Abstract
Here we address the impact nuclear architecture has on molecular flow within the mitotic nucleus of live cells as compared to interphase by the pair correlation function method. The mitotic chromatin is found to allow delayed but continuous molecular flow of EGFP in and out of a high chromatin density region, which, by pair correlation function analysis, is shown as a characteristic arc shape that appears upon entry and exit. This is in contrast to interphase chromatin, which regulates flow between different density chromatin regions by means of a mechanism which turns on and off intermittently, generating discrete bursts of EGFP. We show that the interphase bursts are maintained by metabolic energy, whereas the mitotic mechanism of regulation responsible for the arc is not sensitive to ATP depletion. These two distinct routes of molecular flow were concomitantly measured in the Caenorhabditis elegans germ line, which indicates a conservation of mechanism on a scale more widespread than cell type or organism.
Introduction
A key emerging contributor to genome function is the architectural organization of the cell nucleus. The cell nucleus is a functionally and spatially ordered organelle (1), in which constitutive, passive, and nondirected diffusion is the mode of motion for molecules (2–4). Transit by diffusion constitutes an energetically economical scanning mechanism for molecules to find their intranuclear targets, with no need for a specific signal or signal recognition machinery (5). Molecular diffusion throughout the nucleus is thus mainly regulated by the steric constraints imposed by structural components, such as chromatin (5). Chromatin is the binding target for many nuclear proteins involved in functions such as chromatin remodeling and repair (6), epigenetic regulation (7), or gene transcription (8). Because chromatin fills up to 12% of the cell nucleus (9), it must also be considered a major static obstacle even for nonbinding (inert) molecules (e.g., untagged EGFP (10)).
The regulation imparted by chromatin on the diffusion of nonbinding molecules can be central to their specific mechanism of action as well as to the understanding of nuclear architecture and function in general. Much effort has been conducted in studying the accessibility of the nuclear landscape to inert molecules in interphase cells. The results of these studies have mainly been derived from fluorescence recovery after photobleaching (3,11), single particle tracking (12), and fluorescence correlation spectroscopy (FCS) (13) experiments. We recently addressed the diffusion of untagged monomeric EGFP in interphase nuclei of live cells by the pair correlation function (pCF) method (14). We showed that there are two partially disconnected molecular flows throughout the nucleus associated with high and low chromatin density. Rare and sudden (300 ms) bursts of molecules are observed to travel across the change in density, which predominately behaves as a barrier. We proposed these bursts to be the result of intrinsic localized changes in chromatin structure. This observation, in turn, brings up the important question: How do different chromatin architectures taking place during the cell cycle affect the diffusion of small molecules?
To address this issue here, we apply the pCF analysis to EGFP intranuclear diffusion during mitosis, and compare it to interphase. We use Chinese hamster ovary cells (CHO-K1) stably expressing monomeric EGFP and transiently transfected with mCherry-tagged Histone 2B (H2B), to have a reference for the local chromatin density and position. Notably we find the mitotic chromatin to behave differently from the interphase chromatin in the way it affects molecular flow. During mitosis, chromatin represents a zone that can be freely crossed, although with some delay; a characteristic arc-shaped correlation appears in the pCF carpet upon entry and exit of the chromatin. We show that this delay in flow is not dependent on metabolic energy, and therefore the mode of regulation imparted by chromatin as likely to behave as a physical barrier. On the contrary, the intermittent regulation in interphase is completely turned off under energy-depleted conditions. The distinct paths of EGFP molecular flow observed in interphase and mitotic nuclei of the CHO-K1 cells were also concomitantly measured for both cell cycle stages in the nuclei of the Caenorhabditis elegans adult germ line.
Based on our results, we can thus conclude that the regulation of molecular flow of inert molecules in the nucleus strictly depends on the spatial organization of chromatin, and the characteristic mechanisms defined for each of the cell cycle phases tested are retained on a more widespread level than cell type or organism.
Materials and Methods
Cell culture and treatments
CHO-K1 cells stably transfected with EGFP were grown in Ham's F12K medium supplemented with 10% of Fetal Bovine Serum at 37°C and in 5% CO2. Freshly split cells were plated onto 35-mm glass bottom dishes and transiently transfected with the H2B-mCherry plasmid using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to manufacturer's protocol. The H2B-mCherry plasmid (20972) was purchased from Addgene (www.addgene.com) and provided to Addgene by Robert Benezra. All measurements were performed at 37°C and 5% CO2. Energy-depletion experiments were conducted by using sodium azide and 2-deoxy-d-glucose, as described elsewhere (15). The samples of C. elegans expressing monomeric EGFP in the germ line (JK4461 unc-119(ed3) III; Ppie-1:gfp:fem-3(qIs156)) and stained with Hoechst 33342 (Invitrogen) were kindly prepared and provided by Amanda Cinquin, University of California, Irvine.
