Abstract
Bacterial biofilms are structured multicellular communities that are responsible for a broad range of infections. Knowing how free-swimming bacteria adapt their motility mechanisms near a surface is crucial for understanding the transition from the planktonic to the biofilm phenotype. By translating microscopy movies into searchable databases of bacterial behavior and developing image-based search engines, we were able to identify fundamental appendage-specific mechanisms for the surface motility of Pseudomonas aeruginosa. Type IV pili mediate two surface motility mechanisms: horizontally oriented crawling, by which the bacterium moves lengthwise with high directional persistence, and vertically oriented walking, by which the bacterium moves with low directional persistence and high instantaneous velocity, allowing it to rapidly explore microenvironments. The flagellum mediates two additional motility mechanisms: near-surface swimming and surface-anchored spinning, which often precedes detachment from a surface. Flagella and pili interact cooperatively in a launch sequence whereby bacteria change orientation from horizontal to vertical and then detach. Vertical orientation facilitates detachment from surfaces and thereby influences biofilm morphology.
Introduction
Bacterial biofilms are multicellular structured surface-bound communities that cause a broad range of infections and are notoriously resistant to antibiotics (1–4). Biofilm formation, development, and growth depend critically on how planktonic bacteria adapt their motility mechanisms near a surface (5–11). For Pseudomonas aeruginosa, a commonly studied organism for biofilm formation (12), motility is driven by two types of appendages: a single polar flagellum and multiple type IV pili (TFP). The flagellum operates as a rotor and generates force via the hydrodynamic drag opposing its rotation (13). TFP operate as linear actuators that pull the bacterium along a surface (14,15).
Bacteria can move collectively on surfaces (16) using distinct appendage-specific motility modes. Flagella mediate swarming, a motility mode used for colony expansion along a semisolid surface (17), and TFP mediate twitching, a motility mode commonly observed in dense aggregates with cell-to-cell contact (8,18,19). These surface motility modes are coupled to signaling networks and nutritional sources, and enable exploration of newly colonized surface environments (10,20,21). Investigators have identified additional factors that can influence collective motility modes, such as biosurfactants (10,22–24) that play a key role in swarming. Swarming and twitching are predominantly studied with the use of plate assays, in which an increase in agar concentration drives the transition from swarming to twitching (17,21,25,26). However, bulk assays are ill-suited for observing the cooperation between distinct motility appendages that must occur when multiple appendages are available, and thus do not allow the general conditions that promote the identification of specific motility modes.
At present, little is known about the initial stages of biofilm formation, during which bacteria transition from a free-swimming planktonic state to a surface-associated state and subsequently form microcolonies. Both flagella and TFP influence these developmental steps (6,9,27), as deleting or altering either appendage leads to variations or deficiencies in cell attachment and growth. To understand surface motility in the low-density transition regime, it is necessary to correlate the spatiotemporally resolved motion of individual bacteria to life-cycle events such as attachment, detachment, and division. Genetic techniques can identify specific appendages that mediate collective motility modes; however, these methods do not probe single-bacterium behavior. Conversely, single-cell techniques can measure forces exerted by motility appendages (11,15,28) but do not probe collective behavior or interbacterium interactions. To quantify the initial stages of biofilm formation, during which individual bacteria may rapidly change their behavior in response to their environment (9,12), we need to develop new techniques that can couple single-cell resolution with large sample populations.
In this work, we were able to identify single-bacterium surface motility mechanisms by tracking every cell in a library of microscopy movies, and to show how these mechanisms influence life-cycle events and biofilm morphology. We extracted the motility histories of individual surface-associated P. aeruginosa cells by translating video microscopy movies into searchable databases of bacterial behavior using automated tracking algorithms (29,30). Automated searches of bacteria trajectories were designed to identify life-cycle events (e.g., division) that were then correlated to patterns of surface motility. Using this search-engine strategy, we quantitatively characterized four fundamental appendage-specific surface motility mechanisms of P. aeruginosa that imply different strategies for surface exploration (Fig. 1). TFP mediate two surface motility mechanisms: crawling, by which the bacterium moves lengthwise with high directional persistence, and upright walking perpendicular to the surface, by which the bacterium moves with low directional persistence to rapidly explore microenvironments. The flagellum mediates two additional mechanisms: swimming and surface-anchored spinning, which often precedes detachment from a surface. Cooperation between different appendages influences motility after division and before detachment. These motility mechanisms have striking implications for early biofilm formation. We show that by enabling vertical orientations that facilitate detachment from a surface, TFP contribute to uniform biofilm morphologies.
Figure 1.
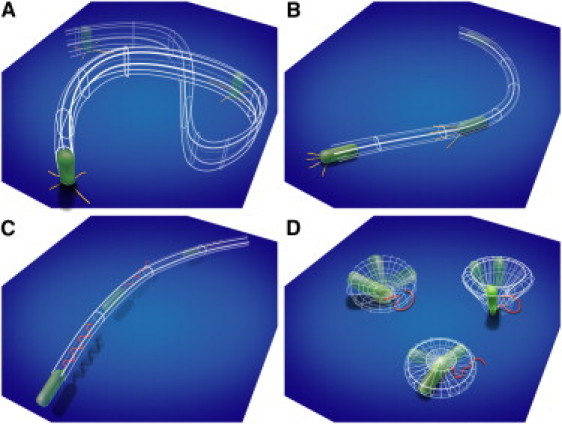
Surface motility mechanisms observed for P. aeruginosa: TFP-driven (A) vertical walking and (B) horizontal crawling, and flagellum-driven (C) near-surface swimming and (D) surface-bound spinning.
Materials and Methods
Flow-cell experiments
P. aeruginosa strain ATCC 15692 wild-type (WT) 1C and ΔpilA or ΔfliM isogenic mutants of this strain were used for all experiments (21). The motility of surface-attached cells was monitored in sterilized flow cells containing FAB medium (31) with 0.6 mM glucose, glutamate, or succinate as the sole carbon source. The medium flow rate was ∼3.75 mL h−1. We prepared an inoculum by growing strains in test tubes containing FAB medium with 30 mM carbon with shaking at 30°C to OD600 ≈ 0.3. We diluted the cultures by adding 50 μL of the bacterial suspension into 950 μL of sterilized FAB (1:20). All strains contained mini-Tn7 chromosomal, constitutive, GFP-expressing insertions that allowed the cells to be visualized by fluorescence microscopy (32). Long time-lapse movies containing 750–2000 images of fluorescent cells were collected at 12 frames per minute with EZC1 software (Nikon, Tokyo, Japan) on a Nikon C1 confocal laser microscope equipped with a 60× objective. Brightfield movies containing up to 20,000 images were collected using an Olympus (Tokyo, Japan) microscope equipped with a 100× objective at 10 frames per second (short time) or 20 frames per minute (long time).
