Abstract
Objective
Multipotent hematopoietic cell line EML can differentiate into myeloid, erythroid, megakaryocytic, and B-lymphoid lineages, but it remained unknown whether EML cells have T-cell developmental potential as well. The goal of this study was to determine whether the coculture with OP9 stromal cells expressing Notch ligand Delta-like 1 (OP9-DL1) could induce differentiation of EML cells into T-cell lineage.
Materials and Methods
EML cells were cocultured with control OP9 or OP9-DL1 stromal cells in the presence of cytokines (stem cell factor, interleukin-7, and Fms-like tyrosine kinase 3 ligand). Their T-cell lineage differentiation was assessed through flow cytometry and reverse transcription polymerase chain reaction expression analysis of cell surface markers and genes characterizing and associated with specific stages of T-cell development.
Results
The phenotypic, molecular, and functional analysis has revealed that in EML/OP9-DL1 cocultures with cytokines, but not in control EML/OP9 cocultures, EML cell line undergoes T-cell lineage commitment and differentiation. In OP9-DL1 cocultures, EML cell line has differentiated into cells that 1) resembled double-negative, double-positive, and single-positive stages of T-cell development; 2) initiated expression of GATA-3, Pre-Tα, RAG-1, and T-cell receptor – Vβ genes; and 3) produced interferon-γ in response to T-cell receptor stimulation.
Conclusions
These results support the notion that EML cell line has the capacity for T-cell differentiation. Remarkably, induction of T-lineage gene expression and differentiation of EML cells into distinct stages of T-cell development were very similar to previously described T-cell differentiation of adult hematopoietic stem cells and progenitors in OP9-DL1 cocultures. Thus, EML/OP9-DL1 coculture could be a useful experimental system to study the role of particular genes in T-cell lineage specification, commitment, and differentiation.
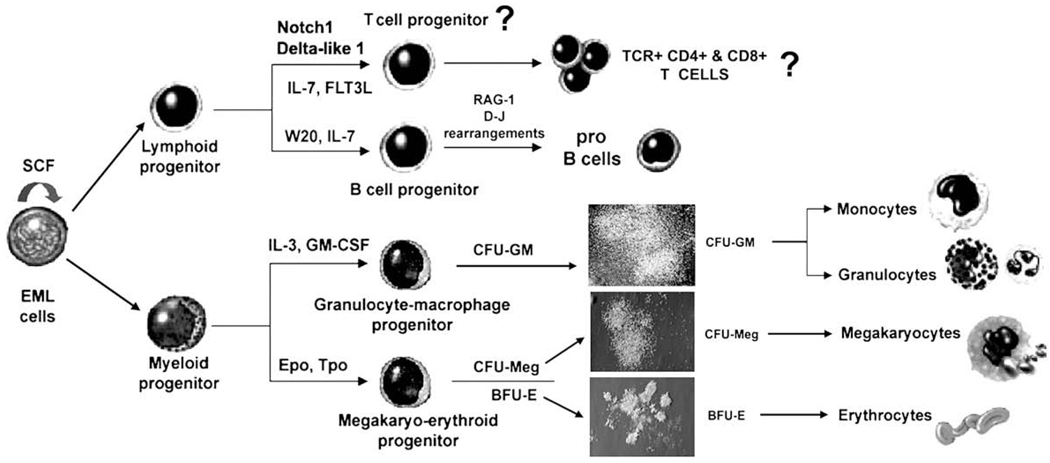
Together with purified hematopoietic stem cells (HSC) and multipotent progenitors (MPPs), several murine hematopoietic cell lines that can differentiate into various blood cell lineages are being used extensively to study molecular mechanisms that regulate lineage commitment and differentiation of hematopoietic progenitors. The stem cell factor (SCF)–dependent, multipotent hematopoietic cell line EML has the capacity for multilineage (erythroid, myeloid, lymphoid) differentiation in vitro [1]. In the presence of SCF, EML cells undergo proliferative self-renewal and remain undifferentiated. In response to cytokines, EML cells differentiate into erythroid, granulocyte-macrophage, and megakaryocytic progenitors in colony-forming assays, and generate colony-forming unit granulocyte-macrophage, burst-forming unit erythroid, and colony-forming unit megakaryocytic colonies (Fig. 1) [1–3]. Moreover, in coculture with W20 stromal cell line and the presence of interleukin-7 (IL-7), EML cells give rise to pro-B cells that express RAG-1 and undergo D-J rearrangements (Fig. 1) [1].
Figure 1.
The model of maintenance and multilineage differentiation of a multipotent hematopoietic cell line EML. In the presence of stem cell factor (SCF), EML cells undergo proliferative self-renewal and remain undifferentiated, whereas in the presence of cytokines and/or stroma EML cells differentiate into erythroid, myeloid, and lymphoid lineages. BFU-E = burst-forming unit erythroid; CFU-GM = colony-forming unit granulocyte macrophage; CFU-Meg = colony-forming unit megakaryocyte; Epo = erythropoietin; GM-CSF = granulocyte macrophage colony-stimulating factor; IL = interleukin; SCF = stem cell factor; TCR = T-cell receptor; Tpo = thrombopoietin.
Based on their multilineage differentiation capacity, expression of HSC and MPP-related cell surface markers (almost all EML cells have the Sca-1+ c-kit+ Flk2− phenotype), and expression of genes relevant for hematopoiesis [3], EML cells resemble Lin−Sca-1+ c-kit+ and Lin−Sca-1+ c-kit+ Flk2− bone marrow cells, highly enriched for long-term and short-term repopulating HSC and MPPs [4–7].
Thus, EML cell line represents a unique and very useful in vitro model for studying molecular mechanisms of lineage commitment and differentiation of multipotent and lineage-committed progenitors for most hematopoietic lineages [8–14]. However, it remained unknown whether EML cells have the potential to differentiate into T-cell lineage as well (Fig. 1).
The Notch pathway is one of the key molecular mechanisms that regulate T-cell development [15,16]. To date, four mammalian Notch receptors (Notch1 – 4) have been identified, along with five ligands that belong to Delta (Delta-like 1, 3, 4) and Jagged (Jagged-1, 2) family. Notch receptors and their ligands are expressed on HSC, progenitors, and stromal cells, and Notch pathway plays an important role in lineage commitment and differentiation of hematopoietic progenitors [17–19]. Conditional inactivation of Notch1 leads to an early block in T-cell development and accumulation of immature B cells in the thymus. In contrast, overexpression of the activated form of Notch1 blocks B-cell development and induces T-cell development in the bone marrow [20–25]. In summary, activation of Notch1 pathway regulates the earliest stages of commitment into the T-cell lineage, while inhibiting B-cell development.
