Abstract
MicroRNAs (miRNAs) are a class of small RNAs that regulate gene expression. Expression profiles of specific miRNAs have improved cancer diagnosis and classification and even provided prognostic information in many human cancers, including lung cancer. Tumor suppressive and oncogenic miRNAs were uncovered in lung carcinogenesis. The biological functions of these miRNAs in lung cancer were recently validated in well characterized cellular, murine transgenic as well as transplantable lung cancer models and in human paired normal-malignant lung tissue banks and tissue arrays. Tumor suppressive and oncogenic miRNAs that were identified in lung cancer will be reviewed here. Emphasis is placed on highlighting those functionally validated miRNAs that are not only biomarkers of lung carcinogenesis, but also candidate pharmacologic targets. How these miRNA findings advance an understanding of lung cancer biology and could improve lung cancer therapy are discussed in this article.
Introduction
MicroRNAs (miRNAs) were discovered as a class of small RNAs that can regulate gene expression.1–3 Long primary miRNA transcripts (pri-miRNAs) are transcribed by RNA polymerase II and are often several kilobases (Kb) in size.4 Excision of a pri-miRNA by the microprocessor, a complex of the RNase III enzyme Drosha and DGCR8, generates in the nucleus a ~70 nucleotide long stem loop miRNA precursor hairpin (pre-miRNA), which is transported into the cytoplasm by nuclear export factor exportin 5.4–7 Mature miRNAs are generated from processing of pre-miRNAs by Dicer, another RNase III enzyme.4 Mature miRNAs can stably associate with Argonaute-containing RNA-induced silencing complex (RISC), while the opposite strand of the pre-miRNA duplex, miRNA*, is typically degraded.4, 8
An important functional role of miRNAs is to regulate target mRNA expression at the post-transcriptional level.1 In combination with RISC, miRNAs bind to partially complementary sequences in the 3′-untranslated region (UTR) of target mRNAs. The seed region (2–8 nucleotides) of miRNAs is critical for target sequence recognition and binding.2, 3 This miRNA-mRNA binding causes mRNA destabilization and degradation9, 10 as well as translational repression.11 Each miRNA is able to bind multiple mRNA sequences.10 Given this, there can be diverse effects on gene expression by changes in expression of a single miRNA.12, 13 An miRNA can bind different mRNAs that share similar target sequences. Yet, consequences of miRNA-mediated changes in gene expression depend on the cell and tissue context as well as the precise physiological and pathophysiological states under study. For example, miR-31 functions as an oncogenic miRNA in lung cancer as well as in head and neck cancer, but also plays a key role in regulating breast cancer metastasis.14– 17 Different effects are engaged through distinct sets of regulated target genes in the respective affected tissues. How miRNAs exert tumor suppressive or oncogenic effects in lung carcinogenesis is the subject of this review.
MiRNAs and Lung Cancer
Lung cancer is the leading cause of cancer mortality for men and women in the United States.18 There are two major types of lung cancer: small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC).18 Since NSCLC is much more prevalent than SCLC, it is the focus of this review. Despite advances made in surgery, radiation therapy and chemotherapy, the five year survival for lung cancer remains only about 16%.18 This underscores the need for an improved understanding of lung cancer biology and for the identification of novel molecular pharmacologic targets to reduce the clinical consequences of lung carcinogenesis.
MiRNAs are important in many cellular processes including proliferation, differentiation and apoptosis by regulating mRNA target gene expression.19 Some components of the miRNA machinery are affected by the carcinogenesis process. For example, down-regulation of Dicer is reported in lung cancer and this predicts a poor clinical outcome.20 Knock-down of Dicer1, the key protein that processes pre-miRNA, can impair global miRNA processing and enhance tumorigenicity in a K-ras-driven murine model of lung cancer.21 Argonaute, a key component of the RISC complex, directs small interfering RNA (siRNA) as well as miRNA-mediated gene regulation and is linked to the biology of Wilms’ tumor and to neuroectodermal tumors.22
The functions of only a few miRNAs have been comprehensively elucidated. Yet, miRNA profiles have already proven useful to improve lung cancer diagnosis and classification as well as to provide prognostic information. Detection of early changes in lung carcinogenesis, including key changes in miRNA expression, offers the prospect to intervene to improve clinical outcomes when the potential for substantial clinical benefits exist. In this regard, it is intriguing that altered miRNA expression is detected in sputum. This can uncover early stages of squamous cell carcinoma23, adenocarcinoma24 and other types of NSCLC.25