Bacteria tracking
We modified particle-tracking algorithms (29) that automatically locate circular features to identify individual bacteria, which are typically elongated. We designated pixels that were local maxima in at least three directions, or were brighter than a predetermined threshold as backbone pixels, and calculated the position and orientation of each bacterium from the moments of its backbone distribution (30). To determine the length and width of the bacterium, we first rotated each feature by its orientation angle and then calculated the maximum x and y distances between the pixels comprising the bacterium.
Bacteria positions were joined into tracks with the use of code written in IDL (ITT VIS, White Plains, NY). From the bacteria trajectories, we calculated the mean-squared displacement (MSD), where the angled brackets denote an average over all times t, and the instantaneous velocity of a bacterium , where is the position at time i. The angle deviation was defined as the difference between velocity angle and orientation, limited to the range . We classified the bacteria as horizontal or vertical using a cutoff generated from the histogram of bacterial lengths. The directional persistence length P of a single bacterium track is the length scale over which correlation in the direction of motion is lost, and is defined via , where Lij is the integrated track length between time i and time j, and θij is the angle between the velocity vectors at times i and j.
Results and Discussion
Pili drive vertical orientation in P. aeruginosa
We first examine the postdivision motility of horizontally oriented, surface-bound WT bacteria immediately after attachment to the surface. Surprisingly, the two sibling daughter cells exhibit a marked asymmetry in motility after division. In >99% of 214 WT division events, one sibling remains horizontally attached to the substrate. However, the other sibling may detach, move away horizontally, or exhibit a motility mechanism whereby it tilts away from the surface and then walks upright away from the division site on one pole (Fig. 2, A–C).
Figure 2.
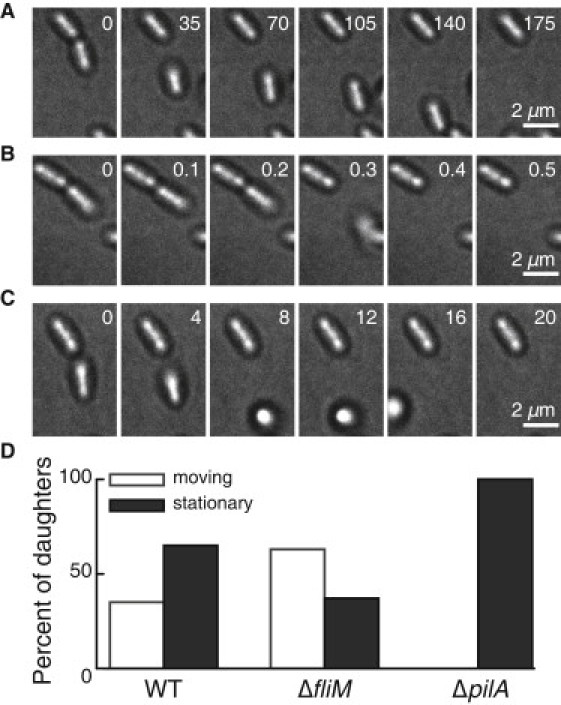
TFP drive postdivision motility in P. aeruginosa. (A–C) Representative time series micrographs of postdivision behavior of daughter cells, showing (A) crawling, (B) detachment, and (C) walking. Timestamps in seconds are shown. (D) Percentage of daughter cells that exhibit motion or remain stationary after division, for the WT strain (N = 214 divisions), ΔfliM strain (N = 105 divisions), and ΔpilA strain (N = 131 divisions). No ΔpilA daughter cells move after division, indicating that postdivision surface motility must depend on TFP.
To identify the motility appendage responsible for walking, we compare the postdivision motility of surface-associated WT bacteria with that of TFP-deficient (ΔpilA) and flagellum-deficient (ΔfliM) mutants. In WT, ∼40% of daughter cells move after division and 60% remain stationary (Fig. 2 D). ΔpilA bacteria do not move after division, indicating that the observed postdivision surface motility mechanisms must depend on TFP. ΔfliM bacteria are more likely to move away after division than the WT (∼60% motile), consistent with reports indicating that flagella are implicated in surface attachment (6).
Pili govern distinct walking and crawling surface motility mechanisms
To investigate TFP-dependent motility, we first examine ΔfliM mutants immediately after surface attachment occurs and then compare the results with those obtained in the WT and ΔpilA strains. These bacteria, whose movement is strictly TFP-driven, exhibit two distinct orientations: horizontal, in which bacteria are oriented parallel to the surface, and vertical, in which bacteria attached to the surface at one end are oriented normal to the surface (33). The histogram of bacterial lengths projected onto the surface is bimodal, with distinct peaks corresponding to vertical and horizontal orientations (Fig. 3 A). The probability distribution of time spent vertical exhibits local maxima at 0 and 1, indicating that the system is bistable, i.e., the bacteria prefer to remain either horizontal or vertical (Fig. 3 B). However, bacteria can change orientations as frequently as once every 10 s, and a very small fraction (<1%) were observed to cartwheel by rapidly changing from vertical to horizontal to vertical, switching the end adhered to the surface. Because ΔfliM bacteria do not possess flagella, TFP can thus mediate attachment at nonflagellated poles. Most ΔfliM bacteria spend time in both orientations, indicating that they can readily switch between TFP-driven mechanisms.
Figure 3.
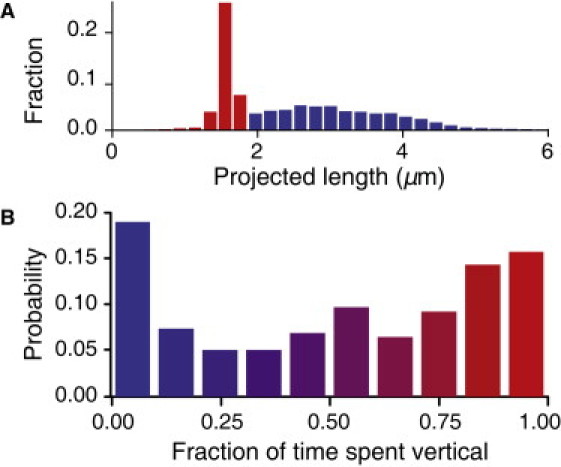
(A) Histogram of projected length and (B) probability of time spent oriented vertically for ΔfliM bacteria (N = 70,073 individual bacteria images).
