Abstract
Myosin II motor proteins play important roles in cell migration. Although myosin II filament assembly plays a key role in stabilization of focal contacts at the leading edge of migrating cells, the mechanisms and signaling pathways regulating localized assembly of lamellipodial myosin II filaments are poorly understood. We performed proteomic analysis of myosin IIA heavy chain (MHC) phosphorylation sites in MDA-MB 231 breast cancer cells to identify MHC phosphorylation sites activated during integrin engagement and lamellar extension on fibronectin. Fibronectin-activated MHC phosphorylation was identified on novel and previously recognized consensus sites for phosphorylation by Protein Kinase C (PKC) and Casein Kinase II (CK II). S1943, a CK-II consensus site, was highly phosphorylated in response to matrix engagement, and phosphoantibody staining revealed phosphorylation on myosin II assembled into leading edge lamellae. Surprisingly, neither pharmacological nor siRNA reduction in CKII activity reduced this stimulated S1943 phosphorylation. Our data demonstrate that S1943 phosphorylation is upregulated during lamellar protrusion, and that CKII does not appear to be the kinase responsible for this matrix-induced phosphorylation event.
INTRODUCTION
The motor protein myosin II plays critical roles in a variety of settings in nonmuscle cells (1). Myosin II is recruited to different locations during cell migration of different cell types, for example, to the posterior in migrating Dictyostelium cells, and to the posterior as well as anterior lamellar zones in epithelial cells and fibroblasts migrating in 2-D (2). In cancer cells, it is now generally accepted that cells can activate multiple distinct modes of cell migration depending on their mechanical environment and nature of the extracellular matrix, with integrin-matrix adhesions being critical in some settings, and more amoeboid or low-adhesion mechanisms of migration dominating in other environments (3, 4). Myosin II has complex and distinct roles in each type of migration. During formation of leading edge protrusions, assembly of myosin II filaments into the cell cortex plays a critical role in stabilizing nascent focal adhesion complexes that are necessary for efficient integrin-based migration (5). Despite the importance of myosin II in leading edge cytoskeletal functions, the mechanisms regulating its recruitment remain poorly understood.
Myosin II molecules consist of two myosin heavy chains (MHC) that form a globular head domain that interacts with F-actin and produces force, and an extended coiled-coil tail that mediates bipolar filament assembly. Two pairs of light chains associate with the neck region, the essential light chains (ELCs) and the regulatory light chains (RLCs). The three mammalian nonmuscle myosin II isoforms (IIA, IIB, and IIC) carry heavy chains encoded by distinct genes (MYH9, MYH10, and MYH14, respectively) (1). While it is well established that RLC phosphorylation has critical roles in regulating nonmuscle myosin II filament assembly, there is also emerging evidence that myosin II heavy chain phosphorylation may be an important modulator of filament assembly. For example, myosin IIB is phosphorylated near the C-terminus in prostate cancer cells via a pathway that involves PAK1 and PKC isoforms during responses to EGF (6). Myosin IIA can be phosphorylated on several C-terminal residues, including a consensus PKC target site at position S1916 (7), and on a consensus casein kinase II (CK-II) site at position S1943, which lies within a 30 residue “non-helical tailpiece” at the very C-terminus of the myosin IIA tail. Phosphorylation at S1943 is triggered when MDA-MB 231 cancer cell responses to EGF, and this phosphorylation was shown to be important for filament assembly responses to EGF (8). Phosphorylation of purified tail domain fragments of myosin IIA on S1943 favors filament disassembly (9), and in live cells mutation of this site to a nonphosphorylatable alanine residue leads to cortical oversassembly (8, 10). Given that myosin IIA is enriched in leading edge lamella in many cell types during persistent polarized migration, MHC phosphorylation at S1943 may represent a mechanism to drive filament turnover, helping disassemble anterior myosin II filaments to replenish the soluble pool of myosin II, available to assemble further forward as the leading edge extends.
Although the involvement of myosin IIA MHC phosphorylation on S1943 in assembly control is recognized, there have been few studies to date which address the identity of the upstream kinase or kinases in live cells. Based upon a consensus motif for CK-II, early studies showed that purified CK-II could phosphorylate purified smooth muscle myosin II (11), and that CK-II could phosphorylated purified myosin IIB (12, 13). Dulyaninova and colleagues also showed that purified CK-II can phosphorylate recombinant myosin IIA tail fragments in vitro (9). However, to our knowledge no studies have ever been performed to directly ask whether CK-II phosphorylates myosin IIA in live cells.
To further investigate the pathways that regulate myosin IIA heavy chain phosphorylation during migration on extracellular matrix as well as to unambiguously characterize the sites of phosphorylation on the MHC regions and understand their effects, we have performed proteomic and related cell biology analyses of MHC IIA phosphorylation events. We find that when cells are seeded on fibronectin to stimulate integrin-based signaling pathways, myosin IIA displays substantial upregulation of phosphorylation on both S1916 and S1943. To test the importance of CK-II in this pathway, we performed both pharmacological inhibition and siRNA knockdown of CK-II. Surprisingly, we find that depletion of CK-II activity has no effect on fibronectin-induced S1943 phosphorylation. These results support the model that matrix-induced signaling may enhance myosin II filament turnover at the leading edge, facilitating dynamic relocalization of myosin II to advancing protrusions during migration. Our data further suggest that CK-II is not a major MHC kinase mediating S1943 phosphorylation in this setting.
METHODS AND MATERIALS
Cell Culture
The highly metastatic MDA-MB-231 human breast cancer cell line that is derived from epithelium of human mammary gland was obtained from the American Type Culture Collection (Rockville, MD). COS-7 cells were a gift from Dr. Cathy Carlin from Case Western Reserve University, Cleveland, OH. MEM and DMEM supplemented with 10% heat-inactivated fetal bovine serum, penicillin/streptomycin, and 1% L-glutamine (Life Technologies, Gaithersburg, MD) to culture MDA-MB 231 cells, and Cos-7 cells, respectively at 37 °C with 6% CO2 and humidity. The cells used for performing experiments were not more than 10 to 15 passages after the initiation of cultures.
Chemicals and Antibodies
Cell dissociation reagent and fibronectin were purchased from Sigma-Aldrich (St. Louis, MO). Cell lysis buffer was purchased from Cell Signaling (Danvers, MA). SiRNA oligonucleotides specific to non-muscle MHC-IIA, MHC-IIB, and CK-II catalytic subunits were purchased from Dharmacon RNA Technologies, USA. Transfection reagents were purchased from Amaxa Biosystems, USA. Alexa Fluor® 568 Phalloidin and DAPI nucleic acid stain and Alexa-conjugated secondary antibodies were from Molecular Probes, Eugene, OR. SuperFemto Western blot reagents were from Pierce, Rockford, IL. Antibodies purchased from Sigma-Aldrich (St. Louis, MO) include non-muscle myosin IIA (catalog #M8064), myosin IIB (catalog #M7939), and actin. Antibodies against CK II are from Santa Cruz Biotechnology, Inc., CA. For double staining of total MHC IIA and phospho-S1943, an anti-MHC IIA monoclonal antibody was used (Abnova Biologicals, CO, catalog #H00004627-M06).
Plasmids
The plasmid pEGFP-NMHC-IIA-C3 expresses the full length corrected human non-muscle myosin heavy chain under the control of a constitutive CMV promoter, with EGFP fused at the amino terminus of the myosin IIA coding region. This plasmid, and the S1943A phosphorylation site mutant derived from it, were both gifts from the lab of Dr. Anne Bresnick (for full details see (8, 10)).
Transfection of cells
siRNAs and plasmid DNA transfections. Cells (1X106 to 1X107) growing in tissue culture plates for not more than 20 hour (60–70% confluent) were collected using cell dissociation reagent followed by washing with PBS and resuspending in 90 μl of transfection solution (Amaxa Biosystems, USA, kit “T” for MDA-MB 231 cells and kit “R” for COS-7 cells). The cell suspension was mixed with 3 μg of plasmid DNA or 5-20 pmol of Smart Pool siRNA oligonucleotides (Dharmacon RNA Technologies, Lafayette, CO). This suspension was transferred to a cuvette and electroporated using program A-023 and W-001 (Amaxa Biosystems) for MDA-MB 231 and COS-7 cells, respectively. Cells were then diluted in growth medium, seeded in tissue culture plates (60-mm or 100-mm), and incubated for 48–72 hour before collection for functional assays.
