Abstract
The phytochrome B (PHYB) gene of Arabidopsis thaliana was introduced into cotton through Agrobacterium tumefaciens. Integration and expression of PHYB gene in cotton plants were confirmed by molecular evidence. Messenger RNA (mRNA) expression in one of the transgenic lines, QCC11, was much higher than those of control and other transgenic lines. Transgenic cotton plants showed more than a two-fold increase in photosynthetic rate and more than a four-fold increase in transpiration rate and stomatal conductance. The increase in photosynthetic rate led to a 46% increase in relative growth rate and an 18% increase in net assimilation rate. Data recorded up to two generations, both in the greenhouse and in the field, revealed that overexpression of Arabidopsis thaliana PHYB gene in transgenic cotton plants resulted in an increase in the production of cotton by improving the cotton plant growth, with 35% more yield. Moreover, the presence of the Arabidopsis thaliana PHYB gene caused pleiotropic effects like semi-dwarfism, decrease in apical dominance, and increase in boll size.
Keywords: Transformation, Gossypium hirsutum, Phytochrome B, Overexpression, Plant growth, Yield
1. Introduction
Phytochromes were first discovered in the 1950s, when it was found that a brief pulse with red light results in initiation of seed germination in dark, and causes de-etiolation, inhibition of leaf elongation and regulation of flowering, extended growth of hypocotyls and stems, and entrainment of circadian clock (Kendrick and Kronenberg, 1994; Guo et al., 1998; Somers et al., 1998). Phytochrome B (PHYB) protein has different domains that act differently to show their roles, which in turn depend on conditions of light as well as on the specific responses of physiology (Usami et al., 2007). The unique feature of phytochromes among photoreceptors is their capacity to interconvert between a red light absorbing form of phytochrome, i.e., Pr (absorption maximum of 660 nm) and a far-red light absorbing form of phytochrome, i.e., Pfr (absorption maximum of 730 nm). The presence of phytochromes provides plants with a superior system for detecting their positions relative to other plants (Mathews, 2006). Modulation from vegetative growth to reproductive growth, establishment of the seedling, and switching of the circadian clock are controlled by phytochrome. Chen et al. (2004) have determined the molecular characterizations and light-regulated subcellular localizations of photoreceptors in plants. Thiele et al. (1999) determined the semi-dwarfism, decrease in apical dominance, smaller but a higher number of thicker leaves, and increase in pigmentation in PHYB overexpressing potato plants. Schittenhelm et al. (2004) determined enhanced photosynthesis in DARA5 leaves, which increased the yield of potato in high irradiation. PHYA and PHYB transgenic lines also showed overexpression in dark grown seedling. The reduction of elongation, increase of anthocyanin pigments, and high amplification of red light irradiance in PHYB transgenic lines were also reported by Said et al. (2007).
In the present study, an attempt was made to increase the yield of cotton crop by using an alternative approach of transforming the PHYB gene to improve the physiology of this cash crop. A lot of work has been done on transformation of PHYB in potato (Thiele et al., 1999; Boccalendo et al., 2003; Schittenhelm et al., 2004), tomato (Said et al., 2007), and Arabidopsis (Wagner et al., 1991). The present investigation was carried out in cotton to determine the effect of overexpression of Arabidopsis thaliana PHYB gene on cotton growth and yield.
2. Materials and methods
2.1. Plant transformation
The mature embryos (kernels from mature seeds) of cotton variety CIM-482 were transformed with PHYB gene through Agrobacterium LBA-4404 and after two months of kanamycin selection the putative transgenic plants were shifted to loamy soil in pots. The cotton seed surface was sterilized by 1 mg/ml HgCl2 and 1 mg/ml sodium dodecyl sulfate (SDS). Mature embryos were isolated from germinating seeds and a cut was made at the apex of the shoot with a sterilized blade. Afterwards, the embryos were cocultivated for 1 h with an Agrobacterium strain containing PHYB gene. The embryos were than dried on sterilized filter paper and cultured on MS medium (Murashige and Skoog, 1962) for 3 d at 28 °C. After 3 d, embryos were subcultured on MS medium containing kanamycin 50 mg/ml optimized for cotton plants for selection. After two months of selection on kanamycin medium, putative transgenic plants were shifted in shoot and root regeneration media without kanamycin, as determined by Rao et al. (2009). The healthy putative transgenic cotton plants were shifted to pots containing loamy soil. The stable putative transgenic plants were subjected to molecular analysis after 15–20 d following shifting.
