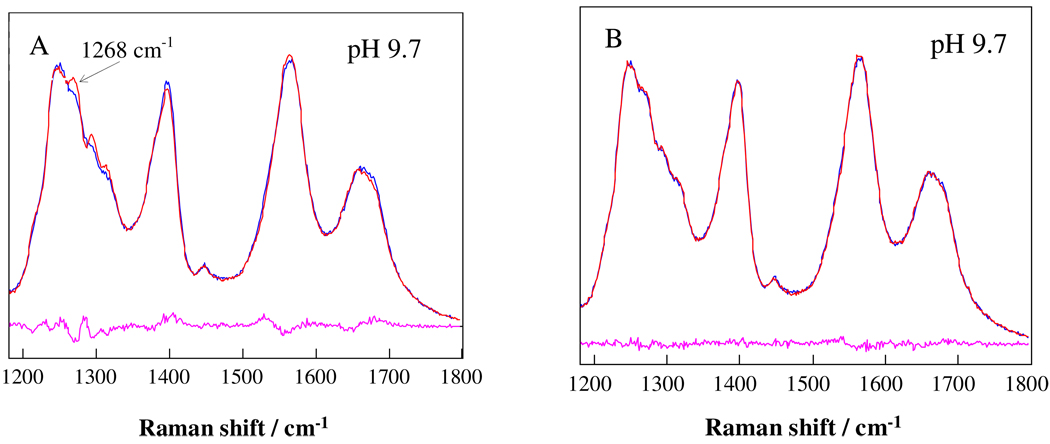
We examined this pH dependence by modeling the observed Raman spectra by linearly adding the two PLL Raman spectra at the extreme pH values using a least-squares method.32 The pH 9.1 to 10.5 spectra cannot be fit well by the use of the pH 11.7 and pH 7.1 (or at any pH < 8.4) basis spectra (Fig. 3A). The modeled results using the pH 7.1 basis spectrum always shows a stronger 2.51-helix band at ~ 1268 cm−1 than is observed between pH 9.1 and 10.5, indicating that the 2.51-helix content decreases relative to that of the PPII as the pH increases. However, the entire pH range between 9.1 to 11.7 can be fit well by using the pH 9.1 and pH 11.7 basis spectra, as shown in Fig. 3B. These results indicate that the pH induced conformational transitions between pH 7.1 to 11.7 involve more than two states, whereas the conformational transition between pH 9.1 to 11.7 involves only two conformation or an equilibrium between two state of multiple conformations whose stoichiometry is constant within the sets.
Figure 3.
Comparison between observed (blue) and modeled (red) 204 nm UVRR of PLL at pH 9.7. (A) Using PLL UVRR pH 7.1 and 11.7 basis spectra. (B) Using PLL UVRR pH 9.1 and 11.7 basis spectra. The residuals show the differences between the modeled and observed spectra.
Our UVRR two-state fitting of the PLL spectra demonstrates the high sensitivity of the AmIII3 band region to peptide conformation, which allows us to discover that the relative fraction of 2.51-helix conformation to the PPII conformation decreases somewhat as the pH increases. These results also demonstrate that the appearance of isosbestic points is not always a reliable indicator of a two-state transition.
2D correlation spectroscopy
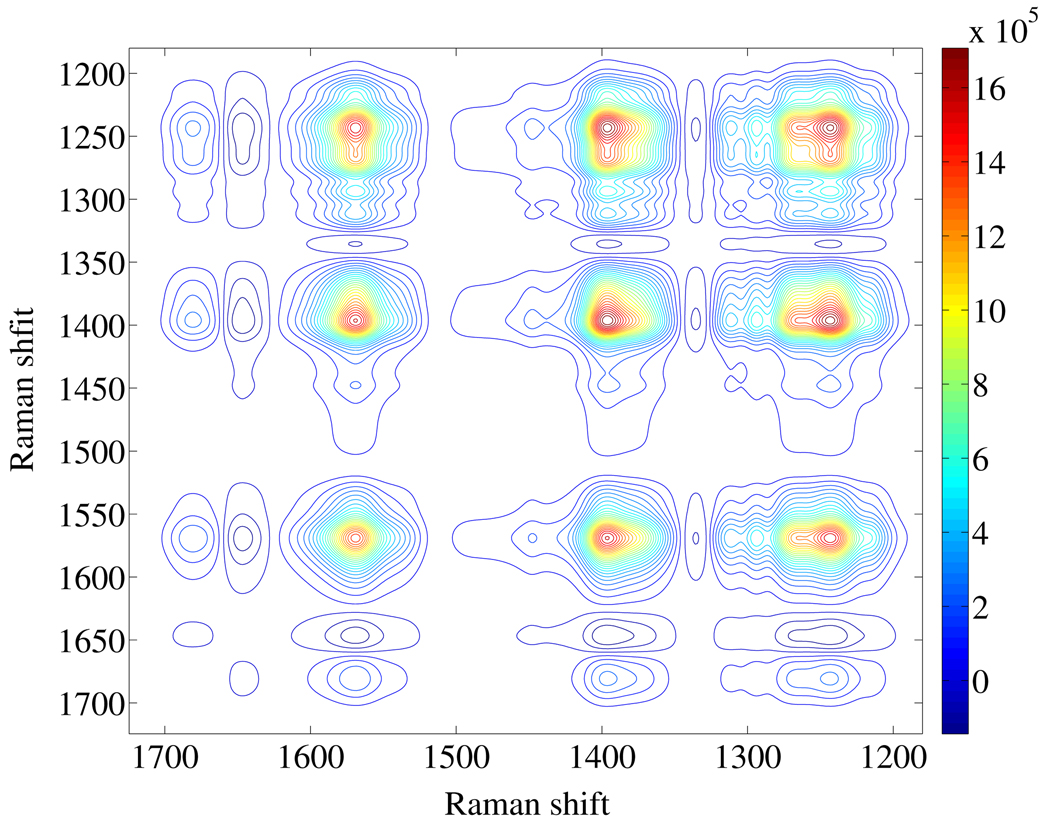
