Abstract
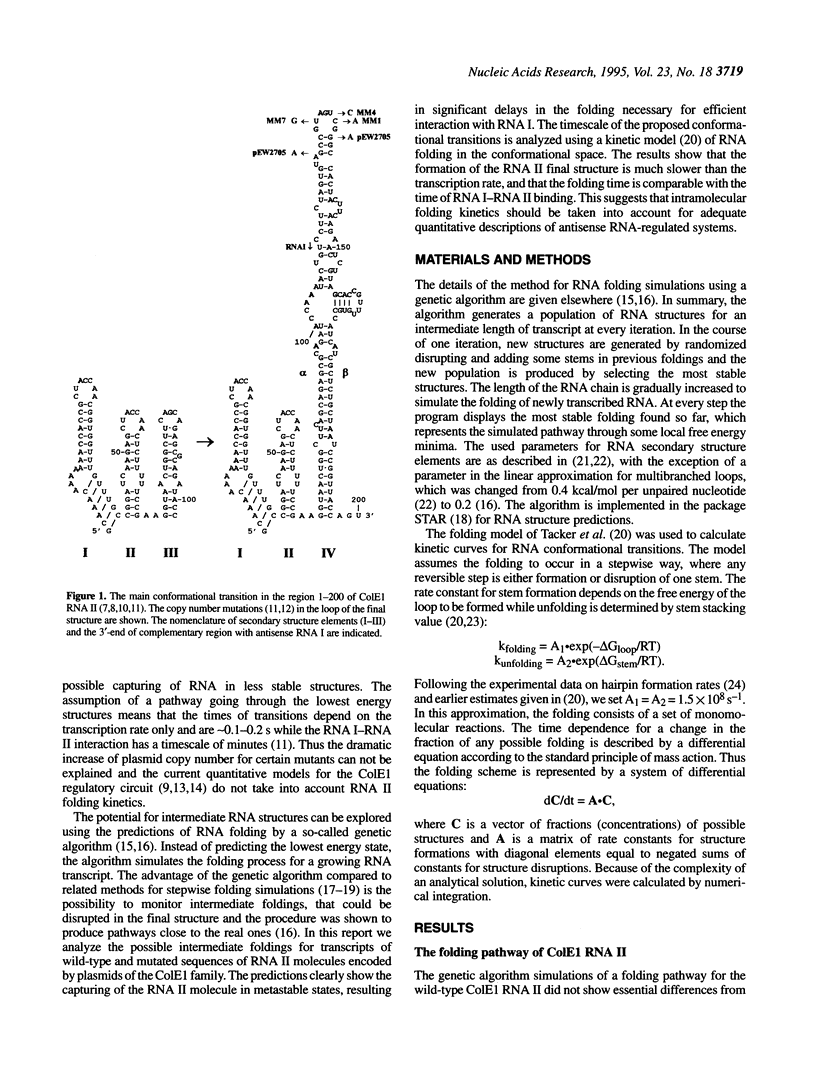
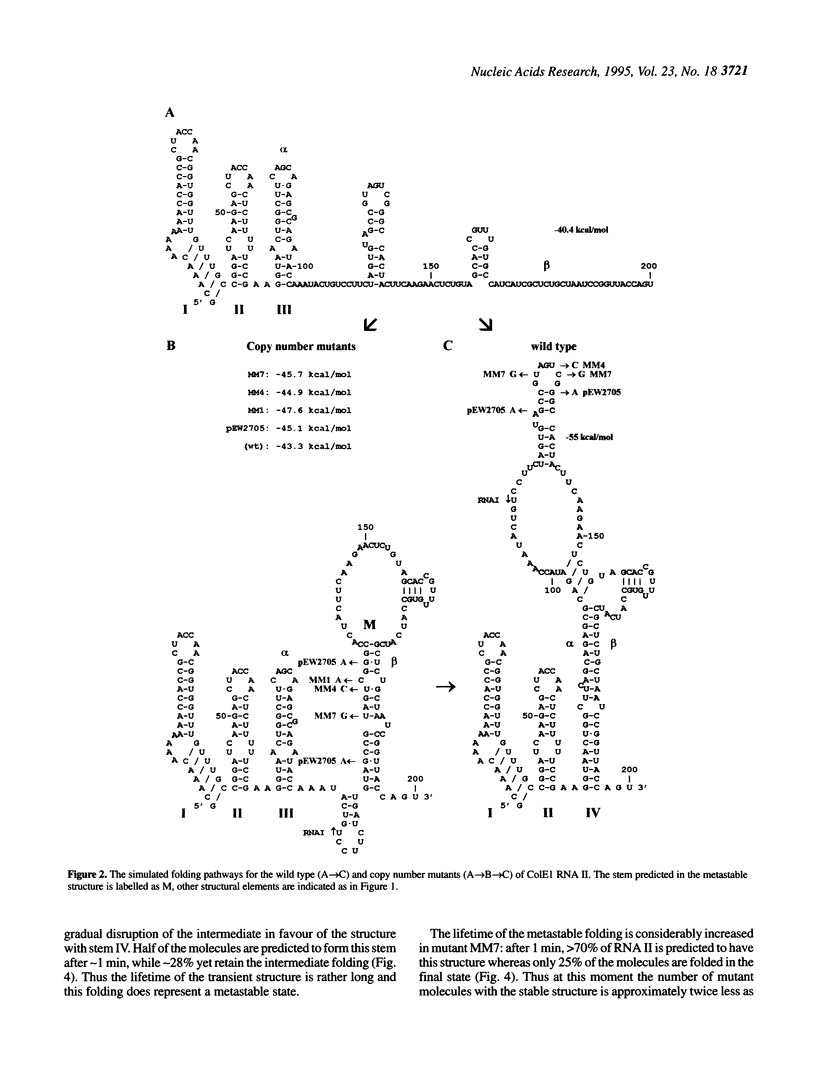
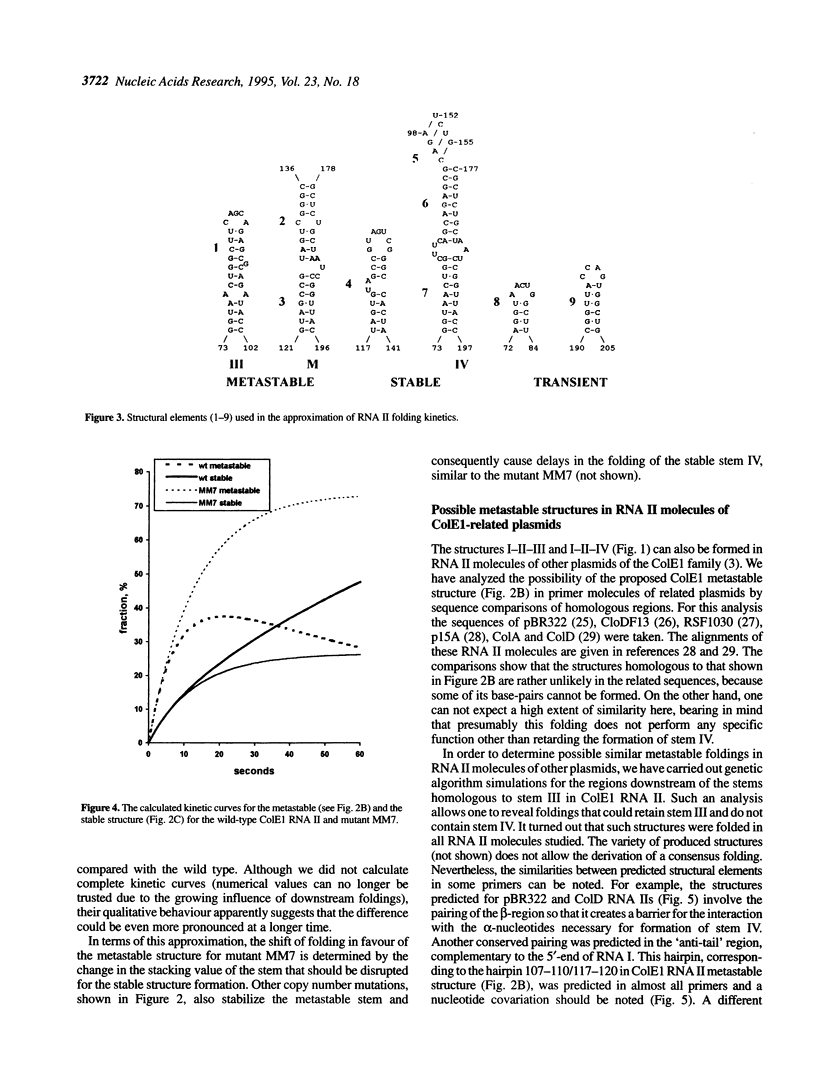
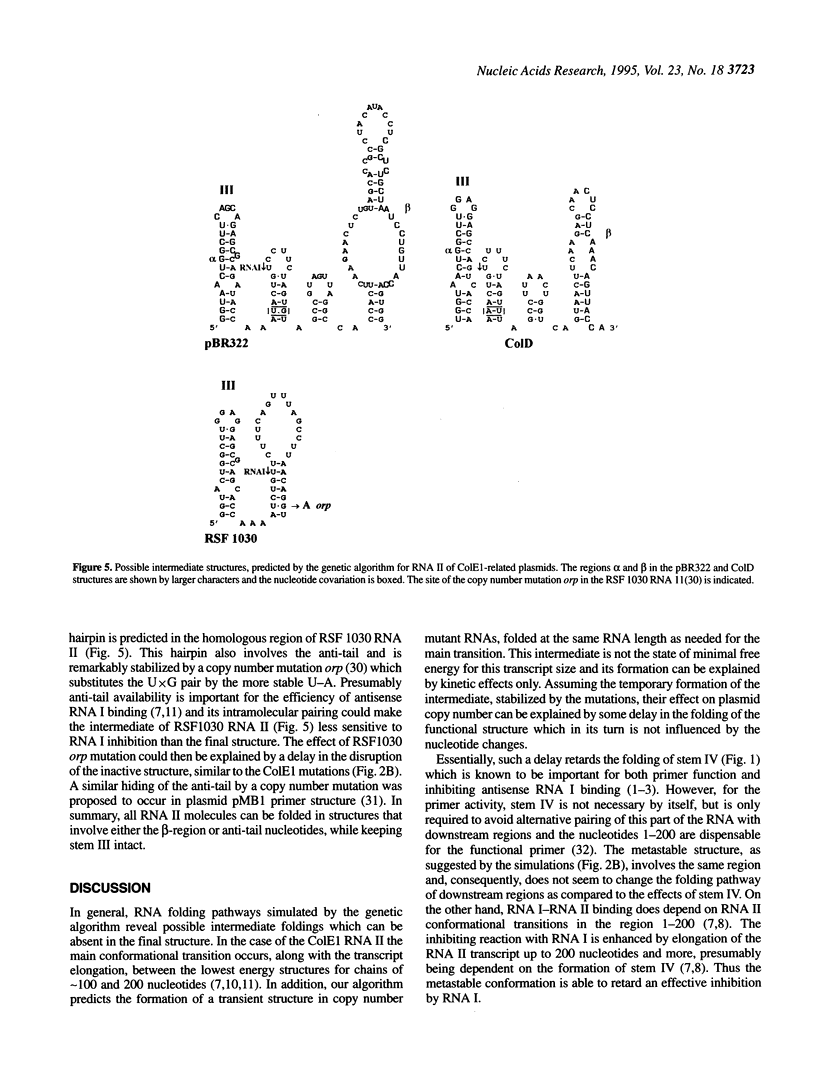
Replication of the ColE1 group plasmids is kinetically regulated by the interaction between plasmid-encoded primer RNA II and antisense RNA I. The binding is dependent on alternative RNA II conformations, formed during the transcription, and effectively inhibits the primer function within some time interval. In this paper, the folding pathways for the wild type and copy number mutants of ColE1 RNA II are studied using simulations by a genetic algorithm. The simulated pathways reveal a transient formation of a metastable structure, which is stabilized by copy number mutations. The folding kinetics of the proposed conformational transitions is calculated using a model of a multistep refolding process with elementary steps of double-helical stem formation or disruption. The approximation shows that the lifetime of the metastable structure is relatively long and is considerably increased in the mutants, resulting in a delay of the formation of the stable RNA II structure, which is the most sensitive to the inhibition by the antisense RNA I. Thus the effect of copy number mutations can be interpreted as a compression of the time window of effective inhibition due to an increased time spent by the RNA II in the metastable state. The implications of metastable foldings in RNA functioning are discussed.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abrahams J. P., van den Berg M., van Batenburg E., Pleij C. Prediction of RNA secondary structure, including pseudoknotting, by computer simulation. Nucleic Acids Res. 1990 May 25;18(10):3035–3044. doi: 10.1093/nar/18.10.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biebricher C. K., Luce R. In vitro recombination and terminal elongation of RNA by Q beta replicase. EMBO J. 1992 Dec;11(13):5129–5135. doi: 10.1002/j.1460-2075.1992.tb05620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brendel V., Perelson A. S. Quantitative model of ColE1 plasmid copy number control. J Mol Biol. 1993 Feb 20;229(4):860–872. doi: 10.1006/jmbi.1993.1092. [DOI] [PubMed] [Google Scholar]