2.2. Molecular analyses of transgenic plants
2.2.1. Polymerase chain reaction (PCR)
Genomic DNA isolated from apical leaves of transgenic and control cotton plants (growing in the greenhouse as well as in the field) was analyzed by PCR for detection of PHYB by amplifying internal fragments of PHYB genes with the method of Saiki et al. (1988) with a little modification. The sequence of PHYB forward primer was 5′-TAGGGCTCCTCATGGTTGTC-3′ and the sequence of reverse primer was 5′-TCGCAGTGTGAGATCGAAAC-3′. The PCR was carried out with the two primers to amplify 646 bp fragment. DNA extracted from untransformed plants was used as negative control and that of plasmid pBinPhyB as positive control. The PCR was performed at 94 °C for 3 min, then 35 cycles of 94 °C for 45 s, 52 °C for 45 s, and 72 °C for 1 min, followed by 72 °C for 7 min. The amplified PCR fragments were resolved on 0.01 g/ml agarose gel and observed under ultraviolet (UV) light.
2.2.2. Southern blot analysis
Southern blot analysis was performed as described by Southern (1975) by extracting genomic DNA from apical leaves of putative transgenic plants and untransformed control plants. DNA was extracted as described in Section 2.2.1. Genomic DNA (20 μg) was digested with Kpn1 for PHYB according to the supplier’s instructions (Enzyme Production Lab of the National Centre of Excellence in Molecular Biology (CEMB), Pakistan). The color was detected by 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium (BCIP/NBT) tablets (Sigma B5655) dissolved in water according to the manufacturer’s instruction.
2.2.3. Quantitative real-time reverse transcription-polymerase chain reaction (RT-PCR)
Primers used for real-time PCR were as follows: forward primer, 5′-CTCCTGGCTGAGTTTCTGCT-3′; reverse primer, 5′-GCTTGTCCACCTGCTGCTAT-3′. Real-time PCR reactions were carried out in an iQ5 cycler with a 96-well plate using the IQTM SYBR_Green Super mix (Bio-Rad, USA). As an internal control, the glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used to normalize the data. We used 50 ng of complementary DNA in each PCR reaction. RT-PCR conditions were as follows: initial denaturation at 95 °C for 3 min followed by 40 cycles of denaturation at 94 °C for 30 s, annealing at 60 °C for 30 s, extension at 72 °C for 45 s, and final elongation step at 72 °C for 10 min. By continuous monitoring fluorescence between 60 and 95 °C with 0.5 °C increments every 30 s, a melting curve analysis was carried out. Statistical analysis of the results of real-time PCR was performed using iQ5 software (Bio-Rad) Version 1.0 on the basis of threshold curve (C T) values of the gene in different samples converted to their linear forms, normalized with GAPDH gene. Analysis of variance (ANOVA) was done to analyze significant differences in transcript expression in leaves of control and PHYB transgenic cotton plants.
2.2.4. Enzyme-linked immunosorbant assay (ELISA)
ELISA was used to screen the plants for expression of PHYB. The total proteins of seven plants were isolated from apical leaves, which were named as QCC2, QCC5, QCC7, QCC10, QCC11, QCC14, and QCC15. These protein samples were bound to microtitre plate wells. After treatment with specific PHYB (aR-19) antibody sc-12704 from Santa Cruz Biotechnology Inc., the presence of PHYB protein was detected by color reaction. Substrate used in this case was BCIP/NBT.
2.3. Measurement of physiological parameters
2.3.1. Photosynthetic activity
The uptake of CO2 per leaf area was determined with the help of infrared (IR) spectroscopy having a transportable gas exchange porometer (model LCA-3, Analytical Development Co. (ADC), Hoddesdon, UK), in order to determine the photosynthetic activities of transgenic and control cotton plants. The terminal leaflets of leaves 6 to 8 of 30-d to 37-d old plants were used. Before measurement was made, the leaflet was fixed in the chamber and exposed to 50 to 500 mmol/(m2∙s) white light provided by a 150-lx lamp (Flexilux, Scholly Fiberoptic, Denzlingen, Germany) at 22 to 25 °C until CO2 assimilation reached a maximum steady-state level (10–15 min).
2.3.2. Stomatal conductance
PHYB transgenic and control cotton plants were also analyzed for their stomatal conductance by LCA-3 analyzer, applying the same method as mentioned above, and the data were recorded from the computer attached to the LCA-3 analyzer (ADC, Hoddesdon, UK).