Several studies have reported that interaction of Notch1 receptor with the Delta-like 1 (DL1) ligand or ectopic expression of active form of Notch1 can induce in vitro differentiation of hematopoietic progenitors (HPCs) into a T-cell lineage in the absence of thymic microenvironment [26–28]. Jaleco et al. [26] have described coculture of human cord blood CD34+ cells with stromal cell line S17 expressing human DL1 or Jagged-1 ligands [26]. In that system, DL1 completely inhibited differentiation of HPCs into B-cell lineage and promoted development of cells with a phenotype of T-cell/natural killer precursors and cells coexpressing CD4 and CD8 markers [26]. Furthermore, coculture with macrophage colony-stimulating factor–deficient bone marrow stromal cell line OP9 expressing Notch ligand DL1 (OP9-DL1 cells) and cytokines (interleukin [IL-7] and Fms-like tyrosine kinase 3 ligand [Flt3L]) induced and supported T-cell development from mouse fetal HPCs [27]. Similarly, Hozumi et al. [28] demonstrated that ectopic expression of activated form of Notch receptors in mouse fetal HPCs and coculture with OP9 stromal cells induce development of Thy-1+CD25+ and CD4+CD8+ cells expressing T-cell receptor (TCR) [28]. The OP9-DL1 coculture system was also used to induce T-cell development from mouse embryonic stem cells, mouse adult HSC, and human cord blood HSC [29–32]. Collectively, these results indicate that in vitro T lymphopoiesis is inducible by activation of the Notch1 pathway in a simple coculture system [31,32].
Thus, in view of these studies, we sought to determine whether EML cells have the capacity for T-cell development and whether the coculture with OP9-DL1 stromal cells could induce their differentiation into T-cell lineage.
We report here that in OP9-DL1 cocultures supplemented with cytokines (SCF, IL-7, and Flt3L), EML cells undergo T-cell lineage commitment and differentiation. In OP9-DL1 cocultures, EML cell line has differentiated into cells that 1) resembled double-negative precursors, and CD4+CD8+ and CD4+, and CD8+ T-lineage cells; 2) initiated expression of GATA-3, pre-Tα, RAG-1, and TCR-Vβ genes, and 3) produced interferon-γ (IFN-γ) in response to TCR stimulation.
Materials and methods
Coculture of EML cells with OP9 stromal cells
SCF-dependent EML cell line was obtained from Dr. Tsai, and was maintained in Iscove’s modified Dulbecco’s medium (Gibco, Gaithersburg, MD, USA) with 20% equine serum (HyClone, Logan, UT, USA) and 10% SCF-conditioned medium from BHK/MKL cell line [1,3]. Green fluorescent protein (GFP)–expressing OP9-DL1 and control OP9 stromal cell lines were obtained from Dr. Zúñiga-Pflücker, and were maintained as described previously [27]. EML/OP9 cocultures were initiated by seeding 4 × 104 EML cells/well into six-well plates containing a confluent monolayer of OP9-DL1 or control OP9 stromal cells. Cocultures were maintained in Iscove’s modified Dulbecco’s medium supplemented with 10% fetal bovine serum (HyClone), recombinant IL-7, and Flt3L (5 ng/mL; R&D Systems, Minneapolis, MN, USA) and 10% v/v conditioned BHK/MKL media containing SCF [1]. After 7 days of coculture, nonadherent EML-derived cells were carefully harvested to avoid dislodging OP9 and adherent EMLderived cells, and transferred into six-well plates (4 × 104 cells/ well) with new OP9-DL1 or OP9 monolayers. Nonadherent EML-derived cells were again transferred onto new OP9 monolayers (4 × 104 cells/well) after 12 and 17 days of coculture.
Antibodies and flow cytometry
Monoclonal antibodies (Abs) α-B220 (RA3-6B2), α-CD3 (145-2C11), α-CD4 (RM4-5), α-CD8a (53-6.7), α-CD24 (M1/69), α-CD25 (7D4), α-CD44 (IM7), α-TCR-β chain (H57-597), α-TCR-γδ chain (GL3), α-TCR-Vβ4 (CTVB4), α-TCR-Vβ5.1 (MR9-4), α-TCR-Vβ6 (RR4-7), α-TCR-Vβ8.1, 8.2 (1B3.3), α-TCR-Vβ14 (14-2), α-Thy-1.2 (53-2.1), α-IFN-γ (XMG1.2), α-c-kit-allophycocyanin, α-Sca-1-phycoerythrin (PE)-Cy7, α-Flk-2-PE, and α-CD34-PE were purchased from BD Pharmingen (San Diego, CA, USA) or eBioscience (San Diego, CA, USA). Isotype control (R35-39) and α-IL-7Rα/CD127 (A7R34) Abs were purchased from eBioscience. Anti – CD3-TCR-complex-fluorescein isothiocyanate (FITC) (17A2) and α-CD44 (IM7) allophycocyanin-conjugated Abs were purchased from BioLegend (San Diego, CA, USA). Unconjugated monoclonal α-Notch1 A6 Ab and secondary PE-conjugated Ab were purchased from Abcam (Cambridge, MA, USA) and Novus Biologicals (Littleton, CO, USA). Cells were analyzed on BD-LSR flow cytometer and sorted on FACSVantage and FACSAria cell sorters (Becton Dickinson, Mountain View, CA, USA).
Western analysis
Cells were lysed with buffer (2 mM TrisCl [pH 8.0], 0.14 M NaCl, 1 mM phenylmethylsulphonyl fluoride, 0.1% Triton X-100 and protease inhibitors) at 4°C for 1 hour. Up to 50 µg protein per lane was resolved on 12% Tris-glycine ready gel (Bio-Rad, Richmond, CA, USA) and electrotransferred to Immuno-Blot polyvinylidene difluoride membrane (Bio-Rad). The polyvinylidene difluoride membranes were incubated overnight at 4°C with anti-Notch1 A6 Ab (Abcam) at 1:500 dilution, washed with TBS-T and incubated with secondary horseradish peroxidase–conjugated antibody (Zymed, San Francisco, CA, USA). Proteins were detected with ECL Western blotting analysis detection system (Amersham, Arlington Heights, IL, USA) or SuperSignal West Chemiluminescent substrate for detection of horseradish peroxidase (Pierce, Rockford, IL, USA). Where indicated, membranes were stripped and reprobed with α-actin Ab (Sigma, St Louis, MO, USA).