Specific miRNA expression profiles can improve lung cancer classification and identify new pharmacological targets. Prior miRNA signatures have been linked to prognosis of clinical subsets of lung cancers.26 This can help classify this malignancy by precisely defining the miRNA expression profiles that are characteristic of different histopathologic types of lung cancer.27
Expression profiles of miRNAs can provide prognostic information in lung cancer. Specific miRNA expression patterns can assess survival outcomes for lung cancer patients. For example, a four-miRNA (miR-486, miR-30d, miR-1 and miR-499) serum signature in NSCLC can predict overall survival. 28 Increased miR-155 and reduced let-7a-2 expression defines an unfavorable survival in pulmonary adenocarcinomas.29 Lung cancer patients whose tumors had a five-miRNA signature (low miR-221 and let-7a, high miR-137, miR-372, and miR-182*) exhibit unfavorable overall and disease-free survivals.26 Recurrence of stage I NSCLC after surgical resection was predicted by miRNA expression profiles.30
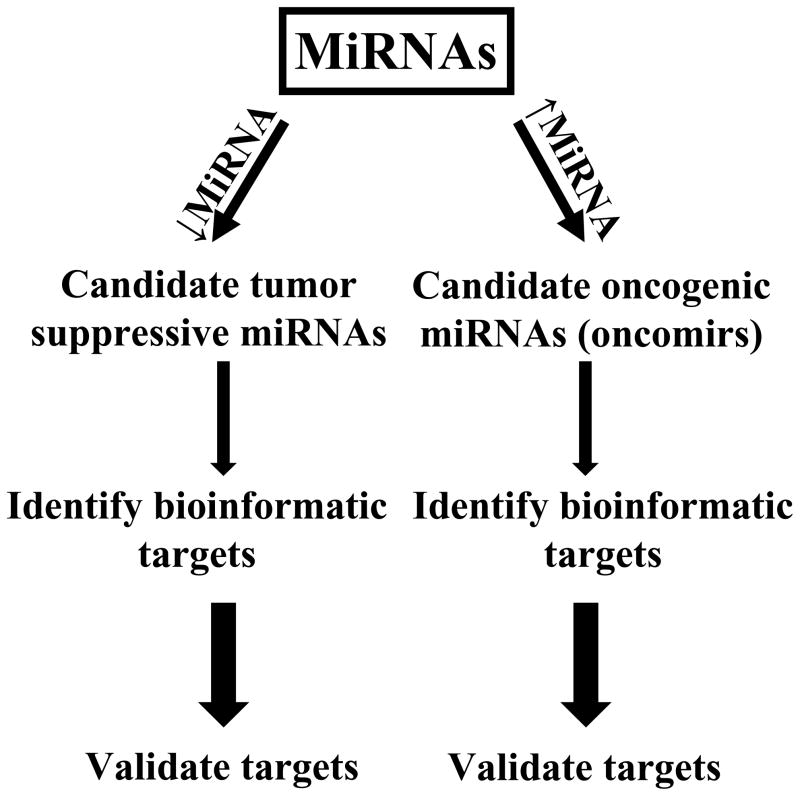
There is a need to identify lung cancer-associated miRNAs, which are biomarkers of treatment response or pharmacological targets. Given this, searches were undertaken to find those miRNAs that would exert tumor suppressive or oncogenic (oncomir) effects in the lung. Candidate tumor suppressive miRNAs are those that exhibit reduced expression in malignant versus adjacent normal lung tissues, as shown in Figure 1. Potential oncomirs are defined by overexpression in the malignant versus adjacent normal lung tissues, as summarized in Figure 1. Consequences of this include changes in expression of critical target genes that could confer tumor suppressive or oncogenic effects of specific miRNAs. Changes in these direct target genes can be used to improve the diagnosis or classification of lung cancers. Some of these species are also therapeutic or chemopreventive targets in the lung, as in the example of miR-31.14
Figure 1.
Identification and functional characterization of oncogenic and tumor suppressive miRNAs in lung cancer. Candidate tumor suppressive miRNAs are repressed (↓miRNA) while candidate oncogenic (oncomir) miRNAs are increased (↑miRNA) in their expression within lung cancers (relative to adjacent normal lung tissues). Bioinformatic analysis is used to prioritize potential target genes before functional validation, as described in the text. The increased arrow sizes are meant to convey an increased stringency of selection for the highlighted miRNAs.
Oncomirs in Lung Cancer
Oncomirs have higher basal expression in malignant as compared to adjacent normal lung tissues. Some are potential prognostic biomarkers as in the case of miR-92a-2* in SCLC31 or the examples of miR-155, miR-130a, let-7f and miR-30e-3p in NSCLC.29, 32, 33 To date, few candidate oncomirs have had mechanistic validation or identification of the target genes that would exert their oncogenic effects.
One of these oncomirs is miR-21 that plays a key functional role in several cancers34–37 including lung cancer.38 Prior work revealed that miR-21 promotes cellular growth and augments tumor invasion and metastasis39 by reducing expression of specific sets of target genes that exert tumor suppressive effects. For example, in K-ras-dependent mouse lung cancers there is an increase in miR-21 expression; miR-21 targets multiple negative regulators of the Ras/MEK/ERK pathway to promote proliferation by regulating expression of Spry1, Spry2, Btg2 and Pdcd4.39–41 Also, miR-21 inhibits apoptosis by reducing expression of pro-apoptotic gene products that include Apaf1, Faslg, Pdcd4 and RhoB.39, 40 Reduction of Pdcd4 expression can increase invasion and metastasis.41
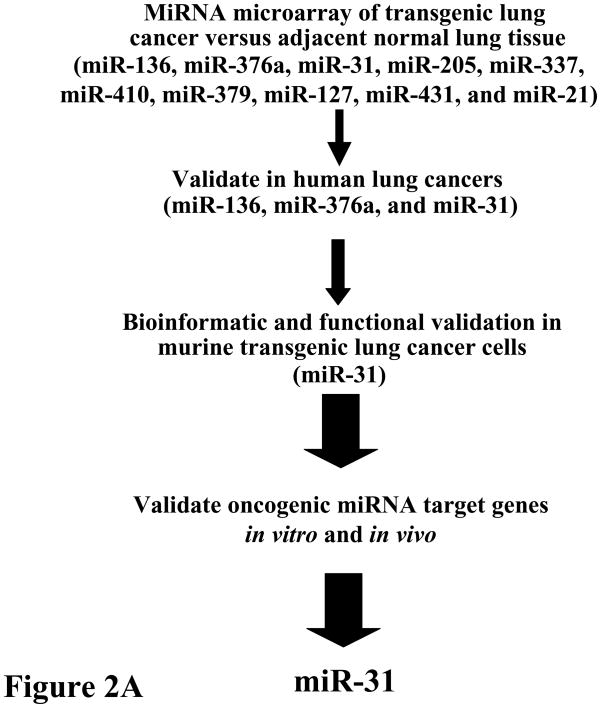
Our team reported that miR-31 acts as an oncomir in murine and human lung cancers.14 The strategy that was taken to uncover oncomirs and their target genes in lung cancers appears in Figure 2A. That miR-31 and other miRNAs were overexpressed in transgenic cyclin E-driven murine lung cancers implied that a similar miRNA expression profile would occur in human lung cancers.42 This was the case when human lung cancers (versus adjacent normal lung tissues) were found to have a similar expression pattern for several of the miRNAs that were highlighted in previously described transgenic lung cancers.14, 42 Functional validation was sought for the candidate oncomirs that were over-expressed in the majority of examined murine and human lung cancers.