Generalized two-dimensional (2D) correlation spectroscopy was developed by Noda in 1986 to study the response of a system to an applied external perturbation.23 The 2D correlation analysis produces two resulting plots where the correlation intensity is plotted as a function of two independent spectral axes: a 2D synchronous spectrum and a 2D asynchronous spectrum which represent the similarity and dissimilarity of spectral variations as a function of the perturbation. These spectra enhance spectral resolution. This increased resolution may enable discrimination of highly overlapped spectral peaks.
We performed a 2D correlation analysis on the pH dependent UVRR spectra of PLL. The 2D synchronous spectrum shown in Fig. 4 is characterized by strong and broad autopeaks and cross peaks for the AmIII band, the Cα-H b band and the AmII band. The color scale shows the correlated intensity variation spanned by the 2D contours. The large correlation intensities in the 2D synchronous plot indicate that the AmIII band, the Cα-H b band and the AmII band intensities of PLL vary with pH. The AmI band clearly resolves into two components at ~1650 cm−1 and 1665 cm−1. The correlation magnitudes for the AmI band are smaller than for the other peaks, because its pH induced intensity variations are much smaller than for the other bands. Essentially, no additional information is gained from the synchronous spectrum compared to the conventional analysis of UVRR spectra described above.
Figure 4.
Synchronous pH dependence 2D UVRR spectrum of PLL.
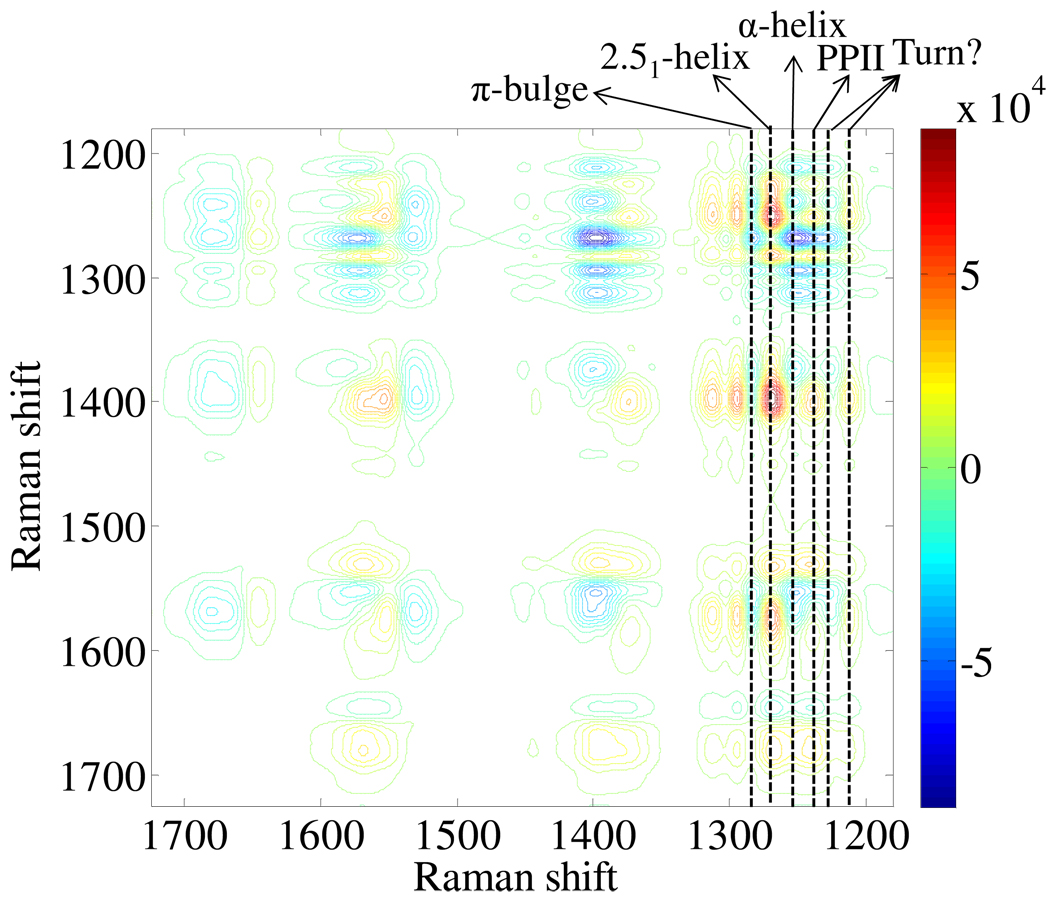
Fig. 5 shows the 2D asynchronous correlation spectrum of PLL. The cross peaks occur when the intensities of two bands change at different rates or vary out of phase with respect to each other. No asynchronous cross peak occurs in the AmI region (1620–1700 cm−1), which indicates that the correlation intensities in this region are weak. However, the cross peaks between the AmI and other bands resolve into two AmI band components that derive from extended and α-helix-like conformations.