TCR stimulation and IFN-γ production
To measure IFN-γ production 106 nonadherent EML cells from day 17 OP9-DL1 cocultures, and thymocytes from C57BL/6 J mice as a positive control, were stimulated for 3 days with immobilized anti-CD3 (5 µg/mL) and anti-CD28 (0.1 µg/mL). Cells were then restimulated with anti-CD3/CD28 for 5 hours in the presence of 2 µM Monesin as a Golgi-Stop. For intracellular staining with α-IFN-γ Ab (XMG1.2), cells were permeabilized with 0.3% saponin (Sigma) and fixed in 4% paraformaldehyde (Sigma).
RNA isolation, reverse transcription, and PCR analysis
Total RNA was isolated from 1) undifferentiated EML cells; 2) nonadherent EML-derived cells from day 7, 12, and 17 EML/OP9-DL1 and EML/OP9 cocultures; 3) EML cells cultured with cytokines only (IL-7 and Flt3L at 5 ng/mL) for 72 hours; 4) sorted CD25+CD44− nonadherent cells from day 17 EML/OP9-DL1 cocultures; 5) OP9-DL1 and OP9 stromal cells; and 6) C57BL6/J thymocytes. Equal amount of RNA (5 µg) from each cell sample was reverse-transcribed using Superscript II kit (Invitrogen, Carlsbad, CA, USA). Polymerase chain reactions (PCR) were performed in an Eppendorf Mastercyler for 35 cycles (95°C for 30 seconds, 57 – 62°C for 45 seconds, and 72°C for 30 seconds), using primers for following genes: Ikaros (sense-CACTACCTCTGGAGCAGCAGAA, antisense-CATAGGGCATGTCTGACAGGCACT); VAV1 (sense-GACGAAGATATTTACAGTGG, antisense-GCTTATCATACTCTGTCATC); GATA-3 (sense-GCGGTCCTCAACGGTCAGCAC, antisense-TCGGGCACATAGGGCGGATAG); PU.1 (sense-CGGATGACTTGGTTACTTACG, antisense-TTGGACGAGAACTGGAAGGTA); IL-7Rα (sense-GTTCTCTGTGTTTTTGTTGG, antisense-GGCTCTGTGTCCCTGTGTCT); pre-Tα (sense-CTGGCTCCACCCATCACACT, antisense-TGCCATTGCCAGCTGAGA), and RAG-1 (sense-TGCAGACATTCTAGCACTCTGG, antisense-ACATCTGCCTTCACGTCGAT). Primers for Notch1, Hes-1, Manic, Lunatic, and Radical Fringe, TCR-Vβ chains and hypoxanthine phosphoribosyltransferase were described previously [2,17,33–40]. For semi-quantitative reverse transcription (RT)-PCR analysis, PCR reactions were performed for 23, 27, 31, and 35 cycles. PCR products were run on 1% agarose gels and visualized with ethidium bromide staining.
Results
Coculture with OP9-DL1 stromal cells induces differentiation of EML cells into double-negative T-cell precursors
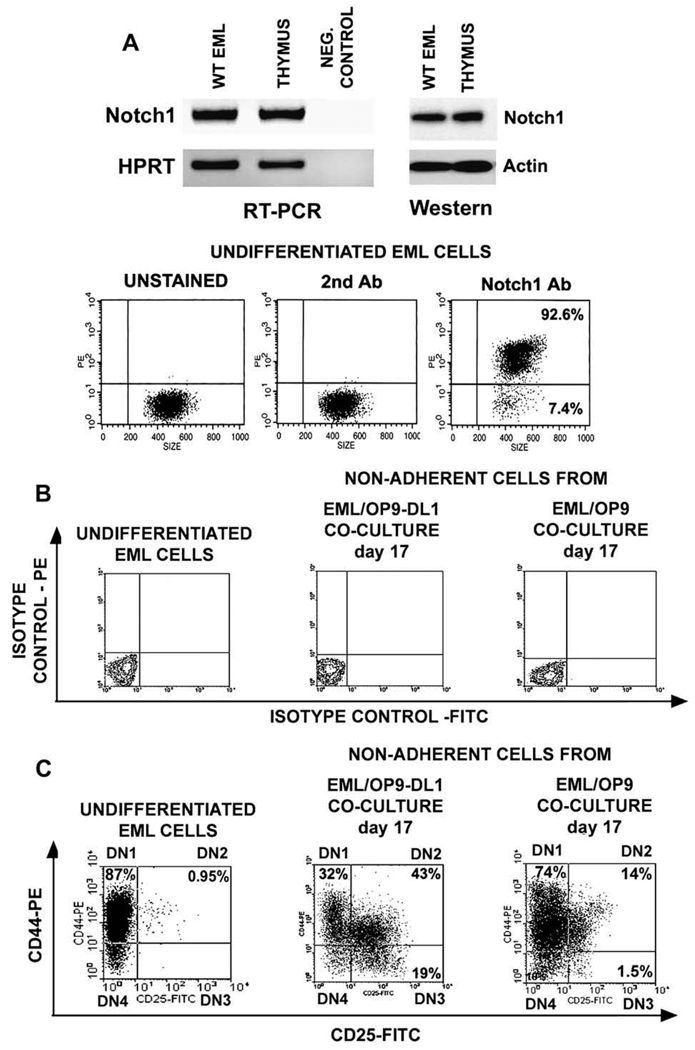
Prior to coculture with OP9-DL1 stromal cells, we analyzed expression of Notch1 by EML cells. The RT-PCR, Western, and flow cytometry analyses have shown that undifferentiated EML cells express Notch1 transcript and protein, and that the majority of EML cells express Notch1 on their cell surface (Fig. 2A).