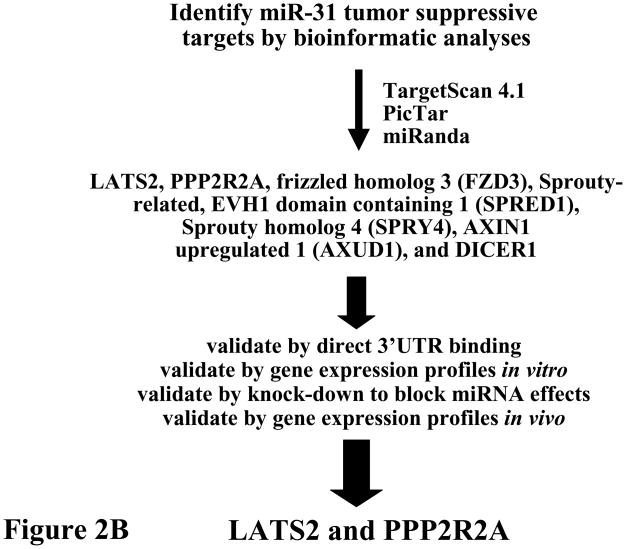
Figure 2.
Comprehensive analysis of known miRNAs was performed to uncover those that were increased in lung cancers relative to adjacent normal lung.14 (A) Initial analysis was performed in transgenic cyclin E-driven mouse models that recapitulated frequent features of human lung cancer such as presence of chromosome instability, hedgehog pathway activation, single or multiple pre-malignant and malignant (adenocarcinoma) lung lesions and even the onset of metastases. Results were confirmed and extended for a subset of highlighted miRNAs (see figure) within a panel of paired normal-malignant human lung tissues. Gain and loss of function studies were performed in murine and human lung cancer cells to explore the anti-neoplastic effects of each highlighted miRNA. This led to finding miR-31 as a key oncomir in the lung. This conclusion was made based on engineered loss of miR-31 that caused a significant reduction in lung cancer formation after transplantation of these cells into syngeneic mice, as discussed in the text.14 (B) Bioinformatic analysis was performed using the indicated programs shown in this panel. This uncovered the tumor suppressors (LATS2 and PPP2R2A) as regulators of the oncogenic effects of miR-31. The increase in arrow size conveys the increased stringency of selection for the highlighted miRNAs.
Of a panel of over-expressed miRNAs in murine lung cancers, only a small subset was also augmented in human lung cancers.14 Of those shown in Figure 2A, basal miR-31 expression level was enhanced in a large number of lung cancer cell lines studied and engineered knock-down of miR-31 significantly reduced both cellular growth and colony formation.14 Among potential target genes uncovered by bioinformatic analysis, several tumor suppressive species were identified as possible direct miR-31 targets.14 Of these, two were functionally validated as direct miR-31 targets: large tumor suppressor 2 (LATS2) and PP2A regulatory subunit B alpha isoform (PPP2R2A); engineered knockdown of LATS2 or PPP2R2A reversed the growth inhibitory effect by miR-31 knock-down.14 This revealed a direct link between miRNA-31 and that of LATS2 and PPP2R2A expression, as shown in Figure 2B. Prior work of others highlighted miR-31 as a candidate oncomir in head and neck cancer.15 Intriguingly, regulating miR-31 expression also affects breast cancer metastasis by regulating expression of other target genes.16, 17 Taken together, these findings underscore that miRNAs exert their functions in cell, tissue and disease specific contexts. Pharmacologic knock-down of a critical oncogenic miRNA, such as miR-31 in lung cancer, might exert anti-neoplastic effects.14
Tumor Suppressive MiRNAs in Lung Cancer
In contrast to oncomirs, expression profiles of candidate tumor suppressive miRNAs are repressed in cancers versus adjacent normal lung tissues. For example, reduced expression of the miRNA let-7 was reported to occur in bronchioloalveolar carcinoma43 and lung adenocarcinoma.29, 44 The miR-146b expression patterns in squamous cell lung cancer were found to predict a poor clinical outcome and miR-34a expression was found as a biomarker for clinical relapse in surgically resected NSCLC.45,46 Also, miR-34a was inactivated by CpG methylation and this caused transcriptional silencing in lung cancers.47 The precise mechanisms through which these miRNAs exert their tumor suppressive effects remain to be determined.