Figure 5.
Asynchronous pH dependence 2D UVRR spectrum of PLL.
The asynchronous contour in the AmII region (1500–1600 cm−1) shows three out-of-phase cross peaks appearing at ~1530, 1553, 1573 cm−1 indicating that there are three components in the AmII region. As seen in Fig. 1B, the AmII band of PLL at pH 7.1 contains two subbands at ~ 1549 and 1567 cm−1. The third AmII subband at ~1530 cm−1, resolved in the 2D asynchronous spectrum, derives from the high pH α-helix-like PLL conformation. The Cα-H b band resolves into two components at ~1373 and 1396 cm−1, respectively, whose origin is not yet clear.
The asynchronous contour in the AmIII region (1200–1320 cm−1) is very complex. The complexity comes, in part, from the occurrence of the multiple conformations of PLL as the pH changes. It is very difficult to carry out band assignments based on just the AmIII cross peaks. Fortunately, the cross correlation regions of the AmIII band with the AmII and Cα-H b bands shed light on the analysis of the existence of multiple conformations.
As seen in Fig. 5, five AmIII subbands are observed at ~1316, 1294, 1270, 1241 and 1213 cm−1,that correlate to the Cα-H b 1396 cm−1 subband, and the AmII 1530 and 1573 cm−1 subbands; three AmIII peaks at ~1283, 1252, and 1225 cm−1 correlate with the 1373 cm−1 Cα-H b subband and the 1553 cm−1 AmII subbands. Therefore, 8 subbands resolve in the AmIII region at ~1316, 1294, 1283, 1270, 1252, 1241, 1225 and 1213 cm−1.
The 1316 and 1394 cm−1 subbands come from the AmIII2 and AmIII1 bands, while the other six are associated with the the AmIII3 bands that derive from PLL different secondary structures. The subbands at 1270 and 1241 cm−1 derive from the 2.51-helix and PPII conformation, respectively. The subband at 1213 cm−1 may result from the 1212 cm−1 shoulder observed in the Fig. 1 low pH PLL spectra. The other three subbands at 1283, 1252, and 1225 cm−1 result from high pH induced conformations. By using the method of Mikhonin et al.,30 we calculate that the subband at 1283 cm−1 most likely derives from π-helix/bulge conformations with an average Ramanchandran Ψ angle of −72°. The subband at 1252 cm−1 derives from an α-helix conformation with an average Ψ = −35°, and the 1225 cm−1 subband may derive from a β-turn structure.
Thus, the asynchronous spectrum shows greatly enhanced spectral resolution. It reveals two components in the Cα-H b band and three components in the AmII band. The cross peaks in asynchronous spectrum, between the AmIII3 band and the other bands, resolves into six AmIII3 subbands which represents six PLL conformations including three new structures induced by increasing pH: the π-helix, α-helix and some turn structure.
A widely used application of 2D correlation spectroscopy is to determine the sequential order of reaction events based on the analysis of the sign of the peaks in the synchronous and asynchronous spectra. Generally, this analysis utilizes the sequential order rule proposed by Noda, which was originally developed from periodic perturbation 2D correlation spectroscopy.33,34 The reliability of this sequential order rule for analyzing nonperiodic data set was recently questioned.35,36 The work showed that synchronous and asynchronous spectra in the generalized 2D correlation spectroscopy of nonperiodic data cannot be simply interpreted identically to that of 2D correlation spectroscopy of periodic data. We conclude that we cannot determine the pH sequence of PLL conformational changes directly from these 2D synchronous and asynchronous results.