Microscope
The microscopy measurements were performed on a model No. LSM710 META laser scanning microscope, using a 63× water immersion objective 1.2 NA (Carl Zeiss, Jena, Germany). EGFP was excited with the 488-nm emission line of an Argon laser. mCherry was excited with the 561-nm emission line of a diode pump solid-state laser. EGFP and mCherry were measured sequentially using the 492–560 nm and 580–696 nm collection ranges, respectively. For each channel, the pinhole was set to 1 Airy unit. The potential cross talk, bleed-through, and FRET effects between the two fluorophores were tested and found to be nonexistent in the acquisition settings used. The average laser power at the sample was always maintained at the milliWatt level. The volume of the 488-nm laser point spread function (PSF) was calibrated by measuring the autocorrelation curve for 20 nM fluorescein in 0.01 M NaOH, which has a known diffusion coefficient of 400 μm2/s. The measured values of ω0 (that defines the PSF) varied in the range of 0.26 ± 0.04 μm.
Experimental
A detailed description of the experimental settings used for the line-scan measurement is present in a previous publication (14). We acquire data by rapidly scanning a diffraction-limited laser beam (488 nm) along a line drawn inside the nucleus that traverses a high density chromatin region. Measuring a line of 32 points at maximum zoom, we achieve 100-nm pixel dimensions, resulting in a line length of 3.2 μm. The maximum scanning speed for these settings was selected (pixel dwell time 6.3 μs, line time 0.472 ms) so that the EGFP molecules could be correlated in time between lines measured. In general for each experiment, 2 × 105 consecutive lines (with no intervals between lines) were acquired. Time regions within each experiment (∼6.4 × 104 lines, corresponding to ∼30 s) with no average change in fluorescence intensity (e.g., photobleaching) were then selected for the correlation analysis.
Data analysis and simulations
Calculation of the auto- and pair-correlation functions was done using the SimFCS software developed at the Laboratory for Fluorescence Dynamics (www.lfd.uci.edu). Details about the mathematical derivation of the pCF for diffusing particles can be found in previously published articles (14,16,17). Briefly, intensity data are presented using a carpet representation in which the x coordinate corresponds to the point along the line (pixels) and the y coordinate corresponds to the time. The ACF is calculated according to the expression
where F(t) is the fluorescence intensity at time t and τ is the time shift. The fit of the ACF was done using the diffusion model for two species according to the expression
with
where Di is the diffusion coefficient of species i, γ = 0.3536 depends on the illumination profile which is assumed to be Gaussian, and w0 and wz are the radial and axial waists of the illumination profile, respectively.
The pair correlation function pCF(n) is a cross correlation function of the time trace at two different points that differ by a distance of n pixels. The pCF(n) function at a given pixel distance n is given by the expression
where F(t,n) is the fluorescence intensity at time t at the pixel n along the scan line, τ is the time shift, and δr is the distance between pixels. For the experiments reported here, the distance between adjacent pixels is 100 nm. For example, pCF(10) indicates that the correlation function was calculated at a distance of 10 pixels (1000 nm). An analytical form of the pCF(n) cannot be obtained except in very simple cases of isotropic diffusion because the positions of the boundaries are not known. Instead, we use the maximum of the pCF(n) to determine the average time a molecule takes to travel a given distance. This time is different if there is an obstacle or barrier along the line of measurement with respect to the absence of the obstacle. Bleaching is not affecting the calculation of the pair correlation function because we only use the pair correlation function to determine the time a molecule take to go from one pixel to the other. If the molecule bleaches, then it will not appear at the pixel at a given distance. This will decrease the amplitude of the correlation, but not the time. We note that a molecule can take any three-dimensional path between the two points in space so that the molecule is not continuously illuminated. In all our experiments, overall bleaching was very small.
We display the ACF and pCF pixels in pseudo colors in a image in which the x coordinate corresponds to the point along the line where the function is calculated (or the first point along the line used for the pair correlation) and the vertical coordinate corresponds to the autocorrelation time in a log-scale. Because we have 32 points in a line, when we calculate pCF (10), line 1 is cross correlated with line 11, line 2 with line 12, and so on. This operation is repeated only up to line 22. The rest of the lines are not calculated and they are shown in black in the pCF(n) display. The distances at which pCF analysis was carried out were not fixed across all experiments, but instead determined on an individual basis by the chromatin density variation along each line measured.
Simulations were performed using a random walk in a grid using the simulation engine of the SimFCS software. Each point in the grid is associated with a characteristic diffusion coefficient. The size of the grid element can be scaled arbitrarily. The barrier is simulated by a squared region that has different diffusion properties compared to the surrounding environment. In the simulation of the impenetrable barrier to diffusion, we set the square region to impart a slower diffusion coefficient, with a probability of zero for molecules to get in and out of this zone. In the simulation of the penetrable barrier to diffusion, we set the square region to impart a slower diffusion coefficient, with a probability of one for molecules to get in and out of this zone.