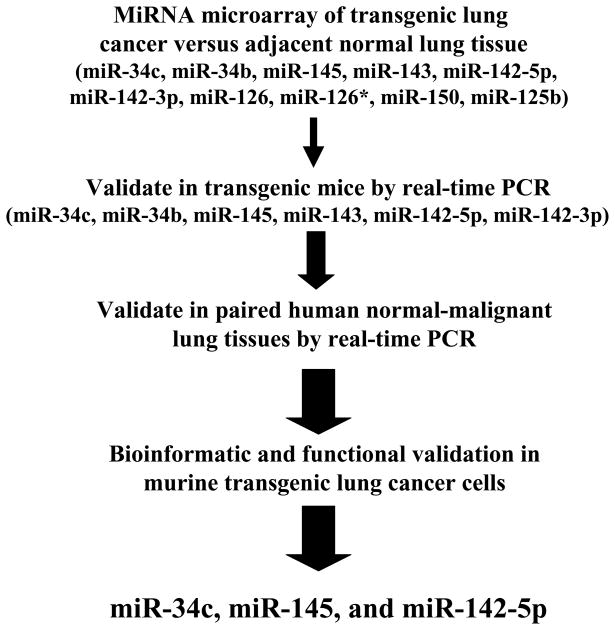
The miR-34 family is reported as a p53-induced tumor suppressive miRNA family in diverse types of cancers.48–53 As a transcription factor, p53 directly induces miR-34 family transcription. Ectopic miR-34 expression can augment apoptosis, cell-cycle arrest or senescence. The promoter regions of the miR-34 family are often inactivated by CpG methylation.47 Repression of the miR-34 family was linked to resistance to p53 activating agents that can cause apoptotic response to specific chemotherapy treatments.54 Direct miR-34 target genes include: in CKD4/6,55–57 c-Myc,58, 59 CREB,60 SIRT1,61 cyclin E 62, and Bcl-2.53 Our laboratory found that miR-34c expression was repressed in both murine and human lung cancers and that ectopic expression of miR-34c significantly reduced lung cancer cell growth by specifically targeting cyclin E for repression.62 Other highlighted candidate tumor suppressive miRNAs in lung cancer appear in Figure 3. The expression patterns of oncogenic and tumor suppressive miRNAs in lung cancers were confirmed by use of in situ hybridization (ISH) assays. Representative results are shown in Figure 4.
Figure 3.
Comprehensive analysis of known miRNAs was performed to uncover those that were repressed in murine lung cancers relative to adjacent normal lung tissues.62 Results were confirmed and extended for a subset of highlighted miRNAs in a panel of paired normal-malignant human lung tissues. Gain and loss of function studies were independently performed in murine and human lung cancer cells to explore the anti-neoplastic effects of each highlighted miRNA. Of these species, three prominent growth suppressive miRNAs were found and were each functionally validated. These were: miR-34c, miR-145, and miR-142-5p. The increase in arrow size conveys the increased stringency of selection for the highlighted miRNAs.
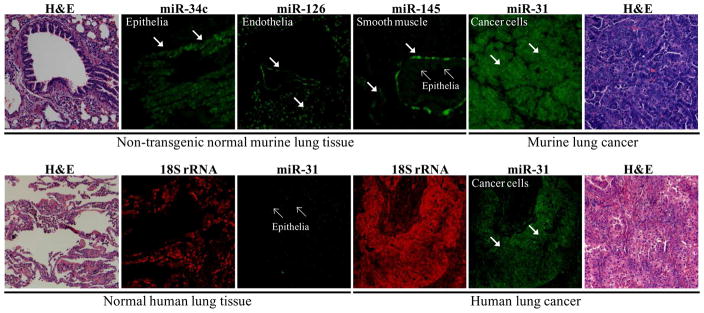
Figure 4.
Spatial characterization of miRNA expression profiles in murine and human lung tissues. Representative in situ hybridization (ISH) assay results are displayed using locked nucleic acid (LNA)-modified probes against candidate tumor suppressive or oncogenic miRNAs. These assays were performed using formalin-fixed paraffin-embedded normal or malignant lung tissues, as previously described.14,62 The upper panel shows the miRNA expression patterns of the indicated miRNAs in age-matched non-transgenic sibling controls (normal lung tissue) and in a cyclin E-driven mouse model of lung cancer, as previously described.42 ISH analyses indicated the predominant bronchial epithelial cell expression pattern for miR-34c (arrows) as well as for expression of miR-145 in epithelial cells (small arrows), though miR-145 was also expressed in the smooth muscle cells of the lamina muscularis of the mucosa and vasculature (arrows). In contrast, miR-126 expression was expressed in endothelial cells (arrows) and by this profile was viewed as having a lower priority for miRNA functional assessment than either miR-34c or miR-145. The lower panel displays the ISH expression patterns for miR-31 and for the 18S ribosomal RNA (as control for RNA integrity) in paired human normal lung tissue versus adjacent lung cancer (adenocarcinoma). This ISH analysis confirmed augmented miR-31 expression (relative to adjacent normal lung) within both murine and human lung cancers. Representative hematoxylin and eosin (H & E) stained lung tissues are displayed.
