Abstract
microRNAs (miRNAs) are newly discovered, small, non-coding ribonucleic acids (RNAs) that play critical roles in the regulation of host genome expression at the post-transcriptional level. During last two decades, miRNAs have emerged as key regulators of various biological processes including immune cell lineage commitment, differentiation, maturation, and maintenance of immune homeostasis and normal function. Thus, it is not surprising that dysregulated miRNA expression patterns have now been documented in a broad range of diseases including cancer, inflammatory, and autoimmune diseases. This rapidly emerging field has revolutionized our understanding of normal immunoregulation and breakdown of self-tolerance. This review focuses on the current understanding of miRNA biogenesis, the role of miRNAs in the regulation of innate and adaptive immunity, and the association of miRNAs with autoimmune diseases. We have discussed miRNA dysregulation and the potential role of miRNAs in systemic lupus erythematosus (SLE), rheumatoid arthritis (RA), and multiple sclerosis (MS). Given that most autoimmune diseases are female-predominant, we have also discussed sex hormone regulation of miRNAs in inflammatory responses, with an emphasis on estrogen, which has now been shown to regulate miRNAs in the immune system. The field of miRNA regulation of mammalian genes has tremendous potential. Identification of specific miRNA expression patterns in autoimmune diseases and further a comprehensive understanding of the role of miRNA in disease pathogenesis offers promise of not only novel molecular diagnostic markers, but also new gene therapy strategies for treating SLE and other inflammatory autoimmune diseases.
1. Introduction
The recent discovery of the gene regulatory role of small non-coding RNAs, miRNAs has led to a paradigm shift in the understanding of expression and function of mammalian genome. The first miRNA, lin-4 was identified in the nematode C. elegans at 1993.1 However, the abundance and ubiquitous nature of miRNAs in different species was not explored until seven years later with the identification of the second miRNA, let-7 in C. elegans,2 which led to large scale screening and identification of new miRNAs in invertebrates and vertebrates.3–6 Currently, by employing a combination of different experimental strategies and robust computational prediction, a total of 17,341 mature miRNAs in 142 species (miRBase database, version 16.0, September 2010) have been identified.7 Thus far, 1,223 human miRNAs and 1,055 mouse miRNAs have been identified in the database, which validate the previous computation-based prediction that miRNA genes constitute about 3% of the human genome and that the total number of human miRNA genes may be in the range of thousands.8, 9 It is estimated that a single miRNA may regulate hundreds to over thousands of target genes, and therefore about 30%–92% of human genes are likely regulated by miRNA.10–13 The powerful gene regulatory role of miRNAs is now well recognized. The expression and function of miRNAs are essential for the development of diverse physiological systems and the maintenance of the cellular homeostasis and normal function.14–18 The field of miRNA research gained widespread attention with the recognition of aberrant expression and/or function of miRNAs in a broad range of human diseases including various types of cancer, metabolic, neurodegenerative, infectious, chronic inflammatory, and autoimmune diseases.19–23 This review will summarize the current knowledge regarding the role of miRNA in inflammatory autoimmune disease, with an emphasis on SLE.
2. miRNA biogenesis and action
Most mammalian miRNA genes (about 80%) have been identified in the intron region of either protein coding or non-protein coding transcripts. Only a small number of miRNA genes (20%) are located in the exon region of non-coding RNAs.24, 25 Some miRNA genes could be either exonic or intronic miRNAs depending on the alternative splicing pattern of host genes.24, 26 The miRNA biogenesis pathways and their regulation have been extensively studied in last decade and well reviewed in recent publications.6, 26–30 As depicted in figure 1, miRNA genes are mainly transcribed by RNA polymerase II into primary miRNA transcripts (pri-miRNAs), which are usually hundreds to thousands of nucleotides (nts) long and contain one or several hairpin loop structures.26, 27, 31 In the canonical miRNA biogenesis pathway, the pri-miRNAs are cleaved in the nucleus by a microprocessor complex that is composed of the nuclear RNase III enzyme, Drosha and the double-stranded-RNA-binding protein, DiGeorge syndrome critical region protein 8 (DGCR8) to produce precursor miRNAs (pre-miRNAs).32, 33 In an alternative, non-canonical miRNA biogenesis pathway, a new class of miRNA precursors, termed “mirtrons”, are produced independent of microprocessor processing.34–36 Unlike pri-miRNAs, the mirtrons do not have the lower stem-loop structure and the flanking single stranded segments that are crucial for the binding and processing by the DGCR8/Drosha microprocessor. Therefore, the mirtrons bypass Drosha processing and are spliced by spliceosome to yield branched pre-mirtrons, which then go through lariat-mediated debranching to generate pre-miRNAs (Figure1).30, 34, 35
Figure 1. miRNA biogenesis and action in animal cells.
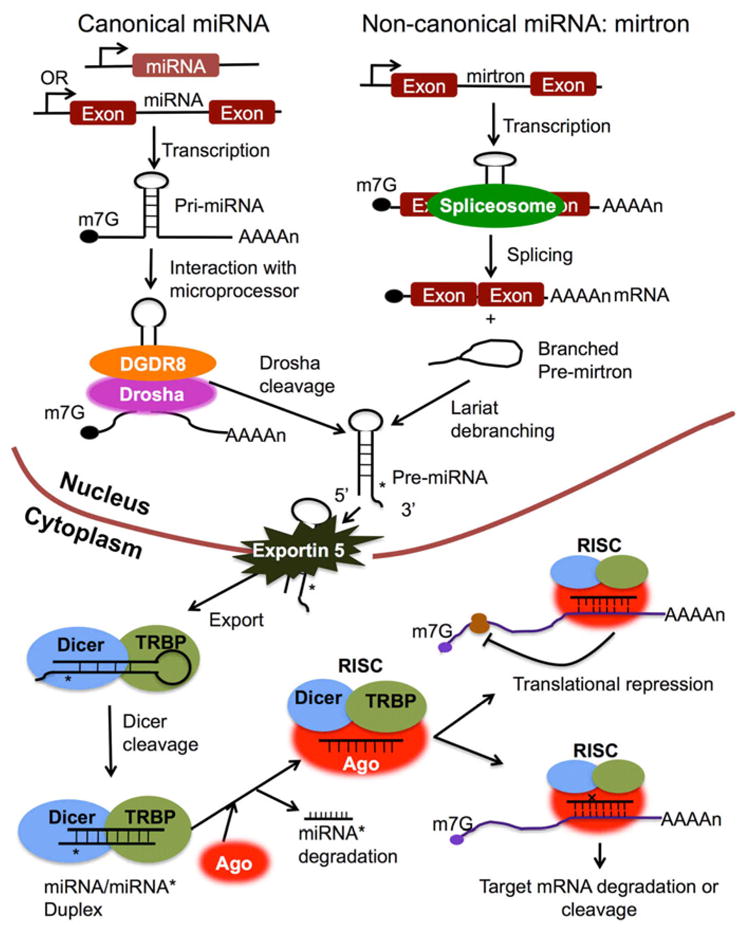
Mature miRNAs are short (with average about 22 nts long), single stranded RNA molecules derived from long primary transcripts, pri-miRNAs through sequential processing in both the nucleus and cytoplasm. In the canonical miRNA biogenesis pathway, the pri-miRNA is processed by DGCR8/Drosha microprocessor to generate pre-miRNA. The 33nts lower stem loop structure, the terminal loop, and the two single stranded segments flanking the hairpin loop in pri-miRNA are together required for the binding of pri-miRNA with DGCR8 and Drosha cleavage at the accurate site . In non-canonical miRNA biogenesis pathway, pre-miRNA is generated from small intronic miRNA, mirtron, through spliceosome splicing and then lariat-mediated debranching. Exportin 5 transports the pre-miRNA that derived from either canonical miRNA or mirtron from the nucleus into the cytoplasm. In the cytoplasm, the pre-miRNA is further processed by Dicer to generate miRNA/miRNA* duplex, which is then loaded into Argonaute protein and forms RISC together with Dicer and TAR RNA binding protein (TRBP). Although both strands of miRNA/miRNA* duplex have the potential to be functional miRNA, only guided strand remains in the RISC as a mature miRNA. Depending on the complementary degree between miRNA and the 3’UTR sequence of target gene, miRNA regulates the expression its target genes through either translation inhibition or mRNA degradation/cleavage. The imperfect sequence complementary between miRNA and target mRNA usually leads to translational inhibition of target gene without affecting the amount of mRNA. While, the perfect complementary match between miRNA and target mRNA usually results in the repression of target genes by mRNA decay or cleavage.
After being exported from the nucleus into cytoplasm by the nucleocytoplasmic shuttle protein Exportin-5, pre-miRNAs are further processed by the RNAIII enzyme, Dicer to yield ~22nts long, imperfect matched miRNA/miRNA* duplexes, which are then loaded into the Argonaute (Ago) protein to generate RNA-induced silencing complex (RISC). The guided strand of the miRNA/miRNA* duplex remains in the RISC as a mature miRNA, and the complementary strand (the passenger strand miRNA*) is degraded.26, 28
Once loaded onto RISC, the mature miRNA will interact with the 3’ untranslated region (UTR) of its target messenger RNA (mRNA) to regulate gene expression. The 2–8 nts seed region of miRNA is crucial for target recognition. The complementary degree between the miRNA seed region and the target mRNA 3’UTR determines the mechanism of miRNA-mediated gene regulation: translation repression or miRNA cleavage and degradation.6, 11, 28 High-throughput proteomic analysis revealed that miRNA-regulated genes are mainly subjected to translational inhibition, and those highly translationally repressed genes also display mRNA destabilization.37–39
3. The role of miRNA in immune system development and normal function
During the last several years, mounting evidence has emerged to show that miRNAs are critical for not only the development of the immune system, but also for the function of both innate and adaptive arms of the immune system.18, 40–45 In the following sections, we will discuss the current relevant findings with regard to the roles of miRNAs in the regulation of innate immunity, adaptive immunity, autoimmunity, and inflammation.
3.1 miRNAs in the regulation of innate immunity
The cells of the innate immune system include granulocytes, monocyte-derived macrophages and dendritic cells. Of the granulocytes, neutrophils constitute the largest percentage of peripheral white blood cells and are regarded as the first cellular line of defense against invading pathogens. The Toll-like receptors (TLRs) on the membrane of monocytes recognize and bind to specific microbial products called pathogen-associated molecular patterns (PAMPs) and then trigger downstream signaling pathway to initiate inflammatory responses.46, 47 The TLR signaling is tightly controlled in vivo by different classes of negative regulators to prevent overwhelming inflammation.48 Recent studies revealed that miRNAs not only regulate innate immune cell development, but also fine-tune the innate immune response, thereby adding a new layer of negative feedback regulation of TLR signaling (Figure 2).40, 42, 43
Figure 2. miRNAs in the regulation of innate immunity.
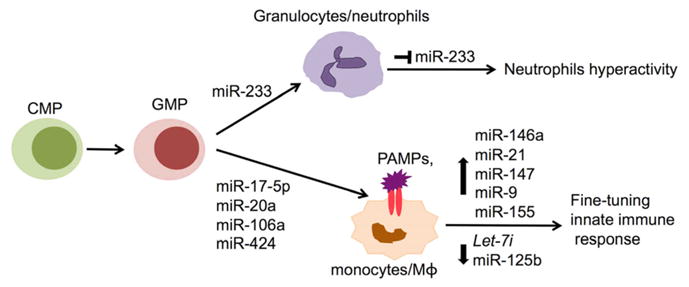
Illustrates the potential role of various miRNAs that have been reported to be involved in the development and function of neutrophils and monocyte/macrophage (Mφ). Please see the text for a detailed explanation. CMP, myeloid progenitor; GMP, granulocyte-monocyte progenitor; PAMPs, pathogen-associated molecular patterns.