2.3.3. Transpiration rate
Transpiration rates of the PHYB transgenic and control cotton plants were measured by utilizing the LCA-3 IR gas analyzer, and the data were recorded from the computer attached to the LCA-3 analyzer.
2.3.4. Derived growth parameters
Relative growth rate (RGR) is the increase in plant material weight per unit of time. This was calculated using the formula as described by Radford (1967): RGR=(logm2−logm1)/(t2−t1), where m 2 and m 1 are the weights of plants after the second and first harvests, respectively, and t 2 and t 1 are the time of the second and first harvests, respectively. Net assimilation rate (NAR) is the productive efficiency of the leaf calculated in relation to leaf area, and was derived as NAR=2(m2−m1)/[(A1−A2)∙(t2−t1)], where A 1 and A 2 are the leaf areas of plants after the first and second harvests, respectively. Specific leaf area (SLA) is the leaf morphology and is determined as the partitioning of biomass between the different organs, further determined (per plant) by expressing the dry weight attributable to a particular organ as a quotient of total plant dry weight. This is termed the weight ratio for that organ. Thus, SLA is equal to the ratio of leaf area (A)/leaf weight (m).
2.3.5. Yield
Yields of transgenic and control plants were calculated in two generations in the greenhouse as well as in the field. Five lines of PHYB transgenic plants were selected for this experiment along with one control line. Each line contained ten plants. Yield increase (∆Y) was calculated by using the formula: ∆Y=(Yt−Yc)×100%/Yc, where Y t and Y c are the yields of transgenic and control plants, respectively.
2.4. Statistical analysis
The data collected were analyzed using the Fisher’s analysis of variance technique and the experimental plants as well as control plants were compared using the least significant difference test (LSD) at 5% probability (Petersen, 1994). Relative growth rate, net assimilation rate, and relative leaf growth rate were calculated according to the classical growth analysis.
3. Results
3.1. Agrobacterium-mediated transformation of CIM-482 with pBinPhyB construct
In order to incorporate the PHYB gene in CIM-482, Agrobacterium-mediated transformation was used. In all, 8 500 embryos were transformed with PHYB gene and selected on MS medium containing 50 mg/ml kanamycin. Ninety-nine putative transgenic plants out of a total of 8 500 transformed embryos were obtained after eight weeks of selection, and shifted to MS medium. Transformation efficiency was 1.1% (Table 1).
Table 1.
Transformation efficiencies of cotton plants
| No. | ntotal | ncon. |
nplant |
TE (%) | |
| Control | Transgenic | ||||
| 1 | 500 | 50 | 35 | 7 | 1.4 |
| 2 | 1000 | 50 | 40 | 15 | 1.5 |
| 3 | 1000 | 50 | 25 | 7 | 0.7 |
| 4 | 1000 | 50 | 32 | 12 | 1.2 |
| 5 | 1000 | 50 | 29 | 15 | 1.5 |
| 6 | 1000 | 50 | 20 | 11 | 1.1 |
| 7 | 1000 | 50 | 27 | 10 | 1.0 |
| 8 | 1000 | 50 | 30 | 9 | 0.9 |
| 9 | 1000 | 50 | 35 | 13 | 1.3 |
| Total | 8500 | 450 | 273 | 99 | 1.1 |
n total: total number of embryos; n con.: number of control (non-transgenic plants); n plant: number of plants obtained after eight weeks; TE: transformation efficiency
3.2. Molecular analyses of putative transgenic plants
The plants which survived the kanamycin selection and which were successfully transferred to the soil in pots, were considered to be putative transgenic. These plants were further analyzed for the presence and expression of Arabidopsis PHYB gene. The molecular analysis included PCR, Southern blot, and ELISA (Table 2).
Table 2.