The let-7 miRNA family is located in genomic regions that are fragile sites or frequently deleted in specific cancers.63 Let-7 expression is often repressed in certain types of lung cancers.29, 43, 44 Engineered let-7 over-expression can inhibit cancer cell proliferation.64, 65 Several let-7 target genes are reported, including: K-ras,65 HMGA2,64, 66 and c-Myc21, 67 as well as the cell cycle regulators CDC25A, CDK6, and cyclin D2.68 Engineered over-expression of let-7 was achieved in the K-ras-driven murine lung cancer model via adenoviral delivery and this reduced lung cancer formation in vivo.21 Forced over-expression of tumor suppressive miRNAs can exert anti-neoplastic effects in the lung.62 Pharmacologic strategies exist that can regulate miRNA expression and these are next discussed.
MiRNAs as Lung Cancer Targets
Specific miRNAs play critical roles in regulating tumorigenicity. It is therefore appealing to consider pharmacologic strategies to target specific miRNAs in lung carcinogenesis. These approaches can use miRNA derivatives as anti-cancer agents. Inhibition of specific oncomirs is achieved by use of optimized antisense derivatives. As an example, locked nucleic acid (LNA) miRNA derivatives represent a class of chemically-modified nucleic acids that inhibit specific miRNAs in mouse models69 and in non-human primates.70 LNAs and related compounds are designed to be stable in serum and to exert limited off-target toxicities.70 Use of LNAs to target oncomirs in lung cancer is an attractive approach. Other chemically-modified nucleotide compounds were shown to antagonize miRNA activities in in vitro and in vivo models. For example, 2′-O-methyl-modified nucleotide derivatives inhibited miR-122 (cholesterol-conjugated antagomir) in mice, which caused a decrease in cholesterol biosynthesis. 71 An analogous approach inhibited miR-10b activity in a mouse mammary tumor model and this caused a decrease in lung metastases. 72
In contrast to suppression of oncomirs as a way to confer anti-tumorigenic effects, restoration of tumor suppressive miRNAs was also examined in pre-clinical models. A chemically synthesized miR-34a analog was used along with a lipid-based formulation.73 This was delivered to murine models by intravenous delivery, which caused anti-turmorigenic effects.73 Other examples were that of intratumoral or intranasal delivery of let-7 to established tumors in mouse models of NSCLC and this conferred anti-neoplastic effects.74
Agents that affect expression of specific miRNAs were shown to overcome tumor suppressive or oncogenic miRNA effects. These agents are in the early phases of pre-clinical testing. Pre-clinical validation would set the stage for conduct of proof of principle clinical trials targeting desired miRNAs. In this regard, the miRNA expression profile within a patient’s lung cancer might need to be discerned before selection of the miRNA-based therapy that would target aberrant miRNA expression within the tumor.
Future Directions
Expression profiles of miRNAs are useful to improve diagnosis, classification and to provide clinical prognostic information in lung cancer. There is a need to assess the miRNA signatures for each histopathologic subtype of lung cancer and to compare any changes relative to the normal lung as well as to the clinical features present at diagnosis such as stage, smoking history, age, and gender. Likewise, miRNA profiles might prove informative to predict response to chemotherapeutic or targeted therapies as well as to predict survival after diagnosis of early or advanced stages of lung cancer. At the same time, miRNA profiles might identify those at high risk for development of lung cancer. This information might guide the use of chemopreventive agents to reduce this clinical risk.
It is known that miRNA profiles exert biological effects by regulating expression of many target genes. Bioinformatic analysis can predict potential target genes for each miRNA.75, 76 Functional validation of any highlighted target gene must be experimentally confirmed by establishing direct complex formation of the miRNA of interest with the expected mRNA sequence. In these analyses, it is important to recognize that any miRNA profile would likely depend on the cell and tissue contexts as well as on the examined physiological or pathophysiological states. It is notable that a change in a single miRNA can lead to compensatory and time-dependent changes in protein (or mRNA) expression profiles within individual cells or tissues.12, 13 Examination of any individual or small series of miRNAs may not reflect the complexity of changes in miRNA expression participating in clinical tumor biology. Comprehensive analysis of the functional role of each highlighted network of miRNAs and their targets is needed.
In the analyses of miRNA expression profiles, it has proven useful to relate changes in genetically-engineered mouse models of lung cancer to those observed in lung cancers of patients. The utility of this approach is summarized in Figure 4 and in prior work.14, 62 In that work, functional validation of candidate tumor suppressive or oncogenic miRNAs was confirmed by gain and loss of miRNA studies in murine lung cancer cells that can be transplanted via tail vein injections into syngeneic mice to assess lung tumor formation.14, 62 This is an active area of investigation. Yet, functional validation of only a few miRNAs has been achieved in lung cancer. Further work is needed to elucidate the precise miRNA target genes and pathways that play critical roles in regulating lung carcinogenesis. Both in vitro and in vivo gain and loss of function experiments are needed to validate the role of any miRNA in lung cancer biology. In these studies, ISH assays are useful to assess the specific cells or tissues that exhibit miRNA deregulation. This deregulation may occur in the cancer cells themselves, in normal cellular elements within a cancer, and in the stroma or tumor microenvironment that can be the site of altered expression of a specific miRNA in lung cancer. 14, 62, 77
Once specific miRNAs are found to play a key role in lung cancer biology, the same species might become targets to reduce lung carcinogenesis. It is intriguing to keep in mind that chemically-modified RNA analogs (such as LNAs) exist. 78 These are candidate therapeutic agents. One potential advantage of using LNAs or other miRNA derivatives is that off-target toxicities might be clinically well tolerated.78 One possible disadvantage is that prolonged regulation of any single miRNA of interest may confer global changes in gene expression that could elicit unwanted compensatory changes in target gene expression. This in turn could confer clinical resistance or even toxicity. Off-target effects of specific miRNA-based therapy might cause adverse clinical side effects. In the near term, pharmacological miRNA delivery approaches should be optimized in pre-clinical models before clinical testing begins.