During granulopoiesis, miR-223 was highly induced by the transcription factor, CCAAT enhancer binding protein α (C/EBPα), which in turn resulted in the repression of transcription factors nuclear factor 1 A-type (NFI-A) and E2F1, and led to enhanced granulocytes differentiation.49, 50 Nevertheless, an in vivo study with miR-223 knockout mice revealed that miR-223 acted as a negative regulator of granulocyte differentiation by targeting another transcription factor, myocyte-specific enhancer factor 2C (mef2c).51 The miR-223 knockout mice surprisingly had increased number of granulocyte progenitors, which led to increased expansion of granulocytes. Furthermore, the miR-223 knockout mice spontaneously develop inflammatory lung pathology due to the hyperactivity of neutrophils. These data suggested that although the role of miR-223 in neutrophils development and function is apparent, further work is warranted to understand the precise function of miR-223 in the regulation of granulopoiesis and granulocyte function at different differentiation stages.
Monocyte-derived macrophages play critical roles in innate immune responses. Monocytopoiesis is controlled by a circuitry loop consisting of miR-17-5p, miR-20a, miR-106a, acute myeloid leukaemia-1 (AML-1; also known as Runt-related transcription factor 1, Runx1), and macrophage colony-stimulating factor receptor (M-CSFR).52 During monocytopoiesis, the expression of miR-17-5p, miR-20a, and miR-106a was decreased. In parallel, their target gene, AML-1, was upregulated to promote the expression of M-CSFR, which plays a pivotal role in monocyte–macrophage differentiation and maturation. On the other hand, AML-1 also negatively regulated the expression of miR-17-5p-20a-106a by binding to the promoter of the miR-17-92 and miR-106a-92 cluster, which suggests a mutual negative feedback regulation loop in monocytopoiesis.52 In addition to the above miRNAs, miR-424 was shown to enhance monocyte differentiation by translational repression of NFI-A, which also led to the activation of M-CSFR.53
The importance of miRNA in the regulation of innate immune responses is evident in a series of studies in which various miRNAs were shown to alter dramatically in response to PAMPs or inflammatory cytokines stimulation.42, 54–57 In a pioneer study, Taganov et al. have shown that miR-146 was rapidly upregulated in response to lipopolysaccharide (LPS) stimulation in human monocytic cells, and acted as a negative feedback regulator of TLR signaling by targeting tumor necrosis factor receptor-associated factor 6 (TRAF6) and IL-1 receptor-associated kinase 1 (IRAK1).57 The upregulation of miR-146a was also observed in monocytes in response to the activation of TLR4, TLR2, and TLR5 signaling pathways or inflammatory cytokine such as tumor necrosis factor (TNF)α and IL-1β stimulation.57 In addition to miR-146a, other miRNAs were identified as negative regulator of TLR signaling. miR-155 was induced in macrophages in response to both bacterial and viral derived antigens that activate TLR4, TLR2, TLR3, or TLR9.54, 55, 57 In a different study, miR-155 was shown to downregulate inflammation by targeting myeloid differentiation primary response protein 88 (MyD88), a key adaptor protein in the TLR signaling pathway.58 miR-21 was found to negatively regulate LPS- activated TLR4 signaling by targeting the tumor suppressor, PDCD4, which in turn decreased nuclear factor-κB (NF-κB) activation and IL-10 production.59 miR-147 was induced following the activation of TLR2, TLR3, or TLR4, and acted as a negative regulator to prevent excessive inflammatory responses in macrophages in the lungs of mice.60 miR-9 was induced by LPS in both neutrophils and macrophages and negatively regulated NF-κB dependent inflammatory responses by fine-tuning the expression of NF-κB.61 Some miRNAs such as Let7i and miR-125b were decreased during innate immune responses.55, 56 LPS stimulation and parasite Cryptosporidium parvum infection of cultured human cholangiocytes decreased let7i expression in a MyD88 and NF-κB- dependent manner, which led to the upregulation of TLR4 protein and enhanced immune responses. Accordingly, there was decreased parasite infection in let7i antisense inhibitor transfected cells and increased parasite burden in let7i precursor-transfected cells when compared to control.56 Decreased miR-125b expression in response to LPS may contribute to elevated TNFα level in activated macrophages as miR-125b was shown to target TNFα mRNA.55
Taken together, these reports clearly demonstrate the importance of miRNA in the regulation of innate immune cell development and in fine-tuning innate immune responses to prevent overwhelming inflammation in the body.
3.2 miRNAs in the regulation of adaptive immunity
T and B lymphocytes are major cellular components of adaptive immunity. The overall significance of miRNA in the regulation of adaptive immunity is clearly evident in studies in which the disruption of miRNA biogenesis in lymphocyte progenitors was found to impair the development of T and B lymphocytes. Disruption of miRNA synthesis by conditional depletion of Dicer in early stages of T lymphocytes impaired T cell development with reduced T cell numbers in thymus and peripheral lymphoid organs, and caused aberrant T helper (Th) cell differentiation and cytokine production.62, 63 Depletion of Dicer in early B cell progenitors resulted in a complete developmental block of B cells at pro- to pre-B transition and affected antibody diversity, which was partially due to the loss of miR-17-92 mediated repression of the proapoptotic protein, Bim.64
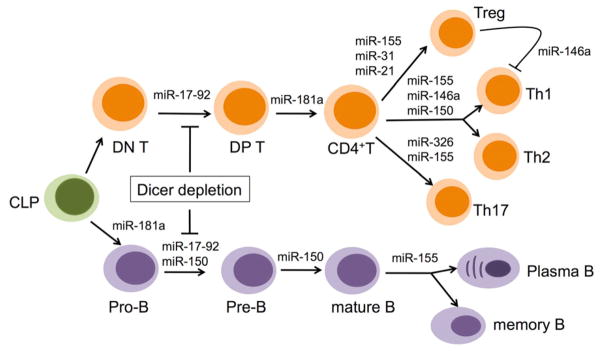
The distinct role of individual miRNA in the regulation of lymphocyte development and function is also documented in recent studies (Figure 3). miR-155 is upregulated in both T and B cells upon activation and is required for maintaining lymphocyte homeostasis and normal immune function by targeting different genes including c-maf, Pu.1, and activation-induced cytidine deaminase (AID).65–68 Although miR-155 knockout mice apparently possess normal lymphocyte development, they have defective T and B immunity including abnormal Th1/Th2 differentiation ratio with increased Th2 polarization, increased Th2 cytokine production, a reduced number of Treg cell, decreased germinal center responses, and low numbers of IgG class-switched plasma cells and memory cells.65–67, 69, 70 miR-181a also plays an important role in the regulation of both of T and B cell development. While ectopic expression of miR-181a in hematopoietic progenitor cells in vitro resulted in an increase of B-lineage cells without affecting T-lineage cells, transfer of bone marrow cells overexpressing miR-181 into irradiated host mice led to a substantial increase of CD19+ B cells accompanied by a significant decrease in T cells.71 miR-181a was fund to target phosphatases SHP-2, PTPN-22, and DUSP5/6, negative regulators of TCR signaling, and therefore plays a critical role in T cell maturation by regulating T cell receptor (TCR) signaling strength and then tuning T cells sensitivity to positive and negative selection.72 Inhibiting of miR-181a expression in immature T cells impaired both positive and negative selection during T cell development.72 miR-150 is selectively expressed in mature resting B and T cells but not in their progenitors.73, 74 Overexpression of miR-150 prematurely in hematopoietic stem cells blocked the B cell development at pro- to pre-B stages without affecting T cells, granulocytes and macrophage maturation.74, 75 Further, Xiao et al. demonstrated that miR-150 controls B cell development by targeting transcription factor, c-myb, in a dose dependent manner. Mice with miR-150 deficiency have increased number of B1 cells in spleen and peritoneal cavity and enhanced humoral responses.74 In contrast to miR-150, miR-17-92 is highly expressed in T and B precursors and decreased following maturation. Overexpression of miR-17-92 in lymphocyte progenitors promoted T and B lymphocyte proliferation and resulted in the development of lymphoma by targeting proapoptotic proteins, Bim and phosphatase and tensin homolog (PTEN). On the other hand, ablation of miR-17-92 inhibited B cell development at the pro-B to pre-B transition, which is attributed to apoptotic death of Pro-B cells due to augmented expression of proapoptotic protein Bim that is targeted by miR-17-92.76, 77
Figure 3. miRNAs in the regulation of adaptive immunity.
This figure depicts various miRNAs that have been reported in the development and differentiation of T cells (upper part of the figure), and B cells (lower part of the figure). Please see the text for a detail explanation. CLP, common lymphoid progenitor; DN, CD4− CD8− double negative; DP, CD4+CD8+ double positive; Treg, regulatory T cell.
Regulatory T cells (Tregs) suppress the activation of effector T cells to maintain immune system homeostasis and tolerance to self-antigens. Various miRNAs have been shown to play critical role in the regulation of Treg development and/or function. miR-155 knockout mice have a reduced number of Tregs in both thymus and peripheral lymphoid tissues. However, the miR-155 deficient Tregs have competent suppressive activity in vivo.70 This suggests that miR-155 is critical for Tregs development, but not essential for Treg suppressive functions. In a different study, miR-155 was shown to play a role in Treg-mediated suppression activity by regulating the sensitivity of effector CD4+ Th cells. miR-155 inhibition in CD4+ Th cells resulted in enhanced natural Treg (nTreg) mediated suppression.78 miR-146a has been shown to be upregulated in Th1 cells and abolished in Th2 cells during T cell differentiation.73 A recent study revealed that miR-146a is highly expressed in Tregs and selectively controls Treg-mediated suppression of interferon (IFN)γ-dependent Th1 responses and inflammation by targeting signal transducer and activator of transcription (STAT)-1.79 The mice with miR-146a ablation in Tregs have elevated production of Th1 cytokine IFNγ and develop severe Th1-mediate lesions in a variety of organs leading to conjunctivitis, blepharitis, and dermatitis.79 In addition, miR-31 and miR-21 have also been shown to regulate Treg development by negatively and positively, respectively, regulating the expression of Foxp3, a critical transcription factor in Treg development.80
Many recent studies have shown that IL-17, a powerful neutrophilic inflammation-inducing cytokine, is involved in autoimmune diseases.81 Two miRNAs, miR-326 and miR-155, have now been shown to be associated with the Th17 differentiation and induction of IL-17. miR-326 was shown to target Ets, a transcription factor that negatively regulates Th17 development, which consequently led to the promotion of Th17 differentiation and IL-17 production.82 miR-155 was required for the production of Th17-promoting cytokines. When immunized with experimental allergic encephalitis (EAE)-inducing myelin oligodendrocyte glycoprotein35–55 (MOG35–55), miR-155 knockout mice had a marked decrease in number of Th-17 cells compared to wild type mice, and displayed remarkably resistance to (MOG35–55)-induced EAE.83
In summary, the above emerging data clearly show that proper expression and function of miRNAs is essential for T and B cell development, differentiation, and function. It is therefore important that the expression of miRNA must be tightly regulated for the maintenance of immune homeostasis. Conceivably, dysregulation of miRNA can promote the induction of autoimmunity.