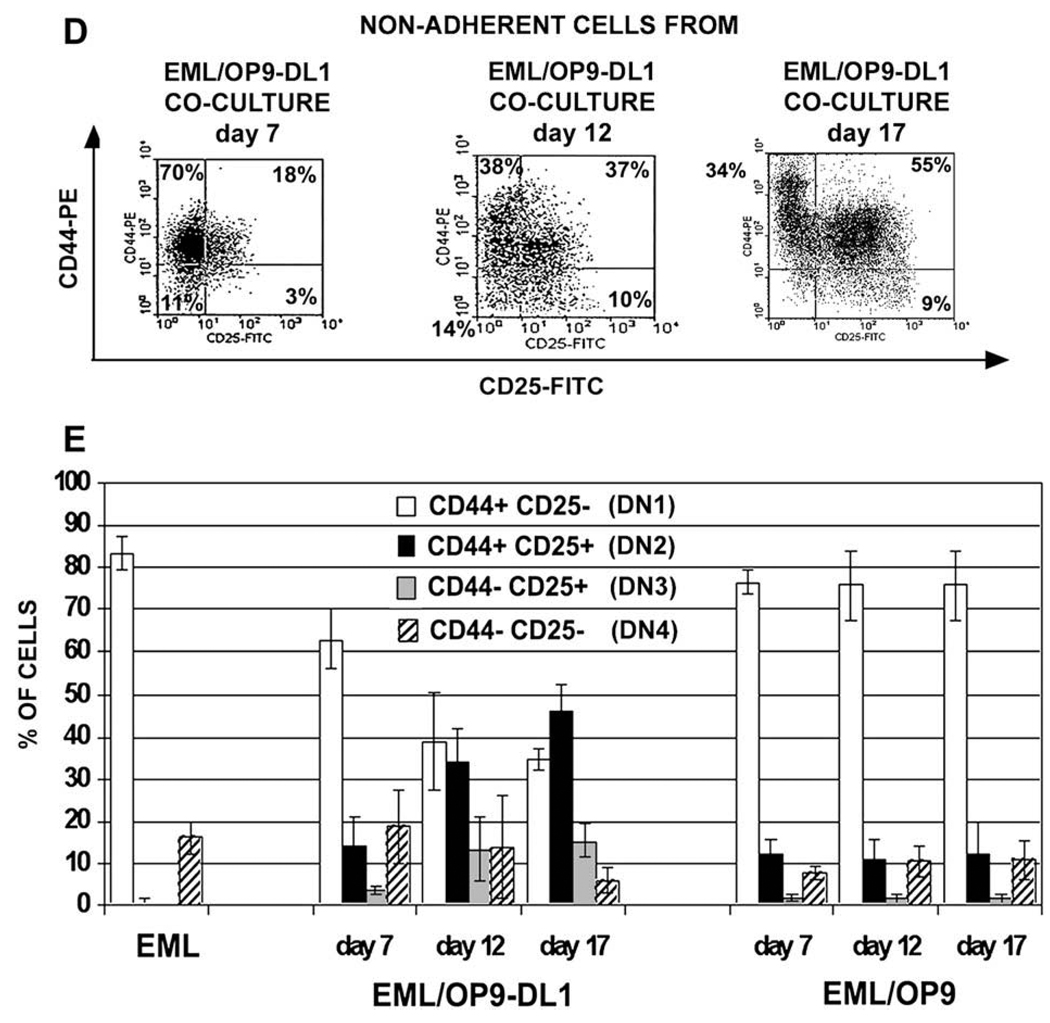
Figure 2.
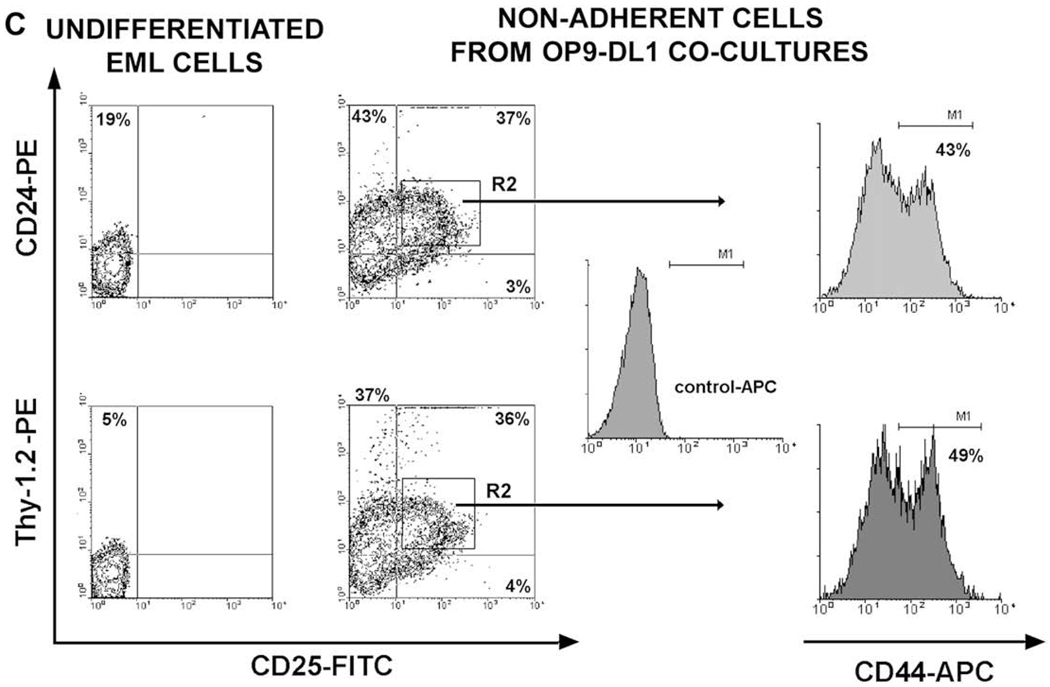
In OP9 stromal cells expressing Notch ligand Delta-like 1 (OP9-DL1) cocultures EML cells differentiate into cells resembling double-negative (DN) T-cell precursors. (A) Reverse transcriptase polymerase chain reaction (RT-PCR), Western, and flow cytometry analyses have shown that EML cells express Notch1 transcript and protein, and that the majority of cells express Notch1 on their cell surface. (B) Flow cytometry analysis of undifferentiated EML cells and nonadherent EML (naEML) cells from day 17 EML/OP9-DL1 and EML/OP9 cocultures after staining with fluorescein isothiocyanate (FITC)- and phycoerythrin (PE)-conjugated isotype controls shows that the harvested naEML cells did not contain dislodged OP9 cells, as evidenced by the absence of green fluorescent protein–positive (GFP+) cells. (C) Flow cytometry analysis of CD44 and CD25 expression on undifferentiated EML and naEML cells from day 17 EML/OP9-DL1 and control EML/OP9 cocultures. Clockwise from the upper left the numbers in each quadrant indicate the percentage of CD44+CD25−, CD44+CD25+, CD44−CD25+ and CD44−CD25− cells, phenotypically resembling double-negative DN1, DN2, DN3, and DN4 stages of T-cell development. (D) Temporal kinetics of DN2, DN3, and DN4 cell development in EML/OP9-DL1 cocultures. The numbers in each quadrant indicate the percentage of cells expressing the CD44 and/or CD25 marker. (E) Comparative analysis of the temporal kinetics of DN cell development in day 7, 12, and 17 EML/OP9-DL1 and EML/OP9 cocultures. The histograms show the percentages of undifferentiated EML cells and naEML cells from OP9-DL1 and OP9 cocultures with the CD44+CD25−, CD44+CD25+, CD44−CD25+, and CD44−CD25− phenotypes. Data are representative of five separate coculture experiments and are shown as mean ± standard error of mean.
