Abstract
The phosphorylation and dephosphorylation of proteins by kinases and phosphatases constitute an essential regulatory network in eukaryotic cells. This network supports the flow of information from sensors through signaling systems to effector molecules, and ultimately drives the phenotype and function of cells, tissues, and organisms. Dysregulation of this process has severe consequences and is one of the main factors in the emergence and progression of diseases, including cancer. Thus, major efforts have been invested in developing specific inhibitors that modulate the activity of individual kinases or phosphatases; however, it has been difficult to assess how such pharmacological interventions would affect the cellular signaling network as a whole. Here, we used label-free, quantitative phosphoproteomics in a systematically perturbed model organism (Saccharomyces cerevisiae) to determine the relationships between 97 kinases, 27 phosphatases, and more than 1000 phosphoproteins. We identified 8814 regulated phosphorylation events, describing the first system-wide protein phosphorylation network in vivo. Our results show that, at steady state, inactivation of most kinases and phosphatases affected large parts of the phosphorylation-modulated signal transduction machinery, and not only the immediate downstream targets. The observed cellular growth phenotype was often well maintained despite the perturbations, arguing for considerable robustness in the system. Our results serve to constrain future models of cellular signaling and reinforce the idea that simple linear representations of signaling pathways might be insufficient for drug development and for describing organismal homeostasis.
INTRODUCTION
Protein kinases, and, to a lesser extent, protein phosphatases, are attractive drug targets (1–5); however, although their respective catalytic activities are well characterized, their functions in vivo remain relatively poorly understood. Despite extensive in vitro (6), in silico (7), or indirect in vivo assays (8), our knowledge of the global relationships between kinases, phosphatases, and their substrates remains fragmented (2). Even less is known about the more downstream, indirect consequences of kinase activity, making rational selection of suitable candidates for therapeutic interventions difficult; consequently, many promising kinase inhibitors are ultimately retired from development (9).
One promising approach for closing this knowledge gap is the organism-wide, quantitative assessment of all phosphorylated proteins, comparing phosphorylation status in wild-type cells to that in cells that have undergone systematic perturbations of their kinases or phosphatases. Progress in phosphoproteomics technology has brought this goal within reach by enabling the reproducible quantification of thousands of phosphorylation sites in a single study (10–12). Although the throughput is not yet sufficient to systematically address all 518 protein kinases and 147 protein phosphatases in human cells (13, 14), simpler organisms, such as yeast, can be addressed. Yeast in particular is frequently used as a model to study human diseases (15), including cancer, mitochondrial diseases, and even neurological disorders caused by protein misfolding (16, 17). Although some signaling systems, such as the apoptotic machinery, are absent in yeast, other parts of its signaling network display substantial similarities to those in human cells (18, 19). Of the 161 kinases and phosphatases in yeast, 136 are conserved in humans at more than 30% amino acid sequence identity (table S1), and some human signaling proteins can even replace their yeast counterparts (20). Here, we used a combination of phosphoproteomics measurements and computational methods (11) to detect and quantify the system-wide responses in the yeast phosphoproteome upon deletion or inhibition of most of its kinases and phosphatases.
RESULTS
Experimental strategy
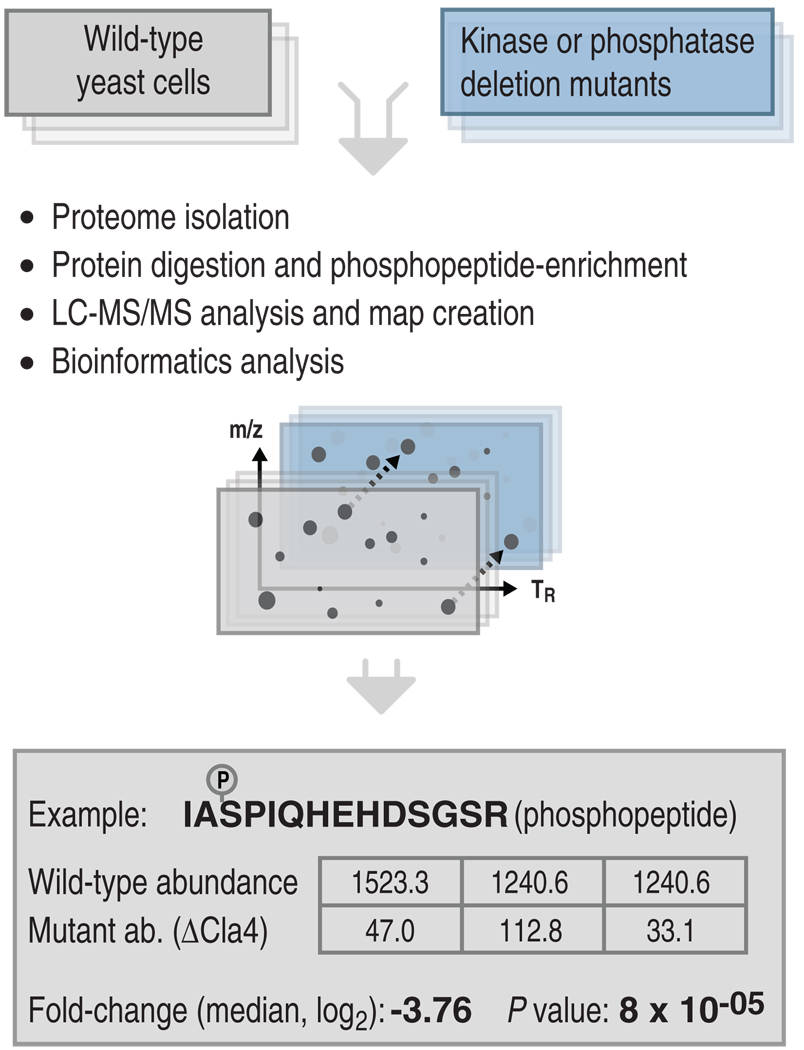
We developed an integrated experimental and computational strategy for high-throughput comparative phosphoproteomic analysis in Saccharomyces cerevisiae (Fig. 1), which consisted of the following steps. First, we systematically perturbed the kinase-substrate and phosphatase-substrate networks by selecting gene deletion mutants of the nonessential kinases or phosphatases or, for some essential kinases, by generating mutants inhibitable by cell-permeable drugs, which are referred to as “analog-sensitive” kinase strains (21). To minimize compensatory mutations that might accumulate over time in the gene deletion strains, we freshly prepared all mutant strains. To enable a statistical characterization of our observations, we always grew, processed, and measured each perturbed strain in three independent replicates, together with three replicates of wild-type, control cells. Phosphopeptides were isolated from each sample (22, 23) and submitted to high-performance mass spectrometry to generate liquid chromatography coupled to mass spectrometry LC-MS/MS phosphoproteome maps. The triplicate phosphoproteome maps generated from each perturbed or wild-type cell sample were annotated with the amino acid sequences of the detected phosphopeptide features and were aligned with the algorithm SuperHirn (24), which was followed by additional postprocessing (see Supplementary Materials for details). The statistical significance of observed changes in the perturbed states was then computed for each phosphopeptide with the Corra software suite (25).