4. miRNAs in the development and prevention of autoimmunity
To maintain tolerance, there are many checkpoints at both central and peripheral lymphoid organ levels. These checkpoints ensure that autoreactive T and B cells that are routinely and randomly generated during lymphogenesis are either deleted or silenced.84, 85 However, in some circumstances, the self-reactive lymphocytes escape the checkpoints and survive in peripheral lymphoid tissues. Once being activated, these autoreactive cells mount a devastating attack on self-tissues, leading to the induction of autoimmune diseases. With the increased recognition that miRNAs are stringently regulated in the cells of the normal immune system to maintain immune homoeostasis and normal immune function, it is conceivable that dysregulated miRNA expression will lead to the immune tolerance breakdown and the development of autoimmune diseases. Disruption of miRNA synthesis by depleting Drosha or Dicer in Tregs led to the complete loss of immune regulatory function of Tregs, which resulted in the development of inflammatory disease and fatal autoimmunity.86–88 Selective ablation of miR-146a in Treg cells resulted in the breakdown of immune tolerance and the development of autoimmune diseases in mice. This was mainly due to a selective loss of Treg mediated suppression of Th1 inflammatory responses in mice with miR-146a deficient Tregs.79 Overexpression of miR-17-92 in lymphocytes induced lymphoproliferative disease and autoimmunity in miR-17-92 transgenic mice as the result of miR-17-92-mediated repression of Bim and PTEN, which play critical roles in immune tolerance mechanism.76, 84 Moreover, the unique dysregulated miRNA expression patterns have been identified in human patients with SLE, RA, and MS. Selected disease-associated miRNAs have been shown to play critical pathogenic roles in the development of autoimmune diseases (Table 1).
Table 1.
| Diseases | miRNAs | Pathogenic contribution | Reference |
|---|---|---|---|
| Systemic lupus erythematosus | miR146a, ↓PBMC | Targets STAT-1 and IRF-5, negative regulator of Type I interferon pathway | 100 |
| miR-148a and miR-21 ↑CD4+ T cells | Target DNA methyltransferase 1 (DNMT1) directly and indirectly, respectively, induces DNA hypomethylation and the expression autoimmune-associated genes. | 99 | |
| miR-125a ↓ T cells | Targets Kruppel-like factor 13 (KLF13), Negative regulator of inflammatory chemokine RANTES | 101 | |
| Other miRNAs dysregulated in SLE including miR-223, miR-31, miR-371-5p, miR-473-5p. | Unknown | 97, 98, 100, 102 | |
| Rheumatoid Arthritis | miR-146a, ↑PBMC, CD4T cells, Th-17 cells, synovial fibroblasts (RASFs) and/or synovial tissues | Targets FAF1, Negative regulator of T cell apoptosis | 106, 116– 119 |
| miR-155, ↑PBMC, Th-17 cell, RASFs and synovial tissues | Targets Matrix metalloproteinase (MMP)-3/1 in RASFs, Regulation of inflammation and potentially involved in RASFs mediated tissue damages | 106, 116, 117 | |
| miR-124a, ↓ synoviocytes | Targets cyclin-dependent kinase 2 (CDK-2) and chemokine MCP-1, Negative regulator of cell proliferation and MCP-1 secretion | 120 | |
| Other miRNAs dysregulated in RA including miR-223, miR-132, and miR-16. | Unknown | 106, 117, 119, 121 | |
| Multiple sclerosis | miR-326, ↑ CD4 T cells | Targets Ets-1, Promotes Th-17 cell differentiation | 82 |
| miR-17-5P, ↓ whole blood and ↑CD4 T cells | Potentially involved in the regulation of T cell activation | 125, 126 | |
| miR-20a, ↓ whole blood | Potentially involved in the regulation of T cell activation | 126 | |
| miR-34a, miR-155 and miR-326, ↑ MS lesion | Targets CD47, promotes phagocytosis of myelin by releasing macrophage from inhibitory signaling. | 122 | |
| Other miRNAs dysregulated in MS including miR-25, niR-18b, and miR-599. | Unknown | 123, 124, 127 |
miRNAs in human inflammatory autoimmune diseases
4.1 miRNA in SLE
SLE is a classical autoimmune disease that is characterized by the production of autoantibodies against a wide-range of nuclear and phospholipid antigens and multi systemic organ injuries. Numerous immunological abnormalities including dysregulated B and T lymphocyte functions, aberrant TLR, type I IFN, NF-κB signaling pathways, and abnormal interactions among immune cells have been identified in human and murine lupus.89–92 In addition to the existence of genetic loci with SLE predisposition, epigenetic regulatory defects such as abnormal DNA methylation, histone modification have also been shown to play important role in lupus pathogenesis.93–95 Recent studies have now revealed a potential role of miRNA in SLE, thereby providing new insights into lupus pathogenesis. A computational prediction analysis of 72 lupus susceptibility genes revealed that most lupus-related genes (71 out of 72 genes) contain at least one miRNA target site for over 140 miRNAs.96 Interestingly, four lupus susceptibility genes (Roquin, B-cell lymphoma 2 (Bcl2), Bim, and PTEN), which are involved in the regulation of T cell co-stimulation, cell survival, apoptosis and/or tolerance control, were each predicted to contain over 50 miRNA target sites. This study emphasized the potential contribution of miRNA in the regulation of autoimmune genes and self-tolerance control in autoimmune lupus.96
Dysregulated miRNA expression pattern in the peripheral blood mononuclear cells (PBMCs) from patients with SLE was first reported by Dai et al. at 2007.97 They identified 16 lupus-related miRNAs that were altered specifically in patients with lupus, but not in patients with idiopathic thrombocytopenic purpura (ITP, an organ specific autoimmune disease).97 Since then, several reports have been published to show dysregulated miRNA expression profiles in PBMC, CD4+ T cells, kidney biopsy, and Epstein-Barr virus (EBV)-transformed B cell lines from human patient with lupus.98–102
Using Taqman PCR array, Tang et al. identified 42 miRNAs that were differentially expressed in PBMC from human patients with SLE when compared to healthy controls.100 Among them, miR-146a, a negative regulator of TLR signaling, was profoundly decreased (more than 6-fold) in patients with SLE versus healthy controls. Further, the authors reported that miR-146a negatively regulated the type I IFN pathway by targeting interferon regulatory factor (IRF)-5 and STAT-1. Overexpression of miR-146a in normal PBMC greatly reduced the induction of IFNα/β, and inhibition of endogenous miR-146a increased the production of IFNα/β in response to the activation of TLR-7. Therefore, decreased expression of miR-146a in PBMC may contribute to the enhanced type I IFN production in human lupus.100 In a separate study from the same group, Pan et al. identified that miR-21 and miR-148a were upregulated in CD4+ T cells from both MRL-lpr mice and human patients with lupus.99 A previous report has shown that miR-148a targets DNA (cytosine-5-)-methyltransferase 3 beta (DNMT3B),103 which led to the discovery of the role of miR-148a and miR-21 in the regulation of DNA methylation in lupus T cells. Pan et al. confirmed that miR-148a targeted DNA methyltransferase 1 (DNMT1) directly by binding to the protein coding region, and that miR-21 indirectly downregulated DNMT1 by targeting its upstream regulator, RAS guanyl nucleotide-releasing protein 1(RASGRP1). Overexpression of miR-148a and miR-21 in CD4+ T cells led to DNA hypomethylation and then increased expression of autoimmune-associated methylation-sensitive genes, CD70 and lymphocyte function-associated antigen 1 (LFA-1).99 In a more recent report, Zhao et al. reported that miR-125a was reduced in PBMCs from patients with lupus when compared to healthy controls. Further experiments revealed that miR-125a, which is mainly expressed in T cells, targeted Kruppel-like factor 13 (KLF13) gene, a critical transcription factor in the regulation of the chemokine RANTES, in a dose dependent manner. Decreased expression of miR-125a resulted in the upregulation of KLF13, which in turn contributes to the elevation of the inflammatory chemokine RANTES in lupus T cells.101 Together, the above three studies have shown that dysregulated miRNAs in lupus contribute to disease pathogenesis by regulating Type I interferon signaling pathway,100 DNA methylation,99 and RANTES expression,101 respectively.
So far, numerous dysregulated miRNAs were identified in human patients with lupus.97–100, 102 Nevertheless, many dysregulated miRNAs in lupus that were determined by microarray assay need further validation by Real-time RT-PCR or Northern blotting. Interestingly, it seems that the dysregulated miRNAs in patients with lupus that were determined in one study were not well reproduced in other studies. For example, the decrease of miR-146a in PBMC from patients with lupus was observed in Tang et al.’s report,100 but not in other studies.97, 98, 102 The discrepancy of the data generated in different human lupus miRNA studies may be due to differences in the type of samples, sensitivity of detection methods, and also the diversity in the medical history, disease severity, length of disease, and race of human patients with lupus.
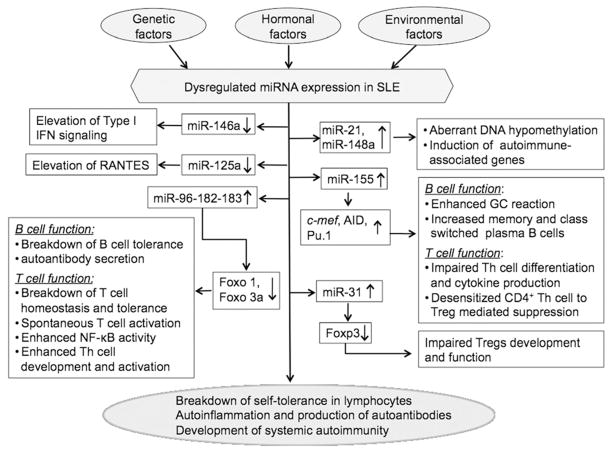
Considering that genetic factors contribute significantly to lupus etiology and miRNA expression, we have recently identified a common lupus disease-associated miRNA expression pattern in three murine autoimmune-prone models (MRL-lpr, C57BL/6-lpr, and NZB/W) with different genetic background.104 By a combination of miRNA microarray and Real-time RT-PCR analysis, we identified that the miR-182-96-183 cluster, miR-31, and miR-155 are commonly upregulated in splenocytes from three strains of lupus prone mice when compared to their respective controls. It is interesting to note that the upregulation of these common lupus-related miRNAs was evident in NZB/W mice only at an age when lupus disease is manifested, which suggested that the dysregulation of these miRNAs are lupus associated. In addition, by Real-time RT-PCR, we identified that miR-146a, miR-17-5p, and miR-101a, which have been shown to be dysregulated in human lupus or implicated the development of autoimmunity,23, 76, 99, 100, 105, 106 were also dysregulated in splenocytes and/or purified splenic lymphocytes subsets from MRL-lpr mice when compared to control MRL mice.104 While the direct contribution of these common lupus disease-associated miRNAs to the pathogenesis of murine lupus is as yet not determined, the identified targets and immune regulatory function of miR-155, miR-182-96-183 and miR-31 strongly suggest the potential contribution of these miRNAs to immune tolerance breakdown and altered B and T lymphocyte function in autoimmune-prone mice (Figure 4). 104 miR-155 plays a critical role in the regulation of B cell and T cell immunity,65–67, 83 therefore, the upregulation of miR-155 in lupus B and T lymphocytes may lead to abnormal B cell activation, enhanced germinal center responses, and abnormal inflammatory T cell development and cytokine production in human and murine lupus. The upregulation of miR-31 in T cells may correlate with the deficiency of Treg cell development/function in lupus since miR-31 targets Foxp3, which is critical for Treg cell development.80 The miR-182-96-183 cluster is primarily expressed in the retina, and rarely detected in the spleen, thymus, and other tissues from normal B6 mice.107 Importantly, miR-182 and miR-96 target forkhead transcription factor O (Foxo)1 and Foxo3a, which are newly emerged regulator of T lymphocyte homeostasis and tolerance, and T cell immune responses.108–113 T cell-specific deletion of Foxo1 gene in mice led to spontaneous T cell activation, an increased number of IFNγ, IL-17 secreting CD4+ T cells, anti-dsDNA autoantibody production, and tissue inflammation.110 Inactivation of Foxo3a in mice led to NF-κB activation, hyperactivation of Th cell with increased Th1/Th2 cytokine production and autoimmune inflammation in salivary gland, lung and kidney.109 The direct involvement of miR-182 in immune regulation is further supported by a recent report in which miR-182 was shown to be induced by IL-2 to suppress the activity of Foxo1. This in turn promoted clonal expansion of activated Th cells and IL-2 driven Th mediated immune responses.114 Extrapolation of these findings, it is therefore conceivable that the upregulation of miR-96-182-83 in lupus lymphocytes could potentially cause the decrease of Foxo1/3a, which in turn may lead to T lymphocyte hyperactivation, self-tolerance breakdown, autoantibody production, and development of autoimmunity. This view is supported by a finding of decreased Foxo1 transcript in human SLE that has been reported.115 Further, we have also observed that Foxo1 and Foxo3a protein levels are decreased in splenocytes from MRL-lpr mice when compared MRL mice (unpublished lab data).