We find that switching from a horizontal to a vertical orientation does not depend on the direction of flow relative to the bacterium, and occurs in the absence of flow. This suggests that motility switching is actively driven and is not due to interactions with flow. ΔpilA mutants do not exhibit this switching, and therefore TFP are necessary for active switching. Pili-driven switching was previously observed in Myxococcus xanthus, which exhibits a slow, vertical, pili-driven jiggling motion before it transitions to a horizontal orientation for conventional crawling lateral motion (34). By contrast, P. aeruginosa can undergo lateral motion while oriented either horizontally or vertically.
The trajectories of vertical and horizontal ΔfliM bacteria exhibit distinct morphological and dynamical signatures. Visually, the tracks of horizontal bacteria appear smoother and straighter than those of vertical bacteria (Fig. 4 A). Quantitatively, the average directional persistence length Lp, which measures the average length over which trajectories appear straight, is longer for tracks of horizontal bacteria ( μm) than for those of vertical bacteria ( μm). The typical Lp of tracks of vertical bacteria is similar to the extension distance of a single TFP (35), suggesting that sequential steps in these tracks are caused by multiple splayed TFP pulling the bacterium in different, uncorrelated directions. By contrast, because TFP are predominantly located at the poles of the bacterium, horizontal pulling can result in significantly more directional persistence. Each morphology is associated with distinct dynamical behavior: horizontal bacteria preferentially move along their body axis, moving faster along their body than they do perpendicular to it, whereas vertical bacteria exhibit no directional preference for motion (Fig. 4 B). These differences indicate that TFP mediate two distinct motility mechanisms: 1), vertically oriented walking with low directional persistence; and 2), horizontally oriented crawling with high directional persistence.
Figure 4.
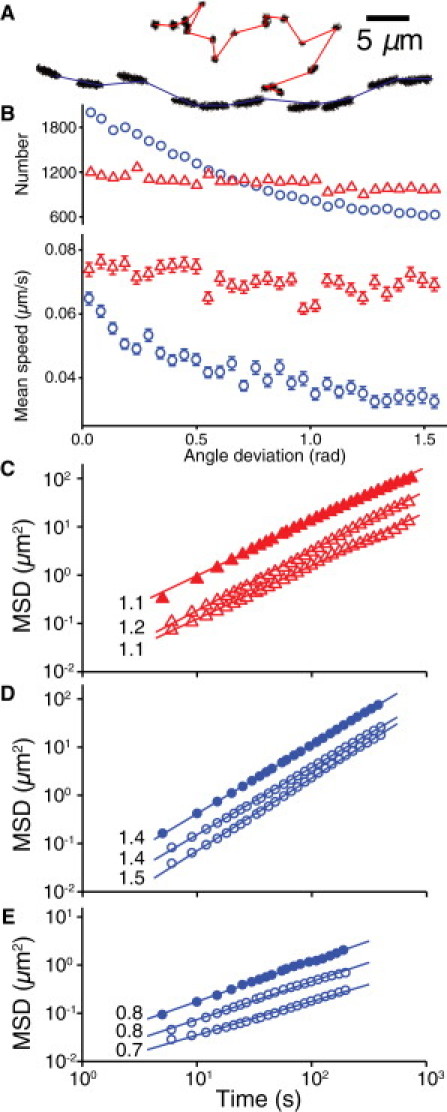
Motility characteristics of the ΔfliM strain. (A) Representative trajectories of walking (top) and crawling (bottom) motility mechanisms, showing morphological differences. (B) Number and mean speed of walking (▵) and crawling (○) bacteria versus angle deviation for ΔfliM bacteria (N = 70,073). Error bars indicate 1 standard deviation (SD). (C) MSD versus time for walking bacteria over a 1-h measurement (▴) and density-limited 4- and 7-h measurements (▵). (D) MSD versus time for superdiffusive crawling bacteria for 1-h (●) and 4- and 7-h (○) measurements. (E) MSD versus time for subdiffusive trapped bacteria for 1-h (●) and 4- and 7-h (○) measurements.
We investigate differences in the area- and distance-covering properties of walking and crawling by calculating the ensemble-averaged MSD as a function of time. The slope of the MSD reflects the shape of the trajectories; by definition, slopes of 1.0 and 2.0 respectively indicate random diffusive motion and geometrically straight motion. Walking bacteria traverse linear distances slightly more efficiently than diffusion (slope = 1.1; Fig. 4 C). Crawling bacteria can be separated into two subpopulations: one with nearly straight, superdiffusive motion (slope = 1.4; Fig. 4 D) and one with subdiffusive motion (slope = 0.8; Fig. 4 E). Walking bacteria exhibit the highest instantaneous velocity, whereas superdiffusive crawling bacteria exhibit a lower instantaneous velocity but move further and more efficiently on long timescales due to the longer Lp. Subdiffusive crawling bacteria have both the lowest instantaneous velocity and a low displacement on long timescales, and often appear locally trapped on length scales (1–2 μm) comparable to those of motility appendages. Each motility mechanism thus confers specific advantages for surface exploration (6,9,36): crawling enables directional motion for efficient coverage of distance, whereas walking enables rapid local exploration of the area. At later times after surface attachment, the magnitude of the MSD decreases for each type of motion, as shown in Fig. 4, C–E, for acquisitions lasting 1, 4, and 7 h after initial attachment. However, the slopes of the MSD are nearly constant, indicating that the characteristic dynamical properties of each mechanism remain the same even though both the density of bacteria and the biofilm characteristics change dramatically over time.