Cell Spreading Assays
Cell spreading assays to study myosin II localization and phosphorylation of the MHC of myosin IIA were carried out as described earlier (14). Briefly, cells growing in tissue culture plates were collected using cell dissociation reagent followed by seeding on fibronectin (20μg/ml) coated tissue culture plates. Cells were allowed to spread in the growth medium for 80–90 min in the tissue culture incubator maintained with humidity and CO2 (5%) at 37° C. For making total cell lysates, cell spreading assays were performed in 10 cm tissue culture plates coated with fibronectin prior to use.
Immunostaining and imaging
Immunostaining and imaging of the cells were performed as described earlier (14) with a few modifications. To perform immunostaining, cells growing or actively spreading on Lab-Tek II Chambered Coverglass were washed with PBS at room temperature followed by incubating with fixing buffer I (1X PBS, 2 mM MgCl2, 2 mM EGTA, and 4% paraformaldehyde) for 30 min at room temperature. After washing with PBS, cells were permeabilized by incubating with buffer II (Fixing buffer carrying 0.5% Triton X-100) for 5–8 min at room temperature. Cells were washed with PBS several times and blocked with blocking buffer (1X TBS with 0.1% Tween and 1% BSA) at room temperature for 10 min followed by incubating with antibodies (diluted in blocking buffer) at room temperature for 2 hour. After washing with washing buffer (blocking buffer with no BSA), cells were incubated with Alexa-conjugated secondary antibodies (Molecular Probes, Eugene, OR) diluted to 1,000-fold in blocking buffer for 1 hour at room temperature. Cells were washed with washing buffer and incubated with DAPI (20 μg/ml) solution for 5–10 min at room temperature. Cells were washed with PBS several times before imaging with a Zeiss LSM510 confocal microscope.
Western blot
Western blot analyses were carried out as described earlier (15) with some modifications. Total cell lysates were prepared from the cells after completely removing growth medium and rinsing with ice-cold PBS. Cells were lysed directly by the addition of 1X cell lysis buffer carrying an array of protease inhibitors (16) and incubated on ice for 3–5 min. Cell lysates were then collected by scraping with a rubber policeman. Samples were then transferred to microfuge tubes and subjected to water bath sonication for four strokes. Clear supernatant obtained by microcentrifugation at high speed at 4° C were resuspended in SDS sample buffer. Protein samples were heated at 95° C for 2 min and stored frozen till use. To ensure equal sample loading for SDS-PAGE and Western blotting, aliquots of samples from each experiment were subjected to SDS-PAGE and Coomassie blue staining. These gels were then used for total lane densitometry, allowing normalization of sample loading for subsequent Western blot analysis and estimating the relative levels of myosin II isoforms. SDS samples were resolved on polyacrylamide gels and then transferred to polyvinylidene difluoride membrane. Membranes were incubated with the primary antibodies and then with horseradish peroxidase–conjugated secondary antibodies. The immunoblotted proteins were visualized using Super-Femto Western blot reagents (Pierce, Rockford, IL).
Pro-Q Diamond staining and densitometry
Stimulation of the phosphorylation of the MHC of myosin II in the spreading cells was studied by performing Pro-Q Diamond staining (Molecular Probes, Inc., Eugene, OR). The protein lysates made from the non-migrating quiescent, suspension and spreading MDA-MB 231 cells were subjected to SDS-PAGE. The gel was immersed in 100 ml of fixation solution (50% methanol, 10% acetic acid, and water) with gentle agitation at room temperature for 30 min. After repeating fixation, the gel was washed with 200 ml distilled water (three changes, 10 min per wash). Then, the gel was incubated with 100 ml Pro-Q Diamond phosphoprotein stain for 60–90 min in the dark, and destained with 250 ml of destaining solution in the dark (20% acetonitrile, 50 mM sodium acetate pH 4.0, two changes, 1 hour per wash). The image was acquired on Phosphorimager (Bio-Rad, Hercules, CA) with a 532 nm laser excitation and a 580 nm bandpass emission filter. Following the image acquisition, densitometry of the phosphorylated band corresponding to the MHC was performed using ImageJ program (NIH).
Development and characterization of phosphospecific polyclonal antibodies
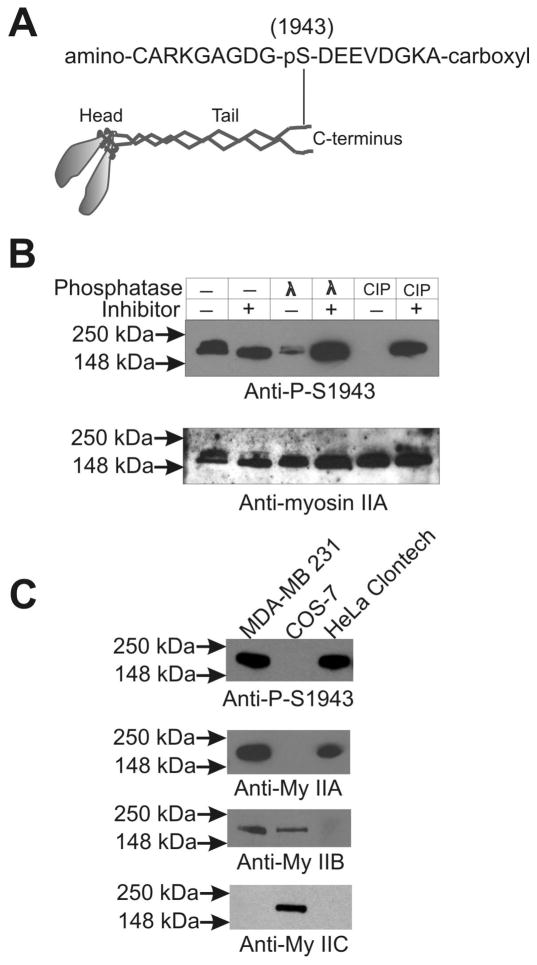
Polyclonal antiserum was raised against a custom-made specific phosphopeptide (amino-CARKGAGDG-pS1943-DEEVDGKA-carboxyl) of the MHC of non-muscle myosin IIA motor protein encoded by MyH9, under contract with Covance Laboratories. Antiserum obtained was subjected to affinity purification using the phosphopeptide to capture specific antibody and non-phosphopeptide to remove cross-reacting antibody. For testing these affinity purified phosphospecific polyclonal antibodies, total lysates made from spreading MDA-MB 231 cells using lysis buffer (50 mM Tris-HCl pH7.8 carrying 1% TritonX-100, 150 mM NaCl, 0.1 mM EGTA, 5 mM dithiothreitol, and 2 mM MnCl2) were treated with λ phosphatase and or calf intestinal phosphatase (New England BioLabs, USA) in the presence and absence of a cocktail of phosphatase inhibitors (50 mM NaF, 20 mM PMSF, 1 mM sodium orthovanadate, 10 μg/ml each of chymostatin, leupeptin, antipain and pepstatin - CLAP) at 30° C for 10 min followed by SDS-PAGE and Western blotting. For testing their cross reactivity with the MHCs of myosin IIB and IIC motor proteins, Western blot analysis of total lysates made from myosin IIA null cells (COS-7), myosin IIB null (HeLa cells from Clontech), and myosin IIC null cells (HeLa and MDA-MB 231) were performed.