NaCl concentration dependence of PLL UVRR
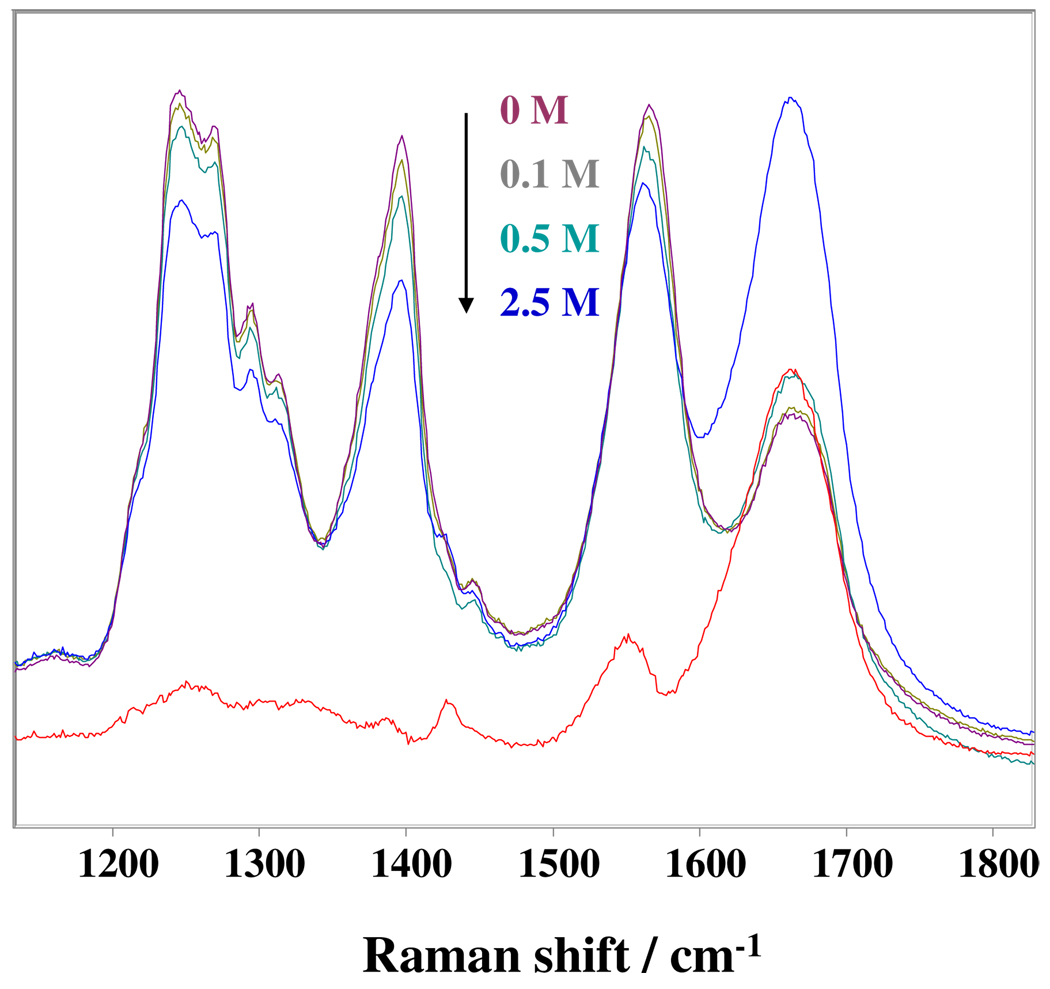
It has long been known that salts can significantly impact peptide and protein conformations mainly due to Debye-Hückel screening, ion binding and ion modulation of water structure.37,38 However, there appears to be little impact of high NaCl concentrations on the PLL conformation. For example, Fig. 6 shows the NaCl concentration dependence of the PLL UVRR spectra. Increasing the NaCl concentration causes the AmII, the Cα-H b and the AmIII band intensities to decrease slightly. The apparently large intensity increase of the AmI band actually results from a dramatic intensity increase of the overlapping water bending band due to the formation of a Cl−-water complex that has a strong charge transfer transition.39
Figure 6.
NaCl concentration dependence of the 204 nm PLL UVRR spectra (0.0, 0.1, 0.5 and 2.5 M NaCl). Also shown is a difference spectrum of PLL in the presence and absence of 2.5 M NaCl.
