Abstract
We have implemented aggregation and correlation toolbox (ACT), an efficient, multifaceted toolbox for analyzing continuous signal and discrete region tracks from high-throughput genomic experiments, such as RNA-seq or ChIP-chip signal profiles from the ENCODE and modENCODE projects, or lists of single nucleotide polymorphisms from the 1000 genomes project. It is able to generate aggregate profiles of a given track around a set of specified anchor points, such as transcription start sites. It is also able to correlate related tracks and analyze them for saturation–i.e. how much of a certain feature is covered with each new succeeding experiment. The ACT site contains downloadable code in a variety of formats, interactive web servers (for use on small quantities of data), example datasets, documentation and a gallery of outputs. Here, we explain the components of the toolbox in more detail and apply them in various contexts.
Availability: ACT is available at http://act.gersteinlab.org
Contact: pi@gersteinlab.org
1 INTRODUCTION
There is now an abundance of genome-sized data from high-throughput genomic experiments. For instance, there are ChIP-chip, ChIP-seq and RNA-seq experiments from the ENCODE (ENCODE Project Consortium, 2007) and modENCODE (modENCODE consortium, 2009) projects. There are also genome sequence data that can be used to generate tracks measuring sequence content, such as the densities of single nucleotide polymorphisms (SNPs) from dbSNP (Sharry et al., 2001) and the 1000 genomes project. In most cases, the representations of these data take the form of either signal tracks that describe a genomic landscape or distinct region tracks that tag portions of the genome as active. The aggregation and correlation toolbox (ACT) provides a powerful set of programs that can be applied to any experiments producing data in these formats. The ability to analyze multiple genomic datasets is important, as demonstrated by tools like Galaxy (Giardine et al., 2005). ACT provides a unique set of functionality that complements existing methods of analysis.
2 THE ACT TOOLBOX: OVERVIEW
ACT facilitates three main types of analysis:
Aggregation: in many scenarios, it is useful to determine the distribution of signals in a signal track relative to certain genomic anchors (Fig. 1, aggregation). For example, it has recently been reported that the contribution of each transcription factor binding site to tissue-specific gene expression depends on its position relative to the transcription start site (TSS) (MacIssac et al., 2010). It is thus useful to aggregate binding signals of transcription factors at a certain distance from the TSSs of all genes (the anchors). In general, this type of aggregation analyses helps identify proximity correlations and functional relationships between the signals and anchors. In the ENCODE pilot study (ENCODE Project Consortium, 2007), it has been used to demonstrate positional relationships between chromatin features and TSSs.
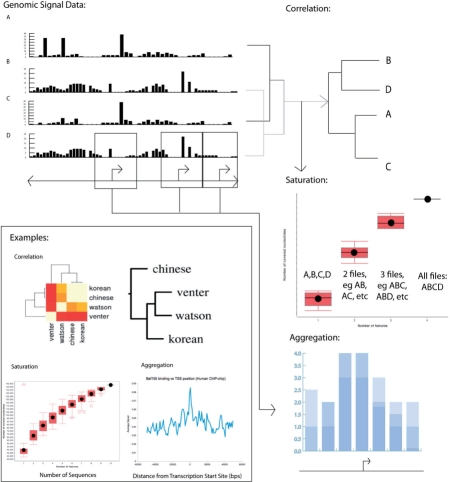
Fig. 1.