Fig. 1.
Integrated experimental and computational pipeline to determine in vivo kinase-substrate and phosphatase-substrate relationships. Yeast kinase and phosphatase genes were systematically deleted one by one and the phosphoproteomes were systematically compared between mutant and wild-type strains. To achieve this, for each mutant strain and wild-type, we grew and processed three independent biological replicates by proteome isolation, protein digestion with trypsin, phosphopeptide enrichment by applying a TiO2 resin, and quantification and identification of the phosphopeptides with LC-MS/MS. Observed phosphopeptide ion features were aligned, quantified, and tested for statistical significance. For the example phosphopeptide shown, IAS*PIQHEHDSGSR, the resulting matrix gives the intensity values measured in the wild-type and mutant samples, as well as the corresponding log2 fold change (here −3.76) with its associated significance. Abbreviations for the amino acids are as follows: A, Ala; D, Asp; E, Glu; G, Gly; H, His; I, Ile; P, Pro; Q, Gln; R, Arg; and S, Ser.
We assessed the reliability of our measurements and computational data processing at two levels. First, we assessed the confidence of the phosphopeptide identifications generated by database searching, and second, we assessed the reproducibility of detecting quantitative phosphopeptide differences between wild-type and mutant strains. For the first check, and to determine the reliability of our phosphopeptide identifications from the peptide fragment ion spectra, we performed statistical analyses with the PeptideProphet tool (26) and a decoy database strategy (27). From these analyses, we found that a PeptideProphet probability cutoff of 0.9 corresponded to a false discovery rate (FDR) of ~0.038 (3.8%) (table S2), which confirms that our chosen cutoff of 0.9 yielded an acceptably low degree of incorrect peptide identifications, in particular because most phosphopeptides were identified repeatedly in the context of this extensive study.
We then used the statistical tool Corra (25), which supports an empirical Bayesian alternative to the t test (28). The test improves the reliability of conclusions in cases of large-scale testing. For each phosphopeptide feature, the test provided a P value of the observed differences between wild-type and mutant replicates. The P values were further corrected for multiple testing according to the Benjamini and Hochberg procedure (29) (see the Supplementary Materials). After this quantitative analysis step, we chose an FDR threshold of 0.015 in conjunction with a minimum fold-change requirement of log2 >1.5, both of which had to be met before we would consider any phosphopeptide as reproducibly regulated. At this threshold, nine comparisons between wild-type and lowest-impact kinase mutants resulted in only a single or no phosphopeptide being designated as regulated, which verified the validity of our selected criteria. On the basis of these results, we concluded that our applied cutoffs ensured that, despite a high sensitivity (fig. S1), only a minimal amount of noise entered our analyses and that we achieved high reproducibility in the observed regulatory events.
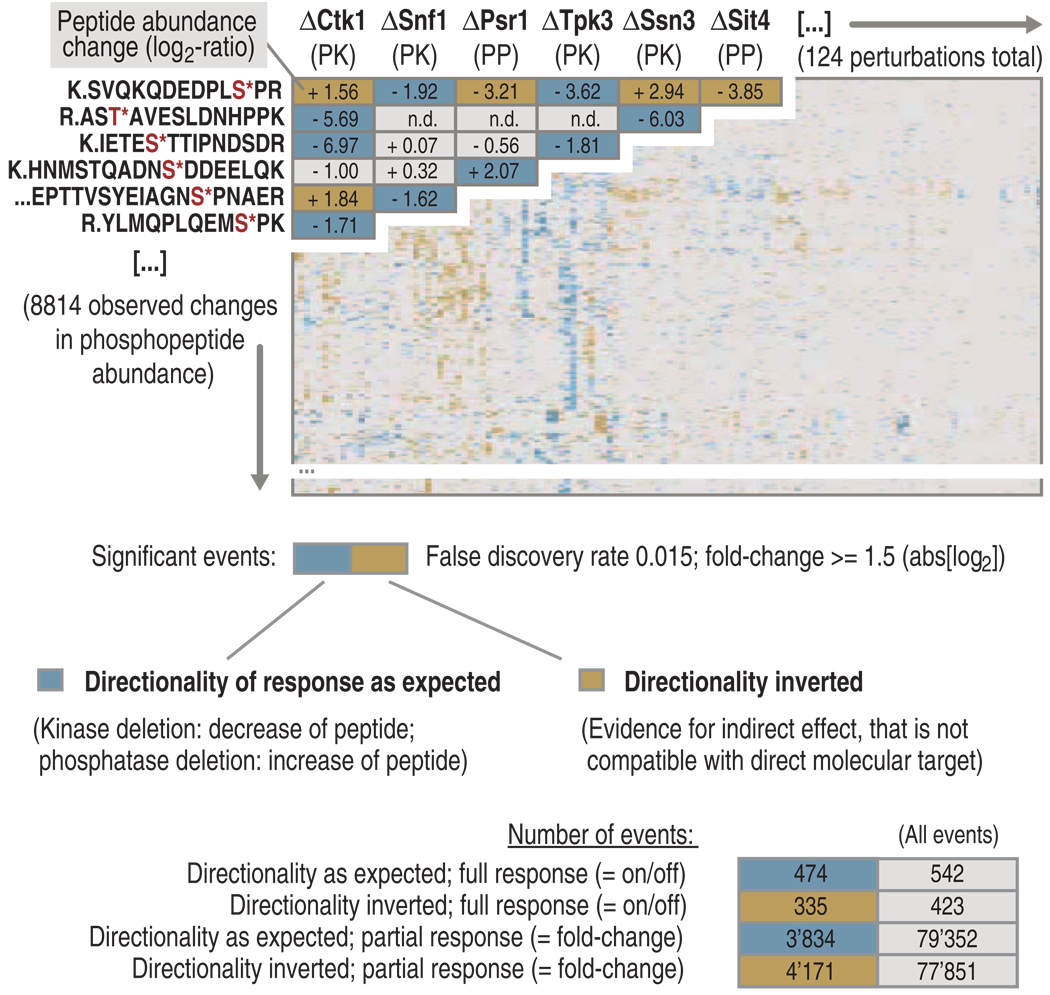
Overall, we attempted the analysis of 161 mutant strains of yeast. Of these, 37 strains could not be analyzed because they were not viable, not inhibitable, or otherwise not amenable to our procedure (table S1). In total, we generated quantitative data for 116 gene deletion mutants and for an additional 8 strains in which analog-sensitive kinases were pharmacologically inhibited (table S1). Together, this corresponds to coverage of 78% of the theoretical kinase and phosphatase space in yeast and covered 77% of those enzymes that show sequence conservation with human enzymes (table S1). A matrix and a network generated from these data related the observed changes in the abundance of a phosphopeptide (measured in triplicate) to the corresponding kinase or phosphatase deletion (Fig. 2 and fig. S2). The matrix contains 8814 reproducible changes in peptide abundance that mapped to 1026 phosphoproteins that were clustered according to the coregulation of the phosphopeptides (tables S3 and S4). Of note, an additional 7550 phosphopeptides were consistently identified but did not exhibit a substantial change in abundance under any of the perturbations tested.