Proteomics methodology and nano-LC-MS/MS analysis
For protein characterization experiments, protein lysates made from the quiescent, suspension and spreading MDA-MB 231 cancer cells were subjected to electrophoresis using 4–12% polyacrylamide gradient gels in SDS running buffer followed by staining with Coomassie blue. Protein concentrations were normalized via Bradford assays and then equal amount of total protein lysates loaded in each gel lane. After destaining the gel, the MHC bands were excised carefully, based on the molecular weight range of interest, and then subjected to in-gel reduction, alkylation, and tryptic digestion in a HEPA-filtered hood to reduce keratin background. All the bands of interest were processed in batches at the same time to ensure the robustness of the in-gel proteomic methodology adopted hereby. The LC-MS/MS analysis was performed using an UltiMate 3000 parallel LC system (Dionex, San Francisco, CA) that was interfaced to a LTQ-Orbitrap XL mass spectrometer (Thermo-Finnigan, Breman, Germany). Equal volumes of the samples were used for all the chromatographic analyses such that systematic errors that may be present will be a common factor for all the samples. The LC/MS platform was operated in the nano-LC mode using the standard nano-ESI API stack fitted with a picotip emitter (uncoated fitting, 10μm spray orifice, New Objective Inc., Woburn, MA). The solvent flow rate through the column was maintained at 300nL/min using a 1:1000 splitter system. The protein digests (5 μL) were injected into a reversed-phase C18 PepMap100 trapping column (0.3 × 5 mm, 5 μm particle size, Dionex) equilibrated with 0.1% Formic Acid (FA)/2% acetonitrile (v/v) and washed for 5 min with the equilibration solvent of 100% water, 0.1% formic acid at a flow rate of 25 μL/min, using an isocratic loading pump operated through an autosampler. After the washing step, the trapping column was switched in-line with a reversed-phase C18 Acclaim PepMap 100 column (0.075 × 150 mm, Dionex) and the peptides were chromatographed using a linear gradient of acetonitrile from 5% to 50% in aqueous/0.1% formic acid over a period of 45 min at the above-mentioned flow rate such that the eluate was directly introduced to the mass spectrometer. The high resolution mass spectrometer was operated in a data-dependent MS to MS/MS switching mode, with the five most intense ions in each MS scan subjected to MS/MS analysis. The full scan (325–1800 m/z range) was performed at 60000 resolution and the followup` fragmentation scans were performed in the CID mode such that the total scan cycle frequency was approximately 1 second. The spray voltage was set at 2.5kV and the threshold intensity for the MS/MS trigger was set at 1000 with the fragmentation carried out using the CID mode, using normalized collision energy (NCE) of 35. The data was collected in the profile mode for the full scan and centroid mode for the MS/MS scans. Dynamic exclusion function for previously selected precursor ions was enabled during the analysis such that the following parameters were applied: repeat count of 2, repeat duration of 45 seconds, and exclusion duration of 60 seconds and exclusion size list of 150. Xcalibur software (version 2.0.5, build 0704), Thermo-Finnigan) was used for instrument control, data acquisition, and data processing.
Phosphopeptide enrichment, when performed, was done using the TiO2 Mono Tip (GL Biosciences) product with usage instructions followed according to manufacturer specifications. A standard volume of the tryptic digest (6 μL) from the protein characterization experiments were used for the enrichment analysis, during the sample loading phase. After washing steps, the phosphopeptides were eluted with 75 μL of 3% ammonium hydroxide such that the volume was further reduced to 10 μL prior to LC injection. Each enrichment analysis run was performed using 5 μL of the reduced volume sample such that a duplicate analysis could be performed for all the relevant samples. The chromatographic and mass spectrometric method parameters were modified for the phosphopeptide analysis, in comparison to the above-mentioned protein characterization method. The LC separation time was extended to 105 minutes such that it is a 0.44% gradient increase during the gradient separation time. The neutral loss chromatograms were also collected such that the characteristic loss of phosphoric acid (H3PO4, loss of 98 Da) from phosho-Ser and phospho-Thr-containing peptides can be identified as well. For the protein identification and post-translational modification (PTM) data analysis steps, all the raw files obtained using the Orbitrap instrumentation platform were processed using the locally licensed version of the MASCOT (Matrix Science Inc., London, UK) search engine (2010, Version 2.3). Nominal peptide score threshold of 20 was applied to filter the peptides from the database search results. Manual analysis and annotation of the CID-based MS/MS files was done especially in relation to PTM reporting such that site-specific modifications can be assigned unambiguously. For relative abundance calculation of phosphorylated vs non-phosphorylated peptides of interest, the raw LC-MS/MS files were processed in the extracted ion chromatogram (EIC) mode such that peptide abundances of phosphorylated and non-phosphosphorylated native precursors can be chromatographically well resolved and accurately analyzed in peak area calculations.
Determination of the relative levels of myosin II motor proteins using mass spectrometry and an extracted ion chromatogram-based analysis
Total lysates made from MDA-MD 231 human breast cancer cells growing in tissue culture plates were subjected to SDS-PAGE and staining with Coomassie blue. After destaining, the MHC bands were excised and then subjected to in-gel reduction, alkylation, and tryptic digestion. The eluted peptide samples of the MHC were subjected to mass spectrometry analysis as described above. Using the known sequences of the MHCs of myosin IIA, IIB, and IIC motor protein complexes, individualized Mascot Search Databases were created. The raw LC-MS/MS data files obtained for the digested MHC protein gel band samples, prior to any phosphopeptide enrichment, were searched against these specific databases and protein coverage was obtained after applying the peptide threshold filter. The relative abundances of myosin IIA, IIB, and IIC motor proteins were preliminarily evaluated based on the sequence coverage obtained from the Mascot search results. Even though the sequence coverage numbers suggest greatest abundance of Myosin IIA amongst the 3 isoforms as a first approximation, we resorted to the more reliable chromatographic peak area-based relative quantitation using extracted ion intensities of a chosen peptide from the sequencing data. The choice of such a peptide was based on a criterion that very close peptide sequence similarity and peptide length match was essential for locating such a peptide amongst the three isoforms. This was essential in order to assume that the respective peptides ionization efficiencies were comparable during analysis in the LTQ Orbitrap XL mass analyzer, when analyzed from the same gel band. Upon careful examination of our datasets, the peptide EQADFAIEALAK (MHC IIA) was employed in relation to the same peptide being detected based on the EQADFAVEALAK sequence in MHC IIB and as EQADFALEALAK in MHC IIC. The chosen peptide was discriminative enough to allow us to comment unambiguously on the myosin isoforms relative abundances as the chromatographic resolution to discriminate all the 3 peptides was obtained under our run conditions. Peak area summation was done after boxcar averaging to smooth the extracted ion chromatogram in each case. Peak definition was done using the peak define tool of the XCalibur (Thermo Finnigan) MS data analysis software.
RESULTS
Myosin II isoform expression in the highly metastatic MDA-MB 231 human breast cancer cells
Although it appears that all three nonmuscle myosin II isoforms can support some contractile processes, such as cytokinesis, in the context of cell migration and cell protrusion, several studies have revealed differential effects of depleting MHC IIA versus MHC IIB. For example, during isotropic spreading of breast cancer cells, depletion of MHC IIA increases spreading, while depletion of MHC IIB reduces protrusion (14, 17). Cai and colleagues observed a similar isoform-specific role for myosin IIA in spreading fibroblasts (18). Even-Ram et al. (19) found that myosin IIA promoted microtubule dynamics, and depletion of MHC IIA increased cell migration rates.
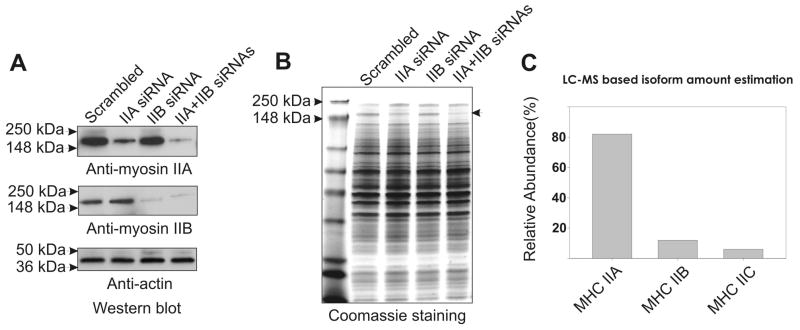
Although depletion studies argue for differential mechanical roles for myosin II isoforms in adhesion and lamellar extension, full understanding of relative mechanical roles of isoforms requires evaluation of the relative molar abundance of each isoform in intact cells. This analysis has been absent in many earlier studies. Western blot analysis of MDA-MB 231 lysates reveals expression of MHC IIA and IIB isoforms (Figure 1A). Each isoform is readily depleted after siRNA transfection of IIA or IIB siRNA oligo pools, and both isoforms can be depleted with simultaneous transfection of these pools (Figure 1A). Examination of corresponding cell lysates by SDS-PAGE and Coomassie Blue staining reveals depletion of the Coomassie-stained band at ~220 kDa that corresponds to the MHC II polypeptide (Figure 1B, arrowhead). As a more quantitative approach to establish isoform expression levels, we performed mass spectrometric evaluation of MHC II in MDA-MB 231 cells, performed on the MHC II band excised from Coomassie Blue stained SDS-PAGE cell lysate samples. The fractional abundance of myosin IIA, IIB, and IIC isoforms was calculated based on sample peptide relative abundance calculations (see Methods). This approach revealed presence of MHC IIC that had not been detected via western blotting, and established relative molar abundance of 82%, 12%, and 6% for MHC IIA, IIB, and IIC, respectively (Fig. 1C). This can be attributed to the greater analytical sensitivity of the nano LC-MS/MS proteomic methodology adopted in our study. Overall, both methods were in good concurrence that MHC IIA is the most abundance isoform in MDA-MB 231 cells. Therefore, the present study focused on understanding the mechanism of myosin IIA recruitment to the leading edge during spreading.