To determine how bacteria use pili-driven motility mechanisms in the presence of flagella, we characterize the motility of WT bacteria using the same metrics. In WT bacteria, which possess both flagella and TFP, the distribution of projected lengths is again bimodal (Fig. 5 A), with distinct populations of vertical and horizontal bacteria, and the distribution of time spent vertical again exhibits local maxima at 0 and 1 (Fig. 5 B). WT bacteria also exhibit walking, superdiffusive crawling, and subdiffusive trapped behavior as distinguished by their characteristic signatures (Fig. 6). However, the prevalence of walking decreases from 36% in ΔfliM to 16% in WT; moreover, the number of vertically oriented bacteria and the percentage of time spent vertical also decrease in WT (Figs. 5 and 6 A). These quantitative differences reflect the availability of additional surface motility mechanisms driven by flagella. Conversely, organisms that do not possess flagella, such as Neisseria gonorrheae (37), N. meningitidis (38), Myxococcus xanthus (39,40), and Synechocystis species (41), should rely even more heavily on TFP-mediated motility mechanisms.
Figure 5.
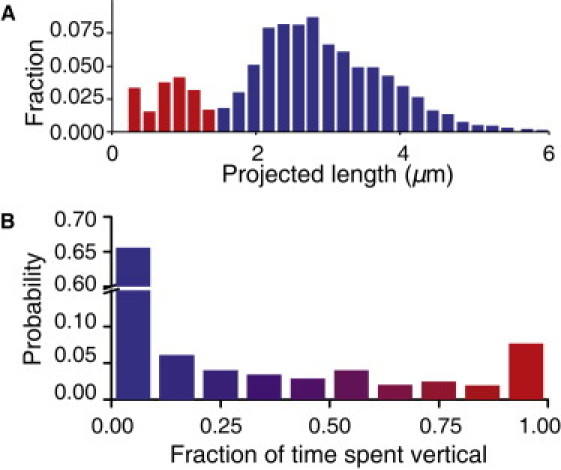
(A) Histogram of projected length and (B) probability of time spent oriented vertically for WT bacteria (N = 170,073 individual bacteria images).
Figure 6.
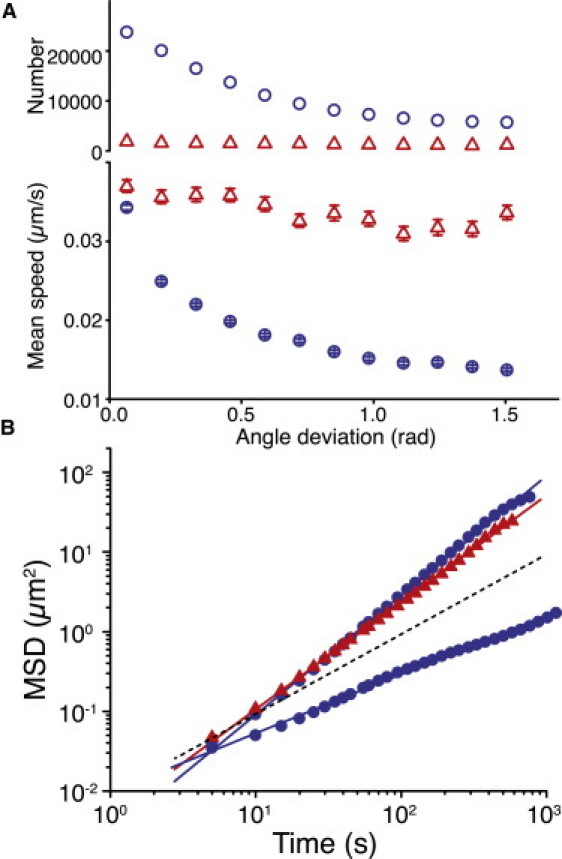
Motility characteristics of the WT strain. (A) Number and mean speed of walking (▵) and crawling (○) bacteria versus angle deviation for WT bacteria (N = 170,073). Error bars indicate 1 SD. (B) MSD versus time for walking (▴) and crawling (●) WT bacteria (N = 170,073); the dotted line indicates a slope of one (diffusive motion). Characteristic slopes match those of ΔfliM bacteria (33).
Flagella mediate swimming and spinning mechanisms
To elucidate the role of flagella in near-surface motility, we examine the ΔpilA strain, whose motility is strictly flagellum-driven, in comparison with the WT. Neither walking nor crawling is present in ΔpilA, because they are governed by TFP. In contrast to ΔfliM and WT, the distribution of projected lengths appears trimodal: the third peak at the longest lengths represents pairs of bacteria that have not moved appreciably after dividing and are thus difficult to distinguish from single long bacteria (Fig. 7 A). Both vertical and horizontal surface-attached bacteria remain immobile over long periods of time. The slope of the MSD is nearly zero, indicating that ΔpilA bacteria move even less than the trapped subdiffusive bacteria in TFP-competent strains (Fig. 7 B).
Figure 7.
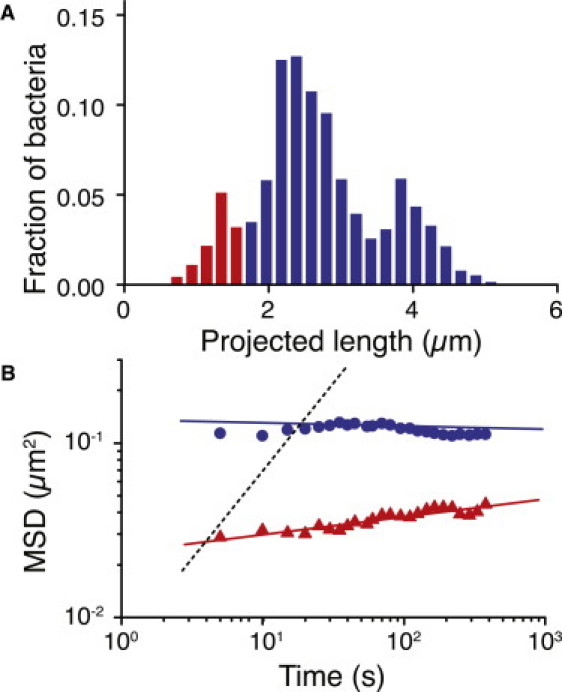
(A) Histogram of projected length for ΔpilA bacteria (N = 17,437 individual bacteria images). (B) MSD versus time for vertical (▴) and horizontal (●) ΔpilA bacteria (N = 17,437); the dotted line indicates a slope of 1 (diffusive motion).