Fig. 2.
Matrix of kinases and phosphatases analyzed in this study and their effects on the phosphoproteome. Overall, 124 kinases and phosphatases were interrogated through our experimental and computational pipeline. Each row (y axis) corresponds to a regulated phosphopeptide and each column (x axis) summarizes the responders of a given kinase or phosphatase. Phosphopeptides with a directionality as expected (that is, kinase deletion resulted in a decrease in peptide abundance, whereas phosphatase deletion resulted in an increase in peptide abundance) are shown in graded blue, and phosphopeptides with an inverted directionality (evidence for indirect effect, not compatible with direct molecular target) are displayed in graded gold, according to the observed fold change for each peptide. Phosphopeptides observed but not regulated or not detected are displayed in gray. At the bottom, the total numbers of events observed in this study are listed. “Full response” corresponds to phosphopeptides that appeared or vanished when wild-type and mutant strains were compared, and “partial response” corresponds to phosphopeptides that showed a statistically significant change in abundance, but were detected in both wild-type and mutant samples. Abbreviations for the amino acids are as follows: A, Ala; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; and V, Val.
Finally, the cellular abundance distribution of detected phosphoproteins (regulated and unregulated) was roughly similar to that of the total yeast proteome; however, the complete phosphoproteome was still not covered (fig. S3), because under our chosen growth conditions, many phosphorylation sites would not be phosphorylated, and because our experimental pipeline had several biases, among them that only tryptic peptides with a mass/charge ratio (m/z) suitable for LC-MS/MS analysis (30) could be identified. Nevertheless, the observed phosphorylation sites covered a reasonably large fraction of the phosphoproteome, and therefore an existing bias should not impair our conclusions (31).
Direct versus indirect phosphorylation events
Because kinases and phosphatases are components of complex, interconnected signaling networks, we fully expected to observe a number of indirect, downstream responses, that is, phosphopeptides whose abundance would change despite their not being a direct molecular target of the kinase or phosphatase in question. Indeed, we found that such events seemed to strongly outnumber direct kinase-substrate interactions, as argued by the following observations. First, we determined for each kinase or phosphatase the number of phosphopeptides whose responses showed the expected directionality (that is, reductions in abundance in the case of kinase deletions and increases in abundance in the case of phosphatase deletions). In general, the number of phosphopeptides that responded in the expected directionality was roughly similar to that of phosphopeptides that responded with “inverted” directionality (Fig. 2 and fig. S4). Exceptions to this finding were analog-sensitive kinases that were inhibited over the short term; for example, in the case of Cdc28, about 76% of the phosphopeptides were regulated in the expected directionality. No difference in the direction of regulation was observed between nonessential kinases or phosphatases (fig. S4). Second, we conservatively assumed that phosphopeptides that changed in abundance in only a single deletion strain might be direct molecular targets of the kinase or phosphatase in question. By this measure, we found that, at most, 32% of the observed regulatory events might have been direct for kinases (that is, that the events mapped to just a single kinase), whereas in the case of phosphatases this number was 53%. The data sets generated by the short-term inhibition of the analog-sensitive kinases showed a higher fraction of potential direct targets (44%) than did the permanent deletion strains.
Third, we tested the overlap of our data with various previously established reference protein-protein interactions in yeast (32–35), such as the STRING database (tables S5 and S6). We observed that the overlap of our data with these direct interactions was small (table S5). This is consistent with the long-held notion that kinase-substrate interactions are too weak and transient to be detectable by typical affinity purification–based protein interaction screens. Reassuringly, however, first, the overlap of the heavily studied kinase Cdc28 with our data set on the level of regulated phosphoproteins was high, showing a 43% overlap with the study of Ubersax et al. (36) and a 76% overlap with the study of Holt et al. (10) (on the phosphorylation site level, the overlap was 46%). Second, all other phosphorylation events that did overlap showed substantial enrichments for the expected directionality. Likewise, we observed substantial enrichment of confirmed interactions, in particular for those phosphopeptides that responded only in a single perturbation (table S7). This indicates that our data included a sizeable fraction of direct enzyme-target interactions; however, from all three tests, we can conclude that indeed a large majority of our observed events were indirect consequences of the deletion. Not a single kinase showed exclusively direct effects, indicating that a focused modulation of a pathway (branch) without system-wide adaptations might not be possible with a single drug.
Changed extents of phosphorylation versus changed protein abundance
As is the case in prolonged pharmacological intervention, our genetic kinase-deletion approach gave the cells ample time to accommodate (and potentially compensate for) the loss of kinase activity. This should not only have led to downstream, indirect consequences on the phosphoproteome, but could have also entailed subsequent changes in gene expression and the amounts of proteins produced. To assess the extent of this effect, we measured not only abundance changes in the phosphoproteome but also abundance changes of the proteins themselves, by observing unphosphorylated peptides in a subset of 16 kinase deletion strains. The kinases selected for this test ranged from those that had a small effect on the phosphoproteome to those that had a large effect. The data indicated that for a total of 467 regulated phosphopeptides that matched to 118 proteins covered in this analysis, 79% of the proteins remained unchanged in abundance, and, in a single case, the directionality of the phosphopeptide regulation was opposite to the protein abundance change (figs. S5 and S6). In 21% of the cases in which a phosphopeptide was regulated, we also observed a change in protein abundance in the same direction.