Figure 1.
Quantification of myosin II isoform expression levels in MDA-MB 231 human breast cancer cells. (A) Western blot indicting depletion of individual MHC II isoforms following siRNA. (B) Depletion of myosin II motor proteins in MDA-MB 231 cells via siRNA, detected via Coomassie Blue staining. Arrowhead denotes position of the MHC II in cell lysates after siRNA knockdown, followed by SDS-PAGE and Coomassie Blue staining. Note that the MHC II band intensity is reduced with either MHC IIA siRNA or MHC IIB siRNA treatment, and treatment with both siRNA pools leads to near-complete loss of Coomassie staining at the position of the MHC II band. (C) Extracted ion chromatogram (EIC)-based estimation of myosin isoform abundance.
Recruitment of myosin IIA to leading edge lamella correlates with upregulation of MHC phosphorylation
Myosin II isoforms are recruited to leading edge protrusions in many cell types including fibroblasts (20) and breast cancer cells (8). While myosin IIA localization is often diffuse in quiescent non-polarized cells, it is specifically recruited to the leading edge lamella in polarized migrating cells, and it is recruited to the analogous lamellar margin when cells are stimulated for spreading and leading edge protrusive activity by plating on fibronectin-coated surfaces (14). Given evidence that RLC phosphorylation is upregulated in the spreading lamellar zone of spreading cells (14), we investigated whether MHC IIA phosphorylation might also be regulated during the cell adhesion and spreading that is triggered upon fibronectin-integrin engagement.
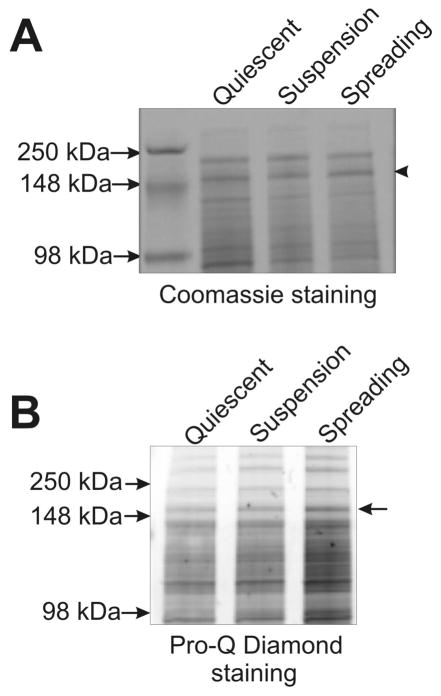
As a first test of cellular phosphorylation responses to fibronectin engagement, we collected cell lysates from actively spreading cells and performed SDS-PAGE followed by treatment with Pro-Q Diamond stain (Molecular Probes, Inc, OR), a dye that preferentially stains phospho-proteins. This analysis indicated increases in phosphorylation of many proteins in MDA-MB 231 lysates, with a notable increase in Pro-Q staining of the MHC II band in lysates of actively spreading cells (Figure 2A & B, arrows).
Figure 2.
Activation of MHC II phosphorylation during spreading on fibronectin. (A) Coomassie staining of total cell lysates separated on SDS-polyacrylamide gel. Total lysates made from the non-migrating quiescent, suspension, and spreading MDA-MB 231 cells were subjected to SDS-PAGE followed staining with Coomassie blue. Arrowhead indicates position of MHC II. (B) Increased MHC II phosphorylation is indicated via staining with the dye Pro-Q Diamond. Note that this Pro-Q staining was performed on the same gel presented in panel A, thus uniform loading is established by the Coomassie staining of the same sample. The Pro-Q staining suggests that spreading on fibronectin enhances phosphorylation on a number of polypeptides in addition to the 220 kDa band containing MHC. Arrow indicates position of the MHC II band.
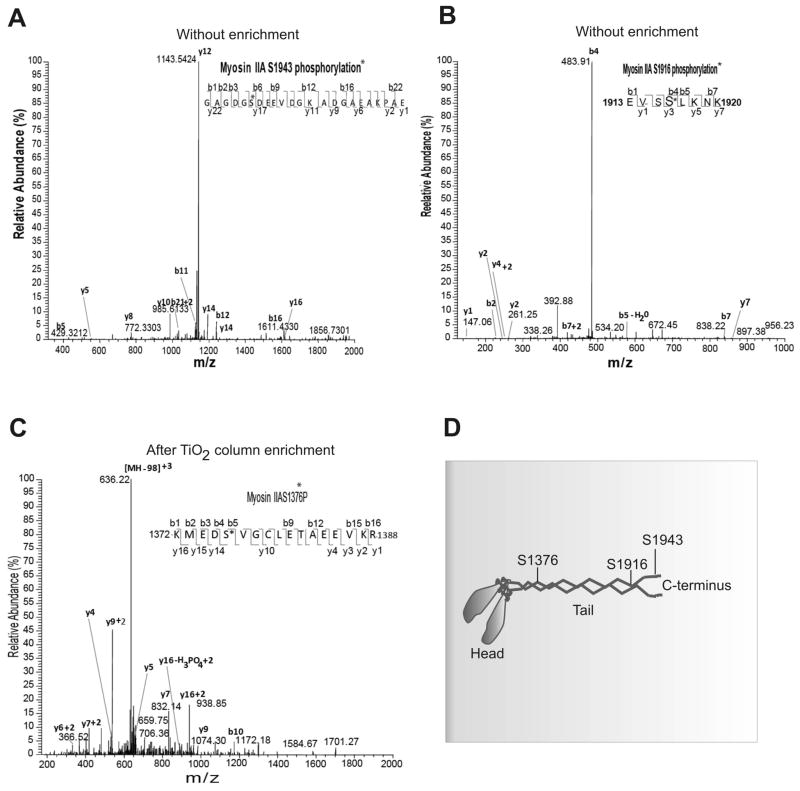
Mass spectrometric approaches were next employed to identify specific phosphorylated sites in the MHC II band isolated from actively spreading cells. Total coverage of MHC IIA peptides in this analysis was typically 60–70% (Figure S1). This analysis revealed multiple phosphorylation events in the MHC IIA polypeptide that were stimulated during active spreading on fibronectin. Three major fibronectin-stimulated MHC IIA phosphorylation events observed are represented by the tandem mass spectrum of monophosphorylated peptides 1372-KMEDpS*(1376)VGCLETAEEVKR-1388, 1913-EVSpS*(1916)LKNK-1920, and 1938-GAGDGpS*(1943)DEEVDGKADGAEAKPAE-1960 (Figure 3). Asterisk indicates amino acids that are potential sites for phosphorylation on MHC. In the manual analysis of the MS/MS CID spectrum related to the identification of S1943 phosphorylation, we observed a series of y-ions and several b-ion fragments that reflect the S1943 phosphorylation modification annotation. Using an expanded scale tandem MS spectra we can clearly observe, for example, 670.34 (b8+80–98) and 768.17(b8+80) ion pair. Similarly 799.3 (b9+80–98) and 897.3 (b9+80) ion pair can be detected as well. In addition to these b series ion pairs, we observed and annotated s few other y-ion pairs and b ion pairs that could be identified and attributed only to the modified peptide 1938-GAGDGpS*(1943)DEEVDGKADGAEAKPAE-1960 with one phosphorylation site on Ser-1943 (Figure 3A). We also identified other phosphorylation sites, Ser-1916 and Ser-1376 on the MHC of myosin IIA during cell spreading by applying similar manual data analyses approach (Figure 3B and 3C). The phosphorylation sites, Ser-1943 and Ser-1916 were identified without enrichment of phosphopeptides based on the TiO2 approach, however, the other phosphorylation site of Ser-1376 on MHC was identified only after the enrichment of phosphopeptides using TiO2 columns due to low abundance of the modified peptide as corroborated by MS peak intensities. This suggests that the Ser-1943 and Ser-1916 on the MHC of myosin IIA undergo a higher level phosphorylation in comparison with the phosphorylation of Ser-1376 in spreading MDA-MB 231 cells.