The AmII band downshifts ~ 3 cm−1 in 2.5 M NaCl. Surprisingly, the presence of 2.5 M NaCl does not change the relative intensity of the PPII to 2.51-helix bands, even though the 2.51-helix conformation is stabilized by electrostatic sidechain repulsions. Further, the expected large NaCl screening does not induce α-helix conformations. It should be noted that Xiong et al. also found that 2 M NaCl and KCl concentrations does not impact the PPII, 2.51-helix and α-helix conformational equilibrium of unfolded poly-L-glutamate (PLG).40
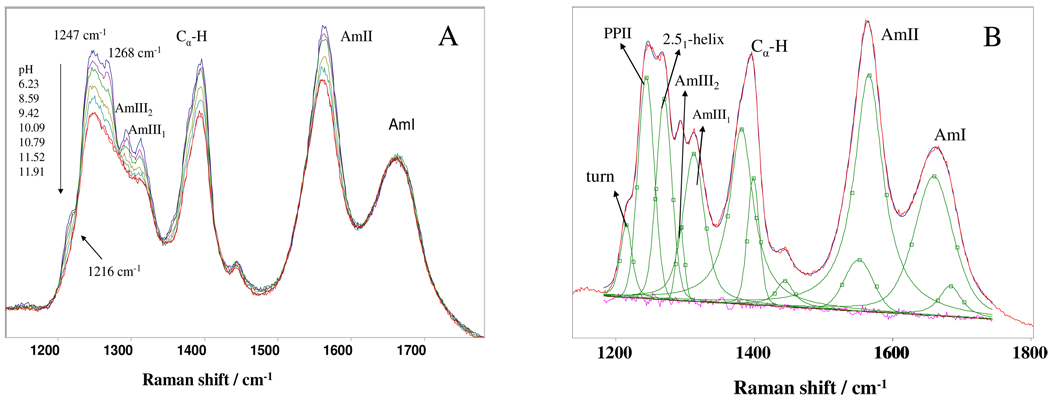
This surprising lack of NaCl impact on PLL and PLG conformations triggered us to reconfirm our assignment of 2.51-helix AmIII3 band assignment. We examined the pH dependence of the 204 nm UVRR spectrum of K10 (Fig. 7A). At neutral or low pH K10 shows UVRR very similar to those of low pH PLL (Fig. 1). Fig. 7B displays three resolved AmIII3 peaks at ~1216 cm−1, 1247 cm−1 and 1268 cm−1, that derive from the turn, the PPII and the 2.51-helix conformations. The AmIII2 band occurs at ~1293 cm−1, the AmIII1 band at ~1313 cm−1, the Cα-H band at ~1395 cm−1, the AmII band at ~1563 cm−1 and the AmI band at ~1665 cm−1. As the pH increases to 11.52, the Cα-H band integrated intensity decreases by ~ 20%. The decrease in K10 intensity is reminiscent of the intensity loss of high pH PLL which results from a conversion to the α-helical conformation. The intensity loss for K10 is much less because the propensity for the α-helical conformation is decreased due to its short length.41,42 K10 remains in ~80% extended conformation even at pH 11.9 where the sidechains are neutral. PLL is mainly α-helical structure at pH 11.7.
Figure 7.
A) pH dependence of 204 nm UVRR spectra of K10 at 10 °C. The spectra were normalized to the AmI band integrated intensity.16 B) Spectral deconvolution of 10 °C 204 nm UVRR K10 spectrum at pH 6.2.
In K10, the 2.51-helix 1268 cm−1 AmIII3 peak intensity significantly decreases relative to the PPII 1247 cm−1 peak as the pH increases, the 1247 cm−1 peak dominates at high pH. Thus, for K10, that 2.51-helix becomes destabilized as the sidechain electrostatic repulsion decrease. This result is consistent with our previous band assignment, that the 1268 cm−1 band derives from the 2.51-helix.
We can estimate the electrostatic repulsion decrease induced by salt screening by calculating the Debye lengths of the 0.1, 0.5, and 2.5 M NaCl solutions which are ~ 9.7, 4.3 and 1.9 Å. In PLL PPII and 2.51-helix conformation, the nearest neighbor sidechain spacings are 9.2 Å and 10.1 Å,13 respectively. Thus, we expect strong charge screening at these salt concentrations. We can estimate the expected solution sidechain electrostatic repulsion decrease induced by NaCl screening from: ψ(l) = ψ0e−l/λD, where ψ(l) is the electrostatic potential at distance l and λD is the Debye length. We estimate that for the 2.51-helix conformation the electrostatic repulsion between the nearest neighboring side chains spaced by 10.1 Å would decrease by about 3-fold, 10-fold and nearly 200-fold for 0.1, 0.5 and 2.5 M NaCl, respectively.
Based on our Raman data, PLL is ~ 50% α-helix-like structure at pH 10.1. The average pKa value of the PLL sidechains determined by Dos et al. using N15 NMR is 9.85±0.2.43 Thus, the degree of ionization of PLL at pH 10.1 is about 36% giving an average sidechain charge of ~ 0.36 esu. Since the electrostatic interaction is proportional to the square of the charge, the resulted average sidechain electrostatic repulsion at pH 10.1 decreases about 8-fold compare to that at low pH if the salt distributed uniformly throughout the peptide solution.
The electrostatic repulsion decrease induced by 2.5 M NaCl screening should be much larger than that induced by pH 10.1 sidechain neutralization. It is surprising that 2.5 M NaCl does not induce α-helix-like structures. Even more surprising is that is does not decrease the 2.51-helix fraction compared to the PPII conformation. At pH 10.1 PLL adopts ~50% α-helix-like content, and the relative fraction of the 2.51-helix conformation to the PPII conformation decreases compared to that at low pH. This result indicates that NaCl does not penetrate the sidechain backbone region of PLL.
α-helix-like conformations of PLL induced by high pH and ClO4−
In contrast, NaClO4 converts unfolded PPII and 2.51-helix charged sidechain PLL to the α-helix conformation.16 Addition of 0.8 M NaClO4 at pH 5.5 2.5 °C converts PLL to an ~86% α-helix-like content. The lack of NaCl impact on PLL conformation indicates specific ClO4− interactions with the PLL sidechains probably involving ion pairing. The ‘law of matching water affinities’ predicts a high ion pairing propensity between the -NH3+ sidechains and ClO4− because both ions are weakly hydrated.