We also performed additional orthogonal, but more indirect, analyses based on the coregulation or antiregulation of phosphorylation sites on the same protein, which we found in more than half of the phosphoproteins. We reasoned that a synchronous change with a similar amplitude and directionality of such phosphopeptides would indicate an abundance change of the corresponding protein. In contrast, a discordant abundance change of the phosphopeptides from such proteins would indicate a change in phosphorylation site occupancy. These data (fig. S7) can be summarized as follows: For about 25% of the observed events, only a single regulated phosphopeptide was detected on the entire length of the phosphoprotein, impeding this type of analysis. The remainder of events fell into three classes: In 49% of the remaining cases, at least two phosphopeptides originating from the same protein were observed to be regulated, and these exhibited identical directionality. In contrast, in 5% of events, the changes were of opposing directionality; the latter pattern was not consistent with a simple protein abundance change. Of note, in a large part of the data, that is, in 46% of cases, a phosphopeptide that had substantially changed in abundance was detected with at least one other phosphopeptide on the same protein, but the other phosphopeptides were not observed to be regulated. The latter two categories indicate that for most events detected in this study, changes in the abundance of a phosphopeptide could not be explained by changes in protein abundance alone.
Effect of a given kinase or phosphatase on the phosphoproteome
The number of phosphopeptides that were affected by the deletion of a given kinase or phosphatase varied considerably (Fig. 2). Therefore, we (i) quantified the impact of each kinase or phosphatase on the phosphoproteome under the growth conditions tested, (ii) assessed whether the kinases and phosphatases were associated with different biological processes according to their effect on the phosphoproteome, and (iii) determined which biological processes were affected by each kinase and phosphatase.
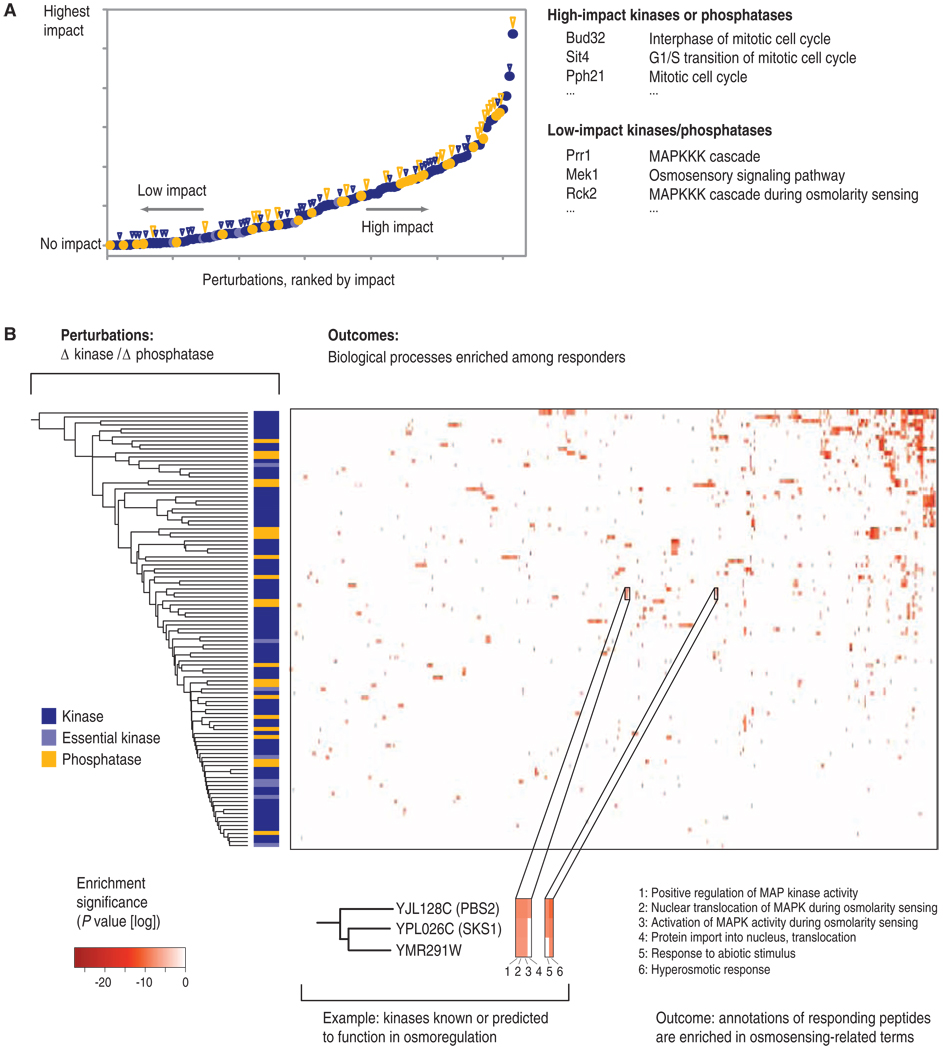
We first computed the fraction of phosphopeptides that were affected by a given kinase or phosphatase relative to the total number of phosphopeptides that were affected by the kinases and phosphatases (Fig. 3A and table S8). We observed that the deletion of 22% of the kinases and phosphatases that we tested resulted in fewer than 10 perturbed phosphopeptides each; therefore, we considered these deletions to have had minimal effects on the fraction of the phosphoproteome detected in this study. These included kinases important in cellular stress response mechanisms, such as Mrk1 (37) and Gcn2 (38). In contrast, for 78% of the kinase and phosphatase deletion strains, distinct changes in the phosphoproteome could be detected. The kinases with the largest effects on the phosphoproteome were Ctk1 (39), a kinase with key roles in the regulation of transcription and translation, and Psk2, which is involved in sugar flux and translational regulation (40). These data show that the loss of most kinases or phosphatases indeed perturbed large parts of the signaling network.
Fig. 3.