However, the flagella drive two distinct motility modes—swimming and spinning—whose characteristic trajectories are readily distinguished (Fig. 8 A). We find that P. aeruginosa, like other flagellated species such as Escherichia coli, Vibrio alginolyticus, and Caulobacter crescentus, can swim at high speeds approximately parallel to a surface for long distances as a result of hydrodynamic surface coupling that generates curved trajectories (42). The typical swimming speed (∼60 μm/s) and curvature (∼0.2) for WT are similar to those measured for other species (43–46) (Fig. 8 B). In addition, both WT and ΔpilA bacteria can spontaneously anchor one pole to the surface by the flagellum and spin either clockwise or counterclockwise about an axis perpendicular to the surface while oriented slightly out of plane. Attachment for flagellum-driven spinning can only occur at the flagellar pole, in contrast to the bipolar attachment seen for TFP-driven walking. The typical angular velocity for both directions of rotation in WT (∼5 rad/s) is comparable to that measured for artificially tethered species (47,48), indicating that the motor speed of the P. aeruginosa flagellum is similar to that in E. coli (Fig. 8 C).
Figure 8.
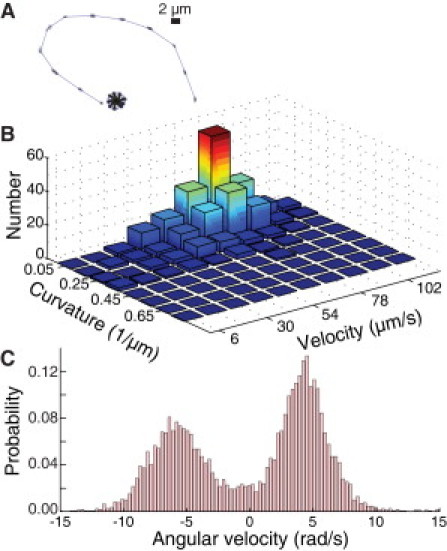
Flagella-driven swimming and spinning motility. (A) Representative trajectories of swimming and spinning motility mechanisms. (B) Two-dimensional histogram of trajectory curvature and instantaneous velocity for swimming WT bacteria. Curvatures are calculated from three consecutive points in each trajectory. (C) Histogram of angular velocity for a representative spinning WT bacterium; positive angular velocity indicates clockwise motion.
Bacteria can switch between motility mechanisms mediated by different appendages
Using single-particle tracking techniques, we identified four distinct near-surface motility mechanisms, with each motility appendage driving two mechanisms. Experiments on N. gonorrheae have shown that bacteria can cooperatively deploy multiple pili to generate lateral crawling motion (11,49), and we also show here (Figs. 3 B and 5 B) that pili-competent bacteria can switch between walking and crawling. To fully exploit the advantages of each motility mechanism, bacteria must switch between motility mechanisms driven by different appendages. The postdivision asymmetry in motility (Fig. 2, A–C) provides one such example: the daughter cell lacking a fully developed flagellum (50,51) is more likely to adopt a TFP-driven mechanism. Instances of motility appendage switching and synergy also occur during cell detachment. In a typical example, a surface-bound WT bacterium initially spins around a fixed center and then ceases to rotate, after which it abruptly tilts away from the surface on one pole and detaches (Fig. 9 A). Spinning bacteria are observed in WT and ΔpilA but not in ΔfliM, consistent with a flagellum-driven mechanism. However, this full launch sequence is only observed for WT, suggesting that TFP can facilitate detachment of spinning bacteria by ceasing their rotation and changing their orientation to out-of-plane. Indeed, ΔpilA bacteria maintain an angle of <30° relative to the surface while spinning, whereas WT bacteria spin at angles of up to 70°. Although both spinning and postdetachment swimming are driven by the flagellum, the transition between them is mediated by the transient attachment of TFP. Ultimately, both the flagellum and the TFP must detach from the surface to allow the bacterium to swim away.
Figure 9.
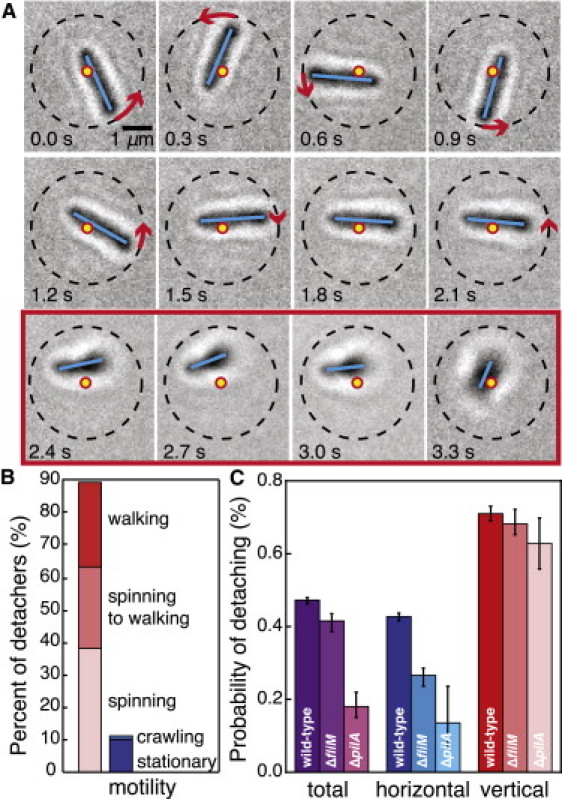
Vertical orientation and appendage cooperation facilitate detachment. (A) Representative image series of a spinning WT bacterium detaching from the surface. Dots indicate the original center of rotation, dashed lines indicate the initial radius of the trajectory, solid lines indicate the bacterial backbone, and arrows indicate the direction and magnitude of rotation between consecutive images. Images inside the box outline (2.4–3.3 s) are those in which the bacterium has tilted off the surface. The bacterium rotates (0–0.9 s), slows (1.2–2.1 s), tilts away from the surface (2.4 s), and then detaches (3.3 s), using both flagella and TFP. (B) Percentage of detaching WT bacteria that exhibit out-of-plane (left) and in-plane (right) motility mechanisms. (C) Detachment probabilities for total, horizontal, and vertical bacteria as a function of strain (ΔfliM (N = 70,073), ΔpilA (N = 17,437), and WT (N = 170,073)). The WT consistently exhibit higher detachment probabilities, showing that both flagella and TFP facilitate detachment. Error bars indicate 1 SD.