Results
Setting the rules to interpret molecular flow upon encountering a barrier from the pCF carpet
The pair correlation function (pCF) can show the diffusive route taken by molecules along a line measured by temporal cross correlation of a pair of points at a given distance (16). Upon encountering a zone that behaves as a barrier, molecular diffusion will be directed either around or through this zone, depending on the degree of penetrability. The chromatin network of a nucleus is known to obstruct molecular diffusion (1–4). We simulate here two different scenarios which result in molecular flow adopting two different diffusive routes. pCF analysis of each scenario gives rise to distinct characteristic shapes in the pCF carpet (see Fig. 1 caption, for explanation on how the pCF carpet is derived). If the chromatin is impenetrable (Fig. 1 A), an absence of molecular flow is observed for diffusion in and out of the chromatin environment, which causes characteristic gaps in the pCF carpet. In contrast, if the chromosome is penetrable (Fig. 1 B) there is a continuity of molecular flow, although with a delay, which causes characteristic arc shapes to appear in the pCF carpet.
Figure 1.
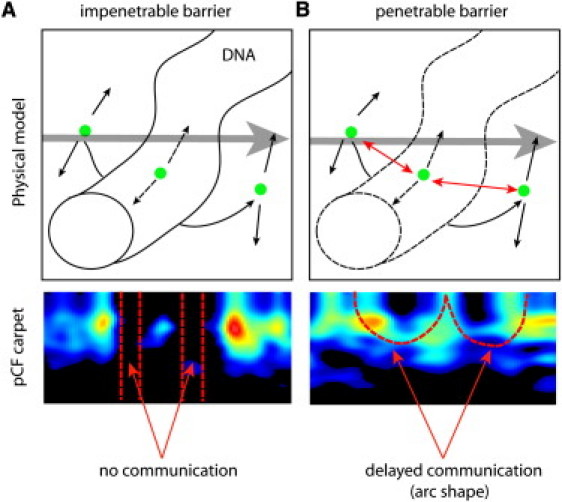
Rules to interpret molecular flow upon encountering a barrier in the pCF carpet. The pCF carpet is obtained by measuring the fluorescence intensity along a line numerous times, and then cross correlating the fluorescence intensity of a pair of points along the line, differing by a selected number of pixels. When this operation is repeated for each pair of points at the selected distance, we represent the cross correlation function under the form of an image that we call the pCF carpet. We simulated diffusion of a particle in a plane in the presence of a region (which represents the chromatin) of a different diffusion coefficient with a barrier to enter and exit the region that can be impenetrable or penetrable. (A) The pCF carpet derived for the impenetrable barrier separating two regions of different diffusion displays characteristic gap regions for this absence of communication. (B) When communication is allowed between the two regions, the pCF carpet displays characteristic arc-shapes due to delayed but positive communication.
Molecular flow of EGFP in a mitotic nucleus
We use CHO-K1 cells stably transfected with monomeric EGFP and transiently transfected with the H2B-mCherry to label the chromatin. In our system, ∼2% of the plated cells are usually undergoing mitosis at any given time, and are selected by visual inspection of the sample. Fig. 2 A depicts a typical CHO-K1 mitotic cell expressing EGFP, with the chromosomes marked by H2B-mCherry. For each mitotic cell tested (N = 5), five line scans were selected within the nucleus, with each line being deliberately positioned across a region of high chromatin density, to test this zone as a barrier to EGFP diffusion (Fig. 2, B and C). The selected line is scanned ∼2 × 105 times (∼90 s) in the EGFP channel over a distance of 32 pixels (∼3.2 μm) and then constructed into an intensity carpet as shown in Fig. 2 D. From the intensity carpet, we can perform pair correlation analysis and investigate molecular flow between any of the columns along the line. Local diffusive information can also be obtained from this method of data representation, if the distance at which pCF analysis is carried out is set to zero pixels. Under these conditions we cross correlate a position with itself and derive the autocorrelation function (ACF) for that location (Fig. 2 E).
Figure 2.