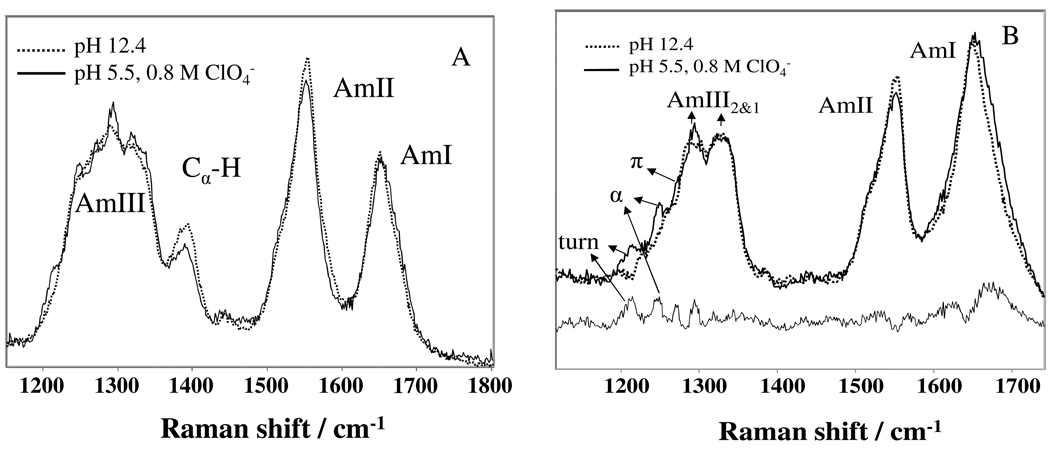
Fig. 8A compares the PLL UVRR spectra at pH 12.4 and 2.5 °C in the absence of ClO4− to that at pH 5.5 in the presence of 0.8 M ClO4−. Since the Cα-H b band only occurs in the PPII, 2.51-helix, β-sheet-like conformations, we can use the Cα-H band intensity to calculate the α-helix-like content. We assume that the unfolded PLL conformation UVRR spectra in ClO4− are identical to the low pH unfolded conformation UVRR in the absence of ClO4−. We also assume identical Cα-H b band cross sections of the PPII and 2.51-helix conformation. This calculation indicates that at pH 5.5 at 2.5 °C PLL contains ~80% α-helix-like content at pH 12.4, almost 6% less than that in 0.8 M ClO4−.
Figure 8.
(A) Comparison of PLL 204 nm UVRR spectra at pH 12.4 in pure water to that at pH 5.5 in the presence of 0.8 M ClO4− at 2.5 °C. The spectra are normalized to the AmI band integrated intensity. (B) Calculated pure α-helix-like PLL UVRR spectra at pH 12.4 in water and at pH 5.5 in the presence of 0.8 M ClO4−. Shown below is their difference spectrum. The α-helix-like UVRR spectra are calculated by subtracting the appropriate amount of the measured unfolded PLL conformation spectra (UVRR PLL spectra at pH 9.1) such that the Cα-H band disappears.
We previously found that NaClO4 increases the α-helix concentration of an ala-based, 21-residue peptide. Molecular dynamic simulation carried by Asciutto et al44 reveals that ClO4− binds strongly to the peptide backbone and excludes water from the peptide surface, thus, stabilizing the α-helix. We expect a similar phenomenon in PLL, where water would be excluded from PLL peptide surface, which results in a stabilization of the α-helix structure.
The spectra of the pure α-helix-like conformations of PLL can be obtained by subtracting appropriate amounts of the extended PLL conformation spectra from the observed UVRR. The calculated α-helix-like conformation spectra of PLL at pH 12.4 versus at pH 5.5 in 0.8 M ClO4− at 2.5 °C are compared in Fig. 8B. At pH 12.4, the AmI band is slightly narrower, the intensity of the AmII band is slightly higher, the AmIII3 peak intensities of turn and α-helix conformation are slightly weaker than that at pH 5.5 in 0.8 M ClO4−.
The narrower pH 12.4 PLL spectrum AmI bandwidth suggests that the α-helix-like conformation hydrogen bonding state is better defined than that of pH 5.5 0.8 M ClO4− conformation. The weaker turn and α-helix AmIII3 bands at pH 12.4 compared to that in 0.8 M ClO4− indicates that the high pH solution has less turn content and longer α-helices than at 0.8 M ClO4−, since a longer α-helix will result in a smaller Raman cross section due to more extensive hypochromism. This longer α-helix would result in more intramolecular hydrogen bonding which would narrow the AmI bandwidth.