Figure 4. An overview of dysregulated miRNAs and the potential pathogenic contribution of dysregulated miRNAs in SLE.
Multiple factors including genetic, hormonal, and environmental factors may contribute to the miRNA dysregulation in SLE. The published reports have shown that miR-146a, miR-125a, miR-21, and miR-148a contribute to lupus pathogenesis by regulating the type I interferon pathway, inflammatory chemokine RANTES expression, and DNA methylation, respectively. The potential pathogenic contribution of miR-155, miR-182-96-183 and miR-31, which are commonly upregulated in three murine lupus models, is suggested by either their well-defined function in immunity or the role of their target genes in the control of immune tolerance to self-antigens. The targets of these specific miRNAs are indicated. Please see the text for a detail explanation.
The current studies have revealed that various miRNAs are dysregulated in human and murine lupus. However, the functional significance of most of lupus-related miRNAs remains elusive, which impedes our comprehensive understanding of the role of miRNA in lupus pathogenesis.
4.2 miRNA in RA
RA is an autoimmune disease that causes chronic inflammation primarily in the joints, as well as in the tissue around the joints and other organs in the body. In reports from different research groups, miR-146a and miR-155 were found consistently to be upregulated in synovial fibroblasts (RASFs),116 PBMC,106 synovial fluid,117 PBMC-derived CD4+ T cells,118 and Th-17 cells from patients with RA when compared to healthy control or patients with osteoarthritis (OA).119 Stanczyk et al. confirmed that miR-155 targets matrix metallopeptidase (MMP)3 and MMP1. This suggests that elevated miR-155 in RA may have a protective function in RASFs-mediated tissue damages by inhibition of MMP1 and MMP3.116 miR-146a has been shown to negatively regulate inflammatory immune responses by targeting TRAF6 and IRAK-1.57 Although miR-146a was upregulated in active RA disease, its target genes, TRAF6 and IRAK-1, had no changes in RA when compared to healthy controls. It is plausible that the increased level of miR-146a in RA does not function effectively as a negative regulator of inflammation, thereby resulting in constitutive production of inflammatory TNFα in RA.106 Recent reports revealed that miR-146a was increased in CD4+ T cells and correlated with the levels of the inflammatory cytokines TNFα and IL-17, and that elevated miR-146a expression may contribute to RA pathogenesis by suppressing T cell apoptosis and enhancing the differentiation of IL-17 cells, respectively.118, 119 By comparing to patients with OA, Nakamachi Y et al. found that miR-124a was significantly decreased in RA synoviocytes.120 Further investigation revealed that miR-124a targeted cyclin-dependent kinase 2 (CDK-2). Overexpression of miR-124a in RA synoviocytes significantly repressed cell proliferation and arrested the cell cycle at G1 phase. In addition, miR-124a was also shown to target monocyte chemoattractant protein 1 (MCP-1), which is elevated in RA synovial fluid and tissues and has been implicated in the RA pathogenesis by recruiting mononuclear phagocytes into joint. Taken together, decreased miR-124a expression may contribute to RA pathogenesis by increasing cell proliferation and MCP-1 production.120
In addition, several other miRNAs including miR-223, miR-16, and miR-132 were found to be dysregulated in human patients with RA.106, 117, 119, 121 However, the potential contribution of these miRNAs in RA inflammation and pathogenesis is not known yet, therefore, further investigation is warranted.
4.3 miRNA in MS
MS is an autoimmune disease with chronic inflammation of myelin sheaths in the central nervous system (CNS). The functional significance of miRNAs in MS pathogenesis was highlighted in a recent study where miR-326 was shown to play a critical role in the pathogenesis of MS by upregulating the differentiation of damaging Th-17 cell.82 By Real-time RT-PCR analysis of miRNA expression in peripheral blood leukocytes from patients with relapsing-remitting MS, neuromyelitis optica (a CNS inflammatory disease with similar signs and symptoms as MS) and also healthy controls, Du, et al. identified that miR-326 was significantly upregulated in patients with relapsing-remitting MS, but not in patients with neuromyelitis optica when compared to healthy controls. The potential role of miR-326 in MS was reinforced in studies with EAE mice, a classical murine model of MS. The authors have shown that while silencing miR-326 in vivo in EAE mice inhibited Th-17 cell production and led to mild EAE disease symptoms, overexpression of miR-326 in vivo led to increased Th-17 cell number and severe EAE symptoms.82 In addition, miR-34a, miR-155, and miR-326 were found upregulated in active MS lesions and contributed to MS pathogenesis by targeting CD47 to release macrophages from the inhibitory control signal, which in turn cause increased phagocytosis of myelin.122 A very recent report also revealed that miR-155 may contribute to EAE by promoting the development of inflammatory Th1 and Th17 cells.83
By using Taqman RT- PCR array and miRNA microarray assay, Otaegui et al. and Kelly, et al. have identified over one hundred miRNAs that were differentially expressed in PBMC from patients with MS when compared to healthy controls.123, 124 Specifically, the expression of miR-18b and miR-599 was related to relapse and miR-96 might be involved in remission.123 In addition, differentially expressed miRNAs have also been identified in different lymphocyte subsets including CD4+ T cells, CD8+ T cells, B cells, and CD4+CD25+ Treg cells from patients with MS.125-127 Intriguingly, while Lindberg et al. demonstrated that miR-17-5p was upregulated in CD4+ T cells,125 Cox et al. found that miR-17-5p was underexpressed in whole blood cells from patients with MS when compared to healthy controls.126 Although the potential function of the MS-associated miRNAs is suggested by the correlation of the miRNA expression level and their candidate genes in some studies, the direct involvement and contribution of dysregulated miRNAs in MS has largely remained unknown and needs further investigation.
5. Estrogen, miRNA and autoimmune diseases
A striking common feature of many autoimmune diseases in both humans and experimental animal models is that females are more susceptible to autoimmune conditions than males.128–130 More than 85 percent of patients with thyroiditis, scleroderma, lupus, and Sjögren’s syndrome are women.131 The female to male ratios for SLE, RA, and MS are 9-13:1, 2-4:1, and 2:1, respectively.128, 130 In addition to genetic factors such as X-chromosome abnormalities, sex hormones such as estrogens and androgens are believed to play a significant role in the sex-based susceptibility to many autoimmune diseases. The studies with murine lupus models, especially with NZB/W mice, revealed that estrogen exacerbated lupus development and that androgens inhibited autoantibody production and exerted protective effect on lupus.132, 133 Moreover, estrogen-treatment can break B cell tolerance even in normal non-autoimmune mice to induce several lupus-related autoantibodies including those against dsDNA, cardiolipin, and other phospholipids.134–137 While in murine lupus models, estrogen has been shown to promote lupus, the role of estrogen in promotion of human lupus is thus far equivocal.138–140 The Nurses’ cohort studies indicated that the use of oral contraceptive pills (OCP) and hormone replacement therapy (HRT) increased the risk for developing SLE.141–143 However, the studies of the Safety of Estrogen in Lupus: National Assessment (SELENA) trials of using OCP and HRT revealed that the use of OCP has no significant effect on global lupus disease activity and incidence of flares, and that the use of HRT is only associated with a small risk for mild and moderate lupus flare, but not severe flare.144–146 In addition to estrogen, other sex hormones such as prolactin, androgen, and testosterone should be considered in the gender bias of SLE and the disease course changes during pregnancy and menopause.139
It is clear that sex hormones have profound influence on immune system development and function. Depending on the cell types, animal models, dose, administration methods and duration, estrogen has been shown to exert both anti-inflammatory pro-inflammatory immune responses in different studies.147–151 We, and others, have reported that in vivo estrogen (17β-estradiol) treatment of non-autoimmune mice promoted the secretion of IFNγ, a major inflammatory cytokine that is implicated in autoimmune diseases.152, 153 In addition, we have recently shown that estrogen upregulated a powerful inflammatory cytokine, IL-17, in non-autoimmune C57BL/6 mice.154 We found that estrogen not only modulated key transcription factors involved in inflammation,155–157 but also regulated miRNA expression to promote inflammatory responses in activated splenocytes from estrogen-treated mice.158 Through the combination of high throughput microarray assay and Real-time RT-PCR confirmation, we identified that miR-451, miR-486, miR-223, miR-148a, miR-18a, and miR-708 were upregulated, and that miR-146a, miR-125a, miR-125b, miR-143, miR-145, let-7e, miR-126, and miR-181a were downregulated in splenocytes from estrogen-treated orchiectomized male C57BL/6 mice when compared to placebo controls. Further, we demonstrated that the downregulation of miR-146a and upregulation of miR-223 contributed to enhanced IFNγ production in LPS activated splenocytes from estrogen-treated mice, which suggested a novel mechanism of immune modulation by estrogen.158 Of relevance, the decreased level of miR-146a and miR-125a and increased level of 148a have been identified in human patients with lupus and reported to contribute to lupus pathogenesis by regulating type I IFN pathway, the inflammatory chemokine RANTES and DNA methylation, respectively.99–101
Recent studies revealed that estrogen receptor (ER)α, rather than ERβ plays a critical role in the regulation estrogen-mediated promotion of inflammatory responses,159 autoimmunity in NZB/W mice,160 and also miRNA induction in MCF-7 cell line.161–163 Interestingly, a subset of estrogen-induced miRNAs also target ERα mRNA or ERα signaling components, therefore forming a negative feedback loop to coordinate and fine-tune cellular responses to estrogen.162
Androgens such as 5-α-dihydrotestosterone and testosterone have been shown to suppress immune responses and inflammation and exert protective effect on a broad range of autoimmune diseases in experimental models of these diseases.133, 164–167 Experiments have shown that androgens regulate miRNA expression in non-lymphoid organs including prostate, muscle and liver, and that the mutual interaction between miRNA and androgen receptor signaling is implicated in prostate cancer progression or suppression.168–171 Although there are no reports so far of androgen regulation of miRNAs in the immune system or in autoimmune diseases, it is conceivable that miRNA may also be regulated by androgen in lymphoid organs and plays a role in androgen-mediated immune suppressive function.