Molecular analyses of PHYB transgenic plants
| Plants | PCR | Southern blot | ELISA |
| QCC2 | + | + | + |
| QCC5 | + | + | + |
| QCC7 | + | + | + |
| QCC10 | + | + | + |
| QCC11 | + | + | + |
| QCC14 | + | + | + |
| QCC15 | + | + | + ve control |
| QCC21 | − | − | − ve control |
3.2.1. Detection and functional integration of Arabidopsis PHYB gene in cotton plants
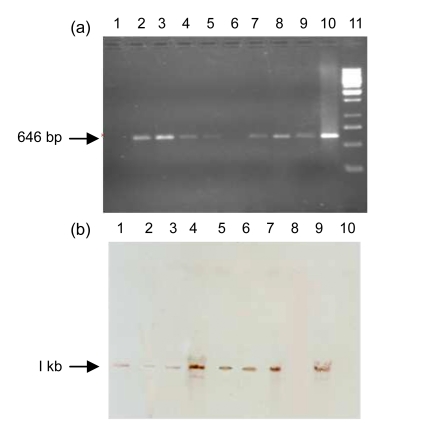
The insertion of the Arabidopsis PHYB gene in cotton plants was detected by PCR. The 646-bp fragment was amplified with internal gene specific primers. Seven of eight plants, namely QCC2, QCC5, QCC7, QCC10, QCC11, QCC14, and QCC15, were detected to be positive putative transgenic cotton plants. No amplification was detected in the negative control and QCC21 plant (Fig. 1a). The stable integration of the Arabidopsis PHYB gene in the cotton plant genome was confirmed by Southern blot analysis. Gene integration was detected with the PHYB specific probe, which highlighted the full cassette of PHYB 3.8 kb after the genomic DNA of the PCR positive plant was digested with Kpn1 restriction enzyme (Fig. 1b). It is clear from Fig. 1b that all the plants which were PCR positive, i.e., QCC2, QCC5, QCC7, QCC10, QCC11, QCC14, and QCC15 show integration.
Fig. 1.
PCR (a) and Southern blot (b) analyses of PHYB transgenic plant DNA
(a) Lanes 3–9: putative transgenic plants QCC2, QCC5, QCC7, QCC10, QCC11, QCC14, and QCC15; Lane 1: negative plant control (QCC21); Lane 10: positive plasmid control; Lane 2: blank; Lane 11: 1 kb Ladder. (b) Lanes 1–7: samples (QCC2, QCC5, QCC7, QCC10, QCC11, QCC14, and QCC15); Lane 8: negative plant control (QCC21); Lane 9: positive plasmid control; Lane 10: λ/HindIII marker
3.2.2. Quantification of PHYB gene RNA in transgenic cotton plants
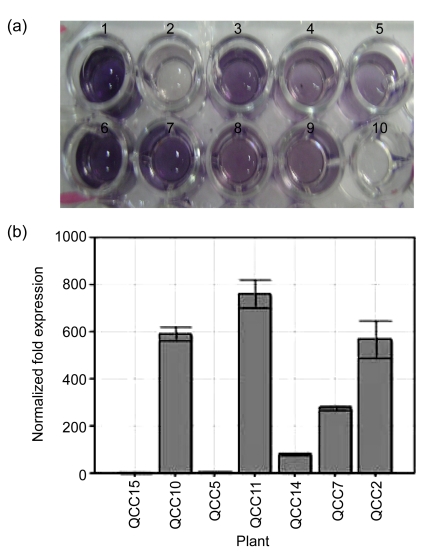
Quantitative real-time RT-PCR was used to check the expression levels of PHYB in leaf samples of seven transgenic lines. GAPDH gene was used as the reference gene to normalize the expression levels. It is obvious from Fig. 2b that all the lines showed different levels of PHYB mRNA expression, but the plant lines QCC2, QCC10, and QCC11 showed much higher levels of overexpression of PHYB as compared to other lines (Fig. 2b) of PHYB plants. The figure also shows the lowest expression of PHYB in lines QCC5 and QCC15.
Fig. 2.
ELISA with anti-goat AtPhyB (a) and quantitative real-time RT-PCR (b) of PHYB plants
Lane 1: +ve control; Lane 2: negative control (QCC21); Lanes 3–9: QCC2, QCC5, QCC7, QCC10, QCC11, QCC14, QCC15; Lane 10: blank
3.2.3. Expression of PHYB gene in cotton plants
The plants which were positive for PCR and Southern blot analyses were further analyzed for the protein expression of the PHYB gene. Total protein was isolated from 10 plants, and subjected to ELISA. Plants QCC2, QCC5, QCC7, QCC10, QCC11, QCC14, and QCC15 gave positive results for the expression of PHYB protein, as detected by ELISA, while no expression was observed in QCC21 negative control plants (Fig. 2a).
3.3. Physiological analysis
3.3.1. Photosynthetic activity
Photosynthetic activity performed by mature control and transgenic leaves is shown in Table 3. The 32–37 d old leaves from non-senescent plants were used. The leaf area photosynthetic rate was higher at photon fluxes of more than 250 mmol/(m2∙s) in the transgenic plants than in the control plants. The increase in the photosynthetic rate per individual plant from 13.10 μmol/(m2∙s) to 42.43 μmol/(m2∙s) was observed.
Table 3.