EML cells were cocultured with OP9-DL1 or control OP9 stromal cells [27] for up to 21 days in the presence of IL-7, Flt3L, and SCF, which is necessary for survival of undifferentiated EML cells [1,3,11]. Cocultures were initiated with 4 × 104 EML cells/well in six-well plates containing a confluent monolayer of OP9-DL1 or OP9 stromal cells. After 7 days, the cocultures consisted of a monolayer of OP9 cells, EML-derived cells that adhered to OP9 cells, and nonadherent EML-derived (naEML) cells. At 7, 12, and 17 days of coculture, naEML cells were harvested and plated at 4 × 104 cells/well into six-well plates with newly established OP9-DL1 or OP9 monolayers. An approximately 30-fold increase in EML cell numbers was observed in EML/OP9-DL1 cocultures by day 7, and an additional 20-fold and 10-fold increase in cell yields by days 12 and 17, respectively.
To ensure that naEML cells are not contaminated with dislodged GFP-expressing OP9 or OP9-DL1 stromal cells [27], harvested naEML cells were analyzed by flow cytometry for GFP+ cells, which were absent in all experiments (Fig. 2B). The naEML cells from OP9-DL1 or OP9 cocultures were first analyzed by flow cytometry for expression of CD44 and CD25 markers (Fig. 2C), which characterize four double-negative (DN) stages of T-cell development [41,42]. The majority (up to 90%) of undifferentiated EML cells are CD44+CD25−, thus having a DN1 phenotype. The remaining EML cells are CD44−CD25− (Fig. 2C). After 17 days of coculture with OP9-DL1 cells, naEML cells reproducibly consisted of CD44+CD25−, CD44+CD25+, CD44−CD25+, and CD44−CD25− cell populations, phenotypically resembling DN1, DN2, DN3, and DN4 stages of T-cell development [41,42] (Fig. 2C). In contrast, the majority of naEML cells from control EML/OP9 cocultures retained CD44+CD25− phenotype, with only up to 15% of cells coexpressing CD44 and CD25, and <3% of cells being CD44−CD25+ on day 17 (Fig. 2C).
In terms of temporal kinetics of DN cell development in EML/OP9-DL1 cocultures, cells with DN2 phenotype were already present on day 7, and their frequency increased through day 17 (Fig. 2D and E). Similarly, the percentage of DN3 cells increased three- to fivefold by days 12 and 17. The frequency of cells with the DN4 phenotype remained similar until day 17, when their numbers decreased. On the other hand, the percentage of DN1 (CD44+CD25−) cells in EML/OP9-DL1 cocultures decreased steadily during the culture period (Fig. 2D and E).
The DN2 cells appeared in EML/OP9 cocultures too, but at a much lower frequency, and their percentage remained similar throughout the coculture. Importantly, DN3 cells never properly developed in EML/OP9 cocultures (Fig. 2E). The percentage of CD44+CD25− (DN1) and CD44−CD25− (DN4) cells remained similar throughout EML/OP9 coculture and did not differ significantly from undifferentiated EML cells (Fig. 2E).
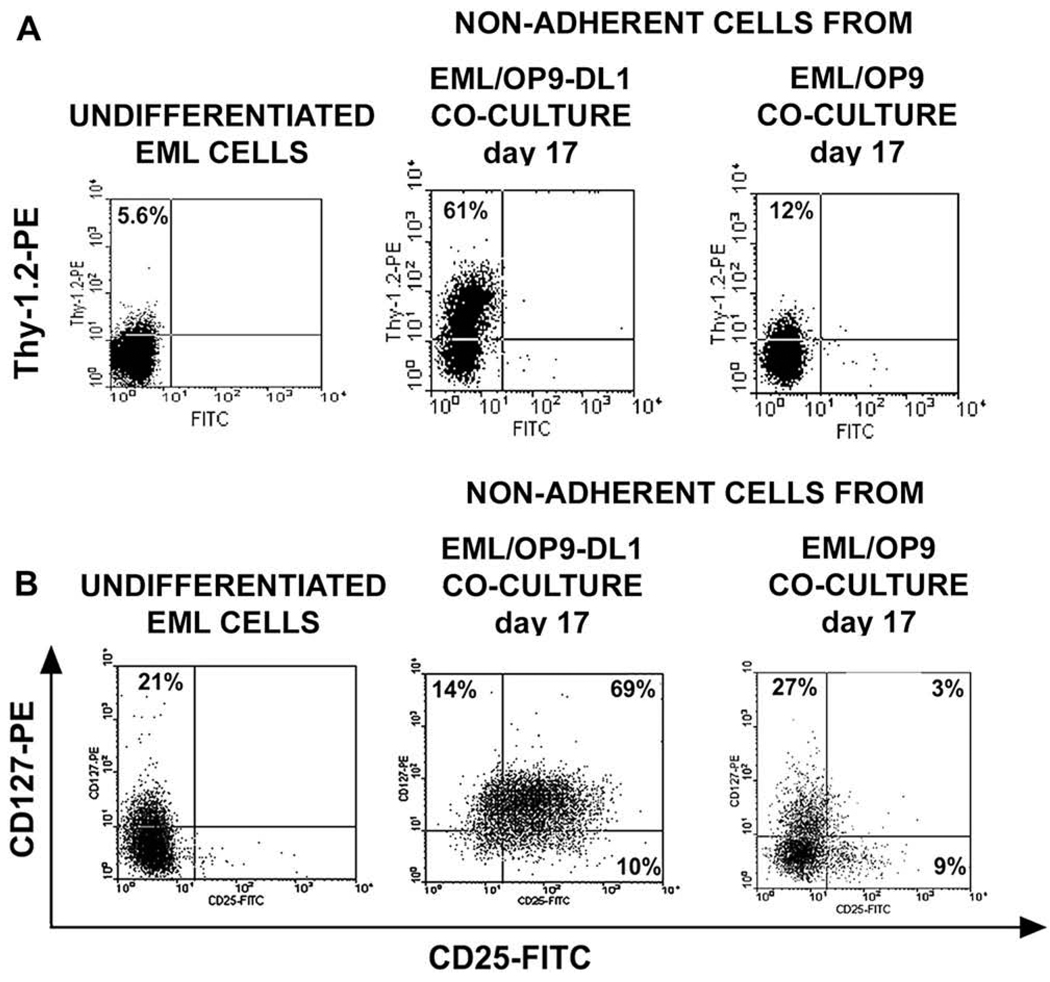
The naEML cells from OP9-DL1 and OP9 cocultures were next examined for expression of Thy-1.2, CD127 (IL-7 receptor α-chain), CD24 (heat stable antigen) and CD25 markers that characterize DN stages of T-cell development [41,43–46]. Both the percentage of Thy-1.2+ cells and the level of Thy-1.2 expression on naEML cells from day 17 EML/OP9-DL1 cocultures increased significantly when compared to Thy-1.2 expression on undifferentiated EML and naEML cells from OP9 cocultures (Fig. 3A). Up to 21% of undifferentiated EML cells express IL-7Rα chain (CD127), but none coexpressed CD25. The frequency of IL-7Rα+ and CD25+ cells among naEML cells from day 17 OP9-DL1 cocultures increased 4-fold and > 25-fold, respectively, and the majority of IL-7Rα+ cells coexpressed CD25, characteristic for DN2 and DN3 stages (Fig. 3B). The percentage of IL-7Rα+ cells in EML/OP9 cocultures remained similar to undifferentiated EML cells, with <3% of cells coexpressing IL-7Rα and CD25 (Fig. 3B).