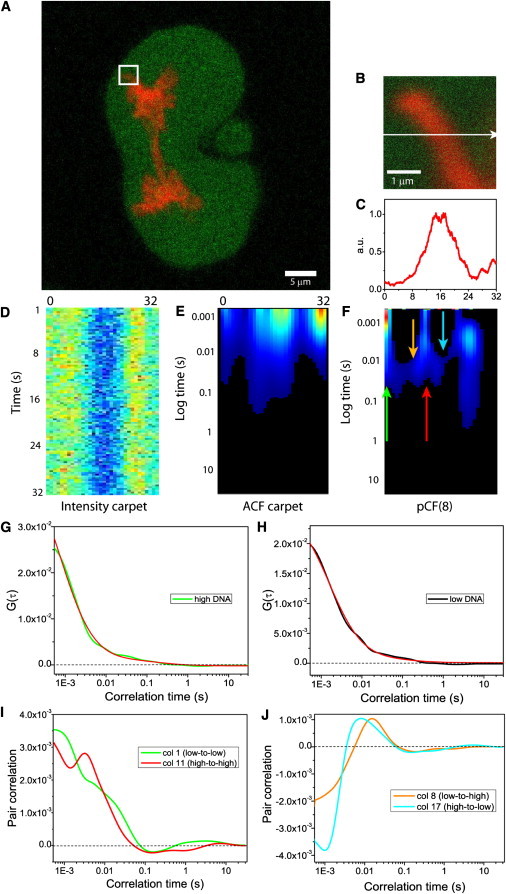
Pair correlation function analysis of intranuclear diffusion of EGFP in a mitotic nucleus. (A) CHO-K1 mitotic cell stably expressing EGFP with the chromosomes marked by H2B-mCherry. (B) Overlay of free EGFP and H2B-mCherry localization in the plane of the 3.3 μm line drawn in the nucleus. (C) Intensity profile of the H2B-mCherry stain across the line measured. (D) Fluorescence intensity carpet of the line drawn across freely diffusing EGFP (200,000 lines). (E) ACF carpet of the line drawn across freely diffusing EGFP (130,000 lines analyzed). (F) The pCF(8) carpet derived for EGFP diffusion between adjacent chromatin density environments across the line measured. The green and red arrows indicate positions where diffusion within the same chromatin density is tested and the orange and cyan arrows indicate positions where diffusion through a change in chromatin density is tested. (G) Fitting of column 15 (which corresponds to high chromatin density) from the ACF carpet in panel E with a two-species model. (H) Fitting of column 30 (which corresponds to low chromatin density) from the ACF carpet in panel E with a one-species model. (I) A plot of the cross correlation function derived from the pCF(8) carpet in panel F for diffusion within a chromatin density environment from the marked arrow positions: low-to-low (column 1 in green) and high-to-high (column 11 in red). (J) A plot of the cross correlation function derived from the pCF(8) carpet in panel F for diffusion through a change in chromatin density from the marked arrow positions: (column 8 in orange) and high-to-low (column 17 in cyan).
In close agreement with the results reported in interphase (14), we find that the high chromatin density region is adequately fitted to a two-component model (D1 = 17.6 ± 6.6 μm2/s, D2 = 0.18 ± 0.1 μm2/s; mean ± SD in five sets of lines in the nuclei of N = 5 observed cells) (Fig. 2 G). This is in contrast to the low chromatin density region which is best fitted to a one-component model (D1 = 19 ± 2 μm2/s, D2 is negligible) (Fig. 2 H). What these results do not tell us is the nature of the obstruction and the manner in which it directs the diffusive path of EGFP between each environment, because the ACF approach is local and only measuring diffusion in a single point. We therefore investigate molecular flow of EGFP by performing the pCF analysis at several distances along the line, determined in each case by the intensity profile of the H2B-mCherry marked chromosomes. The carpet shown in Fig. 2 F is generated at a distance of eight pixels (800 nm), which is no longer a measurement of local diffusion, given that we are now cross correlating points at a distance much larger than the PSF.
Inspection of Fig. 2 C shows that at this distance we are able to analyze three different EGFP flow regimes:
-
1.
Intranuclear (isotropic) flow.
-
2.
Flow from the nucleoplasm into the chromosome.
-
3.
Flow from the chromosome into the nucleoplasm.
The pCF carpet clearly shows that the EGFP flow within a low or high chromatin density environment (e.g., columns 1 and 11) results in communication (positive cross correlation) on the submillisecond timescale, which is indicative of free diffusion (Fig. 2 I). In contrast, flow of EGFP through a change of chromatin density (e.g., columns 8 and 17) results in delayed communication on the tens-of-milliseconds timescale (10–70 ms in N = 5 analyzed cells, five lines per cell), which is indicative of obstructed diffusion (Fig. 2 J).
These two observations cause the pCF carpet to mirror the shape of the chromosome that is being measured as a barrier: we observe a characteristic arc-shape upon entry into the chromosome, and another upon exit. This double-arc feature is analogous to that obtained upon simulation of a penetrable barrier in Fig. 1 B. Closer consideration of how the penetrable barrier was simulated, however, points out an interesting difference between simulation and the in vivo experiments. The penetrable barrier in Fig. 1 B was simulated by imparting a reduced diffusion coefficient within this zone. This in turn, resulted in the diffusing molecules accumulating in the chromatin at steady state. In contrast, in live cells we observe the EGFP molecules to be less concentrated in the chromosome territory. Thus, the chromatin must have an excluded volume for EGFP large enough to circumvent any accumulation of EGFP that would otherwise occur due to reduced diffusion in that region.
Comparison of EGFP intranuclear diffusion in a mitotic versus interphase nucleus
By comparing mitosis (Fig. 3, A–C and D–F) with interphase (Fig. 3, H–J and K–M), we highlight the differences in the regulation of EGFP molecular flow which depend on chromatin conformation. The characteristic mitotic arc-shapes (Fig. 3, C and F) due to delayed molecular flow in (e.g., column 5, Fig. 3 G) and out (e.g., column 17, Fig. 3 G) of the chromosome are replaced by the typical disconnect communication of the interphase nucleus (Fig. 3, J and M). The apparent absence of flow between chromatin environments of different density (e.g., columns 3 and 18 in Fig. 3 N) is analogous to what was obtained upon simulation of an impenetrable barrier in Fig. 1 A. However, as recently demonstrated (14) and shown here in Fig. 3 O, detailed temporal analysis of those pixel positions resulting in an absence of communication, reveals intermittent bursts of EGFP molecules flowing through a change in chromatin density.