Field performances of PHYB transgenic plants
| Plant | PR (µmol/(m2∙s)) | TR (mmol/(m2∙s)) | SC (mol/(m2∙s)) |
| QCC2 | 40.00a | 10.53a | 102.0a |
| QCC7 | 39.01a | 8.19a | 94.0a |
| QCC10 | 33.21b | 6.91a | 69.0a |
| QCC11 | 42.43a | 12.41a | 103.3a |
| QCC14 | 34.13a | 8.20a | 71.0a |
| Control | 13.10b | 3.38b | 20.0b |
PR: photosynthetic rate; TR: transpiration rate; SC: stomatal conductance
3.3.2. Stomatal conductance
Results also indicated that stomatal conductance of QCC11, i.e., 103.3 mol/(m2∙s), is four-fold greater than control, the latter of which has the stomatal conductance of 20.0 mol/(m2∙s). Similarly other PHYB transgenic plants, such as QCC2, QCC10, and QCC7, had stomatal conductances of 102, 69, and 94 mol/(m2∙s), respectively (Table 3).
3.3.3. Transpiration rate
The results of plants analyzed by IR gas analyzer showed that the transpiration rate of PHYB transgenic plant was greater than that of control. The plant QCC2 had 10.53 mmol/(m2∙s). Similarly, other plants like QCC11 had transpiration rate 12.41 mmol/(m2∙s), greater than control; QCC14 and QCC7 showed similar results, except QCC10 whose transpiration rate was (6.91 mmol/(m2∙s)) a little greater than control (3.38 mmol/(m2∙s)) (Table 3).
3.3.4. Growth analysis
Critical examination of data shown in Table 4 shows that PHYB has a significant effect on relative growth rate. Maximum relative growth rate was recorded with PHYB transgenic plants, showing the significant effect of PHYB on relative growth rate. Relative growth rate is strongly and positively correlated with net assimilation rate. The data collected for PHYB transgenic plants showed a significant difference from control plants (Table 4), while data obtained for specific leaf area showed that there was no significant effect of phytochrome on the specific leaf areas of transgenic plants (Table 4).
Table 4.
Growth comparison of PHYB transgenic cotton plants with control cotton plants in the field and greenhouse
| Plant | Relative growth rate |
Net assimilation rate |
Specific leaf area |
||||||
| 10 d | 20 d | 30 d | 10 d | 20 d | 30 d | 10 d | 20 d | 30 d | |
| PHYB transgenic cotton | 0.0053a | 0.0068a | 0.0121a | 0.0046a | 0.0055a | 0.0099a | 6.22a | 5.57a | 5.53a |
| Control cotton | 0.0037b | 0.0043b | 0.0046b | 0.0034b | 0.0037b | 0.0041b | 5.33a | 5.82a | 5.05a |
3.3.5. Yield
Data shown in Table 5 revealed that with increasing photosynthetic rate the plant yield was enhanced. In QCC2, photosynthetic rate was recorded to be 40.00 μmol/(m2∙s) and a 38.79% increase in yield was found, as compared to control (Fig. 3b and Table 5). Similarly, in QCC11, photosynthetic rate was measured to be 42.43 μmol/(m2∙s) and 38.96% increased in yield was recorded as compared to control. In the case of QCC14, with a photosynthetic rate 34.13 μmol/(m2∙s), only a 26.64% increase in yield was found over control. On average, a 34.09% increase in yields of PHYB transgenic plants was recorded during this study as compared to control (Table 5).
Table 5.
Comparisons of physiological and growth performances of PHYB plants with control under the field and greenhouse conditions
| Plant | Field |
Greenhouse |
||||||
| Mean radius (mm) | Mean cotton weight (g/plant) | Mean height (cm) | Yield increase (%) | Mean radius (mm) | Mean cotton weight (g/plant) | Mean height (cm) | Yield increase (%) | |
| QCC2 | 1.69a | 83.30a | 90a | 38.79 | 1.48a | 81.03a | 81a | 39.61 |
| QCC7 | 1.79a | 80.45a | 110a | 33.99 | 1.75a | 78.35a | 95a | 34.99 |
| QCC10 | 1.89a | 79.30a | 100a | 32.07 | 1.76a | 77.41a | 94a | 33.37 |
| QCC11 | 1.89a | 83.43a | 96a | 38.96 | 1.78a | 82.32a | 88a | 41.83 |
| QCC14 | 1.44b | 76.40a | 113a | 26.64 | 1.15a | 72.01a | 87a | 22.39 |
| Control | 1.50b | 60.04b | 140b | 0.99b | 58.04b | 113b | ||
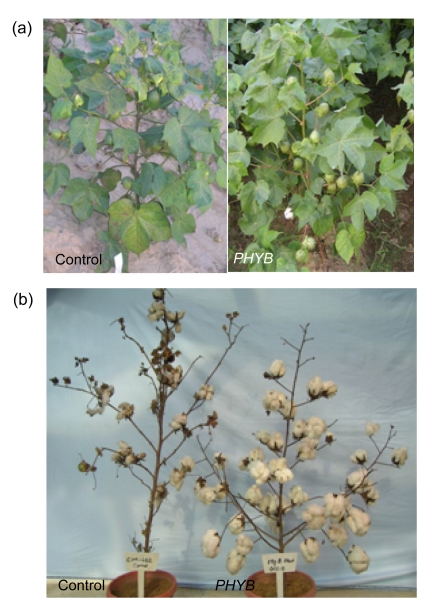
Fig. 3.
