Abstract
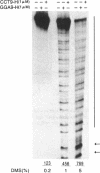
The GGA9-H molecules consisting of a double helical stretch followed by a single-stranded 3'-terminal overhang of nine GGA sequence repeats exhibited a gel mobility-shifted band in a concentration-dependent manner, suggestive of the intermolecular complex formation. The position of the shifted band in a gel was almost identical to that of the Y-shaped dimer marker of the same molecular weight that had the two double-helices at one side. This suggests that GGA9-H dimerizes in a parallel orientation without the formation of four-stranded hairpin structure. Since the GGA9-H homoduplex was stably formed at pH 4, 7 and 9, the formation does not require protonation or deprotonation of the N1 position of adenines. Neither does it require the N7 group of guanines responsible for Hoogsteen base pairing from the methylation interference and modification studies. Modification of the N7 group of guanines with dimethyl sulfate (DMS) did not inhibit the association and also the N7 group in the homoduplex was not protected from DMS. On the other hand, the GAA9-H having the G to A base substitution did not show such an association with either GGA9-H or GAA9-H. These results suggest that the homoduplex formation may be due to G.G base pairing through non-Hoogsteen hydrogen bonds.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beal P. A., Dervan P. B. Second structural motif for recognition of DNA by oligonucleotide-directed triple-helix formation. Science. 1991 Mar 15;251(4999):1360–1363. doi: 10.1126/science.2003222. [DOI] [PubMed] [Google Scholar]
- Beckman J. S., Weber J. L. Survey of human and rat microsatellites. Genomics. 1992 Apr;12(4):627–631. doi: 10.1016/0888-7543(92)90285-z. [DOI] [PubMed] [Google Scholar]
- Bennett K. L., Hill R. E., Pietras D. F., Woodworth-Gutai M., Kane-Haas C., Houston J. M., Heath J. K., Hastie N. D. Most highly repeated dispersed DNA families in the mouse genome. Mol Cell Biol. 1984 Aug;4(8):1561–1571. doi: 10.1128/mcb.4.8.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn E. H. Structure and function of telomeres. Nature. 1991 Apr 18;350(6319):569–573. doi: 10.1038/350569a0. [DOI] [PubMed] [Google Scholar]
- Camerini-Otero R. D., Hsieh P. Parallel DNA triplexes, homologous recombination, and other homology-dependent DNA interactions. Cell. 1993 Apr 23;73(2):217–223. doi: 10.1016/0092-8674(93)90224-e. [DOI] [PubMed] [Google Scholar]
- Cooney M., Czernuszewicz G., Postel E. H., Flint S. J., Hogan M. E. Site-specific oligonucleotide binding represses transcription of the human c-myc gene in vitro. Science. 1988 Jul 22;241(4864):456–459. doi: 10.1126/science.3293213. [DOI] [PubMed] [Google Scholar]
- Han J., Hsu C., Zhu Z., Longshore J. W., Finley W. H. Over-representation of the disease associated (CAG) and (CGG) repeats in the human genome. Nucleic Acids Res. 1994 May 11;22(9):1735–1740. doi: 10.1093/nar/22.9.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanvey J. C., Klysik J., Wells R. D. Influence of DNA sequence on the formation of non-B right-handed helices in oligopurine.oligopyrimidine inserts in plasmids. J Biol Chem. 1988 May 25;263(15):7386–7396. [PubMed] [Google Scholar]
- Hearne C. M., Ghosh S., Todd J. A. Microsatellites for linkage analysis of genetic traits. Trends Genet. 1992 Aug;8(8):288–294. doi: 10.1016/0168-9525(92)90256-4. [DOI] [PubMed] [Google Scholar]
- Hoffman-Liebermann B., Liebermann D., Troutt A., Kedes L. H., Cohen S. N. Human homologs of TU transposon sequences: polypurine/polypyrimidine sequence elements that can alter DNA conformation in vitro and in vivo. Mol Cell Biol. 1986 Nov;6(11):3632–3642. doi: 10.1128/mcb.6.11.3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas D., Bellsolell L., Casasnovas J. M., Coll M., Azorín F. Alternating d(GA)n DNA sequences form antiparallel stranded homoduplexes stabilized by the formation of G.A base pairs. EMBO J. 1993 Oct;12(10):4029–4038. doi: 10.1002/j.1460-2075.1993.tb06081.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohwi-Shigematsu T., Kohwi Y. Poly(dG)-poly(dC) sequences, under torsional stress, induce an altered DNA conformation upon neighboring DNA sequences. Cell. 1985 Nov;43(1):199–206. doi: 10.1016/0092-8674(85)90024-8. [DOI] [PubMed] [Google Scholar]
