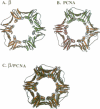Abstract
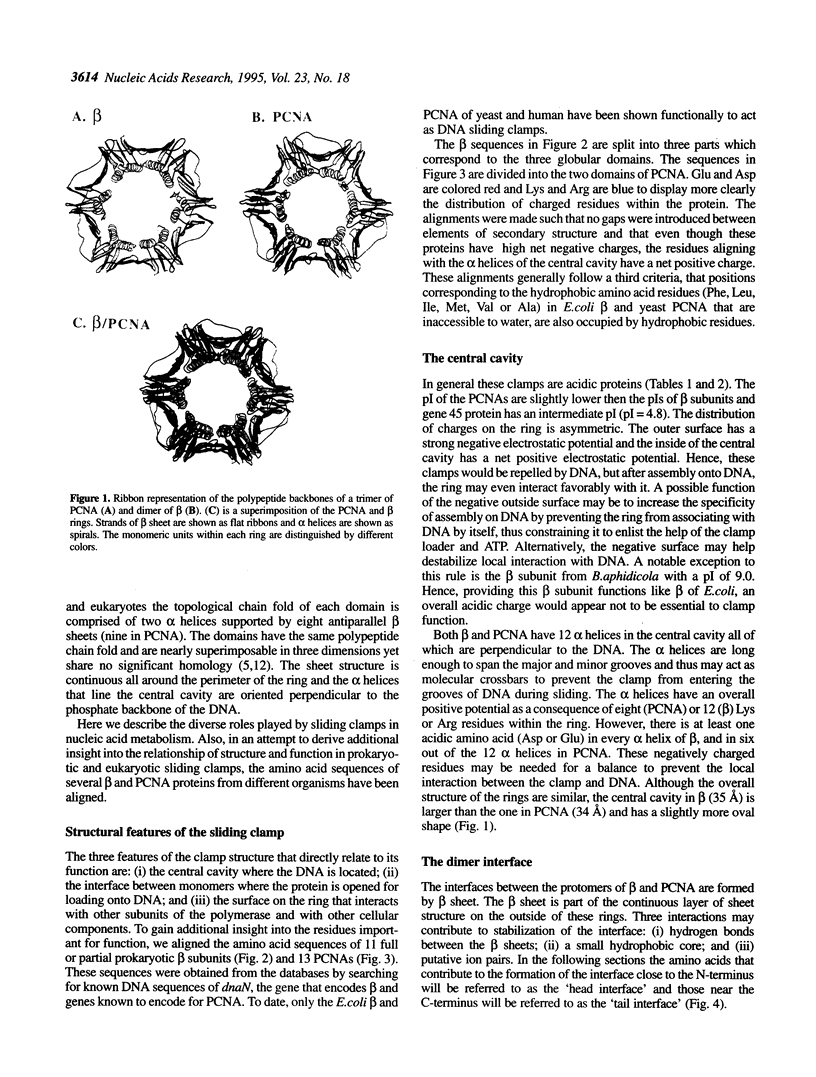
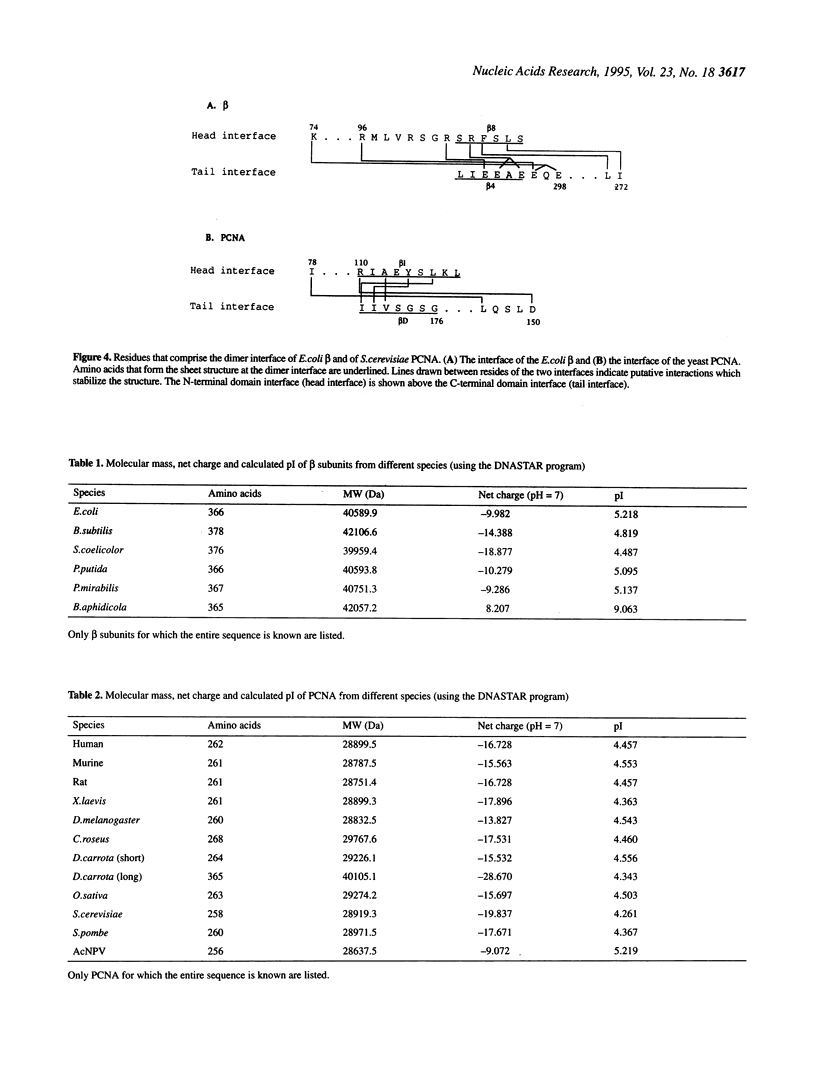
The remarkable processivity of cellular replicative DNA polymerases derive their tight grip to DNA from a ring-shaped protein that encircles DNA and tethers the polymerase to the chromosome. The crystal structures of prototypical 'sliding clamps' of prokaryotes (beta subunit) and eukaryotes (PCNA) are ring shaped proteins for encircling DNA. Although beta is a dimer and PCNA is a trimer, their structures are nearly superimposable. Even though they are not hexamers, the sliding clamps have a pseudo 6-fold symmetry resulting from three globular domains comprising each beta monomer and two domains comprising each PCNA monomer. These domains have the same chain fold and are nearly identical in three-dimensions. The amino acid sequences of 11 beta and 13 PCNA proteins from different organisms have been aligned and studied to gain further insight into the relation between the structure and function of these sliding clamps. Furthermore, a putative embryonic form of PCNA is the size of beta and thus may encircle DNA as a dimer like the prokaryotic clamps.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Almendral J. M., Huebsch D., Blundell P. A., Macdonald-Bravo H., Bravo R. Cloning and sequence of the human nuclear protein cyclin: homology with DNA-binding proteins. Proc Natl Acad Sci U S A. 1987 Mar;84(6):1575–1579. doi: 10.1073/pnas.84.6.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer G. A., Burgers P. M. Molecular cloning, structure and expression of the yeast proliferating cell nuclear antigen gene. Nucleic Acids Res. 1990 Jan 25;18(2):261–265. doi: 10.1093/nar/18.2.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calcutt M. J., Schmidt F. J. Conserved gene arrangement in the origin region of the Streptomyces coelicolor chromosome. J Bacteriol. 1992 May;174(10):3220–3226. doi: 10.1128/jb.174.10.3220-3226.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Rozas H., Kelman Z., Dean F. B., Pan Z. Q., Harper J. W., Elledge S. J., O'Donnell M., Hurwitz J. Cdk-interacting protein 1 directly binds with proliferating cell nuclear antigen and inhibits DNA replication catalyzed by the DNA polymerase delta holoenzyme. Proc Natl Acad Sci U S A. 1994 Aug 30;91(18):8655–8659. doi: 10.1073/pnas.91.18.8655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M. Q., Yoshikawa H., Ogasawara N. Structure of the dnaA and DnaA-box region in the Mycoplasma capricolum chromosome: conservation and variations in the course of evolution. Gene. 1992 Jan 2;110(1):17–23. doi: 10.1016/0378-1119(92)90439-v. [DOI] [PubMed] [Google Scholar]