Together, these data suggest that sex hormones such as estrogen may contribute to the pathogenesis of lupus and other gender biased autoimmune disease via the regulation of miRNA expression.
6. Other potential causes of miRNA dysfunction in SLE
In addition to the hormonal factors we discussed above, genetic and environmental factors, two key factors that are involved in autoimmune disease etiology, may also contribute to lupus pathogenesis by altering the expression and/or function of miRNAs (Figure 4).96, 172 It has been shown that the genetic polymorphism in miRNA genes and the 3’ UTR sequences of its target genes, and translocation, gene amplification or deletion in host genome all contribute to dysregulated miRNA expression and/or function in diseases.96, 173–176 For example, a single-nucleotide polymorphism (SNP) in pre-miR-146a decreased the expression of mature miR-146a ,which has been shown to relate to the genetic predisposition to papillary thyroid carcinoma.177 So far, there is only one study to explore the association of genetic variants of miRNA with SLE susceptibility.178 Although this study did not reveal any significant relationship between the SNP in miR-146a and miR-449 and SLE, it suggested the importance of investigation of miRNA polymorphism in SLE susceptibility. Environmental factor such as EBV viral infections have been implicated in SLE pathogenesis. EBV infection induced the expression of miR-155 and miR-146a in a NF-κB dependent manner.179–181 Although miR-146a was decreased in PBMC from human patients with lupus,100 both of miR-146a and miR-155 were found to be upregulated in CD4+ T cells, PBMC, RASFs from human patient with RA,106, 116, 118 and splenic T cells from murine lupus.104 These data suggest the potential role of these two miRNAs in EBV-mediated autoimmune pathogenesis.
7. Conclusion and future direction
Even though that miRNAs were recognized as key regulators of mammalian genome only a few short years ago, there has been a stunning pace of publications documenting the physiological and pathological roles of miRNA in the immune system. A single miRNA may exert diverse immunoregulatory function, which may be specific to developmental stages and the microenvironment of different types of immune cells. miR-146a and miR-155 are two well studied miRNAs, which have been shown to play critical role in the regulation of lymphocyte development, differentiation, function, and also in the control of both innate and adaptive immune responses.18, 40, 43, 54, 57, 79, 83, 182 Conceivably, dysregulated miR-146a and miR-155 expression and/or function has been associated with various autoimmune diseases including SLE, RA, and MS (table1).100, 106, 116, 122 Although miR-146a was decreased in PBMC from human patients with lupus,100 it was increased in PBMC and RASFs from human patients with RA and in murine lupus T cells.104, 106 Thus, the unique role of individual miRNA must be recognized based on the cell type or disease type and not generalized.
The identification of unique, signature autoimmune disease-related miRNAs provides the foundation for the next phase of studies: 1) to mechanistically understand the pathogenic contribution of these disease-related miRNAs, 2) to determine the cause and mechanism underlying miRNA dysregulation in autoimmune diseases, and 3) to ultimately develop novel, miRNA-based biomarkers for diagnosis and therapeutic strategies for curing and preventing autoimmune diseases. To address these central questions, relevant animal models of autoimmune diseases will be instrumental. Figure 4 summarizes the potential causes of miRNA dysregulation in SLE, and also documents potential pathogenic contribution of selected miRNAs in human and murine lupus. While a single miRNA may target hundreds of genes, the effect of miRNA on individual target protein synthesis is mild and moderate.38, 39 Given that a specific lupus-related gene may contain targeting sites for different miRNAs,96 it is plausible that multiple lupus-associated miRNAs, rather than a single miRNA, synergistically act together to disrupt the tolerance regulatory mechanism and promote lupus. Current studies have indicated a huge potential of using miRNAs as gene therapy targets in vivo to treat cancer.183, 184 We anticipate that in the near future, to treat autoimmune diseases, novel effective miRNA-based gene therapies will be developed to replace the traditional immune suppressive therapies, which are usually life-long with undesirable side effects.
Acknowledgments
This work was supported by a Lupus Foundation of America Novel Pilot Project (208-11-110B-033-918-1) grant, a Virginia-Maryland Regional College of Veterinary Medicine (VMRCVM) Intramural Research Competition Grant (IRC 17385), and in part by National Institutes of Health (RO1AI051880). Dr. Rujuan Dai and Dr. S. Ansar Ahmed have filed a provisional patent application through Virginia Tech Intellectual Properties (VTIP) on the finding of the common lupus disease-associated miRNAs in lupus mice. We thank Mrs. Rebecca Phillip for editing this manuscript.
Abbreviations
- AID
activation-induced cytidine deaminase
- AML-1
acute myeloid leukaemia-1
- Ago
Argonaute
- C/EBPα
CCAAT enhancer binding protein α
- CNS
central nervous system
- DGCR8
DiGeorge syndrome critical region protein 8
- DNMT
DNA methyltransferase
- EAE
experimental autoimmune encephalomyelitis
- EBV
Epstein-Barr virus
- ER
estrogen receptor
- Foxo
forkhead transcription factor O
- HRT
hormone replacement therapy
- IFN
interferon
- IRAK-1
interleukin-1 receptor-associated kinase 1
- IRF
interferon regulatory factor
- KLF13
Kruppel-like factor 13
- LPS
lipopolysaccharide
- MCP-1
monocyte chemotactic protein-1
- M-CSFR
macrophage colony-stimulating factor
- Mef2c
myocyte-specific enhancer factor 2C
- miRNA
microRNA
- MMP
matrix metallopeptidase
- MOG35-55
myelin oligodendrocyte glycoprotein35–55
- mRNA
messenger RNA
- MS
multiple sclerosis
- MyD88
myeloid differentiation primary response protein 88
- NFI-A
nuclear factor 1 A-type
- NF-κ B
nuclear factor-κB
- OA
osteoarthritis
- OCP
oral contraceptive pills
- PAMPs
pathogen-associated molecular patterns
- PBMC
peripheral blood mononuclear cell
- pri-miRNA
primary miRNA transcripts
- pre-miRNA
precursor miRNA
- PTEN
phosphatase and tensin homolog
- RA
rheumatoid arthritis
- RASF
synovial fibroblasts
- RASGRP1
RAS guanyl nucleotide-releasing protein 1
- RISC
RNA-induced silencing complex
- RNA
ribonucleic acid
- SLE
systemic lupus erythematosus
- SNP
single-nucleotide polymorphism
- STAT
signal transducers and activators of transcription
- TCR
T cell receptor
- Th
T helper
- TNF
tumor necrosis factor
- TLR
toll-like receptor
- TRAF-6
tumor necrosis factor receptor associated factor 6
- TRBP
TAR RNA binding protein
- Treg
regulatory T cell
- UTR
untranslated region
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 2.Reinhart BJ, Slack FJ, Basson M, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–6. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 3.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–8. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 4.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–62. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 5.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–4. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 7.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–4. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bentwich I, Avniel A, Karov Y, et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766–70. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 9.Berezikov E, Guryev V, van de Belt J, Wienholds E, Plasterk RH, Cuppen E. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120:21–4. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 10.Engels BM, Hutvagner G. Principles and effects of microRNA–mediated post-transcriptional gene regulation. Oncogene. 2006;25:6163–9. doi: 10.1038/sj.onc.1209909. [DOI] [PubMed] [Google Scholar]
- 11.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 12.Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32:D109–11. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miranda KC, Huynh T, Tay Y, et al. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–17. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 14.Bernstein E, Kim SY, Carmell MA, et al. Dicer is essential for mouse development. Nat Genet. 2003;35:215–7. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 15.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–5. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 16.Neilson JR, Zheng GX, Burge CB, Sharp PA. Dynamic regulation of miRNA expression in ordered stages of cellular development. Genes Dev. 2007;21:578–89. doi: 10.1101/gad.1522907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ambros V. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell. 2003;113:673–6. doi: 10.1016/s0092-8674(03)00428-8. [DOI] [PubMed] [Google Scholar]
- 18.Baltimore D, Boldin MP, O'Connell RM, Rao DS, Taganov KD. MicroRNAs: new regulators of immune cell development and function. Nat Immunol. 2008;9:839–45. doi: 10.1038/ni.f.209. [DOI] [PubMed] [Google Scholar]
- 19.Zhang B, Farwell MA. microRNAs: a new emerging class of players for disease diagnostics and gene therapy. J Cell Mol Med. 2008;12:3–21. doi: 10.1111/j.1582-4934.2007.00196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 21.Krutzfeldt J, Stoffel M. MicroRNAs: a new class of regulatory genes affecting metabolism. Cell Metab. 2006;4:9–12. doi: 10.1016/j.cmet.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 22.Eacker SM, Dawson TM, Dawson VL. Understanding microRNAs in neurodegeneration. Nat Rev Neurosci. 2009;10:837–41. doi: 10.1038/nrn2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pauley KM, Cha S, Chan EK. MicroRNA in autoimmunity and autoimmune diseases. J Autoimmun. 2009;32:189–94. doi: 10.1016/j.jaut.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–10. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim VN, Nam JW. Genomics of microRNA. Trends Genet. 2006;22:165–73. doi: 10.1016/j.tig.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 26.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–39. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 27.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–85. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 28.Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–55. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–34. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 30.Chan SP, Slack FJ. And now introducing mammalian mirtrons. Dev Cell. 2007;13:605–7. doi: 10.1016/j.devcel.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 31.Lee Y, Kim M, Han J, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–60. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gregory RI, Yan KP, Amuthan G, et al. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–40. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 33.Lee Y, Ahn C, Han J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–9. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 34.Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–6. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol Cell. 2007;28:328–36. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakahara K, Kim K, Sciulli C, Dowd SR, Minden JS, Carthew RW. Targets of microRNA regulation in the Drosophila oocyte proteome. Proc Natl Acad Sci U S A. 2005;102:12023–8. doi: 10.1073/pnas.0500053102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 40.Taganov KD, Boldin MP, Baltimore D. MicroRNAs and Immunity: Tiny Players in a Big Field. Immunity. 2007;26:133–7. doi: 10.1016/j.immuni.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 41.Xiao C, Rajewsky K. MicroRNA control in the immune system: basic principles. Cell. 2009;136:26–36. doi: 10.1016/j.cell.2008.12.027. [DOI] [PubMed] [Google Scholar]