- Lee J. S., Evans D. H., Morgan A. R. Polypurine DNAs and RNAs form secondary structures which may be tetra-stranded. Nucleic Acids Res. 1980 Sep 25;8(18):4305–4320. doi: 10.1093/nar/8.18.4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. S. The stability of polypurine tetraplexes in the presence of mono- and divalent cations. Nucleic Acids Res. 1990 Oct 25;18(20):6057–6060. doi: 10.1093/nar/18.20.6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyamichev V. I., Mirkin S. M., Danilevskaya O. N., Voloshin O. N., Balatskaya S. V., Dobrynin V. N., Filippov S. A., Frank-Kamenetskii M. D. An unusual DNA structure detected in a telomeric sequence under superhelical stress and at low pH. Nature. 1989 Jun 22;339(6226):634–637. doi: 10.1038/339634a0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Mergny J. L., Duval-Valentin G., Nguyen C. H., Perrouault L., Faucon B., Rougée M., Montenay-Garestier T., Bisagni E., Hélène C. Triple helix-specific ligands. Science. 1992 Jun 19;256(5064):1681–1684. doi: 10.1126/science.256.5064.1681. [DOI] [PubMed] [Google Scholar]
- Moser H. E., Dervan P. B. Sequence-specific cleavage of double helical DNA by triple helix formation. Science. 1987 Oct 30;238(4827):645–650. doi: 10.1126/science.3118463. [DOI] [PubMed] [Google Scholar]
- Muraiso T., Nomoto S., Yamazaki H., Mishima Y., Kominami R. A single-stranded DNA binding protein from mouse tumor cells specifically recognizes the C-rich strand of the (AGG:CCT)n repeats that can alter DNA conformation. Nucleic Acids Res. 1992 Dec 25;20(24):6631–6635. doi: 10.1093/nar/20.24.6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postel E. H., Flint S. J., Kessler D. J., Hogan M. E. Evidence that a triplex-forming oligodeoxyribonucleotide binds to the c-myc promoter in HeLa cells, thereby reducing c-myc mRNA levels. Proc Natl Acad Sci U S A. 1991 Sep 15;88(18):8227–8231. doi: 10.1073/pnas.88.18.8227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao B. J., Chiu S. K., Radding C. M. Homologous recognition and triplex formation promoted by RecA protein between duplex oligonucleotides and single-stranded DNA. J Mol Biol. 1993 Jan 20;229(2):328–343. doi: 10.1006/jmbi.1993.1038. [DOI] [PubMed] [Google Scholar]
- Rippe K., Fritsch V., Westhof E., Jovin T. M. Alternating d(G-A) sequences form a parallel-stranded DNA homoduplex. EMBO J. 1992 Oct;11(10):3777–3786. doi: 10.1002/j.1460-2075.1992.tb05463.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen D., Gilbert W. A sodium-potassium switch in the formation of four-stranded G4-DNA. Nature. 1990 Mar 29;344(6265):410–414. doi: 10.1038/344410a0. [DOI] [PubMed] [Google Scholar]
- Sen D., Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988 Jul 28;334(6180):364–366. doi: 10.1038/334364a0. [DOI] [PubMed] [Google Scholar]
- Strand M., Prolla T. A., Liskay R. M., Petes T. D. Destabilization of tracts of simple repetitive DNA in yeast by mutations affecting DNA mismatch repair. Nature. 1993 Sep 16;365(6443):274–276. doi: 10.1038/365274a0. [DOI] [PubMed] [Google Scholar]
- Suda T., Oyanagi M., Wakana S., Takahashi Y., Kanada H., Yonekawa H., Miyashita N., Shiroishi T., Moriwaki K., Kominami R. Novel mouse microsatellites: primer sequences and chromosomal location. DNA Res. 1994;1(4):169–174. doi: 10.1093/dnares/1.4.169. [DOI] [PubMed] [Google Scholar]
- Sundquist W. I., Klug A. Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops. Nature. 1989 Dec 14;342(6251):825–829. doi: 10.1038/342825a0. [DOI] [PubMed] [Google Scholar]
- Wang A. H., Quigley G. J., Kolpak F. J., Crawford J. L., van Boom J. H., van der Marel G., Rich A. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979 Dec 13;282(5740):680–686. doi: 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]
- Williamson J. R., Raghuraman M. K., Cech T. R. Monovalent cation-induced structure of telomeric DNA: the G-quartet model. Cell. 1989 Dec 1;59(5):871–880. doi: 10.1016/0092-8674(89)90610-7. [DOI] [PubMed] [Google Scholar]
- Zimmerman S. B., Cohen G. H., Davies D. R. X-ray fiber diffraction and model-building study of polyguanylic acid and polyinosinic acid. J Mol Biol. 1975 Feb 25;92(2):181–192. doi: 10.1016/0022-2836(75)90222-3. [DOI] [PubMed] [Google Scholar]
- de Wachter R., Fiers W. Preparative two-dimensional polyacrylamide gel electrophoresis of 32 P-labeled RNA. Anal Biochem. 1972 Sep;49(1):184–197. doi: 10.1016/0003-2697(72)90257-6. [DOI] [PubMed] [Google Scholar]