Figure 3.
Mapping of phosphorylation sites on MHC IIA by tandem mass spectrometry. Relative abundance of the phosphorylation residues was calculated with total MHC. (A) Tandem mass spectrum of a MHC peptide (amino acids 1938 to 1960) containing pS1943. No peptide enrichment was needed to detect this event. (B) Tandem mass spectrum of a MHC peptide (amino acids 1913 to 1920) containing pS1916. No peptide enrichment was needed to detect this event. (C) Tandem mass spectrum of a MHC peptide (amino acids 1372 to 1388) containing pS1377. Peptide enrichment was performed using TiO2 columns. (D). Schematic location of identified phosphorylation sites in MHC IIA.
We next applied similar mass spectrometry approach to identify phosphorylation sites on the MHC of myosin IIA isolated from non-migrating quiescent and suspension cells. These studies uncovered the same phosphorylation sites, Ser-1376, Ser-1916, and Ser-1943 on the MHC of myosin IIA. Analogous to the spreading cell experimental observations, identification of Ser-1376 as site of phosphorylation needed TiO2 enrichment. The consistently very high phosphorylation levels (Figure 3) during the spreading stage suggests that integrin engagement during spreading activates these phosphorylation events. Therefore, further quantitation of relative phosphorylation levels was needed to establish the level of increase in phosphorylation at these target sites in response to integrin engagement.
The MHC of myosin IIA undergoes a significant phosphorylation on Ser-1943 during lamellipodia extension
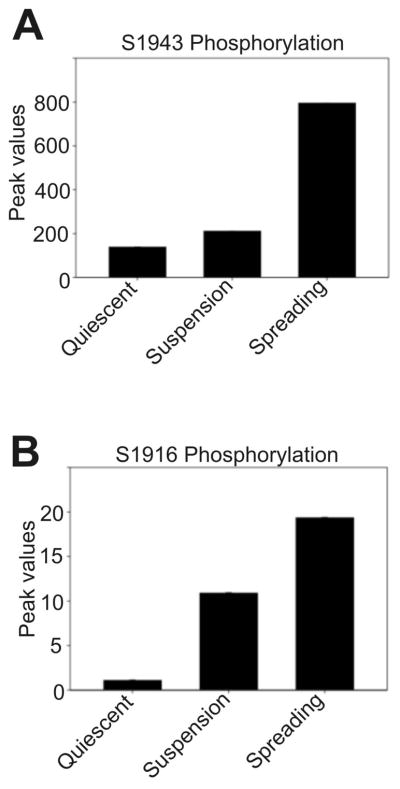
The relative quantitation of phosphorylation levels of the MHC of myosin IIA in quiescent, suspension versus spreading MDA-MB 231 cells was calculated, using duplicate experimental trials, based on well-characterized phosphopeptides from the Ser-1943 and Ser-1916 sites by either using a ratiometric chromatographic peak area measurement with respect to the native unphosphorylated peptide or alternately by ratiometric chromatographic peak area measurements with respect to a native peptide from the protein digest that does not vary in its abundance across all three stages of cells. Specifically, Ser-1943 quantitation was performed in this work using the first-mentioned method as the native peptide for the sequence of interest KGAGDGSDEEVDGKADGAEAKPAE is identified in its +2 or +3 charge state form, depending on the sample (precursor masses 768.3502 and 596.5056, respectively). After evaluating the extracted ion chromatogram (EIC) for this precursor in both the native form as well as phosphorylated form (precursor mass 795.0043), peak area calculations were performed after smoothing the chromatographic profile using a Gaussian curve shape data conversion option within the XCalibur data analysis software. These methods quantify the averaged relative levels to be 138, 211, and 795 for Ser-1943 phosphorylation on MHC IIA in the quiescent, suspension, and spreading populations, respectively, thus revealing a significant increase in the phosphorylation of Ser-1943 on MHC IIA during active spreading on fibronectin (Figure 4A). In the case of quantitation of Ser-1916 phosphorylation, the peak area of a native peptide with the sequence of 15-NFINNPLAQADWAAK-29 (precursor mass 836.9271, +2 charge state) that is unaltered in abundance in quiescent, suspension, and spreading cells, was used as a reference peak for the ratiometric peak area calculations. Upon obtaining the EIC profiles, the peak area calculations were again performed using the Gaussian smoothing option. These studies showed averaged relative levels of 1, 10, and 19.26 for Ser-1916 phosphorylation on MHC IIA in the quiescent, suspension, and spreading states, respectively (Figure 4B). This analysis reveals an increase in the phosphorylation of Ser-1916 on the MHC of myosin IIA during cell spreading as well. However, we routinely observed greater abundance of phosphopeptides carrying Ser-1943 phosphorylation in comparison with the abundance of a peptide with phosphorylated Ser-1916. This observation suggests that in response to integrin engagement, Ser-1943 phosphorylation may have the greater role in regulating myosin IIA filament dynamics.
Figure 4.
Mass spectroscopy based quantitation of MHC IIA phosphorylation changes during cell spreading. (A) Abundance of phospho-S1943 increases significantly during spreading. (B) Abundance of phospho-S1916 also rises, although these peptides were recovered with lower abundance relative to the S1943 peptides.
Leading edge phosphorylation on S1943 of myosin IIA is activated during spreading and regulates myosin IIA localization
We developed phosphospecific polyclonal antibodies using a custom-made specific phosphopeptide (amino-CARKGAGDG-pS1943-DEEVDGKA-carboxyl) of the MHC to understand the role of Ser-1943 phosphorylation in the regulation of myosin II localization to the lamellipodia during spreading and migration (Figure 5A). To test the pS1943-specific antibody, total lysates were made from spreading MDA-MB 231 cells, using lysis buffer without addition of phosphatase inhibitors. These samples were treated with λ phosphatase or calf intestinal phosphatase (CIP) in the presence and absence of a cocktail of phosphatase inhibitors followed by SDS-PAGE and western blotting. As shown in Figure 5B, the anti-pS1943 antibody detects a robust signal in these lysates, but incubation with either λ phosphatase or CIP severely reduces this signal. Presence of a phosphatase inhibitor cocktail blocks this dephosphorylation by λ phosphatase or CIP. These results indicate that the anti-pS1943 antibody is specific to the S1943-phosphorylated MHC. To test for possible cross-reactivity of the anti-pS1943 antibody with the MHCs of the other myosin II isoforms (MHC IIB or MHC IIC), we performed western blot analysis of total lysates made from cells lacking myosin IIA expression (COS-7), and cells lacking myosin IIB expression (HeLa cells from Clontech). The anti-pS1943 antibody reacted only with cells that express MHC IIA, confirming that it is specific to the phosphorylated MHC of myosin IIA motor protein (Figure 5C). Furthermore, western blot analysis of a GFP-MHC-IIA reporter carrying an S1943A substitution revealed a loss of immunoreactivity, confirming epitope specificity (data not shown).
Figure 5.