(A) Phosphoproteome-wide impact of each kinase and phosphatase. For all kinases and phosphatases, we computed the fraction of phosphopeptides affected relative to the total number of phosphopeptides affected by all kinases and phosphatases. The kinases and phosphatases were then ranked accordingly. Blue circles represent kinases, light blue circles represent essential kinases, and golden circles represent phosphatases. A large golden triangle indicates a strong growth or morphological phenotype of a given mutant, whereas a small blue triangle represents a weak growth or morphological phenotype of a given mutant. Right side: examples of kinases that showed either a low or a high effect on the phosphoproteome regions, together with their known cellular functions. (B) For each kinase and phosphatase, the biological processes enriched among their regulated phosphoproteins were computed. Each column corresponds to a biological process, whereas each row corresponds to a given kinase or phosphatase (kinases are depicted in blue, essential kinases in light blue, and phosphatases in gold). The color scale denotes the statistical significance of the observed enrichment. Magnified inset: an example for three clustered kinases, for which a related set of processes is observed enriched among their substrates.
We next determined the distribution of biological processes represented by the phosphoproteins affected by the lower-impact (bottom half) and higher-impact (top half) kinases and phosphatases, respectively. We found that the enzymes with the smallest effect showed a strong enrichment in processes associated with mitogen-activated protein kinase (MAPK) cascade signaling [“MAPKKK (MAPK kinase kinase) cascade,” P = 3.9−10; “response to pheromone,” P = 4.2−6], whereas the enzymes with the largest effects showed a strong enrichment in processes related to the mitotic cell cycle (“interphase of mitotic cell cycle,” P = 3.1−9; “mitotic cell cycle,” P = 1.4−6) (tables S9 and S10). These data showed that under the tested conditions, even stress- or mating-related kinases showed a measurable impact on the phosphoproteome, albeit lower than that of growth- and cell cycle–related kinases or phosphatases. Lastly, we also computed those biological processes that were enriched among the responders of each individual kinase or phosphatase. We found that 575 biological processes were enriched (Fig. 3B and table S11), an average of five processes for each active kinase or phosphatase. The most frequently enriched functions were “endocytosis” (39 times) and “cell morphogenesis” (38 times). Together, these data illustrate that the effects of most kinases and phosphatases on the signal transduction network, and thereby on controlled biological processes, were broad, perhaps broader than expected (2).
Correlation with yeast phenotypes
We next tested the phenotypic consequences of deletion of kinases and phosphatases, which are relevant in particular with regard to effects (side effects) of potential drugs that inhibit kinases or phosphatases. For each deletion strain, we assessed changes in growth speed (41) and morphological features (table S8) (42). Despite 97 of the deletion strains showing reproducible responses in the phosphorylation network, only 9 mutants showed a strong effect on growth speed, and the total was 23 if strong changes in morphological features were also included (Fig. 3A). Conversely, 11 of the 27 kinases and phosphatases that had an undetectable, or only minimal, effect on the section of the phosphoproteome measured in this study showed a phenotype, among them, the kinase Elm1 (43), which showed a strong morphological phenotype. However, many strong morphological phenotypes were indeed observed in mutants that showed a strong change in the phosphoproteome, but the results were nevertheless surprising because they indicated that strong phenotypes were not necessarily reflected in the status of the phosphoproteome, as exemplified by Elm1 and other enzymes. Perhaps, in some cases, compensatory effects (visible at the level of the phosphoproteome) were precisely what prevented the occurrence of strong phenotypic consequences, as exemplified by the lack of correlation between the growth phenotypes and the changes in the phosphoproteome. This observation is particularly relevant because, first, cancer cells might display in some regards increased compensatory power, and second, kinase inhibitors that are specific for a target in vivo might not necessarily result in a cellular phenotype.
DISCUSSION
Our study delineates the responses of the system-wide cellular phosphorylation network upon systematic inactivation of individual kinases or phosphatases. Because the phosphorylation network is one of the main cellular backbones for the processing of information and the implementation of cellular responses, it is highly dynamic. Our measured behavior is only a single snapshot of a large number of possible outcomes, which were constrained by the growth and experimental conditions that we chose.
The first surprising observation that we made was that 7550 phosphopeptides were consistently identified but did not show a substantial amount of regulation. This may be due to, first, our cutoffs being conservative; thus, many putative regulatory events may not have been reproducible or strong enough to be deemed substantial. Second, 22% of the kinase and phosphatase mutants could not be analyzed, mainly because the corresponding genes are essential for cellular viability. Perhaps their essentiality is at least partly due to a generally higher impact on the phosphoproteome, as indicated recently (10), or because their substrates need to be phosphorylated constitutively. Third, in yeast, a large number of paralogous kinase isoforms exist (for example, Tpk1, Tpk2, and Tpk3). Given this, it is reasonable to expect some overlap or redundancy in substrates, which could lead to a considerable number of phosphorylation sites that would appear unregulated as long as only one of the paralogous duplicates was deleted. Fourth, the yeast populations that we analyzed consisted in a strict sense of many mixed subpopulations (for example, cells in different cell cycle states), and it can be assumed that an identical phosphorylation site can become phosphorylated by different kinases during the cell cycle. Therefore, analyzing deletions of single kinases or phosphatases would only manifest in slight, if any, regulation for such sites; for example, a cell cycle phase–specific regulation is masked by all cells that are not in that particular phase at any given time point. Fifth, we also analyzed whether the regulated and nonregulated phosphopeptides fell into different protein abundance classes (for example, the nonregulated are of low abundance and therefore regulation is more difficult to observe), but this was not the case. Overall, it is likely that all five possible explanations contribute to the observed result.
Another finding of this study was the unexpectedly strong dominance of indirect effects (as opposed to direct molecular target effects), which were often without a resulting strong cellular phenotype. To some extent, this observation fits with a view of signaling networks having to be highly flexible and redundant to respond to an ever-changing environment while maintaining stable cellular states (44). This constrains the architecture of the system, as described by the “law of requisite variety” (45, 46), a fundamental law in systems control theory. It states that stable systems have to encode a number of control states that is higher than or equal to the number of states to be controlled. Considering that for each cell the space of “environmental states” is enormous, consequently, also the cellular “control variable space” must have an equal or greater size. The combinatorial possibilities of the phosphoproteome seem to ideally fulfill this demand (44).
An alternative explanation for this observation might also be found in the theory of Neutral Evolution (47). It is possible that only a small number of the observed phosphorylation events are actually relevant for the function and survival of the cell, whereas most phosphorylation events would simply have no effect, or at least have no negative effect, on the cell. As a result, such phosphorylation sites would not be counterselected during evolution. The data generated in this study do not, by themselves, support or refute this hypothesis. Finally, the low correlation between phenotype and the degree of change in the phosphoproteome may have been affected by the growth conditions chosen here, the lack of sensitivity of the phenotypic assays, or the possibility that the phosphoproteomics data were not sampled deeply enough to find such correlations.