- Fujita M. Q., Yoshikawa H., Ogasawara N. Structure of the dnaA region of Micrococcus luteus: conservation and variations among eubacteria. Gene. 1990 Sep 1;93(1):73–78. doi: 10.1016/0378-1119(90)90138-h. [DOI] [PubMed] [Google Scholar]
- Fujita M. Q., Yoshikawa H., Ogasawara N. Structure of the dnaA region of Pseudomonas putida: conservation among three bacteria, Bacillus subtilis, Escherichia coli and P. putida. Mol Gen Genet. 1989 Feb;215(3):381–387. doi: 10.1007/BF00427033. [DOI] [PubMed] [Google Scholar]
- Gogol E. P., Young M. C., Kubasek W. L., Jarvis T. C., von Hippel P. H. Cryoelectron microscopic visualization of functional subassemblies of the bacteriophage T4 DNA replication complex. J Mol Biol. 1992 Mar 20;224(2):395–412. doi: 10.1016/0022-2836(92)91003-8. [DOI] [PubMed] [Google Scholar]
- Hata S., Kouchi H., Tanaka Y., Minami E., Matsumoto T., Suzuka I., Hashimoto J. Identification of carrot cDNA clones encoding a second putative proliferating cell-nuclear antigen, DNA polymerase delta auxiliary protein. Eur J Biochem. 1992 Feb 1;203(3):367–371. doi: 10.1111/j.1432-1033.1992.tb16559.x. [DOI] [PubMed] [Google Scholar]
- Herendeen D. R., Kassavetis G. A., Geiduschek E. P. A transcriptional enhancer whose function imposes a requirement that proteins track along DNA. Science. 1992 May 29;256(5061):1298–1303. doi: 10.1126/science.1598572. [DOI] [PubMed] [Google Scholar]
- Kaboord B. F., Benkovic S. J. Accessory proteins function as matchmakers in the assembly of the T4 DNA polymerase holoenzyme. Curr Biol. 1995 Feb 1;5(2):149–157. doi: 10.1016/s0960-9822(95)00036-4. [DOI] [PubMed] [Google Scholar]
- Kelman Z., O'Donnell M. DNA polymerase III holoenzyme: structure and function of a chromosomal replicating machine. Annu Rev Biochem. 1995;64:171–200. doi: 10.1146/annurev.bi.64.070195.001131. [DOI] [PubMed] [Google Scholar]
- Kelman Z., O'Donnell M. DNA replication: enzymology and mechanisms. Curr Opin Genet Dev. 1994 Apr;4(2):185–195. doi: 10.1016/s0959-437x(05)80044-9. [DOI] [PubMed] [Google Scholar]
- Kelman Z., O'Donnell M. Embryonic PCNA: a missing link? Curr Biol. 1995 Jul 1;5(7):814–814. doi: 10.1016/s0960-9822(95)00159-x. [DOI] [PubMed] [Google Scholar]
- Kodama H., Ito M., Ohnishi N., Suzuka I., Komamine A. Molecular cloning of the gene for plant proliferating-cell nuclear antigen and expression of this gene during the cell cycle in synchronized cultures of Catharanthus roseus cells. Eur J Biochem. 1991 Apr 23;197(2):495–503. doi: 10.1111/j.1432-1033.1991.tb15937.x. [DOI] [PubMed] [Google Scholar]
- Kong X. P., Onrust R., O'Donnell M., Kuriyan J. Three-dimensional structure of the beta subunit of E. coli DNA polymerase III holoenzyme: a sliding DNA clamp. Cell. 1992 May 1;69(3):425–437. doi: 10.1016/0092-8674(92)90445-i. [DOI] [PubMed] [Google Scholar]