Conclusions
Lung cancer is the most common cause of cancer mortality for women and men in the United States18. Given this, there is a pressing need to develop new ways to combat this cancer. One way to begin to address this is by taking advantage of what has already been learnt about miRNAs, which play a key role in regulating gene expression in normal and neoplastic cells or tissues. Expression profiles uncovered miRNAs that are either repressed or increased in lung cancers relative to the adjacent normal lung tissues. These would respectively serve roles as tumor suppressive or oncogenic miRNAs. Expression profiles are also useful to improve the diagnosis or classification of lung cancers. These provide prognostic information and might even guide lung cancer treatments. Intriguingly, miRNAs are also candidate anti-neoplastic agents. Current studies will determine whether chemically-modified miRNAs are suitable for clinical use against lung cancer.
Acknowledgments
This work was supported by National Institutes of Health (NIH) and National Cancer Institute (NCI) grants R01-CA087546 (E. D.), R01-CA111422 (E.D.), R03-CA130102 (E.D.), RO3-CA141564 (L.S.), a Samuel Waxman Cancer Research Foundation award (E.D.), a grant from the American Lung Association (X.L.), an American Cancer Society Institutional Grant (S.J. F.), a grant from The Mary Jo’s Fund to Fight Cancer/Uniting Against Lung Cancer (S.J. F.), and an AACR-Pancreatic Cancer Action Network Cancer Development Award (L.S.). The Wilhelm Johannsen Center for Functional Genome Research is established by the Danish National Research Foundation. E. Dmitrovsky is an American Cancer Society Clinical Research Professor supported by a generous gift from the F.M. Kirby Foundation. We thank Dr. Vincent Memoli (Department of Pathology, Dartmouth Medical School and Dartmouth-Hitchcock Medical Center) for his helpful consultations.
Abbreviations
- ISH
in situ hybridization
- Kb
kilobases
- LNA
locked nucleic acid
- miRNA or miR
microRNA
- NSCLC
non-small cell lung cancer
- oncomir
oncogenic microRNA
- pri-miRNA
primary-miRNAs
- RISC
RNA-induced silencing complex
- SCLC
small cell lung cancer
- siRNA
small interfering RNA
- UTR
untranslated region
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–8. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 2.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–62. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 3.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–4. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 4.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–85. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 5.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–6. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mingot JM, Bohnsack MT, Jakle U, Gorlich D. Exportin 7 defines a novel general nuclear export pathway. EMBO J. 2004;23:3227–36. doi: 10.1038/sj.emboj.7600338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–8. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 8.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–40. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 9.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–55. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–5. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 12.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 13.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu X, Sempere LF, Ouyang H, et al. MicroRNA-31 functions as an oncogenic microRNA in mouse and human lung cancer cells by repressing specific tumor suppressors. J Clin Invest. 2010;120:1298–309. doi: 10.1172/JCI39566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu CJ, Tsai MM, Hung PS, et al. miR-31 ablates expression of the HIF regulatory factor FIH to activate the HIF pathway in head and neck carcinoma. Cancer Res. 2010;70:1635–44. doi: 10.1158/0008-5472.CAN-09-2291. [DOI] [PubMed] [Google Scholar]
- 16.Valastyan S, Chang A, Benaich N, Reinhardt F, Weinberg RA. Concurrent suppression of integrin alpha5, radixin, and RhoA phenocopies the effects of miR-31 on metastasis. Cancer Res. 2010;70:5147–54. doi: 10.1158/0008-5472.CAN-10-0410. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Valastyan S, Reinhardt F, Benaich N, et al. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell. 2009;137:1032–46. doi: 10.1016/j.cell.2009.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–49. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 19.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–31. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 20.Karube Y, Tanaka H, Osada H, et al. Reduced expression of Dicer associated with poor prognosis in lung cancer patients. Cancer Sci. 2005;96:111–5. doi: 10.1111/j.1349-7006.2005.00015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39:673–7. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 22.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–69. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 23.Xing L, Todd NW, Yu L, Fang H, Jiang F. Early detection of squamous cell lung cancer in sputum by a panel of microRNA markers. Mod Pathol. 2010;23:1157–64. doi: 10.1038/modpathol.2010.111. [DOI] [PubMed] [Google Scholar]