In addition to revealing insights into the architecture of cellular signaling, our data set also describes the proteome-wide functional states of yeast cells; this might be useful for determining diagnostic markers for stress conditions, functional states of key pathways, or the activity of a given kinase or phosphatase. These markers could be used in conjunction with targeted proteomics approaches to not only study basic biological processes but also determine how a given pharmacological intervention would affect the cellular signaling network.
With targeted proteomics methods, not only can the cellular information flux under many conditions be observed, at high throughput, but this approach also enables us to understand for all phosphorylation sites whether the observed change is a “true” regulation event or simply as a result of a change in protein abundance (48–50) because both the phosphopeptide and several proteotypic peptides corresponding to the protein could be relatively or absolutely quantified, thus determining the phosphorylation site occupancy and regulation. Overall, our data provide global starting points, and constraints, toward understanding the complexity of phosphorylation regulation in yeast and other organisms. In the future, the results should be complemented by similar data for specific cellular conditions, time courses, or small-molecule interventions, thereby sharpening—step by step—our view of the events in the phosphorylation network. The ensuing insights in general design rules and motifs in cellular information processing will be essential for our ability to develop kinase-based drugs in an informed way.
MATERIALS AND METHODS
The generated LC-MS/MS phosphoproteome maps (table S2), an overview of the generated data (table S12), and the statistical methods used for their analysis are explained in detail in the Supplementary Materials. We have made available all kinase/phosphatase-responder relations in a user-friendly way in the recently described PhosphoPep database (30, 51) (http://www.phosphopep.org). All yeast strains used here can be supplied upon request in a 96-well plate format (table S13).
Supplementary Material
Footnotes
SUPPLEMENTARY MATERIALS
www.sciencesignaling.org/cgi/content/full/3/153/rs4/DC1
Materials and Methods
Fig. S1. Power of the analysis approach.
Fig. S2. Topological properties of the protein phosphorylation network.
Fig. S3. Abundance distribution of responder phosphoproteins (proteins that contain “regulated” phosphopeptides).
Fig. S4. Ratio of phosphopeptides that are reduced or increased in abundance.
Fig. S5. Regulation of phosphopeptides versus regulation of protein abundance.
Fig. S6. Regulation of phosphopeptides versus regulation of protein abundance.
Fig. S7. Regulation of phosphopeptides that map to the same protein.
Table S1. List of enzymes.
Table S2. False discovery rate of peptide identification and specificity of phosphopeptide enrichment for each analyzed phosphorylation pattern.
Table S3. Information on phosphopeptides and phosphoproteins.
Table S4. Significant coregulation of kinases and phosphatases.
Table S5. Overlap of data from this study with other data sets.
Table S6. Confirmed STRING interactions.
Table S7. Overlap of possible direct targets with other data sets.
Table S8. Effects of each kinase and phosphatase on the phosphoproteome.
Table S9. Enrichment of biological processes among the low-impact kinases (bottom half).
Table S10. Enrichment of biological processes among the high-impact kinases (top half).
Table S11. GO terms.
Table S12. Overview of the entire data set.
Table S13. Information on yeast strains.
References
REFERENCES AND NOTES
- 1.Cohen P. Protein kinases—the major drug targets of the twenty-first century? Nat. Rev. Drug Discov. 2002;1:309–315. doi: 10.1038/nrd773. [DOI] [PubMed] [Google Scholar]
- 2.Hunter T. Signaling—2000 and beyond. Cell. 2000;100:113–127. doi: 10.1016/s0092-8674(00)81688-8. [DOI] [PubMed] [Google Scholar]
- 3.Irish JM, Hovland R, Krutzik PO, Perez OD, Bruserud Ø, Gjertsen BT, Nolan GP. Single cell profiling of potentiated phospho-protein networks in cancer cells. Cell. 2004;118:217–228. doi: 10.1016/j.cell.2004.06.028. [DOI] [PubMed] [Google Scholar]
- 4.Pelkmans L, Fava E, Grabner H, Hannus M, Habermann B, Krausz E, Zerial M. Genome-wide analysis of human kinases in clathrin- and caveolae/raft-mediated endocytosis. Nature. 2005;436:78–86. doi: 10.1038/nature03571. [DOI] [PubMed] [Google Scholar]
- 5.Savage DG, Antman KH. Imatinib mesylate—a new oral targeted therapy. N. Engl. J. Med. 2002;346:683–693. doi: 10.1056/NEJMra013339. [DOI] [PubMed] [Google Scholar]
- 6.Ptacek J, Devgan G, Michaud G, Zhu H, Zhu X, Fasolo J, Guo H, Jona G, Breitkreutz A, Sopko R, McCartney RR, Schmidt MC, Rachidi N, Lee SJ, Mah AS, Meng L, Stark MJ, Stern DF, De Virgilio C, Tyers M, Andrews B, Gerstein M, Schweitzer B, Predki PF, Snyder M. Global analysis of protein phosphorylation in yeast. Nature. 2005;438:679–684. doi: 10.1038/nature04187. [DOI] [PubMed] [Google Scholar]
- 7.Linding R, Jensen LJ, Ostheimer GJ, van Vugt MA, Jorgensen C, Miron IM, Diella F, Colwill K, Taylor L, Elder K, Metalnikov P, Nguyen V, Pasculescu A, Jin J, Park JG, Samson LD, Woodgett JR, Russell RB, Bork P, Yaffe MB, Pawson T. Systematic discovery of in vivo phosphorylation networks. Cell. 2007;129:1415–1426. doi: 10.1016/j.cell.2007.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fiedler D, Braberg H, Mehta M, Chechik G, Mukherjee P, Silva AC, Shales M, Collins SR, van Wageningen S, Kemmeren P, Holstege FC, Weissman JS, Keogh MC, Koller D, Shokat KM, Krogan NJ. Functional organization of the S. cerevisiae phosphorylation network. Cell. 2009;136:952–963. doi: 10.1016/j.cell.2008.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rishton GM. Failure and success in modern drug discovery: Guiding principles in the establishment of high probability of success drug discovery organizations. Med. Chem. 2005;1:519–527. doi: 10.2174/1573406054864106. [DOI] [PubMed] [Google Scholar]