- Brenner M., Tomizawa J. Quantitation of ColE1-encoded replication elements. Proc Natl Acad Sci U S A. 1991 Jan 15;88(2):405–409. doi: 10.1073/pnas.88.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castagnoli L., Lacatena R. M., Cesareni G. Analysis of dominant copy number mutants of the plasmid pMB1. Nucleic Acids Res. 1985 Jul 25;13(14):5353–5367. doi: 10.1093/nar/13.14.5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesareni G., Helmer-Citterich M., Castagnoli L. Control of ColE1 plasmid replication by antisense RNA. Trends Genet. 1991 Jul;7(7):230–235. doi: 10.1016/0168-9525(91)90370-6. [DOI] [PubMed] [Google Scholar]
- Chao M. Y., Kan M. C., Lin-Chao S. RNAII transcribed by IPTG-induced T7 RNA polymerase is non-functional as a replication primer for ColE1-type plasmids in Escherichia coli. Nucleic Acids Res. 1995 May 25;23(10):1691–1695. doi: 10.1093/nar/23.10.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X. N., Womble D. D., Rownd R. H. Transcriptional pausing in a region important for plasmid NR1 replication control. J Bacteriol. 1987 Dec;169(12):5353–5363. doi: 10.1128/jb.169.12.5353-5363.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguchi Y., Itoh T., Tomizawa J. Antisense RNA. Annu Rev Biochem. 1991;60:631–652. doi: 10.1146/annurev.bi.60.070191.003215. [DOI] [PubMed] [Google Scholar]
- Fernández A, Shakhnovich EI. Activation-energy landscape for metastable RNA folding. Phys Rev A. 1990 Sep 15;42(6):3657–3659. doi: 10.1103/physreva.42.3657. [DOI] [PubMed] [Google Scholar]
- Fitzwater T., Zhang X. Y., Elble R., Polisky B. Conditional high copy number ColE1 mutants: resistance to RNA1 inhibition in vivo and in vitro. EMBO J. 1988 Oct;7(10):3289–3297. doi: 10.1002/j.1460-2075.1988.tb03196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freier S. M., Kierzek R., Jaeger J. A., Sugimoto N., Caruthers M. H., Neilson T., Turner D. H. Improved free-energy parameters for predictions of RNA duplex stability. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gultyaev A. P. The computer simulation of RNA folding involving pseudoknot formation. Nucleic Acids Res. 1991 May 11;19(9):2489–2494. doi: 10.1093/nar/19.9.2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gultyaev A. P., van Batenburg E., Pleij C. W. Similarities between the secondary structure of satellite tobacco mosaic virus and tobamovirus RNAs. J Gen Virol. 1994 Oct;75(Pt 10):2851–2856. doi: 10.1099/0022-1317-75-10-2851. [DOI] [PubMed] [Google Scholar]
- Gultyaev A. P., van Batenburg F. H., Pleij C. W. The computer simulation of RNA folding pathways using a genetic algorithm. J Mol Biol. 1995 Jun 30;250(1):37–51. doi: 10.1006/jmbi.1995.0356. [DOI] [PubMed] [Google Scholar]
- Jaeger J. A., Turner D. H., Zuker M. Improved predictions of secondary structures for RNA. Proc Natl Acad Sci U S A. 1989 Oct;86(20):7706–7710. doi: 10.1073/pnas.86.20.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loss P., Schmitz M., Steger G., Riesner D. Formation of a thermodynamically metastable structure containing hairpin II is critical for infectivity of potato spindle tuber viroid RNA. EMBO J. 1991 Mar;10(3):719–727. doi: 10.1002/j.1460-2075.1991.tb08002.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez H. M. An RNA folding rule. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):323–334. doi: 10.1093/nar/12.1part1.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masukata H., Tomizawa J. Control of primer formation for ColE1 plasmid replication: conformational change of the primer transcript. Cell. 1986 Jan 17;44(1):125–136. doi: 10.1016/0092-8674(86)90491-5. [DOI] [PubMed] [Google Scholar]
- Merlin S., Polisky B. Assessment of quantitative models for plasmid ColE1 copy number control. J Mol Biol. 1995 Apr 28;248(2):211–219. doi: 10.1016/s0022-2836(95)80043-3. [DOI] [PubMed] [Google Scholar]