We do not expect any intermolecular peptide interactions to influence the pH dependence of the PLL conformation at neutral and low pH values due to the repulsion between the charged peptide sidechains. This expectation is confirmed by our measured essentially identical pH 6.2, 204 nm UVRR spectra of PLL at 1 mg/ml and 2 mg/ml concentrations (not shown). In addition, a comparison of the recent Polavarapu et al.45 10 mg/ml pH 8.8 PLL CD spectrum to that of our Fig. 1 0.64 mg/ml, pH 5.5 CD spectrum16 indicates very similar conformations. Although the pH values are not identical, Fig. 1 in the present manuscript shows very modest UVRR changes for increasing pH values up until pH 9.1. Finally, Meyers31 measured 0.44 mg/ml, pH 8.35 CD PLL spectra that are very close to the 10 mg/ml Polavarapu et al.45 CD spectra.
In contrast, Polavarapu et al.45 demonstrated that at the higher pH value of 11.4 where the sidechains are mainly neutral PLL concentration increases from 1 mg/ml to 10 mg/ml induces conformational changes due to α-helix-like to a transition to β-sheet conformations. This conformational change is not observed by Meyer at pH 11.7 for the lower 0.44 mg/ml concentration. Further, we do not see any β-sheet contributions in the pH 11.5, 0.72 mg/ml PLL CD spectra (not shown). Thus, we conclude that there is a negligible interpeptide associations in the pH dependence of the 1 mg/ml PLL samples measured in the study here.
Conclusions
The pH dependence of PLL UVRR spectra between pH 7.1 and 11.7 cannot be described by a two-state model. The AmIII3 region fitting with pH 7.1 and 11.7 basis spectra reveals a small pH induced decrease in the relative fraction of the 2.51-helix conformation compared to the PPII conformation.
We performed a 2D general correlation analysis on the PLL pH dependence UVRR spectra. The asynchronous spectrum shows enhanced spectral resolution. The 2D asynchronous spectrum reveals multiple components in the Cα-H b band and the AmII band whose origins are unclear. The cross peaks in the 2D asynchronous spectrum between the AmIII band and the other bands reveals that increasing pH induces three new structures: π-helix, α-helix and some turn structure.
We examined the salt effect on PLL conformation, and found that 2.5 M NaCl does not change the equilibrium between the PPII and 2.51-helix conformation by screening sidechain electrostatic repulsion. The result indicates that NaCl does not penetrate the region between the sidechain and the peptide backbone.
We also compared PLL conformations induced by high pH to that induced by 0.8M ClO4−. Both conditions induce α-helix-like conformations. 0.8 M ClO4− induces 6% more α-helix-like conformations than at pH 12.4. Higher pH gives rise to longer α-helices and less turn structures.
ACKNOWLEDGMENT
We gratefully acknowledge Bhavya Sharma, Sergei Bykov, Aleksandr Mikhonin, Nataliya Myshakina, Kan Xiong and Zhenmin Hong for useful discussions. This work was supported by NIH grant 1RO1EBB009089-01.
REFERENCES
- 1.Baldwin RL, Rose GD. Trends Biochem. Sci. 1999;24:26. doi: 10.1016/s0968-0004(98)01346-2. [DOI] [PubMed] [Google Scholar]
- 2.Creighton TE, editor. Protein Structure. Second Edition. New York: W.H. Freeman; 1997. [Google Scholar]
- 3.Dill KA. Biochemistry. 1985;24:1501. doi: 10.1021/bi00327a032. [DOI] [PubMed] [Google Scholar]
- 4.Dill KA. Protein Sci. 1999;8:1166. doi: 10.1110/ps.8.6.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dobson CM, Sali A, Karplus M. Angew. Chem. Int. Ed. 1998;37:868. doi: 10.1002/(SICI)1521-3773(19980420)37:7<868::AID-ANIE868>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 6.Anfinsen CB. Science. 1973;181:223. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- 7.Anfinsen CB, Scheraga HA. Adv. Prot. Chem. 1975;29:205. doi: 10.1016/s0065-3233(08)60413-1. [DOI] [PubMed] [Google Scholar]
- 8.Haber E, Anfinsen CB. J. Biol. Chem. 1962;237:1839. [PubMed] [Google Scholar]