- 10.Holt LJ, Tuch BB, Villen J, Johnson AD, Gygi SP, Morgan DO. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science. 2009;325:1682–1686. doi: 10.1126/science.1172867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huber A, Bodenmiller B, Uotila A, Stahl M, Wanka S, Gerrits B, Aebersold R, Loewith R. Characterization of the rapamycin-sensitive phosphoproteome reveals that Sch9 is a central coordinator of protein synthesis. Genes Dev. 2009;23:1929–1943. doi: 10.1101/gad.532109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 13.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Raymond C, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blocker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AF, Stupka E, Szustakowski J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ, de Jong P, Catanese JJ, Osoegawa K, Shizuya H, Choi S, Chen YJ. International Human Genome Sequencing Consortium, Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 14.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 15.Mager WH, Winderickx J. Yeast as a model for medical and medicinal research. Trends Pharmacol. Sci. 2005;26:265–273. doi: 10.1016/j.tips.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 16.Lindquist S, Krobitsch S, Li L, Sondheimer N. Investigating protein conformation-based inheritance and disease in yeast. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2001;356:169–176. doi: 10.1098/rstb.2000.0762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Outeiro TF, Muchowski PJ. Molecular genetics approaches in yeast to study amyloid diseases. J. Mol. Neurosci. 2004;23:49–60. doi: 10.1385/JMN:23:1-2:049. [DOI] [PubMed] [Google Scholar]
- 18.Hunter T, Plowman GD. The protein kinases of budding yeast: Six score and more. Trends Biochem. Sci. 1997;22:18–22. doi: 10.1016/s0968-0004(96)10068-2. [DOI] [PubMed] [Google Scholar]
- 19.Widmann C, Gibson S, Jarpe MB, Johnson GL. Mitogen-activated protein kinase: Conservation of a three-kinase module from yeast to human. Physiol. Rev. 1999;79:143–180. doi: 10.1152/physrev.1999.79.1.143. [DOI] [PubMed] [Google Scholar]
- 20.Simon JA, Bedalov A. Yeast as a model system for anticancer drug discovery. Nat. Rev. Cancer. 2004;4:481–492. doi: 10.1038/nrc1372. [DOI] [PubMed] [Google Scholar]
- 21.Bishop AC, Ubersax JA, Petsch DT, Matheos DP, Gray NS, Blethrow J, Shimizu E, Tsien JZ, Schultz PG, Rose MD, Wood JL, Morgan DO, Shokat KM. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 2000;407:395–401. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- 22.Bodenmiller B, Mueller LN, Mueller M, Domon B, Aebersold R. Reproducible isolation of distinct, overlapping segments of the phosphoproteome. Nat. Methods. 2007;4:231–237. doi: 10.1038/nmeth1005. [DOI] [PubMed] [Google Scholar]
- 23.Larsen MR, Thingholm TE, Jensen ON, Roepstorff P, Jorgensen TJ. Highly selective enrichment of phosphorylated peptides from peptide mixtures using titanium dioxide microcolumns. Mol. Cell. Proteomics. 2005;4:873–886. doi: 10.1074/mcp.T500007-MCP200. [DOI] [PubMed] [Google Scholar]
- 24.Mueller LN, Rinner O, Schmidt A, Letarte S, Bodenmiller B, Brusniak MY, Vitek O, Aebersold R, Muller M. SuperHirn—a novel tool for high resolution LC-MS-based peptide/protein profiling. Proteomics. 2007;7:3470–3480. doi: 10.1002/pmic.200700057. [DOI] [PubMed] [Google Scholar]
- 25.Brusniak MY, Bodenmiller B, Campbell D, Cooke K, Eddes J, Garbutt A, Lau H, Letarte S, Mueller LN, Sharma V, Vitek O, Zhang N, Aebersold R, Watts JD. Corra: Computational framework and tools for LC-MS discovery and targeted mass spectrometry-based proteomics. BMC Bioinformatics. 2008;9:542. doi: 10.1186/1471-2105-9-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 27.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 28.Smyth G. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article 3. [DOI] [PubMed] [Google Scholar]
- 29.Benjamini Y, Hochberg Y. Controlling the false discovery rate—a practical and powerful approach to multiple testing. J. R. Statist. Soc. Series B Stat. Methodol. 1995;57:289–300. [Google Scholar]
- 30.Bodenmiller B, Malmstrom J, Gerrits B, Campbell D, Lam H, Schmidt A, Rinner O, Mueller LN, Shannon PT, Pedrioli PG, Panse C, Lee HK, Schlapbach R, Aebersold R. PhosphoPep—a phosphoproteome resource for systems biology research in Drosophila Kc167 cells. Mol. Syst. Biol. 2007;3:139. doi: 10.1038/msb4100182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwartz D, Chou MF, Church GM. Predicting protein post-translational modifications using meta-analysis of proteome scale data sets. Mol. Cell. Proteomics. 2009;8:365–379. doi: 10.1074/mcp.M800332-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Breitkreutz BJ, Stark C, Reguly T, Boucher L, Breitkreutz A, Livstone M, Oughtred R, Lackner DH, Bahler J, Wood V, Dolinski K, Tyers M. The BioGRID Interaction Database: 2008 update. Nucleic Acids Res. 2008;36:D637–D640. doi: 10.1093/nar/gkm1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, Bork P, von Mering C. STRING 8—a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mok J, Kim PM, Lam HY, Piccirillo S, Zhou X, Jeschke GR, Sheridan DL, Parker SA, Desai V, Jwa M, Cameroni E, Niu H, Good M, Remenyi A, Ma JL, Sheu YJ, Sassi HE, Sopko R, Chan CS, De Virgilio C, Hollingsworth NM, Lim WA, Stern DF, Stillman B, Andrews BJ, Gerstein MB, Snyder M, Turk BE. Deciphering protein kinase specificity through large-scale analysis of yeast phosphorylation site motifs. Sci. Signal. 2010;3:ra12. doi: 10.1126/scisignal.2000482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stark C, Su TC, Breitkreutz A, Lourenco P, Dahabieh M, Breitkreutz BJ, Tyers M, Sadowski I. PhosphoGRID: A database of experimentally verified in vivo protein phosphorylation sites from the budding yeast Saccharomyces cerevisiae. Database (Oxford ) 2010;2010 doi: 10.1093/database/bap026. bap026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ubersax JA, Woodbury EL, Quang PN, Paraz M, Blethrow JD, Shah K, Shokat KM, Morgan DO. Targets of the cyclin-dependent kinase Cdk1. Nature. 2003;425:859–864. doi: 10.1038/nature02062. [DOI] [PubMed] [Google Scholar]