Comparisons of morphologies and yields of PHYB transgenic plants with the control
(a) PHYB plants showed a greater number of shorter but thicker leaves than the control; (b) Yield of PHYB increased compared to the control
4. Discussion
4.1. Comparison of mRNA overexpression in different PHYB transgenic lines
Transgenic approaches to photosynthetic enhancement and yield increase might be more successful than conventional breeding methods (Dunwell, 2000). Being focused on the aim to utilize the transgenic approach, we produced cotton plants overexpressing PHYB. Molecular analysis of the data indicated that seven out of total 99 putative transgenic cotton lines were PCR and Southern blot positive. Similarly these lines were found to be expressing PHYB protein as shown by ELISA.
The results of quantitative real-time RT-PCR have determined that QCC11 exhibit higher levels of mRNA expression as compared to other plants where mRNA expression is quite low. The results clearly show that the expression level is variable among different lines of PHYB. Expression in QCC2, QCC10, and QCC11 was much higher as compared to other lines (Fig. 2b). These results are similar to Dong and Li (2007), who determined the variation in expression level of transgenic plants, which might be due to nucleotide sequence of the gene, promotor, insertion point of the gene in the DNA of the transgenic variety, transgene copy number, and internal cell environment, as well as several external factors in the environment (Hobbs et al., 1993; Guo et al., 2001; Rao, 2005; Warren, 2007).
4.2. Effects of PHYB overexpression on phenotype
Arabidopsis PHYB DNA, when transformed in cotton lines, led to overexpression of gene under the control of CaMV 35S promoter, supporting the assumption that the plants having overexpression of the phytochrome may be used as agriculturally important plants in the future. A comparison has been made between the already reported data (Husaineid et al., 2007) of different lines that have overexpression of PHYA or PHYB. Overexpressing Arabidopsis PHYB transgenic cotton lines showed agriculturally important characteristics like increase in time span and photosynthetic rate, exhibiting more apical dominance, and increase in thickness, but decrease in size of leaves. This small phenotype of cotton plant in the field is the same as the phytochrome expression in potato (Thiele et al., 1999).
The morphological data indicated that morphology of the plant is directly related to its physiology with increasing photosynthetic rate. This supports the finding of overexpressing PHYA in transgenic tobacco plants studied by Robson et al. (1996). The PHYB lines displayed higher photosynthesis rates (Table 3). This effect may be proportional to the increased chlorophyll levels in transgenic potato leaves as compared to control potato leaves (Thiele et al., 1999). Recognizing the phytochrome-mediated responses of plants to their light environments is an important goal in providing an overall understanding of light-regulated growth and development (Quail, 2007). The transgenic plants showed the higher response to white light due to increase in leaf thickness which results in increase in photosynthetic rate and biomass production and then leads to more supply of assimilates to the leaves by decreases in the stem.
4.3. Effect of PHYB overexpression on physiology and growth parameters
The data reviewed so far indicate the increase in transpiration rate, stomatal conductance, and photosynthetic rate. Improved stomatal conductance was observed because of an increase in transpiration rate. This is because there is a close correlation between stomatal aperture and photosynthetic rate (Jiang et al., 1988; Hirasawa et al., 1992). Importantly 90% of the water loss takes place through stomata which cover only 1% area of the leaf. There is a direct relationship between stoma opening and transpiration rate: if stomata are open more, there will be more transpiration, and vice versa. Transpiration plays a very critical role in plant life. It is not only responsible for water loss, but also has a key role in transportation of sap (sugar, minerals, and plant chemicals), protecting the plant from burning and maintaining the turgidity of the plant.