- 24.Yu L, Todd NW, Xing L, et al. Early detection of lung adenocarcinoma in sputum by a panel of microRNA markers. Int J Cancer. 2010;127:2870–8. doi: 10.1002/ijc.25289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie Y, Todd NW, Liu Z, et al. Altered miRNA expression in sputum for diagnosis of non-small cell lung cancer. Lung Cancer. 2010;67:170–6. doi: 10.1016/j.lungcan.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu SL, Chen HY, Chang GC, et al. MicroRNA signature predicts survival and relapse in lung cancer. Cancer Cell. 2008;13:48–57. doi: 10.1016/j.ccr.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 27.Landi MT, Zhao Y, Rotunno M, et al. MicroRNA expression differentiates histology and predicts survival of lung cancer. Clin Cancer Res. 2010;16:430–41. doi: 10.1158/1078-0432.CCR-09-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu Z, Chen X, Zhao Y, et al. Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J Clin Oncol. 2010;28:1721–6. doi: 10.1200/JCO.2009.24.9342. [DOI] [PubMed] [Google Scholar]
- 29.Yanaihara N, Caplen N, Bowman E, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–98. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 30.Patnaik SK, Kannisto E, Knudsen S, Yendamuri S. Evaluation of microRNA expression profiles that may predict recurrence of localized stage I non-small cell lung cancer after surgical resection. Cancer Res. 2010;70:36–45. doi: 10.1158/0008-5472.CAN-09-3153. [DOI] [PubMed] [Google Scholar]
- 31.Ranade AR, Cherba D, Sridhar S, et al. MicroRNA 92a-2*: a biomarker predictive for chemoresistance and prognostic for survival in patients with small cell lung cancer. J Thorac Oncol. 2010;5:1273–8. doi: 10.1097/JTO.0b013e3181dea6be. [DOI] [PubMed] [Google Scholar]
- 32.Wang XC, Tian LL, Wu HL, et al. Expression of miRNA-130a in nonsmall cell lung cancer. Am J Med Sci. 2010;5:385–8. doi: 10.1097/MAJ.0b013e3181e892a0. [DOI] [PubMed] [Google Scholar]
- 33.Silva J, Garcia V, Zaballos A, et al. Vesicle-related microRNAs in plasma of NSCLC patients and correlation with survival. Eur Respir J. doi: 10.1183/09031936.00029610. in press. [DOI] [PubMed] [Google Scholar]
- 34.Corsten MF, Miranda R, Kasmieh R, Krichevsky AM, Weissleder R, Shah K. MicroRNA-21 knockdown disrupts glioma growth in vivo and displays synergistic cytotoxicity with neural precursor cell delivered S-TRAIL in human gliomas. Cancer Res. 2007;67:8994–9000. doi: 10.1158/0008-5472.CAN-07-1045. [DOI] [PubMed] [Google Scholar]
- 35.Asangani IA, Rasheed SA, Nikolova DA, et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–36. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 36.Wang P, Zou F, Zhang X, et al. MicroRNA-21 negatively regulates Cdc25A and cell cycle progression in colon cancer cells. Cancer Res. 2009;69:8157–65. doi: 10.1158/0008-5472.CAN-09-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ribas J, Ni X, Haffner M, et al. MiR-21: an androgen receptor-regulated microRNA that promotes hormone-dependent and hormone-independent prostate cancer growth. Cancer Res. 2009;69:7165–9. doi: 10.1158/0008-5472.CAN-09-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seike M, Goto A, Okano T, et al. MiR-21 is an EGFR-regulated anti-apoptotic factor in lung cancer in never-smokers. Proc Natl Acad Sci U S A. 2009;106:12085–90. doi: 10.1073/pnas.0905234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu S, Wu H, Wu F, Nie D, Sheng S, Mo YY. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 2008;18:350–9. doi: 10.1038/cr.2008.24. [DOI] [PubMed] [Google Scholar]
- 40.Hatley ME, Patrick DM, Garcia MR, et al. Modulation of K-Ras-dependent lung tumorigenesis by microRNA-21. Cancer Cell. 2010;18:282–93. doi: 10.1016/j.ccr.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu Z, Liu M, Stribinskis V, et al. MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene. 2008;27:4373–9. doi: 10.1038/onc.2008.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ma Y, Fiering S, Black C, et al. Transgenic cyclin E triggers dysplasia and multiple pulmonary adenocarcinomas. Proc Natl Acad Sci U S A. 2007;104:4089–94. doi: 10.1073/pnas.0606537104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Inamura K, Togashi Y, Nomura K, et al. let-7 microRNA expression is reduced in bronchioloalveolar carcinoma, a non-invasive carcinoma, and is not correlated with prognosis. Lung Cancer. 2007;58:392–6. doi: 10.1016/j.lungcan.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 44.Takamizawa J, Konishi H, Yanagisawa K, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–6. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 45.Raponi M, Dossey L, Jatkoe T, et al. MicroRNA classifiers for predicting prognosis of squamous cell lung cancer. Cancer Res. 2009;69:5776–83. doi: 10.1158/0008-5472.CAN-09-0587. [DOI] [PubMed] [Google Scholar]
- 46.Gallardo E, Navarro A, Vinolas N, et al. MiR-34a as a prognostic marker of relapse in surgically resected non-small-cell lung cancer. Carcinogenesis. 2009;30:1903–9. doi: 10.1093/carcin/bgp219. [DOI] [PubMed] [Google Scholar]
- 47.Lodygin D, Tarasov V, Epanchintsev A, et al. Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle. 2008;7:2591–600. doi: 10.4161/cc.7.16.6533. [DOI] [PubMed] [Google Scholar]
- 48.He L, Thomson JM, Hemann MT, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–33. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chang TC, Yu D, Lee YS, et al. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Corney DC, Flesken-Nikitin A, Godwin AK, Wang W, Nikitin AY. MicroRNA-34b and microRNA-34c are targets of p53 and cooperate in control of cell proliferation and adhesion-independent growth. Cancer Res. 2007;67:8433–8. doi: 10.1158/0008-5472.CAN-07-1585. [DOI] [PubMed] [Google Scholar]