Figure 3.
In OP9 stromal cells expressing Notch ligand Delta-like 1 (OP9-DL1) cocultures EML cells differentiate into double-negative (DN) cells that coexpress CD25, Thy-1.2, interleukin-7 receptor-α (IL-7Rα), CD24, and CD44 markers. Flow cytometry analysis of (A) Thy-1.2 expression and (B) CD127 (IL-7Rα) and CD25 coexpression on undifferentiated EML cells, and nonadherent EML (naEML) cells from day 17 EML/OP9-DL1 and control EML/OP9 cocultures. (C) Flow cytometry analysis of CD24, Thy-1.2, CD25, and CD44 coexpression on undifferentiated EML cells and naEML cells from day 17 EML/OP9-DL1 cocultures. The contour plots on the left show the analysis of CD24, Thy-1.2, and CD25 coexpression on undifferentiated EML cells and naEML cells from EML/OP9-DL1 cocultures. CD44 expression was analyzed by three-color flow cytometry among CD24+CD25+ and Thy-1.2+CD25+ naEML cells in the upper right quadrant (R2 gate) of contour plots. Histograms on the right show the expression of CD44 on R2 gated CD24+CD25+ and Thy-1.2+CD25+ naEML cells. The numbers in histograms represent percentage of CD44-positive cells based on the allophycocyanin (APC) control staining.
Notably, the population of naEML cells from EML/OP9-DL1 cocultures also contained cells that coexpressed CD24 and CD25, and Thy-1 and CD25 (Fig. 3C), thus resembling T-lineage cells [41,43–46]. In addition, 40% to 50% of CD24+CD25+ and Thy-1.2+CD25+ naEML cells from EML/OP9-DL1 cocultures coexpressed CD44 as well (Fig. 3C). Taken together, these results support the notion that in OP9-DL1 cocultures EML cells differentiate into cells that coexpress CD44, CD25, Thy-1, IL-7Rα, and CD24, and phenotypically resemble DN stages of T-cell development [41–46].
DL1 ligand and cytokines are both necessary for differentiation of EML cell line into DN T -lineage cells
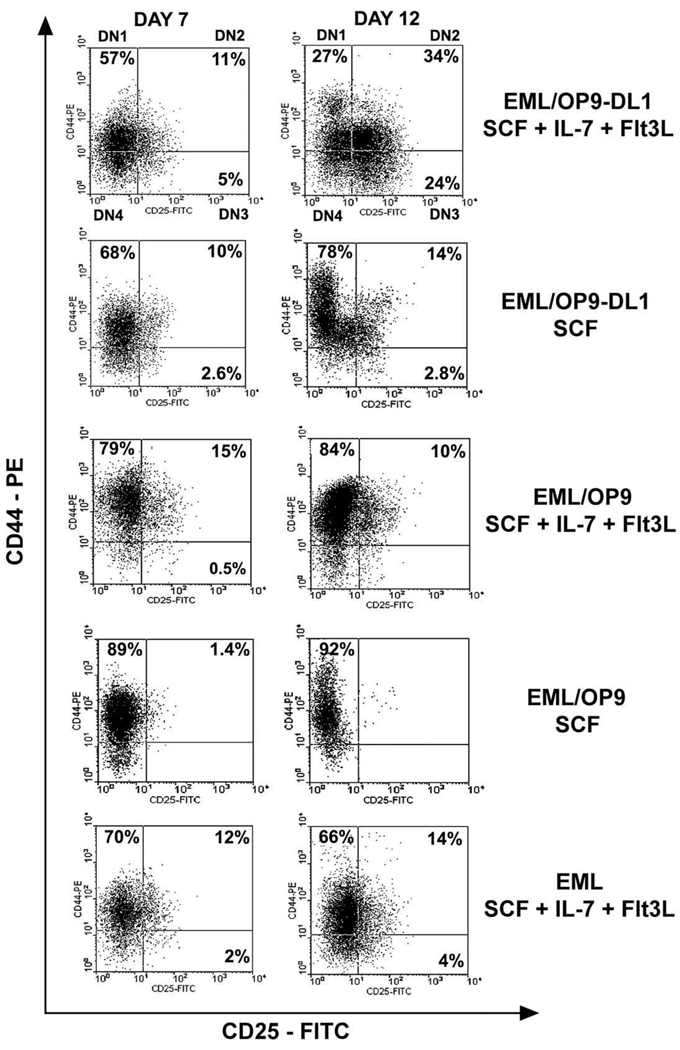
The appearance of CD44+CD25+ DN2 cells in the absence of DL1 ligand in control EML/OP9 cocultures (Fig. 2D) raised the question about the role of cytokines IL-7 and Flt3L in differentiation of EML cells into cells expressing CD25. Hence, we comparatively analyzed development of DN2 (CD44+CD25+) and DN3 (CD44−CD25+) cells in 1) EML/OP9-DL1 and EML/OP9 cocultures supplemented with SCF, IL-7, and Flt3L; 2) EML/OP9-DL1 and EML/ OP9 cocultures supplemented with SCF only; and 3) EML cell cultures supplemented with the cytokines (SCF, IL-7, and Flt3L), but in the absence of OP9 cells. Generation of DN2 cells in EML/OP9-DL1 + SCF cocultures was far less efficient than in EML/OP9-DL1 cultures supplemented with all three cytokines, and development of DN3 cells was minimal as well (Fig. 4). The EML/OP9 + SCF cultures did not support development of CD44+CD25+ cells (Fig. 4). Notably, when EML cells were cultured in the presence of cytokines alone (SCF, IL-7, and Flt3L), up to 15% of EML cells acquired CD44+CD25+ phenotype, but significant development of DN3 CD44−CD25+ cells was never observed in these cultures (Fig. 4). These findings indicate that DL1 ligand and cytokines IL-7 and Flt3L are both necessary although not sufficient for differentiation of EML cell line into cells resembling DN stages of T-cell development.