Relative growth rate and net assimilation rate of PHYB lines were much higher than those of control. Relative growth rate is strongly and positively correlated with net assimilation rate (Tan and Tan, 1981; Shipley, 2002). Thus, there were ultimately a higher photosynthetic rate, growth rate, and shade avoidance response in transgenic lines. PHYB lines have shown 35% more yield as compared to controls. This is the same as the observation by Thiele et al. (1999), in which PHYB potato had more yields as compared to control.
5. Conclusions
The results support that higher photosynthetic rate lead to more relative growth rate and greater yield in cotton plants. It is clearly shown that transgenic plants are potentially more productive. As these plants can take in more CO2 due to a decrease in velocity of chlorophyll breakdown, they might be of considerable worth in those regions which have long growing seasons.
Acknowledgments
We are grateful to Director Central Cotton Research Institute for providing us seeds of cotton variety CIM-482, and to Dr. M. ASHRAF, Chairman of the Botany Department, University of Agriculture Faisalabad, Pakistan for equipment use.
References
- 1.Boccalendo HE, Polschuk EL, Yanovsky MJ, Sanchez RA, Gatz C, Casal JJ. Increased phytochrome B alleviates density effect on tuber yield of field potato crops. Plant Physiol. 2003;133(4):1539–1546. doi: 10.1104/pp.103.029579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen M, Chory J, Fankhauser C. Light signal transduction in higher plants. Ann Rev Genetics. 2004;38(1):87–117. doi: 10.1146/annurev.genet.38.072902.092259. [DOI] [PubMed] [Google Scholar]
- 3.Dong HZ, Li WJ. Variability of endotoxin expression in Bt transgenic cotton. J Agron Crop Sci. 2007;193(1):21–29. doi: 10.1111/j.1439-037X.2006.00240.x. [DOI] [Google Scholar]
- 4.Dunwell JM. Transgenic approaches to crop improvement. J Exp Biol. 2000;51(Suppl. 1):487–496. doi: 10.1093/jexbot/51.suppl_1.487. [DOI] [PubMed] [Google Scholar]
- 5.Guo H, Yang H, Mockler TC, Lin C. Regulation of flowering time by Arabidopsis photoreceptors. Science. 1998;279(5355):1360–1363. doi: 10.1126/science.279.5355.1360. [DOI] [PubMed] [Google Scholar]
- 6.Guo WZ, Sun J, Guo YF, Zhang TZ. Investigation of different dosages of inserted Bt genes and their insect-resistance in transgenic Bt cotton. Acta Genet Sin. 2001;28(7):668–676. (in Chinese) [PubMed] [Google Scholar]
- 7.Hirasawa T, Tsuchida M, Ishihara K. Relationship between resistance to water transport and exudation rate and the effect of the resistance on the midday depression of stomatal aperture in rice plants. Jpn J Crop Sci. 1992;61:145–152. [Google Scholar]
- 8.Hobbs SLA, Warkentin TD, Delong CMO. Transgene copy number can be positively or negatively associated with transgene expression. Plant Mol Biol. 1993;21(1):17–26. doi: 10.1007/BF00039614. [DOI] [PubMed] [Google Scholar]
- 9.Husaineid SS, Kok RA, Schreuder ME, Hanumappa M, Cordonnier-Pratt MM, Pratt LH, van der Plas LH, van der Krol AR. Overexpression of homologus phytochrome genes in tomato exploring the limits in photoreception. J Exp Bot. 2007;58(3):615–626. doi: 10.1093/jxb/erl253. [DOI] [PubMed] [Google Scholar]
- 10.Jiang CZ, Hirasawa T, Ishihara K. Physiological and ecological characteristics of high yielding varieties in rice plants. II. Leaf photosynthetic rates. J Crop Sci. 1988;57:139–145. [Google Scholar]
- 11.Kendrick RE, Kronenberg GHM. Photomorphogenesis in Plants. Dordrecht: Kluwer Academic Publishers; 1994. p. 828. [Google Scholar]
- 12.Mathews S. Phytochrome-mediated development in land plants: red light sensing evolves to meet the challenges of changing light environments. Mol Ecol. 2006;15(12):3483–3503. doi: 10.1111/j.1365-294X.2006.03051.x. [DOI] [PubMed] [Google Scholar]
- 13.Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultivars. Plant Physiol. 1962;15:473–497. [Google Scholar]
- 14.Petersen RG. Agricultural Field Experiments: Design and Analysis. NY, USA: Marcel Dekker Inc.; 1994. pp. 205–260. [Google Scholar]
- 15.Quail PH. Phytochrome-regulated gene expression. J Integr Plant Biol. 2007;49(1):11–20. doi: 10.1111/j.1744-7909.2006.00422.x. [DOI] [Google Scholar]
- 16.Radford PJ. Growth analysis formulae-their use and abuse. Crop Sci. 1967;7(3):171–175. doi: 10.2135/cropsci1967.0011183X000700030001x. [DOI] [Google Scholar]
- 17.Rao AQ, Bakhsh A, Kiani S, Shahzad K, Shahid AA, Husnain T, Riazuddin S. The myth of plant transformation. Biotechnol Adv. 2009;27(6):753–763. doi: 10.1016/j.biotechadv.2009.04.028. [DOI] [PubMed] [Google Scholar]
- 18.Rao CK. Transgenic Bt Technology: 3. Expression of Transgenes. 2005. Available from http://www.monsanto.co.uk/news/ukshowlib.phtml?uid¼9304.