- 51.Welch C, Chen Y, Stallings RL. MicroRNA-34a functions as a potential tumor suppressor by inducing apoptosis in neuroblastoma cells. Oncogene. 2007;26:5017–22. doi: 10.1038/sj.onc.1210293. [DOI] [PubMed] [Google Scholar]
- 52.Tazawa H, Tsuchiya N, Izumiya M, Nakagama H. Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc Natl Acad Sci U S A. 2007;104:15472–7. doi: 10.1073/pnas.0707351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bommer GT, Gerin I, Feng Y, et al. P53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr Biol. 2007;17:1298–307. doi: 10.1016/j.cub.2007.06.068. [DOI] [PubMed] [Google Scholar]
- 54.Zenz T, Mohr J, Eldering E, et al. MiR-34a as part of the resistance network in chronic lymphocytic leukemia. Blood. 2009;113:3801–8. doi: 10.1182/blood-2008-08-172254. [DOI] [PubMed] [Google Scholar]
- 55.Fujita Y, Kojima K, Hamada N, et al. Effects of miR-34a on cell growth and chemoresistance in prostate cancer PC3 cells. Biochem Biophys Res Commun. 2008;377:114–9. doi: 10.1016/j.bbrc.2008.09.086. [DOI] [PubMed] [Google Scholar]
- 56.Sun F, Fu H, Liu Q, et al. Downregulation of CCND1 and CDK6 by miR-34a induces cell cycle arrest. FEBS Lett. 2008;582:1564–8. doi: 10.1016/j.febslet.2008.03.057. [DOI] [PubMed] [Google Scholar]
- 57.He L, He X, Lim LP, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–4. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kong YW, Cannell IG, de Moor CH, et al. The mechanism of micro-RNA-mediated translation repression is determined by the promoter of the target gene. Proc Natl Acad Sci U S A. 2008;105:8866–71. doi: 10.1073/pnas.0800650105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leucci E, Cocco M, Onnis A, et al. MYC translocation-negative classical Burkitt lymphoma cases: an alternative pathogenetic mechanism involving miRNA deregulation. J Pathol. 2008;216:440–50. doi: 10.1002/path.2410. [DOI] [PubMed] [Google Scholar]
- 60.Pigazzi M, Manara E, Baron E, Basso G. MiR-34b targets cyclic AMP-responsive element binding protein in acute myeloid leukemia. Cancer Res. 2009;69:2471–8. doi: 10.1158/0008-5472.CAN-08-3404. [DOI] [PubMed] [Google Scholar]
- 61.Yamakuchi M, Ferlito M, Lowenstein CJ. MiR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci U S A. 2008;105:13421–6. doi: 10.1073/pnas.0801613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu X, Sempere LF, Galimberti F, et al. Uncovering growth-suppressive MicroRNAs in lung cancer. Clin Cancer Res. 2009;15:1177–83. doi: 10.1158/1078-0432.CCR-08-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Calin GA, Sevignani C, Dumitru CD, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007;21:1025–30. doi: 10.1101/gad.1540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Johnson SM, Grosshans H, Shingara J, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–47. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 66.Hebert C, Norris K, Scheper MA, Nikitakis N, Sauk JJ. High mobility group A2 is a target for miRNA-98 in head and neck squamous cell carcinoma. Mol Cancer. 2007;6:5. doi: 10.1186/1476-4598-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Koscianska E, Baev V, Skreka K, et al. Prediction and preliminary validation of oncogene regulation by miRNAs. BMC Mol Biol. 2007;8:79. doi: 10.1186/1471-2199-8-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Johnson CD, Esquela-Kerscher A, Stefani G, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67:7713–22. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- 69.Elmen J, Lindow M, Silahtaroglu A, et al. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res. 2008;36:1153–62. doi: 10.1093/nar/gkm1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Elmen J, Lindow M, Schutz S, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–9. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 71.Krutzfeldt J, Rajewsky N, Braich R, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–9. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 72.Ma L, Reinhardt F, Pan E, et al. Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nat Biotechnol. 2010;28:341–7. doi: 10.1038/nbt.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wiggins JF, Ruffino L, Kelnar K, et al. Development of a lung cancer therapeutic based on the tumor suppressor microRNA-34. Cancer Res. 2010;70:5923–30. doi: 10.1158/0008-5472.CAN-10-0655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Trang P, Medina PP, Wiggins JF, et al. Regression of murine lung tumors by the let-7 microRNA. Oncogene. 2010;29:1580–7. doi: 10.1038/onc.2009.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–98. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 76.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 77.Sempere LF, Preis M, Yezefski T, et al. Fluorescence-based codetection with protein markers reveals distinct cellular compartments for altered microRNA expression in solid tumors. Clin Cancer Res. 2010;16:4246–55. doi: 10.1158/1078-0432.CCR-10-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sempere LF, Kauppinen S. Translational implications of microRNAs in clinical diagnostics and therapeutics. In: Bradshaw RA, Dennis EA, editors. Handbook of Cell Signaling. 2. Oxford: Academic Press; 2009. pp. 2965–81. [Google Scholar]