- Moser D. R., Moser C. D., Sinn E., Campbell J. L. Suppressors of a temperature-sensitive copy-number mutation in plasmid NTP1. Mol Gen Genet. 1983;192(1-2):95–100. doi: 10.1007/BF00327652. [DOI] [PubMed] [Google Scholar]
- Nordström K., Wagner E. G. Kinetic aspects of control of plasmid replication by antisense RNA. Trends Biochem Sci. 1994 Jul;19(7):294–300. doi: 10.1016/0968-0004(94)90008-6. [DOI] [PubMed] [Google Scholar]
- Novick R. P., Iordanescu S., Projan S. J., Kornblum J., Edelman I. pT181 plasmid replication is regulated by a countertranscript-driven transcriptional attenuator. Cell. 1989 Oct 20;59(2):395–404. doi: 10.1016/0092-8674(89)90300-0. [DOI] [PubMed] [Google Scholar]
- Perelson A. S., Brendel V. Kinetics of complementary RNA-RNA interaction involved in plasmid ColE1 copy number control. J Mol Biol. 1989 Jul 20;208(2):245–255. doi: 10.1016/0022-2836(89)90386-0. [DOI] [PubMed] [Google Scholar]
- Polisky B., Zhang X. Y., Fitzwater T. Mutations affecting primer RNA interaction with the replication repressor RNA I in plasmid CoIE1: potential RNA folding pathway mutants. EMBO J. 1990 Jan;9(1):295–304. doi: 10.1002/j.1460-2075.1990.tb08108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu F., Heinrich C., Loss P., Steger G., Tien P., Riesner D. Multiple pathways of reversion in viroids for conservation of structural elements. EMBO J. 1993 May;12(5):2129–2139. doi: 10.1002/j.1460-2075.1993.tb05861.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selzer G., Som T., Itoh T., Tomizawa J. The origin of replication of plasmid p15A and comparative studies on the nucleotide sequences around the origin of related plasmids. Cell. 1983 Jan;32(1):119–129. doi: 10.1016/0092-8674(83)90502-0. [DOI] [PubMed] [Google Scholar]
- Som T., Tomizawa J. Origin of replication of Escherichia coli plasmid RSF 1030. Mol Gen Genet. 1982;187(3):375–383. doi: 10.1007/BF00332615. [DOI] [PubMed] [Google Scholar]