- 42.Gantier MP, Sadler AJ, Williams BR. Fine-tuning of the innate immune response by microRNAs. Immunol Cell Biol. 2007;85:458–62. doi: 10.1038/sj.icb.7100091. [DOI] [PubMed] [Google Scholar]
- 43.O'Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat Rev Immunol. 2010;10:111–22. doi: 10.1038/nri2708. [DOI] [PubMed] [Google Scholar]
- 44.Lodish HF, Zhou B, Liu G, Chen CZ. Micromanagement of the immune system by microRNAs. Nat Rev Immunol. 2008;8:120–30. doi: 10.1038/nri2252. [DOI] [PubMed] [Google Scholar]
- 45.Sonkoly E, Stahle M, Pivarcsi A. MicroRNAs and immunity: novel players in the regulation of normal immune function and inflammation. Semin Cancer Biol. 2008;18:131–40. doi: 10.1016/j.semcancer.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 46.Kawai T, Akira S. TLR signaling. Cell Death Differ. 2006;13:816–25. doi: 10.1038/sj.cdd.4401850. [DOI] [PubMed] [Google Scholar]
- 47.Takeda K, Akira S. Toll-like receptors in innate immunity. Int Immunol. 2005;17:1–14. doi: 10.1093/intimm/dxh186. [DOI] [PubMed] [Google Scholar]
- 48.Liew FY, Xu D, Brint EK, O'Neill LA. Negative regulation of toll-like receptor-mediated immune responses. Nat Rev Immunol. 2005;5:446–58. doi: 10.1038/nri1630. [DOI] [PubMed] [Google Scholar]
- 49.Fazi F, Rosa A, Fatica A, et al. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell. 2005;123:819–31. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 50.Pulikkan JA, Dengler V, Peramangalam PS, et al. Cell-cycle regulator E2F1 and microRNA-223 comprise an autoregulatory negative feedback loop in acute myeloid leukemia. Blood. 2010;115:1768–78. doi: 10.1182/blood-2009-08-240101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Johnnidis JB, Harris MH, Wheeler RT, et al. Regulation of progenitor cell proliferation and granulocyte function by microRNA–223. Nature. 2008;451:1125–9. doi: 10.1038/nature06607. [DOI] [PubMed] [Google Scholar]
- 52.Fontana L, Pelosi E, Greco P, et al. MicroRNAs 17-5p-20a-106a control monocytopoiesis through AML1 targeting and M-CSF receptor upregulation. Nat Cell Biol. 2007;9:775–87. doi: 10.1038/ncb1613. [DOI] [PubMed] [Google Scholar]
- 53.Rosa A, Ballarino M, Sorrentino A, et al. The interplay between the master transcription factor PU.1 and miR-424 regulates human monocyte/macrophage differentiation. Proc Natl Acad Sci U S A. 2007;104:19849–54. doi: 10.1073/pnas.0706963104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.O'Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci U S A. 2007;104:1604–9. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tili E, Michaille JJ, Cimino A, et al. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J Immunol. 2007;179:5082–9. doi: 10.4049/jimmunol.179.8.5082. [DOI] [PubMed] [Google Scholar]
- 56.Chen XM, Splinter PL, O'Hara SP, LaRusso NF. A cellular micro-RNA, let-7i, regulates Toll-like receptor 4 expression and contributes to cholangiocyte immune responses against Cryptosporidium parvum infection. J Biol Chem. 2007;282:28929–38. doi: 10.1074/jbc.M702633200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006;103:12481–6. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tang B, Xiao B, Liu Z, et al. Identification of MyD88 as a novel target of miR-155, involved in negative regulation of Helicobacter pylori-induced inflammation. FEBS Lett. 2010;584:1481–6. doi: 10.1016/j.febslet.2010.02.063. [DOI] [PubMed] [Google Scholar]
- 59.Sheedy FJ, Palsson-McDermott E, Hennessy EJ, et al. Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat Immunol. 11:141–7. doi: 10.1038/ni.1828. [DOI] [PubMed] [Google Scholar]
- 60.Liu G, Friggeri A, Yang Y, Park YJ, Tsuruta Y, Abraham E. miR-147, a microRNA that is induced upon Toll-like receptor stimulation, regulates murine macrophage inflammatory responses. Proc Natl Acad Sci U S A. 2009;106:15819–24. doi: 10.1073/pnas.0901216106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bazzoni F, Rossato M, Fabbri M, et al. Induction and regulatory function of miR-9 in human monocytes and neutrophils exposed to proinflammatory signals. Proc Natl Acad Sci U S A. 2009;106:5282–7. doi: 10.1073/pnas.0810909106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cobb BS, Nesterova TB, Thompson E, et al. T cell lineage choice and differentiation in the absence of the RNase III enzyme Dicer. J Exp Med. 2005;201:1367–73. doi: 10.1084/jem.20050572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Muljo SA, Ansel KM, Kanellopoulou C, Livingston DM, Rao A, Rajewsky K. Aberrant T cell differentiation in the absence of Dicer. J Exp Med. 2005;202:261–9. doi: 10.1084/jem.20050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Koralov SB, Muljo SA, Galler GR, et al. Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell. 2008;132:860–74. doi: 10.1016/j.cell.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 65.Rodriguez A, Vigorito E, Clare S, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–11. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thai TH, Calado DP, Casola S, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–8. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 67.Vigorito E, Perks KL, Abreu-Goodger C, et al. microRNA-155 Regulates the Generation of Immunoglobulin Class-Switched Plasma Cells. Immunity. 2007;27:847–59. doi: 10.1016/j.immuni.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dorsett Y, McBride KM, Jankovic M, et al. MicroRNA-155 suppresses activation–induced cytidine deaminase-mediated Myc-Igh translocation. Immunity. 2008;28:630–8. doi: 10.1016/j.immuni.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Calame K. MicroRNA-155 function in B Cells. Immunity. 2007;27:825–7. doi: 10.1016/j.immuni.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 70.Kohlhaas S, Garden OA, Scudamore C, Turner M, Okkenhaug K, Vigorito E. Cutting edge: the Foxp3 target miR-155 contributes to the development of regulatory T cells. J Immunol. 2009;182:2578–82. doi: 10.4049/jimmunol.0803162. [DOI] [PubMed] [Google Scholar]
- 71.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–6. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 72.Li QJ, Chau J, Ebert PJ, et al. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129:147–61. doi: 10.1016/j.cell.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 73.Monticelli S, Ansel KM, Xiao C, et al. MicroRNA profiling of the murine hematopoietic system. Genome Biol. 2005;6:R71. doi: 10.1186/gb-2005-6-8-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xiao C, Calado DP, Galler G, et al. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131:146–59. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 75.Zhou B, Wang S, Mayr C, Bartel DP, Lodish HF. miR-150, a microRNA expressed in mature B and T cells, blocks early B cell development when expressed prematurely. Proc Natl Acad Sci U S A. 2007;104:7080–5. doi: 10.1073/pnas.0702409104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xiao C, Srinivasan L, Calado DP, et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. 2008;9:405–14. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ventura A, Young AG, Winslow MM, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–86. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stahl HF, Fauti T, Ullrich N, et al. miR-155 inhibition sensitizes CD4+ Th cells for TREG mediated suppression. PLoS ONE. 2009;4:e7158. doi: 10.1371/journal.pone.0007158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lu LF, Boldin MP, Chaudhry A, et al. Function of miR-146a in Controlling Treg Cell-Mediated Regulation of Th1 Responses. Cell. 2010;142:914–29. doi: 10.1016/j.cell.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rouas R, Fayyad-Kazan H, El Zein N, et al. Human natural Treg microRNA signature: role of microRNA-31 and microRNA-21 in FOXP3 expression. Eur J Immunol. 2009;39:1608–18. doi: 10.1002/eji.200838509. [DOI] [PubMed] [Google Scholar]
- 81.Pernis AB. Th17 cells in rheumatoid arthritis and systemic lupus erythematosus. J Intern Med. 2009;265:644–52. doi: 10.1111/j.1365-2796.2009.02099.x. [DOI] [PubMed] [Google Scholar]
- 82.Du C, Liu C, Kang J, et al. MicroRNA miR-326 regulates TH-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nat Immunol. 2009;10:1252–9. doi: 10.1038/ni.1798. [DOI] [PubMed] [Google Scholar]
- 83.O'Connell RM, Kahn D, Gibson WS, et al. MicroRNA-155 Promotes Autoimmune Inflammation by Enhancing Inflammatory T Cell Development. Immunity. 2010;33:607–19. doi: 10.1016/j.immuni.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Goodnow CC. Multistep pathogenesis of autoimmune disease. Cell. 2007;130:25–35. doi: 10.1016/j.cell.2007.06.033. [DOI] [PubMed] [Google Scholar]
- 85.Shlomchik MJ, Craft JE, Mamula MJ. From T to B and back again: positive feedback in systemic autoimmune disease. Nat Rev Immunol. 2001;1:147–53. doi: 10.1038/35100573. [DOI] [PubMed] [Google Scholar]
- 86.Zhou X, Jeker LT, Fife BT, et al. Selective miRNA disruption in T reg cells leads to uncontrolled autoimmunity. J Exp Med. 2008;205:1983–91. doi: 10.1084/jem.20080707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chong MM, Rasmussen JP, Rudensky AY, Littman DR. The RNAseIII enzyme Drosha is critical in T cells for preventing lethal inflammatory disease. J Exp Med. 2008;205:2005–17. doi: 10.1084/jem.20081219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liston A, Lu LF, O'Carroll D, Tarakhovsky A, Rudensky AY. Dicer-dependent microRNA pathway safeguards regulatory T cell function. J Exp Med. 2008;205:1993–2004. doi: 10.1084/jem.20081062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fernandez D, Bonilla E, Phillips P, Perl A. Signaling abnormalities in systemic lupus erythematosus as potential drug targets. Endocr Metab Immune Disord Drug Targets. 2006;6:305–11. doi: 10.2174/187153006779025748. [DOI] [PubMed] [Google Scholar]
- 90.Rahman AH, Eisenberg RA. The role of toll-like receptors in systemic lupus erythematosus. Springer Semin Immunopathol. 2006;28:131–43. doi: 10.1007/s00281-006-0034-3. [DOI] [PubMed] [Google Scholar]