Figure 4.
DL1 ligand and cytokines are both necessary for differentiation of EML cells into double-negative (DN) T-cell precursors. Differentiation of EML cells into DN2 and DN3 cells was examined in (A) day 7 and day 12 EML/OP9 stromal cells expressing Notch ligand Delta-like 1 (OP9-DL1) cocultures supplemented with all cytokines (stem cell factor [SCF], interleukin-7 [IL-7], and Fms-like tyrosine kinase 3 ligand [Flt3L]) or with SCF alone, (B) day 7 and 12 EML/OP9 cocultures supplemented with all cytokines or SCF alone, and (C) day 7 and 12 EML cell cultures with all cytokines in the absence of OP9 stromal cells. The numbers in each quadrant indicate the percentage of cells expressing the CD44 and/or CD25 marker.
In OP9-DL1 cocultures EML cells differentiate into DP and SP T cells and give rise to cells that produce IFN-γ in response to TCR activation
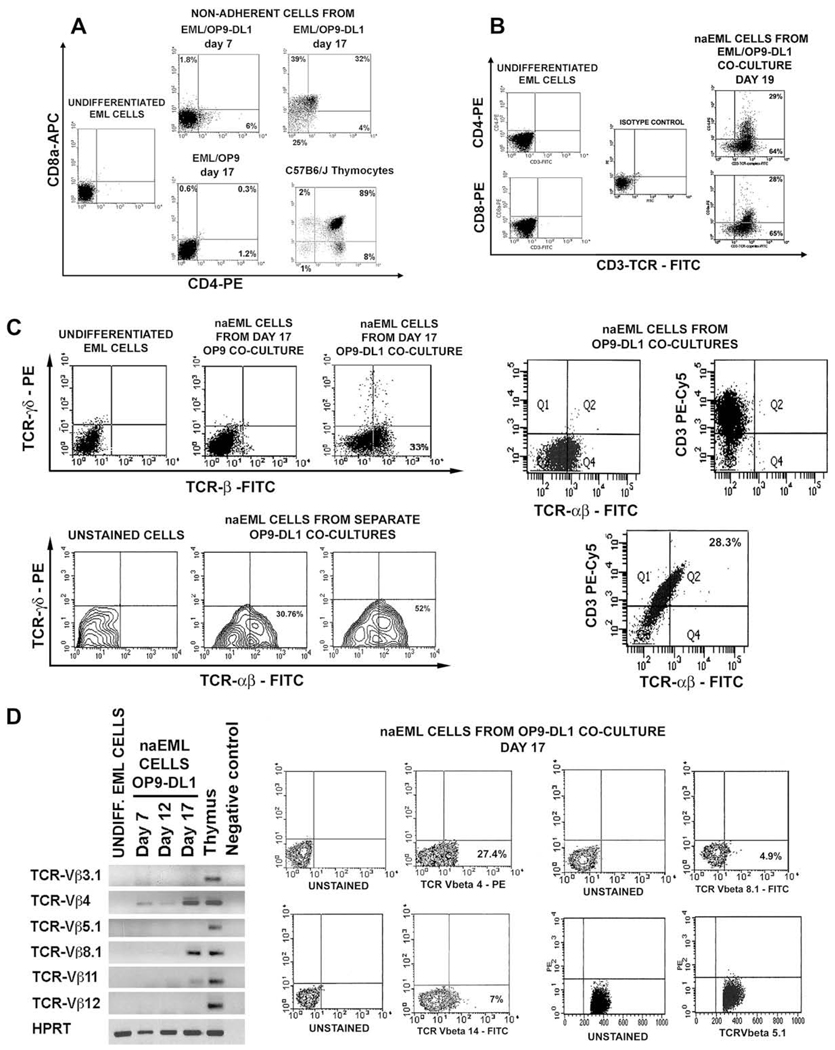
Because EML cells gave rise to DN cells in OP9-DL1 cocultures, we examined their capacity to differentiate into cells resembling more mature stages of T-cell development. Flow cytometry analysis has shown that in OP9-DL1 cocultures EML cells differentiated into CD4+ CD8+ double-positive (DP) cells and CD4+ CD8− and CD8+ CD4− single-positive (SP) cells (Fig. 5A and B). Although CD4+ CD8− cells appeared first on day 7 of coculture (Fig. 5A), by day 17 naEML cell population contained CD4+ CD8+, CD8+ CD4−, and CD4+ CD8− cells (Fig. 5A and B). In contrast, cells with DP and SP phenotypes were never observed in control EML/OP9 cocultures (Fig. 5A).
Figure 5.
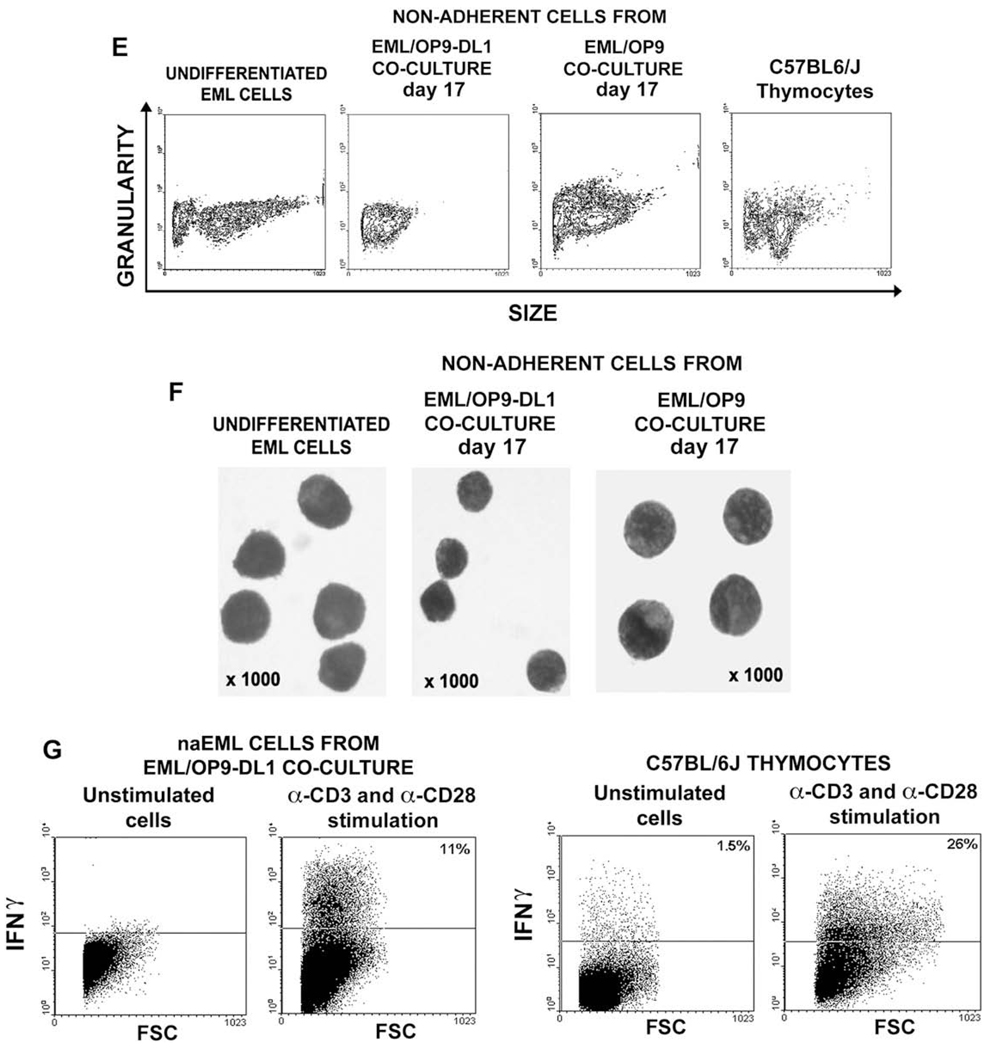
Development of CD4+ and CD8+ T-cell receptor (TCR)-expressing cells in EML/OP9 stromal cells expressing Notch ligand Delta-like 1 (OP9-DL1) cocultures. (A) In cocultures with OP9-DL1 stroma EML cells give rise to CD4+CD8+ double-positive (DP) cells and CD4+CD8− and CD8+CD4− single-positive (SP) cells, which were completely absent in EML/OP9 cocultures. (B) The majority of CD4+ and CD8+ cells from EML/OP9-DL1 cocultures expressed CD3-TCR-complex. (C) Analysis of TCR-αβ and TCRγδ expression on undifferentiated EML cells, and nonadherent EML (naEML) from EML/OP9 and EML/OP9-DL1 cocultures has revealed that EML cells in OP9-DL1 cocultures differentiate into cells resembling TCR-expressing T cells. The contour plots on the lower left show the percentage of TCR-αβ–expressing naEML cells from two separate EML/OP9-DL1 cocultures. Also, all TCRαβ–expressing naEML cells coexpress CD3 as well. (D) Reverse transcription polymerase chain reaction (RT-PCR) and flow cytometry analysis of expression pattern of TCR-Vβ3.1, 4, 5.1, 8-1, 11, 12, and 14 chains in undifferentiated EML cells, thymocytes and naEML from EML/OP9-DL1 co-cultures. The naEML cells from OP9-DL1 cocultures start to transcribe TCR-Vβ4 chain on day 7 of coculture, and TCR-Vβ8-1.2 and 11 chains on day 17 of coculture. Flow cytometry analysis has revealed that naEML cells from day 17 EML/OP9-DL1 cocultures express TCR-Vβ4, TCR-Vβ8-1.2, and TCR-Vβ14 chains, but do not express TCR-Vβ5.1 chain, thus corroborating RT-PCR results. (E) Flow