To estimate the force exerted by TFP to change bacterial orientation, we equate the torque required to effect rotation at an angular velocity ω, , where is the rotational drag on a bacterium of half-width a and length L in a medium with viscosity η (52), to , where θ is the angle between a lever arm of length r and an applied force of magnitude F. In this simple estimate, we ignore the effects of adhesins that are present on the bacterial surface and may increase the adhesive force. Although the bacterium tilts 0.86 radians away from the surface in 1.8 s, approximately half of this angular change (0.42 rad) occurs in < 0.1 s, when the initial angle between the bacterium and the surface is θ = 0.31 rad. We thus estimate the maximum force exerted by TFP during the tilting process in water as pN. Because this value is significantly smaller than the stall force of a single TFP (∼70 pN (28)), even a single pilus can exert sufficient force to change a bacterium's orientation.
We further test the relationship between vertical orientation and detachment by identifying the orientation and motility mechanism of detaching WT bacteria. Detaching bacteria are overwhelmingly more likely to exhibit out-of-plane motility mechanisms (spinning or walking) and rarely crawl immediately before they detach (Fig. 9 B). As a result, the detachment probability of vertical bacteria is roughly twice that of horizontal bacteria (Fig. 9 C). To quantify the effects of appendage cooperativity on detachment, we calculate the detachment probabilities for the ΔfliM and ΔpilA strains. The difference between the horizontal and vertical detachment probabilities is least pronounced in the WT strain; moreover, the total detachment probability for ΔfliM and ΔpilA is significantly smaller than that of WT, indicating that appendage deficiency decreases detachment. This decrease results primarily from the pronounced decrease in detachment probability for horizontal bacteria; once the bacteria are vertical, the detachment probabilities are similar. This supports our observation that both flagella and TFP facilitate surface detachment via a launch sequence that includes a change from a horizontal to a vertical orientation; furthermore, this launch sequence requires the TFP to pull horizontal cells to a vertical orientation. Together, these observations suggest a physical mechanism for the onset of biofilm formation that is signaled by the transition from reversible polar (vertical) attachment to irreversible longitudinal (horizontal) attachment (53).
The motility defects that reduce bacteria's ability to detach strongly influence biofilm morphology (54). In the first 6 h of ΔpilA biofilm formation, the proportion of spinning bacteria increases dramatically with time (Fig. 10). However, these bacteria lack TFP to achieve the near-vertical orientations that facilitate detachment. Consequently, the launch sequence is impaired and these cells detach less frequently than WT cells. The resultant ΔpilA biofilm contains a heterogeneous distribution of bacterial clusters (9) whose positions are governed by the sites of initial attachment, because the bacteria do not walk or crawl away from the attachment sites. Division events () significantly outnumber attachment events () during a 1-h period of cluster formation, indicating that clusters grow primarily via division. By contrast, TFP-competent WT cells actively redistribute and detach, and the numbers of spinning and vertical bacteria decrease over time. Despite a similar number of division events () and a similar surface cell density, the WT biofilm does not contain clusters, indicating that the ΔpilA biofilm morphology is caused by the motility defects in the ΔpilA strain that impede cluster dispersion via pili-driven motility behaviors such as detaching, walking, and crawling. This is consistent with the strong observed likelihood that a daughter cell will deploy TFP to walk or detach after division occurs (Fig. 2). The active regulation of cell density via TFP in WT results in a uniform initial surface coverage that precedes WT biofilm formation. The morphology of a ΔfliM biofilm is less uniform than that of WT, as the bacteria still have a small detachment defect (Fig. 9 C); however, the ΔfliM biofilm does not exhibit the large aggregates seen in the ΔpilA biofilm.
Figure 10.

Motility defects influence biofilm morphology. (A) Percentage of bacteria that exhibit the spinning motility mechanism or vertical orientation for ΔpilA (left, N = 376 and 270 at 0 h and 5.5 h) and WT (right, N = 355 and 257 at 0 h and 5.5 h) bacteria. Error bars indicate 1 SD. The percentage of spinning bacteria increases with time for ΔpilA because they cannot tilt up and detach. (B) Representative micrographs of ΔpilA (left), WT (right), and ΔfliM (right, inset) biofilms 5.5 h after inoculation. The presence of clusters in the ΔpilA biofilm compared with the uniform WT biofilm indicates that only TFP-competent bacteria are able to redistribute and detach. The ΔfliM strain, which has a small detachment defect and no motility defect, is nonuniform but contains no large clusters.
Conclusion
By using an efficient search-engine-based approach to analyze bacterial motility, we were able to identify four fundamental near-surface motility mechanisms in P. aeruginosa. The flagellum mediates near-surface swimming and surface-bound spinning. TFP mediate crawling, the higher directional persistence of which enables efficient directional motion, and walking, the lower directional persistence of which enables efficient local exploration. Tracking the motility of thousands of bacteria over many hours allows us to identify specific mechanisms that make up the TFP-driven motility modes investigated in earlier studies (9,55). Moreover, the improved spatiotemporal resolution of our study will enable fundamentally new investigations of the cooperative deployment of motility appendages. We have shown that appendage switching and synergy allow bacteria to exploit the advantages conferred by different motility mechanisms. For example, we have demonstrated how motility and detachment defects in the ΔpilA strain engender a biofilm morphology distinct from that of WT. Our technique of total analysis, in which every cell in a movie is automatically and individually examined, can be applied to a broad range of microorganisms to quantitatively characterize motility mechanisms that are inaccessible by traditional microscopy methods.
Acknowledgments
This research was funded by the National Institutes of Health (NIH 1RO1HL087920 and No. UL1RR025761 through the Indiana Clinical and Translational Science Institute) and the National Science Foundation (DMR08-04363, CBET08-27293, and Water CAMPWS).