- 91.Christensen SR, Shupe J, Nickerson K, Kashgarian M, Flavell RA, Shlomchik MJ. Toll-like receptor 7 and TLR9 dictate autoantibody specificity and have opposing inflammatory and regulatory roles in a murine model of lupus. Immunity. 2006;25:417–28. doi: 10.1016/j.immuni.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 92.Peng SL. Altered T and B lymphocyte signaling pathways in lupus. Autoimmun Rev. 2009;8:179–83. doi: 10.1016/j.autrev.2008.07.040. [DOI] [PubMed] [Google Scholar]
- 93.Crispin JC, Liossis SN, Kis-Toth K, et al. Pathogenesis of human systemic lupus erythematosus: recent advances. Trends Mol Med. 16:47–57. doi: 10.1016/j.molmed.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Patel DR, Richardson BC. Epigenetic mechanisms in lupus. Curr Opin Rheumatol. 2010;22:478–82. doi: 10.1097/BOR.0b013e32833ae915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pan Y, Sawalha AH. Epigenetic regulation and the pathogenesis of systemic lupus erythematosus. Transl Res. 2009;153:4–10. doi: 10.1016/j.trsl.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 96.Vinuesa CG, Rigby RJ, Yu D. Logic and extent of miRNA-mediated control of autoimmune gene expression. Int Rev Immunol. 2009;28:112–38. doi: 10.1080/08830180902934909. [DOI] [PubMed] [Google Scholar]
- 97.Dai Y, Huang YS, Tang M, et al. Microarray analysis of microRNA expression in peripheral blood cells of systemic lupus erythematosus patients. Lupus. 2007;16:939–46. doi: 10.1177/0961203307084158. [DOI] [PubMed] [Google Scholar]
- 98.Dai Y, Sui W, Lan H, Yan Q, Huang H, Huang Y. Comprehensive analysis of microRNA expression patterns in renal biopsies of lupus nephritis patients. Rheumatol Int. 2009;29:749–54. doi: 10.1007/s00296-008-0758-6. [DOI] [PubMed] [Google Scholar]
- 99.Pan W, Zhu S, Yuan M, et al. MicroRNA-21 and microRNA-148a contribute to DNA hypomethylation in lupus CD4+ T cells by directly and indirectly targeting DNA methyltransferase 1. J Immunol. 2010;184:6773–81. doi: 10.4049/jimmunol.0904060. [DOI] [PubMed] [Google Scholar]
- 100.Tang Y, Luo X, Cui H, et al. MicroRNA-146A contributes to abnormal activation of the type I interferon pathway in human lupus by targeting the key signaling proteins. Arthritis Rheum. 2009;60:1065–75. doi: 10.1002/art.24436. [DOI] [PubMed] [Google Scholar]
- 101.Zhao X, Tang Y, Qu B, et al. MicroRNA-125a contributes to elevated inflammatory chemokine RANTES via targeting KLF13 in systemic lupus erythematosus. Arthritis Rheum. 2010;62:3425–35. doi: 10.1002/art.27632. [DOI] [PubMed] [Google Scholar]
- 102.Te JL, Dozmorov IM, Guthridge JM, et al. Identification of unique microRNA signature associated with lupus nephritis. PLoS ONE. 2010;5:e10344. doi: 10.1371/journal.pone.0010344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Duursma AM, Kedde M, Schrier M, le Sage C, Agami R. miR–148 targets human DNMT3b protein coding region. RNA. 2008;14:872–7. doi: 10.1261/rna.972008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dai R, Zhang Y, Khan D, et al. Identification of a common lupus disease-associated microRNA expression pattern in three different murine models of lupus. PLoS ONE. 2010;5:e14302. doi: 10.1371/journal.pone.0014302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yu D, Tan AH, Hu X, et al. Roquin represses autoimmunity by limiting inducible T-cell co-stimulator messenger RNA. Nature. 2007;450:299–303. doi: 10.1038/nature06253. [DOI] [PubMed] [Google Scholar]
- 106.Pauley KM, Satoh M, Chan AL, Bubb MR, Reeves WH, Chan EK. Upregulated miR-146a expression in peripheral blood mononuclear cells from rheumatoid arthritis patients. Arthritis Res Ther. 2008;10:R101. doi: 10.1186/ar2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Xu S, Witmer PD, Lumayag S, Kovacs B, Valle D. MicroRNA (miRNA) transcriptome of mouse retina and identification of a sensory organ-specific miRNA cluster. J Biol Chem. 2007;282:25053–66. doi: 10.1074/jbc.M700501200. [DOI] [PubMed] [Google Scholar]
- 108.Ouyang W, Li MO. Foxo: in command of T lymphocyte homeostasis and tolerance. Trends Immunol. 2011;32:26–33. doi: 10.1016/j.it.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lin L, Hron JD, Peng SL. Regulation of NF-kappaB, Th activation, and autoinflammation by the forkhead transcription factor Foxo3a. Immunity. 2004;21:203–13. doi: 10.1016/j.immuni.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 110.Ouyang W, Beckett O, Flavell RA, Li MO. An essential role of the Forkhead-box transcription factor Foxo1 in control of T cell homeostasis and tolerance. Immunity. 2009;30:358–71. doi: 10.1016/j.immuni.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Segura MF, Hanniford D, Menendez S, et al. Aberrant miR-182 expression promotes melanoma metastasis by repressing FOXO3 and microphthalmia-associated transcription factor. Proc Natl Acad Sci U S A. 2009;106:1814–9. doi: 10.1073/pnas.0808263106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Guttilla IK, White BA. Coordinate regulation of FOXO1 by miR-27a, miR-96, and miR-182 in breast cancer cells. J Biol Chem. 2009;284:23204–16. doi: 10.1074/jbc.M109.031427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Dejean AS, Beisner DR, Ch'en IL, et al. Transcription factor Foxo3 controls the magnitude of T cell immune responses by modulating the function of dendritic cells. Nat Immunol. 2009;10:504–13. doi: 10.1038/ni.1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Stittrich AB, Haftmann C, Sgouroudis E, et al. The microRNA miR-182 is induced by IL–2 and promotes clonal expansion of activated helper T lymphocytes. Nat Immunol. 2010;11:1057–62. doi: 10.1038/ni.1945. [DOI] [PubMed] [Google Scholar]
- 115.Kuo CC, Lin SC. Altered FOXO1 transcript levels in peripheral blood mononuclear cells of systemic lupus erythematosus and rheumatoid arthritis patients. Mol Med. 2007;13:561–6. doi: 10.2119/2007-00021.Kuo. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Stanczyk J, Pedrioli DM, Brentano F, et al. Altered expression of MicroRNA in synovial fibroblasts and synovial tissue in rheumatoid arthritis. Arthritis Rheum. 2008;58:1001–9. doi: 10.1002/art.23386. [DOI] [PubMed] [Google Scholar]
- 117.Murata K, Yoshitomi H, Tanida S, et al. Plasma and synovial fluid microRNAs as potential biomarkers of rheumatoid arthritis and osteoarthritis. Arthritis Res Ther. 2010;12:R86. doi: 10.1186/ar3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Li J, Wan Y, Guo Q, et al. Altered microRNA expression profile with miR-146a upregulation in CD4+ T cells from patients with rheumatoid arthritis. Arthritis Res Ther. 2010;12:R81. doi: 10.1186/ar3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Niimoto T, Nakasa T, Ishikawa M, et al. MicroRNA-146a expresses in interleukin-17 producing T cells in rheumatoid arthritis patients. BMC Musculoskelet Disord. 2010;11:209. doi: 10.1186/1471-2474-11-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Nakamachi Y, Kawano S, Takenokuchi M, et al. MicroRNA-124a is a key regulator of proliferation and monocyte chemoattractant protein 1 secretion in fibroblast-like synoviocytes from patients with rheumatoid arthritis. Arthritis Rheum. 2009;60:1294–304. doi: 10.1002/art.24475. [DOI] [PubMed] [Google Scholar]
- 121.Fulci V, Scappucci G, Sebastiani GD, et al. miR-223 is overexpressed in T-lymphocytes of patients affected by rheumatoid arthritis. Hum Immunol. 2010;71:206–11. doi: 10.1016/j.humimm.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 122.Junker A, Krumbholz M, Eisele S, et al. MicroRNA profiling of multiple sclerosis lesions identifies modulators of the regulatory protein CD47. Brain. 2009;132:3342–52. doi: 10.1093/brain/awp300. [DOI] [PubMed] [Google Scholar]
- 123.Otaegui D, Baranzini SE, Armananzas R, et al. Differential micro RNA expression in PBMC from multiple sclerosis patients. PLoS ONE. 2009;4:e6309. doi: 10.1371/journal.pone.0006309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Keller A, Leidinger P, Lange J, et al. Multiple sclerosis: microRNA expression profiles accurately differentiate patients with relapsing-remitting disease from healthy controls. PLoS ONE. 2009;4:e7440. doi: 10.1371/journal.pone.0007440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lindberg RL, Hoffmann F, Mehling M, Kuhle J, Kappos L. Altered expression of miR-17-5p in CD4+ lymphocytes of relapsing-remitting multiple sclerosis patients. Eur J Immunol. 2010;40:888–98. doi: 10.1002/eji.200940032. [DOI] [PubMed] [Google Scholar]
- 126.Cox MB, Cairns MJ, Gandhi KS, et al. MicroRNAs miR-17 and miR-20a inhibit T cell activation genes and are under-expressed in MS whole blood. PLoS ONE. 2010;5:e12132. doi: 10.1371/journal.pone.0012132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.De Santis G, Ferracin M, Biondani A, et al. Altered miRNA expression in T regulatory cells in course of multiple sclerosis. J Neuroimmunol. 2010;226:165–71. doi: 10.1016/j.jneuroim.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 128.Whitacre CC. Sex differences in autoimmune disease. Nat Immunol. 2001;2:777–80. doi: 10.1038/ni0901-777. [DOI] [PubMed] [Google Scholar]
- 129.Ansar Ahmed S, Penhale WJ, Talal N. Sex hormones, immune responses, and autoimmune diseases. Mechanisms of sex hormone action. Am J Pathol. 1985;121:531–51. [PMC free article] [PubMed] [Google Scholar]
- 130.Ansar Ahmed S, Hissong BD, Verthelyi D, Donner K, Becker K, Karpuzoglu-Sahin E. Gender and risk of autoimmune diseases: possible role of estrogenic compounds. Environ Health Perspect. 1999;107 (Suppl 5):681–6. doi: 10.1289/ehp.99107s5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.NIH. Progress in Autoimmune Diseases Research. 2005. [Google Scholar]
- 132.Roubinian J, Talal N, Siiteri PK, Sadakian JA. Sex hormone modulation of autoimmunity in NZB/NZW mice. Arthritis Rheum. 1979;22:1162–9. doi: 10.1002/art.1780221102. [DOI] [PubMed] [Google Scholar]
- 133.Roubinian JR, Talal N, Greenspan JS, Goodman JR, Siiteri PK. Effect of castration and sex hormone treatment on survival, anti-nucleic acid antibodies, and glomerulonephritis in NZB/NZW F1 mice. J Exp Med. 1978;147:1568–83. doi: 10.1084/jem.147.6.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Grimaldi CM, Cleary J, Dagtas AS, Moussai D, Diamond B. Estrogen alters thresholds for B cell apoptosis and activation. J Clin Invest. 2002;109:1625–33. doi: 10.1172/JCI14873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Bynoe MS, Grimaldi CM, Diamond B. Estrogen up-regulates Bcl-2 and blocks tolerance induction of naive B cells. Proc Natl Acad Sci U S A. 2000;97:2703–8. doi: 10.1073/pnas.040577497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ansar Ahmed S, Dauphinee MJ, Montoya AI, Talal N. Estrogen induces normal murine CD5+ B cells to produce autoantibodies. J Immunol. 1989;142:2647–53. [PubMed] [Google Scholar]
- 137.Verthelyi DI, Ansar Ahmed S. Estrogen increases the number of plasma cells and enhances their autoantibody production in nonautoimmune C57BL/6 mice. Cell Immunol. 1998;189:125–34. doi: 10.1006/cimm.1998.1372. [DOI] [PubMed] [Google Scholar]
- 138.Petri M, Buyon JP. Oral contraceptives in systemic lupus erythematosus: the case for (and against) Lupus. 2008;17:708–10. doi: 10.1177/0961203308092518. [DOI] [PubMed] [Google Scholar]
- 139.Petri M. Sex hormones and systemic lupus erythematosus. Lupus. 2008;17:412–5. doi: 10.1177/0961203308090026. [DOI] [PubMed] [Google Scholar]