Figure 3.
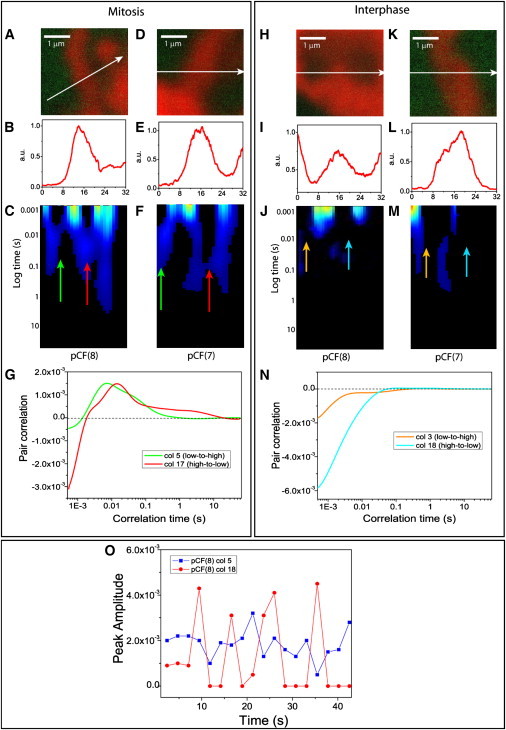
Comparison of EGFP molecular flow through a change in chromatin density (high-to-low or low-to-high) in a mitotic versus interphase nucleus. (A) Overlay of free EGFP and H2B-mCherry localization in the plane of the 3.3 μm line drawn in a mitotic nucleus. (B) Intensity profile of the H2B-mCherry stain across the line measured in a mitotic nucleus. (C) The pCF(8) carpet derived for intranuclear diffusion between adjacent chromatin density environments in a mitotic nucleus. The green and red arrows indicate positions where diffusion through a change in chromatin density environment is tested. (D–F) A second example of molecular flow in a mitotic nucleus. (G) A plot of the cross correlation function derived from the pCF(8) carpet in panel C for delayed diffusion through a change in chromatin density from the marked arrow positions: in (column 5 in green) and out (column 17 in red) of the chromosome of a mitotic nucleus. (H) Overlay of free EGFP and H2B-mCherry localization in the plane of the 3.3 μm line drawn in an interphase nucleus. (I) Intensity profile of the H2B-mCherry stain across the line measured in an interphase nucleus. (J) The pCF(8) carpet derived for intranuclear diffusion between adjacent chromatin density environments in an interphase nucleus. The orange and cyan arrows indicate positions where diffusion through a change in chromatin density environment is tested. (K–M) A second example of molecular flow in an interphase nucleus. (N) A plot of a cross correlation function derived from the pCF(8) carpet in panel J for the absence of diffusion through a change in chromatin density from the marked arrow positions: in (column 3 in orange) and out (column 18 in cyan) of a high density chromatin bundle. (O) Decomposition of a region corresponding to flow through a change in chromatin density into 5000 line segments for the mitotic and interphase nuclei analyzed, in panels C and F, respectively. As can be seen in the case of interphase (red curve), column 18 which originally appeared as having zero correlation for flow through a change in chromatin density now shows the presence of intermittent periods of communication.
This difference prompted us to test the energy dependence of each mechanism (Fig. 4). In the case of the mitotic nucleus (Fig. 4, A and B), we show that ATP depletion has no obvious effect on the appearance of the pCF carpet (Fig. 4 C), i.e., on the diffusion of EGFP. Continuous positive cross correlation is observed with characteristic arc-shapes appearing upon entry (column 8) and exit (column 20) of the chromosome (Fig. 4 G). In contrast, the pCF carpet of the interphase experiment (Fig. 4, D–F) reveals that ATP depletion abolishes the typical flow discontinuity. Continuous flow from a low-to-high (column 3) or high-to-low (column 19) chromatin density region is now possible on the millisecond timescale (Fig. 4 H). As a result, under ATP-depleted conditions, the pCF carpet measured at interphase is no longer disconnected and shows continuous molecular flow through a change in chromatin density.
Figure 4.