- 9.Dill KA, Ozkan SB, Shell MS, Weikl TR. Annu. Rev. Biophys. 2008;37:289. doi: 10.1146/annurev.biophys.37.092707.153558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suarez M, Jaramillo A. J. R. Soc. Interface. 2009;6:S477. doi: 10.1098/rsif.2008.0508.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morra G, Meli M, Colombo G. Curr. Protein Pept. Sci. 2008;9:181. doi: 10.2174/138920308783955234. [DOI] [PubMed] [Google Scholar]
- 12.Thirumalai D, O'Brien EP, Morrison G, Hyeon C. Annu. Rev. Biophys. 39:159. doi: 10.1146/annurev-biophys-051309-103835. [DOI] [PubMed] [Google Scholar]
- 13.Mikhonin AV, Myshakina NS, Bykov SV, Asher SA. J. Am. Chem. Soc. 2005;127:7712. doi: 10.1021/ja044636s. [DOI] [PubMed] [Google Scholar]
- 14.Shi Z, Chen K, Liu Z, Kallenbach NR. Chem. Rev. 2006;106:1877. doi: 10.1021/cr040433a. [DOI] [PubMed] [Google Scholar]
- 15.Krimm S, Mark JE. Proc. Nat. Acad. Sci. U. S. 1968;60:1122. doi: 10.1073/pnas.60.4.1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma L, Ahmed Z, Mikhonin AV, Asher SA. J. Phys. Chem. B. 2007;111:7675. doi: 10.1021/jp0703758. [DOI] [PubMed] [Google Scholar]
- 17.Davidson B, Fasman GD. Biochemistry. 1967;6:1616. doi: 10.1021/bi00858a008. [DOI] [PubMed] [Google Scholar]
- 18.JiJi RD, Balakrishnan G, Hu Y, Spiro TG. Biochemistry. 2006;45:34. doi: 10.1021/bi051507v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bykov S, Lednev I, Ianoul A, Mikhonin A, Munro C, Asher SA. Appl. Spectrosc. 2005;59:1541. doi: 10.1366/000370205775142511. [DOI] [PubMed] [Google Scholar]
- 20.Lednev IK, Karnoup AS, Sparrow MC, Asher SA. J. Am. Chem. Soc. 1999;121:8074. doi: 10.1021/ja003381p. [DOI] [PubMed] [Google Scholar]
- 21.Chi Z, Chen XG, Holtz JSW, Asher SA. Biochemistry. 1998;37:2854. doi: 10.1021/bi971160z. [DOI] [PubMed] [Google Scholar]
- 22.Noda I. Appl. Spectrosc. 2000;54:994. [Google Scholar]
- 23.Noda I, Ozaki Y. Two dimensional correlation spectroscopy-applicationa in vibrational and optical spectroscopy. Chichester, UK: John Wiley & Sons Ltd; 2004. [Google Scholar]
- 24.Imahori K, Tanaka J. J. Mol. Biol. 1959;1:359. [Google Scholar]
- 25.Tinoco IJ, Halpern A, Simpson WT. Polyamino Acids, Polypeptides, and Proteins. Madison: University of Wisconsin Press; 1962. [Google Scholar]
- 26.Jordan T, Spiro TG. J. Raman Spectrosc. 1995;26:867. [Google Scholar]
- 27.Mikhonin AV, Ahmed Z, Ianoul A, Asher SA. J. Phys. Chem. B. 2004;108:19020. [Google Scholar]
- 28.Myshakina NS, Ahmed Z, Asher SA. J. Phys. Chem. B. 2008;112:11873. doi: 10.1021/jp8057355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Triggs NE, Valentini JJ. J. Phys. Chem. 1992;96:6922. [Google Scholar]
- 30.Mikhonin AV, Bykov SV, Myshakina NS, Asher SA. J. Phys. Chem. B. 2006;110:1928. doi: 10.1021/jp054593h. [DOI] [PubMed] [Google Scholar]
- 31.Myer YP. Macromolecules. 1969;2:624. [Google Scholar]
- 32.Chi Z, Chen XG, Holtz JSW, Asher SA. Biochemistry. 1998;37:2854. doi: 10.1021/bi971160z. [DOI] [PubMed] [Google Scholar]
- 33.Noda I. J. Am. Chem. Soc. 1989;111:8116. [Google Scholar]
- 34.Noda I. Appl. Spectrosc. 1993;47:1329. [Google Scholar]
- 35.Huang H. Anal. Chem. 2007;79:8281. doi: 10.1021/ac0708590. [DOI] [PubMed] [Google Scholar]
- 36.Jia Q, Wang N-N, Yu Z-W. Appl. Spectrosc. 2009;63:344. doi: 10.1366/000370209787598861. [DOI] [PubMed] [Google Scholar]
- 37.Goto Y, Takahashi N, Fink AL. Biochemistry. 1990;29:3480. doi: 10.1021/bi00466a009. [DOI] [PubMed] [Google Scholar]
- 38.Lund M, Vacha R, Jungwirth P. Langmuir. 2008;24:3387. doi: 10.1021/la7034104. [DOI] [PubMed] [Google Scholar]
- 39.Xiong K, Asher SA. J. Phys. Chem. A. 2010 in press. [Google Scholar]
- 40.Xiong K, Ma L, Asher SA. submitted to Angew. Chem. Int. Ed [Google Scholar]
- 41.Lifson S, Roig A. J. Chem. Phys. 1961;34:1963. [Google Scholar]
- 42.Zimm BH, Bragg JK. J. Chem. Phys. 1959;31:526. [Google Scholar]
- 43.Dos A, Schimming V, Tosoni S, Limbach H-H. J. Phys. Chem. B. 2008;112:15604. doi: 10.1021/jp806551u. [DOI] [PubMed] [Google Scholar]
- 44.Asciutto EK, General IJ, Xiong K, Asher SA, Madura JD. Biophys. J. 2010;98:186. doi: 10.1016/j.bpj.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shanmugam G, Polavarapu PL. J. Mol. Struct. 2008;890:144. [Google Scholar]