- Stuitje A. R., Spelt C. E., Veltkamp E., Nijkamp H. J. Identification of mutations affecting replication control of plasmid Clo DF13. Nature. 1981 Mar 19;290(5803):264–267. doi: 10.1038/290264a0. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Tacker M., Fontana W., Stadler P. F., Schuster P. Statistics of RNA melting kinetics. Eur Biophys J. 1994;23(1):29–38. doi: 10.1007/BF00192203. [DOI] [PubMed] [Google Scholar]
- Tomizawa J. Control of ColE1 plasmid replication. Intermediates in the binding of RNA I and RNA II. J Mol Biol. 1990 Apr 20;212(4):683–694. doi: 10.1016/0022-2836(90)90230-j. [DOI] [PubMed] [Google Scholar]
- Tomizawa J. Control of ColE1 plasmid replication: binding of RNA I to RNA II and inhibition of primer formation. Cell. 1986 Oct 10;47(1):89–97. doi: 10.1016/0092-8674(86)90369-7. [DOI] [PubMed] [Google Scholar]
- Tomizawa J., Som T. Control of ColE1 plasmid replication: enhancement of binding of RNA I to the primer transcript by the Rom protein. Cell. 1984 Oct;38(3):871–878. doi: 10.1016/0092-8674(84)90282-4. [DOI] [PubMed] [Google Scholar]
- Wagner E. G., Simons R. W. Antisense RNA control in bacteria, phages, and plasmids. Annu Rev Microbiol. 1994;48:713–742. doi: 10.1146/annurev.mi.48.100194.003433. [DOI] [PubMed] [Google Scholar]
- Wong E. M., Polisky B. Alternative conformations of the ColE1 replication primer modulate its interaction with RNA I. Cell. 1985 Oct;42(3):959–966. doi: 10.1016/0092-8674(85)90292-2. [DOI] [PubMed] [Google Scholar]
- Zuker M., Jaeger J. A., Turner D. H. A comparison of optimal and suboptimal RNA secondary structures predicted by free energy minimization with structures determined by phylogenetic comparison. Nucleic Acids Res. 1991 May 25;19(10):2707–2714. doi: 10.1093/nar/19.10.2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zverev V. V., Khmel I. A. The nucleotide sequences of the replication origins of plasmids ColA and ColD. Plasmid. 1985 Nov;14(3):192–199. doi: 10.1016/0147-619x(85)90002-2. [DOI] [PubMed] [Google Scholar]
- van Batenburg F. H., Gultyaev A. P., Pleij C. W. An APL-programmed genetic algorithm for the prediction of RNA secondary structure. J Theor Biol. 1995 Jun 7;174(3):269–280. doi: 10.1006/jtbi.1995.0098. [DOI] [PubMed] [Google Scholar]