Development and characterization of antibodies specific to myosin IIA-pS1943. (A) Sequence of a peptide of myosin IIA carrying the phosphorylated S-1943 residue. (B) Western blot analysis of the specificity of the MHC IIA-pS1943 antibody. Total cell lysates made from spreading MDA-MB 231 cells were treated with λ and calf intestinal phosphatases in the absence or presence of 1 mM Na3VO4 and a cocktail of protease inhibitors (16) at room temperature for 15 min. The dephosphorylation step was stopped by adding equal volume of 2X Laemmli sample buffer followed by heating at 95°C for two min. Following probing for with the MHC IIA-pS1943 antibody, PVDF membranes were stripped and reprobed with myosin IIA Ab. (C) The MHC IIA-pS1943 antibody does not recognize MHC IIB or MHC IIC. Total lysates made from MDA-MB 231 cells (which express myosin IIC at very low level), COS-7 cells (which express no MHC IIA), and HeLa- Clontech cells (which express neither MHC IIB nor IIC) were subjected to western blot analysis using antibodies to MHC IIA, IIB, IIC, and the custom MHC IIA-pS1943 phospho-specific antibody.
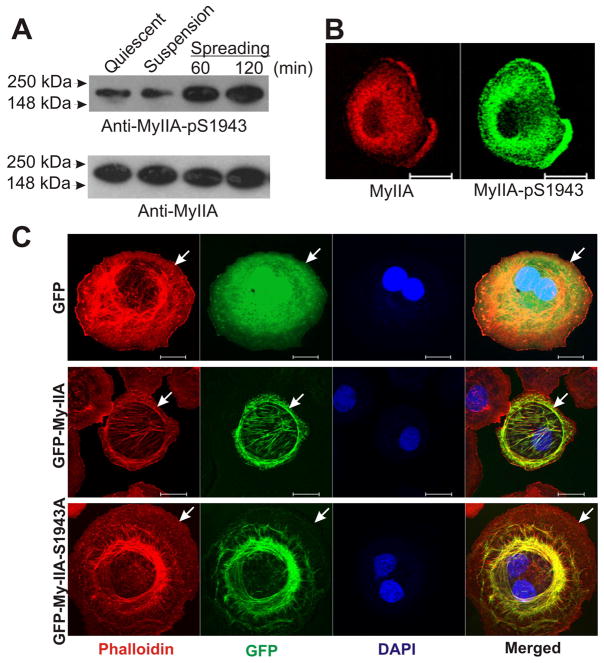
To further examine the timing and spatial distribution of MHC IIA phosphorylation during spreading, we performed western blot analysis and immunohistochemical analysis of MDA-MB 231 cells during spreading with the anti-pS1943 antibody. Western blot analysis revealed a significant increase in the phosphorylation of the MHC of myosin II in 60 min. spreading MDA-MB 231 cells as compared to either undisturbed cells plated overnight (“quiescent”), or as compared to cells kept in suspension for 60 min (Figure 6A). These results support the patterns observed with Pro-Q staining and the elevated phospho-S1943 signal reported by comparative mass spectroscopic approaches. Immunostain analysis of spreading MDA-MB 231 cells revealed significant accumulation of S1943 signal in the spreading margin (Figure 6B). These results provide the first evidence to date that myosin IIA heavy chain phosphorylation is upregulated during integrin-fibronectin engagement, and the first evidence to date that this phosphorylation is specifically enriched in the leading edge lamellar zone. This enrichment of S1943 phosphorylation in leading edge protrusions correlates with elevation of regulatory light chain (RLC) phosphorylation in the same zones, as shown in our earlier work (14). This dual regulation of myosin IIA suggests that that myosin IIA recruited to lamellar protrusions is both activated on RLC to assembly and produce force, but also activated for filament assembly turnover via MHC phosphorylation.
Figure 6.
Phosphorylation and localization of the MHC IIA in MDA-MB 231 cells extending lamellipodia during spreading on fibronectin. (A) Myosin IIA undergoes phosphorylation on Ser-1943 of MHC during spreading. Total lysates made from the non-migrating quiescent, suspension, and spreading MDA-MB 231 cells were subjected to western blot analysis using the anti-pS1943 antibody. (B) Immunostaining of the spreading cell to study colocalization of myosin IIA-pS1943 and endogenous myosin IIA. (C) Abrogation of Ser-1943 phosphorylation on myosin IIA impairs localization to the leading edge during cell spreading. COS-7 cells transiently expressing GFP, GFP-MHC IIA, or the mutant GFP-MHC IIA-S1943A were allowed to spread on fibronectin for 60 min, then processed for imaging. Arrows mark cell edge in each series. Scale bars = 20 μM.
We next performed localization studies by transiently expressing the full length GFP-MHC conjugated protein (GFP-My-IIA) carrying Ser-1943 substituted with alanine (S1943A). Earlier studies have examined behavior of the GFP-myosin IIA-S1943A mutant in MDA-MB 231 or HeLa cells that express endogenous MHC IIA (8, 10), which may ameliorate potential assembly defects of the mutant protein through co-assembly effects. We therefore transfected constructs into COS-7 cells that express no detectable myosin IIA protein, to evaluate GFP-myosin IIA-S1943A behavior in cells lacking an endogenous pool of myosin IIA. During spreading on fibronectin the control GFP-myosin IIA construct displayed lamellar localization corresponding to that of endogenous myosin IIA in MDA-MB 231 cells (14). However, the GFP-myosin IIA-S1943 mutant, while capable of assembly, displayed consistent localization that was more central, with substantial fibrillar assembly in the more central cell body of the spreading cells, and little GFP-myosin IIA assembly at the spreading margin (Figure 6C). Collectively, this data supports a model that leading edge phosphorylation of myosin IIA on S1943 has a role in upregulating filament dynamics at the leading edge, and that the relevant kinase responsible may be specifically activated in response to matrix engagement.
Casein kinase II is not involved in the regulation of the phosphorylation of myosin IIA during cell spreading
We hypothesized involvement of CK II in the regulation of MHC phosphorylation based upon early studies showing that the S1943 phosphorylation site in myosin IIA displays a CK-II consensus motif (21), and that this site can be phosphorylated by CK-II in purified biochemical assays (9, 12). Dulyaninova and colleagues further observed in MDA-MB 231 cells an EGF-stimulated co-immunoprecipitation of CK-II with myosin IIA (8).
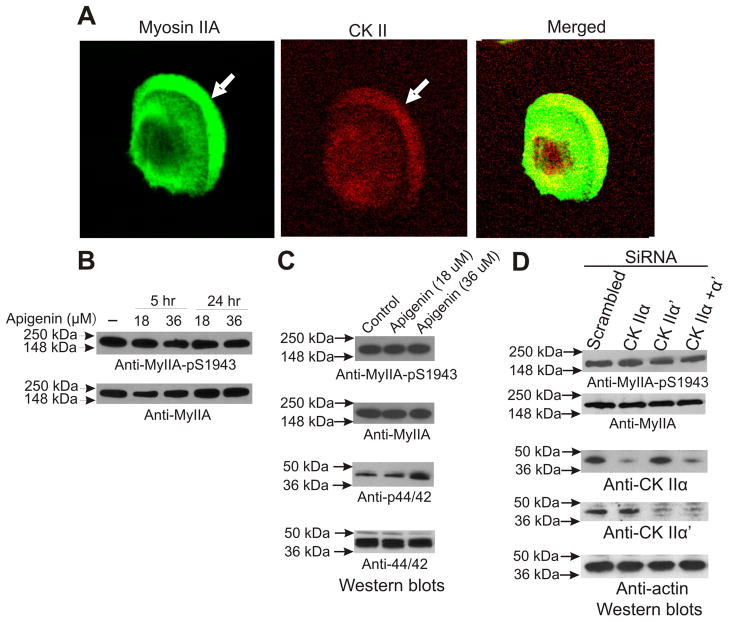
Two distinct genes encode CK-II catalytic subunits, referred to as α and α′ . These associate into a tetrameric holoenzyme carrying two catalytic subunits, believed to intermix so that the active tetramer can contain aa, aα′, or α′α′ forms (22). Via western blot analysis we found that MDA-MB 231 cells express both types of catalytic subunits quiescent and spreading cells. Immunostaining for CK-II a subunit revealed a modest enrichment of CK-II in the leading lamellar region (Figure 7A). These results indicate that CKII could be involved in the phosphorylation of myosin IIA during cell spreading.