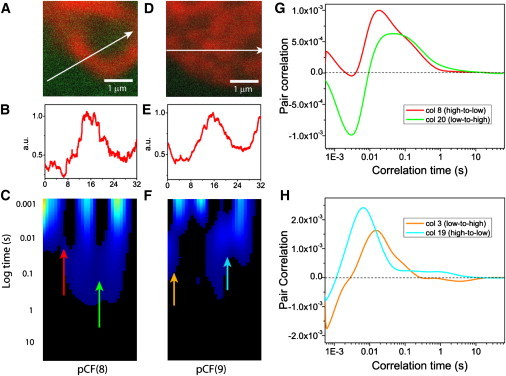
Comparison of EGFP molecular flow through a change in chromatin density (high-to-low or low-to-high) in a mitotic versus interphase nucleus upon ATP depletion. (A) Overlay of free EGFP and H2B-mCherry localization in the plane of the 3.3 μm line drawn in a mitotic nucleus under ATP-depleted conditions. (B) Intensity profile of the H2B-mCherry stain across the line measured in a mitotic nucleus under ATP-depleted conditions. (C) The pCF(8) carpet derived for intranuclear diffusion between adjacent chromatin density environments in a mitotic nucleus under ATP-depleted conditions. The red and green arrows indicate positions where diffusion through a change in chromatin density environment is tested. (D) Overlay of free EGFP and H2B-mCherry localization in the plane of the 3.3 μm line drawn in an interphase nucleus under ATP-depleted conditions. (E) Intensity profile of the H2B-mCherry stain across the line measured in an interphase nucleus under ATP-depleted conditions. (F) The pCF(9) carpet derived for intranuclear diffusion between adjacent chromatin density environments in an interphase nucleus under ATP-depleted conditions. The orange and cyan arrows indicate positions where diffusion through a change in chromatin density environment is tested. (G) A plot of the cross correlation function derived from the pCF(8) carpet in panel C for delayed diffusion through a change in chromatin density from the marked arrow positions: in (column 8 in red) and out (column 20 in green) of the chromosome of a mitotic nucleus under ATP-depleted conditions. (H) A plot of a cross correlation function derived from the pCF(9) carpet in panel F for delayed diffusion through a change in chromatin density from the marked arrow positions: in (column 3 in orange) and out (column 19 in cyan) of a high density chromatin bundle under ATP-depleted conditions.
Intranuclear diffusion of EGFP in the mitotic region of the C. elegans germ line
If the two modes of regulation observed so far are intrinsic rules that govern molecular flow in vivo, then they should be retained at a level that is independent of the organism/cell tested. To address this issue, we used the adult germ line of C. elegans expressing monomeric EGFP: a biological system which enables the concomitant measurement of both cell cycle stages (in one sample!). The distal end of the adult C. elegans gonad contains a stem cell population that is referred to as the mitotic zone (18). This mitotic zone is ∼20 cell diameter in length extending from the distal tip cell. As depicted in Fig. 5 A, when stained with the DNA marker Hoechst 33342, the nuclei actively undergoing mitosis (inside white box) can be easily distinguished from the rest of the cell cycle in the mitotic zone, most of which are in interphase (inside yellow box). Line experiments were carried out measuring EGFP flow in the germ cell nuclei actively undergoing mitosis (Fig. 5 B) and during interphase (Fig. 5 C). The pCF analysis was then performed along the lines measured, at distances determined in each case by the intensity profile of the Hoechst 33342 stain (Fig. 5, D and E). The pCF carpets derived for both cell-cycle stages (Fig. 5, F and G) reveal that the two modes of regulation of molecular flow observed in CHO-K1 cells under physiological conditions are also active in the C. elegans germ line.
Figure 5.
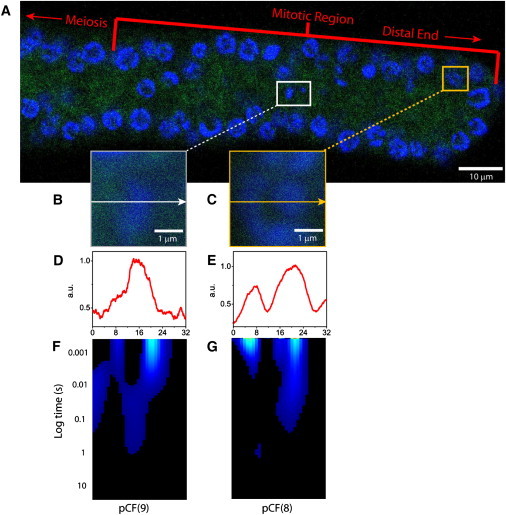
Comparison of EGFP molecular flow through a change in chromatin density (high-to-low or low-to-high) in a mitotic-versus-interphase nucleus in the C. elegans germ line. (A) The adult germ line of C. elegans expressing monomeric EGFP, with the nuclei stained with Hoechst 33342. (B) Overlay of free EGFP and Hoechst 33342 localization in the plane of the 3.3 μm line drawn in a nucleus actively undergoing mitosis in the mitotic zone. (C) Overlay of free EGFP and Hoechst 33342 localization in the plane of the 3.3 μm line drawn in an interphase nucleus in the mitotic zone. (D) Intensity profile of the Hoechst 33342 stain across the line measured in the nucleus actively undergoing mitosis. (E) Intensity profile of the Hoechst 33342 stain across the line measured in the interphase nucleus. (F) The pCF(9) carpet derived for intranuclear diffusion between different adjacent density chromatin environments in a nucleus actively undergoing mitosis. (G) The pCF(8) carpet derived for intranuclear diffusion between different adjacent density chromatin environments in the interphase nucleus.