- 19.Robson PRH, McCormac AC, Irvine AS, Smith H. Genetic engineering of harvest index in tobacco through overexpression of a phytochrome gene. Nat Biotechnol. 1996;14(8):995–998. doi: 10.1038/nbt0896-995. [DOI] [PubMed] [Google Scholar]
- 20.Said SH, Rosan AK, Marielle ELS, Mamatha H, Marie-Michele CP, Pratt LH, Linus HWVDP, Alexander RVDK. Overexpression of homologous phytochrome genes in tomato: exploring the limits in photoperception. J Exp Bot. 2007;58(3):615–626. doi: 10.1093/jxb/erl253. [DOI] [PubMed] [Google Scholar]
- 21.Saiki R, Chang CA, Levenson CH, Warren TC, Boehm CD, Kazazian HHJ, Erlich HA. Diagnosis of sickle cell anemia and β-thalassemia with enzymatically amplified DNA and nonradioactive allele-specific oligonucleotide probes. N Engl J Med. 1988;319(9):537–541. doi: 10.1056/NEJM198809013190903. [DOI] [PubMed] [Google Scholar]
- 22.Schittenhelm S, Hartmann MU, Oldenburg E. Photosynthesis, carbohydrate metabolism, and yield of phytochrome-B-overexpressing potatoes under different light regimes. Crop Sci. 2004;44(1):131–143. doi: 10.2135/cropsci2004.0131. [DOI] [Google Scholar]
- 23.Shipley B. Trade-offs between net assimilation rate and specific leaf area in determining relative growth rate: relationship with daily irradiance. Funct Ecol. 2002;16(5):682–689. doi: 10.1046/j.1365-2435.2002.00672.x. [DOI] [Google Scholar]
- 24.Somers DE, Devlin PF, Kay SA. Phytochromes and cryptochromes in the entrainment of the Arabidopsis Circadian clock. Science. 1998;282(5393):1488–1490. doi: 10.1126/science.282.5393.1488. [DOI] [PubMed] [Google Scholar]
- 25.Southern EM. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98(3):503–517. doi: 10.1016/S0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 26.Tan GY, Tan WK. Net assimilation rate and relative nitrogen assimilation rate in relation to the dry matter production of alfalfa cultivars. Plant Soil. 1981;59(2):185–192. doi: 10.1007/BF02184191. [DOI] [Google Scholar]
- 27.Thiele A, Herold M, Lenk I, Quail PH, Gatz C. Heterologous expression of Arabidopsis phytochrome B in transgenic potato influences photosynthetic performance and tuber. Plant Physiol. 1999;120(1):73–81. doi: 10.1104/pp.120.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Usami T, Matsushita T, Oka Y, Mochizuki N, Nagatani A. Roles for the N- and C-terminal domains of phytochrome B in interactions between phytochrome B and cryptochrome signaling cascades. Plant Cell Physiol. 2007;48(3):424–433. doi: 10.1093/pcp/pcm012. [DOI] [PubMed] [Google Scholar]
- 29.Wagner D, Tepperman JM, Quail PH. Overexpression of phytochrome B induces a short hypocotyls phenotype in transgenic Arabidopsis . Cell. 1991;3(12):1275–1288. doi: 10.1105/tpc.3.12.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Warren AL. Single Cell Gene Expression Analysis by RT PCR. California, USA: California Institute of Technology Pasadena; 2007. pp. 133–135. PhD Thesis. [Google Scholar]