- 37.Hirata Y, Andoh T, Asahara T, Kikuchi A. Yeast glycogen synthase kinase-3 activates Msn2p-dependent transcription of stress responsive genes. Mol. Biol. Cell. 2003;14:302–312. doi: 10.1091/mbc.E02-05-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hinnebusch AG, Natarajan K. Gcn4p, a master regulator of gene expression, is controlled at multiple levels by diverse signals of starvation and stress. Eukaryot. Cell. 2002;1:22–32. doi: 10.1128/EC.01.1.22-32.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sterner DE, Lee JM, Hardin SE, Greenleaf AL. The yeast carboxyl-terminal repeat domain kinase CTDK-I is a divergent cyclin–cyclin-dependent kinase complex. Mol. Cell. Biol. 1995;15:5716–5724. doi: 10.1128/mcb.15.10.5716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grose JH, Smith TL, Sabic H, Rutter J. Yeast PAS kinase coordinates glucose partitioning in response to metabolic and cell integrity signaling. EMBO J. 2007;26:4824–4830. doi: 10.1038/sj.emboj.7601914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hillenmeyer ME, Fung E, Wildenhain J, Pierce SE, Hoon S, Lee W, Proctor M, St Onge RP, Tyers M, Koller D, Altman RB, Davis RW, Nislow C, Giaever G. The chemical genomic portrait of yeast: Uncovering a phenotype for all genes. Science. 2008;320:362–365. doi: 10.1126/science.1150021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ohya Y, Sese J, Yukawa M, Sano F, Nakatani Y, Saito TL, Saka A, Fukuda T, Ishihara S, Oka S, Suzuki G, Watanabe M, Hirata A, Ohtani M, Sawai H, Fraysse N, Latgé JP, François JM, Aebi M, Tanaka S, Muramatsu S, Araki H, Sonoike K, Nogami S, Morishita S. High-dimensional and large-scale phenotyping of yeast mutants. Proc. Natl. Acad. Sci. U.S.A. 2005;102:19015–19020. doi: 10.1073/pnas.0509436102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blacketer MJ, Koehler CM, Coats SG, Myers AM, Madaule P. Regulation of dimorphism in Saccharomyces cerevisiae: Involvement of the novel protein kinase homolog Elm1p and protein phosphatase 2A. Mol. Cell. Biol. 1993;13:5567–5581. doi: 10.1128/mcb.13.9.5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thomson M, Gunawardena J. Unlimited multistability in multisite phosphorylation systems. Nature. 2009;460:274–277. doi: 10.1038/nature08102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ashby WR. An Introduction to Cybernetics. London: Chapman & Hall Ltd.; 1956. [Google Scholar]
- 46.Ashby WR. Requisite variety and its implications for the control of complex systems. Cybernetica. 1958;1:83–99. [Google Scholar]
- 47.Kimura M. Evolutionary rate at the molecular level. Nature. 1968;217:624–626. doi: 10.1038/217624a0. [DOI] [PubMed] [Google Scholar]
- 48.Lange V, Picotti P, Domon B, Aebersold R. Selected reaction monitoring for quantitative proteomics: A tutorial. Mol. Syst. Biol. 2008;4:222. doi: 10.1038/msb.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Picotti P, Aebersold R, Domon B. The implications of proteolytic background for shotgun proteomics. Mol. Cell. Proteomics. 2007;6:1589–1598. doi: 10.1074/mcp.M700029-MCP200. [DOI] [PubMed] [Google Scholar]
- 50.Picotti P, Lam H, Campbell D, Deutsch EW, Mirzaei H, Ranish J, Domon B, Aebersold R. A database of mass spectrometric assays for the yeast proteome. Nat. Methods. 2008;5:913–914. doi: 10.1038/nmeth1108-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bodenmiller B, Campbell D, Gerrits B, Lam H, Jovanovic M, Picotti P, Schlapbach R, Aebersold R. PhosphoPep—a database of protein phosphorylation sites in model organisms. Nat. Biotechnol. 2008;26:1339–1340. doi: 10.1038/nbt1208-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Acknowledgments: We thank the whole team of the Functional Genomics Center Zurich (FGCZ) for fruitful discussions. We thank C. Zheng, Department of Statistics, Purdue University, for help with use and interpretation of the Limma package. Funding: This project was funded in part by ETH Zurich; Federal funds from the National Heart, Lung, and Blood Institute, NIH, under contract no. N01-HV-28179; the PhosphoNetX project of SystemsX.ch, the Swiss initiative for systems biology; and the European Research Council (grant ERC-2008-AdG 233226) to R.A. Work at the FGCZ and at the von Mering laboratory has been supported by the University Research Priority Program Systems Biology and Functional Genomics of the University of Zurich. B.G. is supported by the Bonizzi-Theler Foundation. C.K. is supported by a Marie-Heim Vögtlin fellowship from the Swiss National Science Foundation (SNF). B.B. was the recipient of fellowships by the Boehringer Ingelheim Fonds, the SNF, and the EMBO Long-Term Fellowship. Author contributions: B.B. coordinated the project, conducted most of the experimental work, and contributed to data analysis and writing the manuscript; S.W. and C.v.M. conducted downstream bioinformatics analysis and contributed to writing the manuscript; C.K. and M.P. generated the kinase and phosphatase deletion mutants; J.U. and R.L. generated and characterized a number of the analog-sensitive kinases; D.C. implemented all data into the PhosphoPep database; P.G.P., G.P.N., and A.I.N. contributed to bioinformatics analysis of data; P.P. supported generation of yeast samples; B.G. and B.R. supported LC-MS/MS method optimization and data acquisition; R.S. consulted and coordinated work at the FGCZ; H.L. processed the collected MS/MS spectra of phosphopeptides, validated their identification, and created high-quality spectral libraries of yeast phosphopeptides for distribution; A.C.-L. determined the morphological phenotypes of the yeast cells; M.-Y.B. modified the Corra interface and processed feature-aligned and postprocessed data sets for differentially expressed feature detection and probability assignment; O.V. performed statistical analyses of the data; C.Z. and K.M.S. synthesized kinase inhibitors; and R.A. conceptualized the study and contributed to writing the manuscript. Competing interests: The authors declare that they have no competing interests.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.