- 140.Zen M, Ghirardello A, Iaccarino L, et al. Hormones, immune response, and pregnancy in healthy women and SLE patients. Swiss Med Wkly. 2010;140:187–201. doi: 10.4414/smw.2010.12597. [DOI] [PubMed] [Google Scholar]
- 141.Bernier MO, Mikaeloff Y, Hudson M, Suissa S. Combined oral contraceptive use and the risk of systemic lupus erythematosus. Arthritis Rheum. 2009;61:476–81. doi: 10.1002/art.24398. [DOI] [PubMed] [Google Scholar]
- 142.Sanchez-Guerrero J, Karlson EW, Liang MH, Hunter DJ, Speizer FE, Colditz GA. Past use of oral contraceptives and the risk of developing systemic lupus erythematosus. Arthritis Rheum. 1997;40:804–8. doi: 10.1002/art.1780400505. [DOI] [PubMed] [Google Scholar]
- 143.Sanchez-Guerrero J, Liang MH, Karlson EW, Hunter DJ, Colditz GA. Postmenopausal estrogen therapy and the risk for developing systemic lupus erythematosus. Ann Intern Med. 1995;122:430–3. doi: 10.7326/0003-4819-122-6-199503150-00005. [DOI] [PubMed] [Google Scholar]
- 144.Petri M, Kim MY, Kalunian KC, et al. Combined oral contraceptives in women with systemic lupus erythematosus. N Engl J Med. 2005;353:2550–8. doi: 10.1056/NEJMoa051135. [DOI] [PubMed] [Google Scholar]
- 145.Buyon JP, Petri MA, Kim MY, et al. The effect of combined estrogen and progesterone hormone replacement therapy on disease activity in systemic lupus erythematosus: a randomized trial. Ann Intern Med. 2005;142:953–62. doi: 10.7326/0003-4819-142-12_part_1-200506210-00004. [DOI] [PubMed] [Google Scholar]
- 146.Sanchez-Guerrero J, Uribe AG, Jimenez-Santana L, et al. A trial of contraceptive methods in women with systemic lupus erythematosus. N Engl J Med. 2005;353:2539–49. doi: 10.1056/NEJMoa050817. [DOI] [PubMed] [Google Scholar]
- 147.Straub RH. The complex role of estrogens in inflammation. Endocr Rev. 2007;28:521–74. doi: 10.1210/er.2007-0001. [DOI] [PubMed] [Google Scholar]
- 148.Karpuzoglu E, Ansar Ahmed S. Estrogen regulation of nitric oxide and inducible nitric oxide synthase (iNOS) in immune cells: implications for immunity, autoimmune diseases, and apoptosis. Nitric Oxide. 2006;15:177–86. doi: 10.1016/j.niox.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 149.Calippe B, Douin-Echinard V, Laffargue M, et al. Chronic estradiol administration in vivo promotes the proinflammatory response of macrophages to TLR4 activation: involvement of the phosphatidylinositol 3-kinase pathway. J Immunol. 2008;180:7980–8. doi: 10.4049/jimmunol.180.12.7980. [DOI] [PubMed] [Google Scholar]
- 150.Lang TJ. Estrogen as an immunomodulator. Clin Immunol. 2004;113:224–30. doi: 10.1016/j.clim.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 151.Ghisletti S, Meda C, Maggi A, Vegeto E. 17beta-estradiol inhibits inflammatory gene expression by controlling NF-kappaB intracellular localization. Mol Cell Biol. 2005;25:2957–68. doi: 10.1128/MCB.25.8.2957-2968.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Gourdy P, Araujo LM, Zhu R, et al. Relevance of sexual dimorphism to regulatory T cells: estradiol promotes IFN-gamma production by invariant natural killer T cells. Blood. 2005;105:2415–20. doi: 10.1182/blood-2004-07-2819. [DOI] [PubMed] [Google Scholar]
- 153.Karpuzoglu-Sahin E, Hissong BD, Ansar Ahmed S. Interferon-gamma levels are upregulated by 17-beta-estradiol and diethylstilbestrol. J Reprod Immunol. 2001;52:113–27. doi: 10.1016/s0165-0378(01)00117-6. [DOI] [PubMed] [Google Scholar]
- 154.Khan D, Dai R, Karpuzoglu E, Ahmed SA. Estrogen increases, whereas IL-27 and IFN-gamma decrease, splenocyte IL-17 production in WT mice. Eur J Immunol. 2010;40:2549–56. doi: 10.1002/eji.201040303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Dai R, Phillips RA, Ansar Ahmed S. Despite inhibition of nuclear localization of NF-kappa B p65, c-Rel, and RelB, 17-beta estradiol up-regulates NF-kappa B signaling in mouse splenocytes: the potential role of Bcl-3. J Immunol. 2007;179:1776–83. doi: 10.4049/jimmunol.179.3.1776. [DOI] [PubMed] [Google Scholar]
- 156.Karpuzoglu E, Phillips RA, Dai R, Graniello C, Gogal RM, Jr, Ahmed SA. Signal transducer and activation of transcription (STAT) 4beta, a shorter isoform of interleukin-12-induced STAT4, is preferentially activated by estrogen. Endocrinology. 2009;150:1310–20. doi: 10.1210/en.2008-0832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Dai R, Phillips RA, Karpuzoglu E, Khan D, Ahmed SA. Estrogen Regulates Transcription Factors STAT-1 and NF-{kappa}B to Promote Inducible Nitric Oxide Synthase and Inflammatory Responses. J Immunol. 2009;183:6998–7005. doi: 10.4049/jimmunol.0901737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Dai R, Phillips RA, Zhang Y, Khan D, Crasta O, Ahmed SA. Suppression of LPS-induced IFN{gamma} and nitric oxide in splenic lymphocytes by select estrogen-regulated miRNA: A novel mechanism of immune modulation. Blood. 2008;112:4591–7. doi: 10.1182/blood-2008-04-152488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Calippe B, Douin-Echinard V, Delpy L, et al. 17Beta-estradiol promotes TLR4-triggered proinflammatory mediator production through direct estrogen receptor alpha signaling in macrophages in vivo. J Immunol. 2010;185:1169–76. doi: 10.4049/jimmunol.0902383. [DOI] [PubMed] [Google Scholar]
- 160.Bynote KK, Hackenberg JM, Korach KS, Lubahn DB, Lane PH, Gould KA. Estrogen receptor-alpha deficiency attenuates autoimmune disease in (NZB x NZW)F1 mice. Genes Immun. 2008;9:137–52. doi: 10.1038/sj.gene.6364458. [DOI] [PubMed] [Google Scholar]
- 161.Hossain A, Kuo MT, Saunders GF. Mir-17-5p regulates breast cancer cell proliferation by inhibiting translation of AIB1 mRNA. Mol Cell Biol. 2006;26:8191–201. doi: 10.1128/MCB.00242-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Castellano L, Giamas G, Jacob J, et al. The estrogen receptor-alpha-induced microRNA signature regulates itself and its transcriptional response. Proc Natl Acad Sci U S A. 2009;106:15732–7. doi: 10.1073/pnas.0906947106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Adams BD, Furneaux H, White BA. The micro-ribonucleic acid (miRNA) miR-206 targets the human estrogen receptor-alpha (ERalpha) and represses ERalpha messenger RNA and protein expression in breast cancer cell lines. Mol Endocrinol. 2007;21:1132–47. doi: 10.1210/me.2007-0022. [DOI] [PubMed] [Google Scholar]
- 164.Palaszynski KM, Loo KK, Ashouri JF, Liu HB, Voskuhl RR. Androgens are protective in experimental autoimmune encephalomyelitis: implications for multiple sclerosis. J Neuroimmunol. 2004;146:144–52. doi: 10.1016/j.jneuroim.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 165.Rontzsch A, Thoss K, Petrow PK, Henzgen S, Brauer R. Amelioration of murine antigen-induced arthritis by dehydroepiandrosterone (DHEA) Inflamm Res. 2004;53:189–98. doi: 10.1007/s00011-003-1244-y. [DOI] [PubMed] [Google Scholar]
- 166.Ganesan K, Selvam R, Abhirami R, Raju KV, Manohar BM, Puvanakrishnan R. Gender differences and protective effects of testosterone in collagen induced arthritis in rats. Rheumatol Int. 2008;28:345–53. doi: 10.1007/s00296-007-0446-y. [DOI] [PubMed] [Google Scholar]
- 167.Yokoyama Y, Schwacha MG, Samy TS, Bland KI, Chaudry IH. Gender dimorphism in immune responses following trauma and hemorrhage. Immunol Res. 2002;26:63–76. doi: 10.1385/ir:26:1-3:063. [DOI] [PubMed] [Google Scholar]
- 168.Tessel MA, Krett NL, Rosen ST. Steroid receptor and microRNA regulation in cancer. Curr Opin Oncol. 2010;22:592–7. doi: 10.1097/CCO.0b013e32833ea80c. [DOI] [PubMed] [Google Scholar]
- 169.Narayanan R, Jiang J, Gusev Y, et al. MicroRNAs are mediators of androgen action in prostate and muscle. PLoS ONE. 2010;5:e13637. doi: 10.1371/journal.pone.0013637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Delic D, Grosser C, Dkhil M, Al-Quraishy S, Wunderlich F. Testosterone-induced upregulation of miRNAs in the female mouse liver. Steroids. 2010;75:998–1004. doi: 10.1016/j.steroids.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 171.Shi XB, Xue L, Yang J, et al. An androgen-regulated miRNA suppresses Bak1 expression and induces androgen-independent growth of prostate cancer cells. Proc Natl Acad Sci U S A. 2007;104:19983–8. doi: 10.1073/pnas.0706641104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Sebastiani GD, Galeazzi M. Infection--genetics relationship in systemic lupus erythematosus. Lupus. 2009;18:1169–75. doi: 10.1177/0961203309345737. [DOI] [PubMed] [Google Scholar]
- 173.Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: the implications for cancer research. Nat Rev Cancer. 10:389–402. doi: 10.1038/nrc2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Huppi K, Volfovsky N, Mackiewicz M, et al. MicroRNAs and genomic instability. Semin Cancer Biol. 2007;17:65–73. doi: 10.1016/j.semcancer.2006.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: the implications for cancer research. Nat Rev Cancer. 2010;10:389–402. doi: 10.1038/nrc2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Sethupathy P, Collins FS. MicroRNA target site polymorphisms and human disease. Trends Genet. 2008;24:489–97. doi: 10.1016/j.tig.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 177.Jazdzewski K, Murray EL, Franssila K, Jarzab B, Schoenberg DR, de la Chapelle A. Common SNP in pre-miR-146a decreases mature miR expression and predisposes to papillary thyroid carcinoma. Proc Natl Acad Sci U S A. 2008;105:7269–74. doi: 10.1073/pnas.0802682105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Zhang J, Yang B, Ying B, et al. Association of pre-microRNAs genetic variants with susceptibility in systemic lupus erythematosus. Mol Biol Rep. 2010 doi: 10.1007/s11033-010-0252-6. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 179.Motsch N, Pfuhl T, Mrazek J, Barth S, Grasser FA. Epstein-Barr virus-encoded latent membrane protein 1 (LMP1) induces the expression of the cellular microRNA miR-146a. RNA Biol. 2007;4:131–7. doi: 10.4161/rna.4.3.5206. [DOI] [PubMed] [Google Scholar]
- 180.Gatto G, Rossi A, Rossi D, Kroening S, Bonatti S, Mallardo M. Epstein-Barr virus latent membrane protein 1 trans-activates miR-155 transcription through the NF-kappaB pathway. Nucleic Acids Res. 2008;36:6608–19. doi: 10.1093/nar/gkn666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Cameron JE, Yin Q, Fewell C, et al. Epstein-Barr virus latent membrane protein 1 induces cellular MicroRNA miR-146a, a modulator of lymphocyte signaling pathways. J Virol. 2008;82:1946–58. doi: 10.1128/JVI.02136-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Teng G, Papavasiliou FN. Shhh! Silencing by microRNA-155. Philos Trans R Soc Lond B Biol Sci. 2009;364:631–7. doi: 10.1098/rstb.2008.0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Esquela-Kerscher A, Trang P, Wiggins JF, et al. The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle. 2008;7:759–64. doi: 10.4161/cc.7.6.5834. [DOI] [PubMed] [Google Scholar]
- 184.Wiggins JF, Ruffino L, Kelnar K, et al. Development of a Lung Cancer Therapeutic Based on the Tumor Suppressor MicroRNA–34. Cancer Res. 70:5923–30. doi: 10.1158/0008-5472.CAN-10-0655. [DOI] [PMC free article] [PubMed] [Google Scholar]