Uses of ACT using signal tracks from various sources. Signal around all TSSs is aggregated to give an average signal profile, for example of Baf155 binding around TSSs (Encode Project) (aggregation). Figure made in Excel (correlation). Multiple signal tracks are correlated to show which tracks are more or less related to each other. In the selected example, a heatmap of the SNP track correlation between four individuals (dbSNP) leads to a dendogram of their phylogenetic relationship. Figure made using Web ACT. Each additional signal track increases the number of base pairs covered (saturation). When the addition of signal tracks is considered in all possible combinations, the average increase in coverage, with error bars, can be visualized by a saturation plot. In the example, data are taken from individuals from dbSNP [with additional genomes from Ahn et al. (2009), Bentley et al. (2008), Drmanac et al. (2010), Kim et al. (2009)]. In each box plot, the top and bottom pink bars correspond to the maximum and minimum normal values, the top edge, middle line and bottom edge of the box correspond to the top 25 percentile, median and bottom 25 percentile, the black dot is the mean, and red circles are outliers. Figure made using ACT downloadable saturation program.
Correlation: it is also useful to consider how multiple-related signal tracks are correlated with each other. For example, a previous study (Zhang et al., 2007) demonstrated, using whole-track correlation methods, that there was a consistent relationship among transcription factors as judged by their signal profiles across several ChIP-chip experiments. By providing a means of correlating signal tracks with each other, ACT allows for initial comparison of different experiments to see which are more similar or related than others (Fig. 1, correlation).
Saturation: another important type of analysis is determining the number of experimental conditions required to achieve a high genomic coverage of the biological phenomenon under study. For example, using ChIP-chip or ChIP-seq experiments, one could identify a set of transcription factor binding sites from a human cell line. When the experiment is repeated using another cell line, some additional binding sites could be identified. How many cell lines need to be considered in order to reach the point of saturation, so that few new binding sites would be identified by extra experiments? ACT produces plots that help answer this type of question.
3 DETAILS AND USE CASES
ACT is available as a suite of downloadable scripts corresponding to the aggregation, correlation and saturation components of the toolbox. The tool is intended for Linux/Unix users with Java and Python. In addition, it is useful to have R for output visualization for the aggregation and correlation tools. There is also a compendium of other versions of the tool components written in different languages and with varied functionality. For some types of analysis, there are web components for demonstration purposes on small datasets with built-in visualization features. However, because most whole-genome signal tracks are too large to upload via standard Internet connections, users are recommended to download the toolbox and run it locally. As performing these calculations on whole-genome data can be especially time intensive, the version of the tools presented here has been designed to run efficiently on large datasets.
Aggregation: the aggregation component is designed to take a signal track (.sgr or .wig) and an annotation track (.bed) as input, and compute the average signal over a certain number of base pairs upstream and downstream of (i.e. a fixed radius around) the annotations. In other words, signal values are taken from the region surrounding each annotation, and averaged over the number of annotation anchors provided. The base pair resolution of the aggregation can be specified by the number of bins (narrower bins give more data points and therefore finer granularity). Results of such calculation can be plotted as in Figure 1 (aggregation). ACT also provides features such as computing the standard deviation, median and quartiles that can be viewed as a boxplot, as well as scaling aggregation over regions such as areas between transcription start and end sites or within exons so that all of the aggregate signals within those regions fall into a fixed number of bins. In this case, bin size is dynamically computed for each region so that the same number of bins cover regions of different sizes.
Correlation: the correlation analysis takes a set of active genomic regions (.bed) such as a SNP track or a genomic signal track (.wig). It then divides genomic coordinates into bins and gives each bin a value corresponding to the mean or maximum signal values which fall within the bin, or assigns value based on the number of ‘active regions’ which fall within the bin. A final correlation matrix is created based on either the Spearman's, Pearson's or normal score correlation between each pair of binned datasets. The results can be visualized as a heatmap or as a phylogenetic tree using programs such as PHYLIP (Felsenstein, 1996). One version of the correlation tool uses parallelization to decrease the pro-gram's overall running time. This component was written largely in Java. Examples of correlation output based on SNP tracks and ChIP-chip data are shown in Figure 1 (correlation).