- Krishna T. S., Kong X. P., Gary S., Burgers P. M., Kuriyan J. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell. 1994 Dec 30;79(7):1233–1243. doi: 10.1016/0092-8674(94)90014-0. [DOI] [PubMed] [Google Scholar]
- Kuriyan J., O'Donnell M. Sliding clamps of DNA polymerases. J Mol Biol. 1993 Dec 20;234(4):915–925. doi: 10.1006/jmbi.1993.1644. [DOI] [PubMed] [Google Scholar]
- Lai C. Y., Baumann P. Genetic analysis of an aphid endosymbiont DNA fragment homologous to the rnpA-rpmH-dnaA-dnaN-gyrB region of eubacteria. Gene. 1992 Apr 15;113(2):175–181. doi: 10.1016/0378-1119(92)90393-4. [DOI] [PubMed] [Google Scholar]
- Leibovici M., Gusse M., Bravo R., Méchali M. Characterization and developmental expression of Xenopus proliferating cell nuclear antigen (PCNA). Dev Biol. 1990 Sep;141(1):183–192. doi: 10.1016/0012-1606(90)90113-w. [DOI] [PubMed] [Google Scholar]
- Matsumoto K., Moriuchi T., Koji T., Nakane P. K. Molecular cloning of cDNA coding for rat proliferating cell nuclear antigen (PCNA)/cyclin. EMBO J. 1987 Mar;6(3):637–642. doi: 10.1002/j.1460-2075.1987.tb04802.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya S., Ogasawara N., Yoshikawa H. Structure and function of the region of the replication origin of the Bacillus subtilis chromosome. III. Nucleotide sequence of some 10,000 base pairs in the origin region. Nucleic Acids Res. 1985 Apr 11;13(7):2251–2265. doi: 10.1093/nar/13.7.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell M., Onrust R., Dean F. B., Chen M., Hurwitz J. Homology in accessory proteins of replicative polymerases--E. coli to humans. Nucleic Acids Res. 1993 Jan 11;21(1):1–3. doi: 10.1093/nar/21.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Reilly D. R., Crawford A. M., Miller L. K. Viral proliferating cell nuclear antigen. Nature. 1989 Feb 16;337(6208):606–606. doi: 10.1038/337606a0. [DOI] [PubMed] [Google Scholar]
- Ohmori H., Kimura M., Nagata T., Sakakibara Y. Structural analysis of the dnaA and dnaN genes of Escherichia coli. Gene. 1984 May;28(2):159–170. doi: 10.1016/0378-1119(84)90253-1. [DOI] [PubMed] [Google Scholar]
- Reddy M. K., Weitzel S. E., von Hippel P. H. Assembly of a functional replication complex without ATP hydrolysis: a direct interaction of bacteriophage T4 gp45 with T4 DNA polymerase. Proc Natl Acad Sci U S A. 1993 Apr 15;90(8):3211–3215. doi: 10.1073/pnas.90.8.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders G. M., Kassavetis G. A., Geiduschek E. P. Use of a macromolecular crowding agent to dissect interactions and define functions in transcriptional activation by a DNA-tracking protein: bacteriophage T4 gene 45 protein and late transcription. Proc Natl Acad Sci U S A. 1994 Aug 2;91(16):7703–7707. doi: 10.1073/pnas.91.16.7703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skovgaard O., Hansen F. G. Comparison of dnaA nucleotide sequences of Escherichia coli, Salmonella typhimurium, and Serratia marcescens. J Bacteriol. 