Figure 7.
CK-II is not essential for myosin IIA S1943 phosphorylation during spreading. (A) CK-II colocalizes with myosin IIA in the spreading cells. MDA-MB 231 cells spreading on fibronectin coated plates were fixed staining with antibodies against either MHC IIA and CK- II alpha subunit. Arrows denote cell edge. (B) Pharmacological inhibition of CK-II does not affect S1943 phosphorylation of myosin IIA during cell spreading. Total lysates made from MDA-MB 231 cells spreading in the presence and absence of apigenin for 60 min were subjected to western blot analysis using specific antibodies of myII-pS1943, myIIA, phospho ERK1/ERK2 and ERK1/ERK2. (C) Depletion of CK-II catalytic subunits by siRNA does not reduce S1943 phosphorylation during spreading on fibronectin. MDA-MB 231 cells were transfected with siRNA oligo pools, either scrambled or directed to CK-IIa, CK-IIa′, or with siRNA directed against both subunits. After 48 hrs cells were collected and allowed to spread on fibronectin surfaces for 60 min. Cell lysates were then collected for western blot analysis.
To determine whether CK-II is in fact responsible for phosphorylation of S1943 in live cells, we performed both pharmacological and siRNA inhibition studies. We evaluated quiescent and actively spreading cells, using MDA-MB 231 cancer cells and apigenin, an established pharmacological inhibitor of CK-II (23). Surprisingly, western blot analysis revealed no inhibition of phosphorylation of MHC IIA in cells spreading in the presence of apigenin in either quiescent cells (Figure 7B) or in spreading cells (Figure 7C). Phospho-specific antibodies to measure CK-II activation states are not commercially available. However, earlier studies have reported CK-II inhibition by apigenin results in indirect activation of ERK1/2 (p44/42) (23). We therefore assessed ERK1/2 activation in the apigenin-treated cells as a measure of CK-II inhibition, confirming upregulation of ERK1/2 (Figure 7C), supporting the conclusion that apigenin in our assays is in fact inhibiting CK-II. As a more direct test of the role of CK-II in spreading-induced S1943 phosphorylation, we depleted both CK-IIα and CK-IIα′ subunits via siRNA, and tested MHC IIA phosphorylation behavior during spreading. Despite very strong depletion of either or both subunits, cell lysates from spreading MDA-MB 231 cells displayed no change in S1943 phosphorylation levels on western blots (Figure 7C). Collectively, these studies strongly suggest that in the context of integrin engagement on fibronectin, the activation of MHC IIA phosphorylation on S1943 is not dependent on CK-II in intact cells. Further siRNA studies are clearly merited in other settings where S1943 phosphorylation has been reported in live cells, to determine whether CK-II is in fact involved in phosphorylation of myosin IIA on S1943 with other stimuli.
DISCUSSION
Myosin II heavy chain phosphorylation has long been recognized as a key regulator in the simple amoeba Dictyostelium, where phosphorylation of threonine residues at the distal end of the tail drives filament disassembly, and is the critical determinant of whether the protein is in the monomer versus bipolar filament state in vivo (24). Smooth muscle and nonmuscle myosin II heavy chain phosphorylation events have been reported in a variety of conditions in live cell labeling studies, and via in vitro biochemical assays, events recognized for many years (11, 12, 21). Via examination of target site consensus sequences, combined with phosphorylation analysis performed in vitro with purified myosin II or recombinant myosin II tail fragments, a general conclusion has emerged in the field that the S1916 site on human MHC IIA (and the analogous conserved sites on MHC IIB and on MHC II from other species such as rabbit) is a target for PKC isoforms. This conclusion was validated in one set of studies performed in rat RBL-2H3 mast cells, where myosin IIA is phosphorylated on the corresponding S1917 to drive cytoskeletal reorganization during antigen-stimulated histamine release (25). In that work, pharmacological inhibition of PKC subtypes, together with transfection of PKC isoform constructs was used to demonstrate that PKCβII was responsible for S1917 phosphorylation within intact mast cells. Similar live cell analysis has never been conducted with respect to the putative CK-II target site in the tail of myosin IIA, leaving an open question as to whether CK-II is the bone fide kinase that phosphorylates this site in live cells.
This putative CK-II site at S1943 is phosphorylated in cancer cells responding to EGF (8), and this phosphorylation modulates leading edge myosin IIA assembly both during EGF responses and during cancer cell lamellar spreading (10). Recent work has also suggested that abrogation of S1943 phosphorylation may be responsible for human platelet pathology observed with some alleles of MYH9-related disorders, in which a premature stop codon removes the last ~28 residues of MHC IIA, eliminating the S1943 phosphorylation site (10). Given the recognized role of myosin II lamellar assembly for processes ranging adhesion stabilization during cell migration (26) to platelet activation (27), it is surprising that there have been virtually no studies to date focused on identifying the upstream kinases and signaling events that regulate S1943 phosphorylation in the live cell setting. In view of the strong upregulation of CK-II levels in cancer progression (28), it is an appealing and plausible idea that CK-II could phosphorylate myosin IIA in physiological and pathological cell migration settings. However, in the context of integrin-stimulated S9143 phosphorylation in MDA-MB 231 cells, our data strongly suggests that another kinase or kinases are responsible for phosphorylation this residue. We suggest that CK-II may well participate in S1943 phosphorylation in other settings, but during activation of fibronectin/integrin signaling pathways it does not appear to be the relevant kinase. Future biochemical purification and pull-down studies are needed in the context of fibronectin-stimulated S1943 phosphorylation, and in the context of other stimuli, such as growth factors, to determine what upstream kinases drive this critical phosphorylation event in live cell and tissue settings.
Together with earlier studies documenting roles of S1943 phosphorylation in regulating myosin IIA assembly at the biochemical level (9, 29), the current studies support a model that MHC phosphorylation may be specifically activated during leading edge protrusion formation in response to matrix engagement. We propose that MHC phosphorylation in this setting may serve to destabilize myosin II filaments, facilitating their disassembly, which would be a requisite step necessary for forward monomer diffusion and re-assembly at more anterior zones during ongoing migration. Such a role would be analogous to the accepted models for how MHC phosphorylation regulates filament assembly and recycling during migration in the lower eukaryotic amoeba Dictyostelium (24). However, in the Dictyostelium there is no evidence that substratum engagement can specifically activate MHC phosphorylation. Thus the mammalian regulation of MHC IIA phosphorylation in response to fibronectin engagement may represent a level of control specifically evolved in response to the needs of cells to regulate migration in the context of a complex multicellular, multi-tissue environment.
Cancer cell metastasis involves multiple modes of cell migration, likely all important in different settings such as blood vessel transmigration versus intra-lymphoid migration or target organ penetration. Given the established importance of myosin II for cell migration through tight spaces and the critical role of integrin engagement for transmigration (4, 30), we speculate that regulation of myosin II assembly dynamics in response to integrin engagement could be an important and regulated component of cancer cell migration in the tissue setting. Future studies will be directed at testing this hypothesis.
Supplementary Material
Red Color indicates the protein sequence covered by LC-MS/MS approach with a single-protein specific database searching using Mascot (Matrix Science) Search Engine after applying accepted peptide score filtering. Sample shown represents coverage of myosin IIA taken from quiescent cells.