Discussion
We first set the rules on how the pCF method can distinguish a region that is penetrable from impenetrable, and through in vivo experiment, demonstrate how the intricacies of each type of region can be elucidated. Here we show mitotic chromatin to be penetrable, allowing continuous flow of EGFP through the chromatin network, which is in contrast to the interphase scenario, where the chromatin becomes impenetrable, causing disconnect molecular flow (14). The distinct route of molecular flow measured in mitotic nuclei was first probed in live CHO-K1 cells under physiological conditions. In these experiments, we observe characteristic arcs in the derived pCF carpets, which mirror the shape of the chromosome being measured as a barrier to diffusion.
The continuity of molecular flow caused by the mitotic chromatin is suggestive of a passive mechanism. This is in contrast to the interphase chromatin, where the intermittent bursts of EGFP molecules observed to flow across a high density bundle of chromatin caused us to speculate that the nuclear architecture of this cell cycle stage could be actively regulated. Accordingly, the mitotic mode of regulation of molecular flow (arc) is not sensitive to energy depletion, whereas the intermittent communication (burst) in interphase is completely turned off upon the same treatment. The apparently paradoxical concomitance of energy-dependent but random motion of EGFP molecules in the interphase nucleus can be explained by the fact that chromatin structural dynamics is, in part, the result of default opening and closing events of the chromatin fiber. Most of these events are known to be caused by ATP-dependent local activities on DNA (19,20).
In particular, it has been shown that EGFP-labeled chromatin in CHO-K1 cells exhibit periods of constrained motion alternating with occasional energy-dependent curvilinear leaps that last 0.3–2.0 s (21). The energy-dependence and timing of these leaps are in full agreement with our results in interphase nuclei. Most notably, it has also been established that these activities are turned off during mitosis (20), which is in agreement with the observation reported here for mitotic nuclei, that the characteristic arc-shapes are not affected by energy depletion. Thus, depending on chromatin conformation, the molecular flow of inert molecules (EGFP) throughout the nucleus may be directed by either mere physical obstruction (mitosis) or a higher-order regulation imparted by energy-consuming activities on chromatin (interphase).
Finally, we used the C. elegans germ line as a model to observe concomitantly the distinct flow patterns characteristic of mitosis and interphase within the same organism, as well as to establish whether these two regulatory mechanisms are conserved in a different biological system. We found that the molecular flow and mechanism of regulation imparted by chromatin (i.e., the passive obstruction in mitosis and bursts in interphase), initially observed in CHO-K1 cells, was also found in the C. elegans germ line. This in turn suggests that, despite differences in complexity, chromatin content, and nuclear volume, the structural and functional rearrangements that chromatin must undergo during the cell cycle are retained on a more widespread level than cell type or organism. Fundamental questions remain as to which chromatin structural components (in the case of mitosis) or chromatin-dependent activities/arrangements (in the case of interphase) are responsible for the observed regulation of molecular flow within the nucleus.
An additional contribution can come from the intranuclear ionic environment, given the dramatic conformational changes of chromatin in response to small changes in the ionic strength reported in vitro (22). In terms of in vivo studies, an isolated report on the distribution of cations in nuclei concluded that during the transition from interphase to mitosis, Ca2+ and Mg2+ concentrations increase approximately three- to fourfold (23). The scarcity of experiments conducted in vivo stems from the fact that many of the intranuclear environment parameters (e.g., the ionic strength) are strictly related to cell viability and cannot tolerate change. We thus believe that state-of-art biophysical approaches for in vivo imaging like the pCF method used here should be combined to a back-to-basics approach in vitro, where we are able to dissect the molecular details of the chromatin-driven regulation of diffusion.
Conclusions
In conclusion, we employed the pair correlation method to study the molecular diffusion of EGFP in the nuclei of mitotic cells, and compared the results to the flow pattern previously characterized for nuclei of interphase cells. The mitotic chromatin was found to allow delayed but continuous molecular flow of EGFP in and out of a high chromatin density region. This is in contrast to interphase chromatin, which regulates molecular flow by means of a mechanism which intermittently turns on and off, generating discrete bursts of EGFP. We show that the interphase bursts are maintained by metabolic energy, whereas the mitotic mechanism of regulation responsible for the arc is not sensitive to ATP depletion. These two distinct routes of molecular flow were concomitantly measured in the C. elegans germ line, which indicates a conservation of mechanism on a scale more widespread than cell type or organism.
Acknowledgments
The authors thank Milka Stakic for cultivating and for transfection of the CHO-K1 cells. We thank Amanda Cinquin for preparing the C. elegans germ line samples.
This work was supported by grant Nos. NIH-P41-RRO3155, P50-GM076516 , and NIH-U54 GM064346, Cell Migration Consortium (to M.A.D. and E.G.).