Saturation: we provide an efficient implementation of saturation plot generator. Each input file corresponds to one dataset (e.g. one new individual, in .bed format), and each line in a file specifies a genomic location that has the biological phenomenon under study (e.g. tagged SNPs). The saturation plot shows, with each new dataset (x-axis), what percentage of genomic base pairs are covered (y-axis). The program considers the various combinations in which tracks can be added so that the increase in base pair coverage is a range of values based on all the files in the input. The resulting plot is output in PDF format (Fig. 1, saturation), in which a series of boxplots depicts increasing base pair coverage, where the boxplot at each position m on the x-axis shows the coverage values of all combinations of m conditions. Boxplots that approach a horizontal asymptote indicate that the coverage has reached saturation. Our implementation makes use of special data structures to avoid redundant counting. It normally takes less than a minute to generate the plot for up to 30 input files each with a few thousand lines. To handle more files and files with more lines, the tool also provides an option to compute the coverage of a random sample of the input file combinations.
4 DISCUSSION
There are number of additional analyses that can be done to fine-tune the output of ACT. For instance, it is possible to use the online genomic signal aggregator (GSA), which assigns each genomic position to the nearest anchor in order to reduce the artifacts caused by the subsets of anchors clustering together, to handle tightly clustered anchors. Also, aggregation can be used in conjunction with genome structure correction to determine if the enrichments of a given signal with respect to anchor points are significantly relative to the non-random positioning of the anchors (ENCODE Project Consortium, 2007). This correction takes into account the fact that a ‘random’ distribution of anchors on the genome arises from a distinctly non-uniform distribution. Practically, this could be carried out through ACT by comparing the aggregation over anchors (e.g. TSSs) to that from ‘randomized anchors’, where the latter is generated by shifting anchor coordinates along the chromosome or transferring anchor coordinates from a second chromosome to the one of interest.
Finally, ACT can be used as a starting point for other downstream analyses. In the instance of RNA-seq data tracks, further analysis can be conducted with RseqTools (Habegger et al., 2011) to, for example, determine additional similarities between two or more highly correlated tracks. The results of correlation analysis, for instance, can also be fed into downstream principal component analysis, allowing for grouping of coregulating factors with their coregulated sites. This would simply involve diagonalization of the output correlation matrix from ACT. Saturation analysis can also be used to inform future experimental design.
Funding: National Institute of Health; A.L. Williams Professorship funds.
Conflict of Interest: none declared.
REFERENCES
- Ahn S.M., et al. The first Korean genome sequence and analysis: full genome sequencing for a socio-ethnic group. Genome Res. 2009;19:1622–1629. doi: 10.1101/gr.092197.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley D.R., et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drmanac R., et al. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science. 2010;327:78–81. doi: 10.1126/science.1181498. [DOI] [PubMed] [Google Scholar]
- ENCODE Project Consortium. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Inferring phylogenies from protein sequences by parsimony, distance, and likelihood methods. Methods Enzymol. 1996;266:418–427. doi: 10.1016/s0076-6879(96)66026-1. [DOI] [PubMed] [Google Scholar]
- Giardine B., et al. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 2005;15:1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habegger L., et al. RSEQtools: a modular framework to analyze RNA-Seq data using compact, anonymized data summaries. Bioinformatics. 2011;27:281–283. doi: 10.1093/bioinformatics/btq643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.I., et al. A highly annotated whole-genome sequence of a Korean individual. Nature. 2009;460:1011–1015. doi: 10.1038/nature08211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacIssac K.D., et al. A quantitative model of transcriptional regulation reveals the influence of binding location on expression. PLoS Comput. Biol. 2010;6:e1000773. doi: 10.1371/journal.pcbi.1000773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- modENCODE Consortium. Unlocking the secrets of the genome. Nature. 2009;18:927–930. doi: 10.1038/459927a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharry S.T., et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z.D., et al. Statistical analysis of the genomic distribution and correlation of regulatory elements in the ENCODE regions. Genome Res. 2007;17:787–797. doi: 10.1101/gr.5573107. [DOI] [PMC free article] [PubMed] [Google Scholar]