References
- 1.Costerton J.W., Stewart P.S., Greenberg E.P. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 2.Touhami A., Jericho M.H., Beveridge T.J. Nanoscale characterization and determination of adhesion forces of Pseudomonas aeruginosa pili by using atomic force microscopy. J. Bacteriol. 2006;188:370–377. doi: 10.1128/JB.188.2.370-377.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson G.G., O'Toole G.A. Innate and induced resistance mechanisms of bacterial biofilms. Curr. Top. Microbiol. Immunol. 2008;322:85–105. doi: 10.1007/978-3-540-75418-3_5. [DOI] [PubMed] [Google Scholar]
- 4.Hall-Stoodley L., Stoodley P. Evolving concepts in biofilm infections. Cell. Microbiol. 2009;11:1034–1043. doi: 10.1111/j.1462-5822.2009.01323.x. [DOI] [PubMed] [Google Scholar]
- 5.Nassif X., Marceau M., Taha M.K. Type-4 pili and meningococcal adhesiveness. Gene. 1997;192:149–153. doi: 10.1016/s0378-1119(96)00802-5. [DOI] [PubMed] [Google Scholar]
- 6.O'Toole G.A., Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 1998;30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 7.Merz A.J., Forest K.T. Bacterial surface motility: slime trails, grappling hooks and nozzles. Curr. Biol. 2002;12:R297–R303. doi: 10.1016/s0960-9822(02)00806-0. [DOI] [PubMed] [Google Scholar]
- 8.Mattick J.S. Type IV pili and twitching motility. Annu. Rev. Microbiol. 2002;56:289–314. doi: 10.1146/annurev.micro.56.012302.160938. [DOI] [PubMed] [Google Scholar]
- 9.Klausen M., Heydorn A., Tolker-Nielsen T. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol. Microbiol. 2003;48:1511–1524. doi: 10.1046/j.1365-2958.2003.03525.x. [DOI] [PubMed] [Google Scholar]
- 10.Verstraeten N., Braeken K., Michiels J. Living on a surface: swarming and biofilm formation. Trends Microbiol. 2008;16:496–506. doi: 10.1016/j.tim.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 11.Biais N., Ladoux B., Sheetz M. Cooperative retraction of bundled type IV pili enables nanonewton force generation. PLoS Biol. 2008;6:e87. doi: 10.1371/journal.pbio.0060087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tolker-Nielsen T., Brinch U.C., Molin S. Development and dynamics of Pseudomonas sp. biofilms. J. Bacteriol. 2000;182:6482–6489. doi: 10.1128/jb.182.22.6482-6489.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berg H.C. The rotary motor of bacterial flagella. Annu. Rev. Biochem. 2003;72:19–54. doi: 10.1146/annurev.biochem.72.121801.161737. [DOI] [PubMed] [Google Scholar]
- 14.Craig L., Li J. Type IV pili: paradoxes in form and function. Curr. Opin. Struct. Biol. 2008;18:267–277. doi: 10.1016/j.sbi.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clausen M., Koomey M., Maier B. Dynamics of type IV pili is controlled by switching between multiple states. Biophys. J. 2009;96:1169–1177. doi: 10.1016/j.bpj.2008.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang H.P., Be'er A., Swinney H.L. Collective motion and density fluctuations in bacterial colonies. Proc. Natl. Acad. Sci. USA. 2010;107:13626–13630. doi: 10.1073/pnas.1001651107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Köhler T., Curty L.K., Pechère J.C. Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J. Bacteriol. 2000;182:5990–5996. doi: 10.1128/jb.182.21.5990-5996.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Semmler A.B.T., Whitchurch C.B., Mattick J.S. A re-examination of twitching motility in Pseudomonas aeruginosa. Microbiology. 1999;145:2863–2873. doi: 10.1099/00221287-145-10-2863. [DOI] [PubMed] [Google Scholar]
- 19.Merz A.J., So M., Sheetz M.P. Pilus retraction powers bacterial twitching motility. Nature. 2000;407:98–102. doi: 10.1038/35024105. [DOI] [PubMed] [Google Scholar]
- 20.Harshey R.M. Bacterial motility on a surface: many ways to a common goal. Annu. Rev. Microbiol. 2003;57:249–273. doi: 10.1146/annurev.micro.57.030502.091014. [DOI] [PubMed] [Google Scholar]
- 21.Shrout J.D., Chopp D.L., Parsek M.R. The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol. Microbiol. 2006;62:1264–1277. doi: 10.1111/j.1365-2958.2006.05421.x. [DOI] [PubMed] [Google Scholar]
- 22.Tremblay J., Richardson A.P., Déziel E. Self-produced extracellular stimuli modulate the Pseudomonas aeruginosa swarming motility behaviour. Environ. Microbiol. 2007;9:2622–2630. doi: 10.1111/j.1462-2920.2007.01396.x. [DOI] [PubMed] [Google Scholar]
- 23.Angelini T.E., Roper M., Brenner M.P. Bacillus subtilis spreads by surfing on waves of surfactant. Proc. Natl. Acad. Sci. USA. 2009;106:18109–18113. doi: 10.1073/pnas.0905890106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Be'er A., Smith R.S., Swinney H.L. Paenibacillus dendritiformis bacterial colony growth depends on surfactant but not on bacterial motion. J. Bacteriol. 2009;191:5758–5764. doi: 10.1128/JB.00660-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beatson S.A., Whitchurch C.B., Mattick J.S. Quorum sensing is not required for twitching motility in Pseudomonas aeruginosa. J. Bacteriol. 2002;184:3598–3604. doi: 10.1128/JB.184.13.3598-3604.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chiang P., Habash M., Burrows L.L. Disparate subcellular localization patterns of Pseudomonas aeruginosa type IV pilus ATPases involved in twitching motility. J. Bacteriol. 2005;187:829–839. doi: 10.1128/JB.187.3.829-839.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barken K.B., Pamp S.J., Tolker-Nielsen T. Roles of type IV pili, flagellum-mediated motility and extracellular DNA in the formation of mature multicellular structures in Pseudomonas aeruginosa biofilms. Environ. Microbiol. 2008;10:2331–2343. doi: 10.1111/j.1462-2920.2008.01658.x. [DOI] [PubMed] [Google Scholar]