References
- 1.Misteli T. Concepts in nuclear architecture. Bioessays. 2005;27:477–487. doi: 10.1002/bies.20226. [DOI] [PubMed] [Google Scholar]
- 2.Gorski S.A., Dundr M., Misteli T. The road much traveled: trafficking in the cell nucleus. Curr. Opin. Cell Biol. 2006;18:284–290. doi: 10.1016/j.ceb.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 3.Seksek O., Biwersi J., Verkman A.S. Translational diffusion of macromolecule-sized solutes in cytoplasm and nucleus. J. Cell Biol. 1997;138:131–142. doi: 10.1083/jcb.138.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Phair R.D., Misteli T. High mobility of proteins in the mammalian cell nucleus. Nature. 2000;404:604–609. doi: 10.1038/35007077. [DOI] [PubMed] [Google Scholar]
- 5.Misteli T. Protein dynamics: implications for nuclear architecture and gene expression. Science. 2001;291:843–847. doi: 10.1126/science.291.5505.843. [DOI] [PubMed] [Google Scholar]
- 6.Tini M., Benecke A., Chambon P. Association of CBP/p300 acetylase and thymine DNA glycosylase links DNA repair and transcription. Mol. Cell. 2002;9:265–277. doi: 10.1016/s1097-2765(02)00453-7. [DOI] [PubMed] [Google Scholar]
- 7.Martin C., Zhang Y. The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- 8.Karpova T.S., Kim M.J., McNally J.G. Concurrent fast and slow cycling of a transcriptional activator at an endogenous promoter. Science. 2008;319:466–469. doi: 10.1126/science.1150559. [DOI] [PubMed] [Google Scholar]
- 9.Wachsmuth M., Weidemann T., Langowski J. Analyzing intracellular binding and diffusion with continuous fluorescence photobleaching. Biophys. J. 2003;84:3353–3363. doi: 10.1016/S0006-3495(03)70059-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsien R.Y. The green fluorescent protein. Annu. Rev. Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 11.Braga J., Desterro J.M., Carmo-Fonseca M. Intracellular macromolecular mobility measured by fluorescence recovery after photobleaching with confocal laser scanning microscopes. Mol. Biol. Cell. 2004;15:4749–4760. doi: 10.1091/mbc.E04-06-0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grünwald D., Martin R.M., Cardoso M.C. Probing intranuclear environments at the single-molecule level. Biophys. J. 2008;94:2847–2858. doi: 10.1529/biophysj.107.115014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dross N., Spriet C., Langowski J. Mapping eGFP oligomer mobility in living cell nuclei. PLoS ONE. 2009;4:e5041. doi: 10.1371/journal.pone.0005041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hinde E., Cardarelli F., Gratton E. In vivo pair correlation analysis of EGFP intranuclear diffusion reveals DNA-dependent molecular flow. Proc. Natl. Acad. Sci. USA. 2010;107:16560–16565. doi: 10.1073/pnas.1006731107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cardarelli F., Serresi M., Beltram F. In vivo study of HIV-1 Tat arginine-rich motif unveils its transport properties. Mol. Ther. 2007;15:1313–1322. doi: 10.1038/sj.mt.6300172. [DOI] [PubMed] [Google Scholar]
- 16.Digman M.A., Gratton E. Imaging barriers to diffusion by pair correlation functions. Biophys. J. 2009;97:665–673. doi: 10.1016/j.bpj.2009.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cardarelli F., Gratton E. In vivo imaging of single-molecule translocation through nuclear pore complexes by pair correlation functions. PLoS ONE. 2010;5:e10475. doi: 10.1371/journal.pone.0010475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hubbard, E. J., and D. Greenstein. 2005. Introduction to the germ line. WormBook. DOI:10.1895/wormbook. 1.18.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- 19.Soutoglou E., Misteli T. Mobility and immobility of chromatin in transcription and genome stability. Curr. Opin. Genet. Dev. 2007;17:435–442. doi: 10.1016/j.gde.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Woodcock C.L., Ghosh R.P. Chromatin higher-order structure and dynamics. Cold Spring Harb. Perspect. Biol. 2010;2:a000596. doi: 10.1101/cshperspect.a000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levi V., Ruan Q., Gratton E. Chromatin dynamics in interphase cells revealed by tracking in a two-photon excitation microscope. Biophys. J. 2005;89:4275–4285. doi: 10.1529/biophysj.105.066670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bednar J., Horowitz R.A., Woodcock C.L. Chromatin conformation and salt-induced compaction: three-dimensional structural information from cryoelectron microscopy. J. Cell Biol. 1995;131:1365–1376. doi: 10.1083/jcb.131.6.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Strick R., Strissel P.-L., Levi-Setti R. Cation-chromatin binding as shown by ion microscopy is essential for the structural integrity of chromosomes. J. Cell Biol. 2001;155:899–910. doi: 10.1083/jcb.200105026. [DOI] [PMC free article] [PubMed] [Google Scholar]