1987 Sep;169(9):3976–3981. doi: 10.1128/jb.169.9.3976-3981.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skovgaard O. Nucleotide sequence of a Proteus mirabilis DNA fragment homologous to the 60K-rnpA-rpmH-dnaA-dnaN-recF-gyrB region of Escherichia coli. Gene. 1990 Sep 1;93(1):27–34. doi: 10.1016/0378-1119(90)90131-a. [DOI] [PubMed] [Google Scholar]
- Smith M. L., Chen I. T., Zhan Q., Bae I., Chen C. Y., Gilmer T. M., Kastan M. B., O'Connor P. M., Fornace A. J., Jr Interaction of the p53-regulated protein Gadd45 with proliferating cell nuclear antigen. Science. 1994 Nov 25;266(5189):1376–1380. doi: 10.1126/science.7973727. [DOI] [PubMed] [Google Scholar]
- Stukenberg P. T., Turner J., O'Donnell M. An explanation for lagging strand replication: polymerase hopping among DNA sliding clamps. Cell. 1994 Sep 9;78(5):877–887. doi: 10.1016/s0092-8674(94)90662-9. [DOI] [PubMed] [Google Scholar]
- Suzuka I., Hata S., Matsuoka M., Kosugi S., Hashimoto J. Highly conserved structure of proliferating cell nuclear antigen (DNA polymerase delta auxiliary protein) gene in plants. Eur J Biochem. 1991 Jan 30;195(2):571–575. doi: 10.1111/j.1432-1033.1991.tb15739.x. [DOI] [PubMed] [Google Scholar]
- Tinker R. L., Kassavetis G. A., Geiduschek E. P. Detecting the ability of viral, bacterial and eukaryotic replication proteins to track along DNA. EMBO J. 1994 Nov 15;13(22):5330–5337. doi: 10.1002/j.1460-2075.1994.tb06867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waga S., Hannon G. J., Beach D., Stillman B. The p21 inhibitor of cyclin-dependent kinases controls DNA replication by interaction with PCNA. Nature. 1994 Jun 16;369(6481):574–578. doi: 10.1038/369574a0. [DOI] [PubMed] [Google Scholar]
- Waseem N. H., Labib K., Nurse P., Lane D. P. Isolation and analysis of the fission yeast gene encoding polymerase delta accessory protein PCNA. EMBO J. 1992 Dec;11(13):5111–5120. doi: 10.1002/j.1460-2075.1992.tb05618.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M., Hayashi Y., Hirose F., Matsuoka S., Moriuchi T., Shiroishi T., Moriwaki K., Matsukage A. Molecular cloning and structural analysis of mouse gene and pseudogenes for proliferating cell nuclear antigen. Nucleic Acids Res. 1991 May 11;19(9):2403–2410. doi: 10.1093/nar/19.9.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M., Nishida Y., Moriuchi T., Hirose F., Hui C. C., Suzuki Y., Matsukage A. Drosophila proliferating cell nuclear antigen (cyclin) gene: structure, expression during development, and specific binding of homeodomain proteins to its 5'-flanking region. Mol Cell Biol. 1990 Mar;10(3):872–879. doi: 10.1128/mcb.10.3.872. [DOI] [PMC free article] [PubMed] [Google Scholar]