- 28.Maier B., Potter L., Sheetz M.P. Single pilus motor forces exceed 100 pN. Proc. Natl. Acad. Sci. USA. 2002;99:16012–16017. doi: 10.1073/pnas.242523299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crocker J.C., Grier D.G. Methods of digital video microscopy for colloidal studies. J. Colloid Interface Sci. 1996;179:298–310. [Google Scholar]
- 30.Mohraz A., Solomon M.J. Direct visualization of colloidal rod assembly by confocal microscopy. Langmuir. 2005;21:5298–5306. doi: 10.1021/la046908a. [DOI] [PubMed] [Google Scholar]
- 31.Heydorn A., Ersbøll B.K., Molin S. Experimental reproducibility in flow-chamber biofilms. Microbiology. 2000;146:2409–2415. doi: 10.1099/00221287-146-10-2409. [DOI] [PubMed] [Google Scholar]
- 32.Koch B., Jensen L.E., Nybroe O. A panel of Tn7-based vectors for insertion of the gfp marker gene or for delivery of cloned DNA into Gram-negative bacteria at a neutral chromosomal site. J. Microbiol. Methods. 2001;45:187–195. doi: 10.1016/s0167-7012(01)00246-9. [DOI] [PubMed] [Google Scholar]
- 33.Gibiansky M.L., Conrad J.C., Wong G.C. Bacteria use type IV pili to walk upright and detach from surfaces. Science. 2010;330:197. doi: 10.1126/science.1194238. [DOI] [PubMed] [Google Scholar]
- 34.Sun H., Zusman D.R., Shi W. Type IV pilus of Myxococcus xanthus is a motility apparatus controlled by the frz chemosensory system. Curr. Biol. 2000;10:1143–1146. doi: 10.1016/s0960-9822(00)00705-3. [DOI] [PubMed] [Google Scholar]
- 35.Skerker J.M., Berg H.C. Direct observation of extension and retraction of type IV pili. Proc. Natl. Acad. Sci. USA. 2001;98:6901–6904. doi: 10.1073/pnas.121171698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klausen M., Aaes-Jørgensen A., Tolker-Nielsen T. Involvement of bacterial migration in the development of complex multicellular structures in Pseudomonas aeruginosa biofilms. Mol. Microbiol. 2003;50:61–68. doi: 10.1046/j.1365-2958.2003.03677.x. [DOI] [PubMed] [Google Scholar]
- 37.Holz C., Opitz D., Maier B. Bacterial motility and clustering guided by microcontact printing. Nano Lett. 2009;9:4553–4557. doi: 10.1021/nl903153c. [DOI] [PubMed] [Google Scholar]
- 38.Merz A.J., So M. Interactions of pathogenic Neisseriae with epithelial cell membranes. Annu. Rev. Cell Dev. Biol. 2000;16:423–457. doi: 10.1146/annurev.cellbio.16.1.423. [DOI] [PubMed] [Google Scholar]
- 39.Kaiser D. Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proc. Natl. Acad. Sci. USA. 1979;76:5952–5956. doi: 10.1073/pnas.76.11.5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu S.S., Kaiser D. Genetic and functional evidence that type IV pili are required for social gliding motility in Myxococcus xanthus. Mol. Microbiol. 1995;18:547–558. doi: 10.1111/j.1365-2958.1995.mmi_18030547.x. [DOI] [PubMed] [Google Scholar]
- 41.Bhaya D., Bianco N.R., Grossman A. Type IV pilus biogenesis and motility in the cyanobacterium Synechocystis sp. PCC6803. Mol. Microbiol. 2000;37:941–951. doi: 10.1046/j.1365-2958.2000.02068.x. [DOI] [PubMed] [Google Scholar]
- 42.Lauga E., DiLuzio W.R., Stone H.A. Swimming in circles: motion of bacteria near solid boundaries. Biophys. J. 2006;90:400–412. doi: 10.1529/biophysj.105.069401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Greenberg E.P., Canale-Parola E. Motility of flagellated bacteria in viscous environments. J. Bacteriol. 1977;132:356–358. doi: 10.1128/jb.132.1.356-358.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frymier P.D., Ford R.M., Cummings P.T. Three-dimensional tracking of motile bacteria near a solid planar surface. Proc. Natl. Acad. Sci. USA. 1995;92:6195–6199. doi: 10.1073/pnas.92.13.6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kudo S., Imai N., Magariyama Y. Asymmetric swimming pattern of Vibrio alginolyticus cells with single polar flagella. FEMS Microbiol. Lett. 2005;242:221–225. doi: 10.1016/j.femsle.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 46.Li G., Tam L.-K., Tang J.X. Amplified effect of Brownian motion in bacterial near-surface swimming. Proc. Natl. Acad. Sci. USA. 2008;105:18355–18359. doi: 10.1073/pnas.0807305105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen X., Berg H.C. Torque-speed relationship of the flagellar rotary motor of Escherichia coli. Biophys. J. 2000;78:1036–1041. doi: 10.1016/S0006-3495(00)76662-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Neuman K.C., Chadd E.H., Block S.M. Characterization of photodamage to Escherichia coli in optical traps. Biophys. J. 1999;77:2856–2863. doi: 10.1016/S0006-3495(99)77117-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holz C., Opitz D., Maier B. Multiple pilus motors cooperate for persistent bacterial movement in two dimensions. Phys. Rev. Lett. 2010;104:178104. doi: 10.1103/PhysRevLett.104.178104. [DOI] [PubMed] [Google Scholar]
- 50.Suzuki T., Iino T. Isolation and characterization of multiflagellate mutants of Pseudomonas aeruginosa. J. Bacteriol. 1980;143:1471–1479. doi: 10.1128/jb.143.3.1471-1479.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Amako K., Umeda A. Flagellation of Pseudomonas aeruginosa during the cell division cycle. Microbiol. Immunol. 1982;26:113–117. doi: 10.1111/j.1348-0421.1982.tb00160.x. [DOI] [PubMed] [Google Scholar]
- 52.Berg H. Princeton University Press; Princeton, NJ: 1993. Random Walks in Biology. [Google Scholar]
- 53.O'Toole G.A., Kaplan H.B., Kolter R. Biofilm formation as microbial development. Annu. Rev. Microbiol. 2000;54:49–79. doi: 10.1146/annurev.micro.54.1.49. [DOI] [PubMed] [Google Scholar]
- 54.Wood T.K., González Barrios A.F., Lee J. Motility influences biofilm architecture in Escherichia coli. Appl. Microbiol. Biotechnol. 2006;72:361–367. doi: 10.1007/s00253-005-0263-8. [DOI] [PubMed] [Google Scholar]
- 55.Singh P.K., Parsek M.R., Welsh M.J. A component of innate immunity prevents bacterial biofilm development. Nature. 2002;417:552–555. doi: 10.1038/417552a. [DOI] [PubMed] [Google Scholar]