Acknowledgments
This work was supported by NIH grant GM077224 to T.T.E., a Case Western Proteomics Center pilot grant to V.B., and a National Institutes of Health Grant to the Case Comprehensive Cancer Center, P30-CA-043703 (PI: Gerson, Stanton).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Conti MA, Adelstein RS. Nonmuscle myosin II moves in new directions. J Cell Sci. 2008;121:11–18. doi: 10.1242/jcs.007112. [DOI] [PubMed] [Google Scholar]
- 2.Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR. Cell migration: integrating signals from front to back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 3.Yilmaz M, Christofori G. Mechanisms of motility in metastasizing cells. Mol Cancer Res. 2010;8:629–642. doi: 10.1158/1541-7786.MCR-10-0139. [DOI] [PubMed] [Google Scholar]
- 4.Lammermann T, Bader BL, Monkley SJ, Worbs T, Wedlich-Soldner R, Hirsch K, Keller M, Forster R, Critchley DR, Fassler R, Sixt M. Rapid leukocyte migration by integrin-independent flowing and squeezing. Nature. 2008;453:51–55. doi: 10.1038/nature06887. [DOI] [PubMed] [Google Scholar]
- 5.Vicente-Manzanares M, Ma X, Adelstein RS, Horwitz AR. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat Rev Mol Cell Biol. 2009;10:778–790. doi: 10.1038/nrm2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Even-Faitelson L, Ravid S. PAK1 and aPKCzeta regulate myosin II-B phosphorylation: a novel signaling pathway regulating filament assembly. Mol Biol Cell. 2006;17:2869–2881. doi: 10.1091/mbc.E05-11-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ludowyke RI, Peleg I, Beaven MA, Adelstein RS. Antigen-induced secretion of histamine and the phosphorylation of myosin by protein kinase C in rat basophilic leukemia cells. J Biol Chem. 1989;264:12492–12501. [PubMed] [Google Scholar]
- 8.Dulyaninova NG, House RP, Betapudi V, Bresnick AR. Myosin-IIA heavy-chain phosphorylation regulates the motility of MDA-MB-231 carcinoma cells. Mol Biol Cell. 2007;18:3144–3155. doi: 10.1091/mbc.E06-11-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dulyaninova NG, Malashkevich VN, Almo SC, Bresnick AR. Regulation of myosin-IIA assembly and Mts1 binding by heavy chain phosphorylation. Biochemistry. 2005;44:6867–6876. doi: 10.1021/bi0500776. [DOI] [PubMed] [Google Scholar]
- 10.Breckenridge MT, Dulyaninova NG, Egelhoff TT. Multiple Regulatory Steps Control Mammalian Nonmuscle Myosin II Assembly in Live Cells. Mol Biol Cell. 2009;20:338–347. doi: 10.1091/mbc.E08-04-0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kelley CA, Adelstein RS. The 204-kDa smooth muscle myosin heavy chain is phosphorylated in intact cells by casein kinase II on a serine near the carboxyl terminus. J Biol Chem. 1990;265:17876–17882. [PubMed] [Google Scholar]
- 12.Murakami N, Chauhan VP, Elzinga M. Two nonmuscle myosin II heavy chain isoforms expressed in rabbit brains: filament forming properties, the effects of phosphorylation by protein kinase C and casein kinase II, and location of the phosphorylation sites. Biochemistry. 1998;37:1989–2003. doi: 10.1021/bi971959a. [DOI] [PubMed] [Google Scholar]
- 13.Murakami N, Healy-Louie G, Elzinga M. Amino acid sequence around the serine phosphorylated by casein kinase II in brain myosin heavy chain. J Biol Chem. 1990;265:1041–1047. [PubMed] [Google Scholar]
- 14.Betapudi V, Licate LS, Egelhoff TT. Distinct roles of nonmuscle myosin II isoforms in the regulation of MDA-MB-231 breast cancer cell spreading and migration. Cancer Res. 2006;66:4725–4733. doi: 10.1158/0008-5472.CAN-05-4236. [DOI] [PubMed] [Google Scholar]
- 15.Betapudi V, Mason C, Licate L, Egelhoff TT. Identification and Characterization of a Novel α-Kinase with a von Willebrand Factor A-like Motif Localized to the Contractile Vacuole and Golgi Complex in Dictyostelium discoideum. Mol Biol Cell. 2005;16:2248–2262. doi: 10.1091/mbc.E04-07-0639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steimle PA, Naismith T, Licate L, Egelhoff TT. WD Repeat Domains Target Dictyostelium Myosin Heavy Chain Kinases by Binding Directly to Myosin Filaments. J Biol Chem. 2001;276:6853–6860. doi: 10.1074/jbc.M008992200. [DOI] [PubMed] [Google Scholar]
- 17.Betapudi V. Myosin II motor proteins with different functions determine the fate of lamellipodia extension during cell spreading. PLoS ONE. 2010;5:e8560. doi: 10.1371/journal.pone.0008560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cai Y, Biais N, Giannone G, Tanase M, Jiang G, Hofman JM, Wiggins CH, Silberzan P, Buguin A, Ladoux B, Sheetz MP. Nonmuscle myosin IIA-dependent force inhibits cell spreading and drives F-actin flow. Biophys J. 2006;91:3907–3920. doi: 10.1529/biophysj.106.084806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Even-Ram S, Doyle AD, Conti MA, Matsumoto K, Adelstein RS, Yamada KM. Myosin IIA regulates cell motility and actomyosin-microtubule crosstalk. Nat Cell Biol. 2007;9:299–309. doi: 10.1038/ncb1540. [DOI] [PubMed] [Google Scholar]
- 20.Matsumura F, Ono S, Yamakita Y, Totsukawa G, Yamashiro S. Specific localization of serine 19 phosphorylated myosin II during cell locomotion and mitosis of cultured cells. J Cell Biol. 1998;140:119–129. doi: 10.1083/jcb.140.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moussavi RS, Kelley CA, Adelstein RS. Phosphorylation of vertebrate nonmuscle and smooth muscle myosin heavy chains and light chains. Mol Cell Biochem. 1993;127–128:219–227. doi: 10.1007/BF01076773. [DOI] [PubMed] [Google Scholar]
- 22.Niefind K, Raaf J, Issinger OG. Protein kinase CK2 in health and disease: Protein kinase CK2: from structures to insights. Cell Mol Life Sci. 2009;66:1800–1816. doi: 10.1007/s00018-009-9149-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Llorens F, Miro FA, Casanas A, Roher N, Garcia L, Plana M, Gomez N, Itarte E. Unbalanced activation of ERK1/2 and MEK1/2 in apigenin-induced HeLa cell death. Exp Cell Res. 2004;299:15–26. doi: 10.1016/j.yexcr.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 24.de la Roche MA, Smith JL, Betapudi V, Egelhoff TT, Côté GP. Signaling pathways regulating Dictyostelium myosin II. J Muscle Res Cell Motil. 2002;23:703–718. doi: 10.1023/a:1024467426244. [DOI] [PubMed] [Google Scholar]
- 25.Ludowyke RI, Elgundi Z, Kranenburg T, Stehn JR, Schmitz-Peiffer C, Hughes WE, Biden TJ. Phosphorylation of nonmuscle myosin heavy chain IIA on Ser1917 is mediated by protein kinase C beta II and coincides with the onset of stimulated degranulation of RBL-2H3 mast cells. J Immunol. 2006;177:1492–1499. doi: 10.4049/jimmunol.177.3.1492. [DOI] [PubMed] [Google Scholar]
- 26.Webb DJ, Donais K, Whitmore LA, Thomas SM, Turner CE, Parsons JT, Horwitz AF. FAK-Src signalling through paxillin, ERK and MLCK regulates adhesion disassembly. Nat Cell Biol. 2004;6:154–161. doi: 10.1038/ncb1094. [DOI] [PubMed] [Google Scholar]
- 27.Adelstein RS, Pollard TD. Platelet contractile proteins. Prog Hemost Thromb. 1978;4:37–58. [PubMed] [Google Scholar]
- 28.Trembley JH, Wang G, Unger G, Slaton J, Ahmed K. Protein kinase CK2 in health and disease: CK2: a key player in cancer biology. Cell Mol Life Sci. 2009;66:1858–1867. doi: 10.1007/s00018-009-9154-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murakami N, Kotula L, Hwang YW. Two distinct mechanisms for regulation of nonmuscle myosin assembly via the heavy chain: phosphorylation for MIIB and mts 1 binding for MIIA. Biochemistry. 2000;39:11441–11451. doi: 10.1021/bi000347e. [DOI] [PubMed] [Google Scholar]
- 30.Breckenridge MT, Egelhoff TT, Baskaran H. A microfluidic imaging chamber for the direct observation of chemotactic transmigration. Biomed Microdevices. 2010;12:543–553. doi: 10.1007/s10544-010-9411-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Red Color indicates the protein sequence covered by LC-MS/MS approach with a single-protein specific database searching using Mascot (Matrix Science) Search Engine after applying accepted peptide score filtering. Sample shown represents coverage of myosin IIA taken from quiescent cells.