cytometry analysis of the size of undifferentiated EML cells, naEML cells from EML/OP9-DL1 and EML/OP9 cocultures, and thymocytes from C57BL/6 J mice. (F) Comparison of the morphology of undifferentiated EML cells, and naEML cells from day 17 EML/OP9-DL1 and EML/OP9 cocultures. Cells were deposited on glass slides, stained with May-Grünwald/Giemsa and photographed using a Zeiss Axiovert microscope at × 1000 magnification. (G) The naEML cells from OP9-DL1 cocultures produce interferon-γ in response to TCR activation. The naEML cells from day 17 EML/OP9-DL1 cocultures and C57BL/6 J thymocytes were stimulated with plate-bound α-CD3 and α-CD28 for 72 hours. IFN-γ production was measured by intracellular staining of unstimulated and stimulated cells, followed by flow cytometry analysis. FITC = fluorescein isothiocyanate; PE = phycoerythrin.
Almost all CD4+ and CD8+ cells from day 19 EML/OP9-DL1 cocultures expressed the CD3-TCR-complex (Fig. 5B). Moreover, between 30% and 50% of naEML cells from replicate OP9-DL1 cocultures expressed TCR-αβ, and a very small percentage expressed TCR-γδ (Fig. 5C), indicating that EML cell line can differentiate into cells resembling TCR-expressing T cells [41–50]. Also, all TCR-αβ–expressing naEML cells coexpress CD3 as well (Fig. 5C). On the other hand, the naEML cells from EML/OP9 cocultures did not express TCRαβ and TCRγδ (Fig. 5C).
RT-PCR analysis of TCR-Vβ3.1, 4, 5.1, 8-1.2, 11, and 12 chain expression [40] has revealed that naEML cells from OP9-DL1 cocultures start to express TCR-Vβ4 chain on day 7 of coculture, and TCR-Vβ8-1.2 and 11 chains on day 17 of coculture (Fig. 5D). Undifferentiated EML cells did not express any of the analyzed TCR-Vβ chains (Fig. 5D). In addition, the flow cytometry analysis has shown that naEML cells from day 17 EML/OP9-DL1 cocultures express TCR-Vβ4, TCR-Vβ8-1.2, and TCR-Vβ14 chains, but do not express TCR-Vβ5.1 chain, thus corroborating RT-PCR results (Fig. 5D). These results are comparable with the frequency of cells from HPC/OP9-DL1 cocultures and adult thymus that express various TCR-Vβ chains [27,29]. Thus, naEML cells from OP9-DL1 cocultures express some of the TCR-Vβ chains that are commonly found in T cells from BDF1 (C57BL/6 J X DBA/2) mice, the strain of origin for EML cell line [1,40,47–50].
Notably, at the time of appearance of DP and SP cells in EML/OP9-DL1 cocultures, we also observed changes in the size and morphology of naEML cells (Fig. 5E and F). In comparison to undifferentiated EML cells and naEML cells from day 17 EML/OP9 cocultures, the naEML cells from day 17 OP9-DL1 cocultures were smaller and similar in size to thymocytes (Fig. 5E). This observation was confirmed by May-Grünwald/Giemsa staining (Fig. 5F). Undifferentiated EML cells are blast-like cells with oval shape [1,2]. In contrast, the naEML cells from day 17 OP9-DL1 cocultures were twice smaller, resembling round lymphocytes with high nucleus to cytoplasm ratio. The morphology of naEML cells from control EML/OP9 cocultures remained similar to undifferentiated EML cells (Fig. 5F).
Presence of TCR-expressing cells in EML/OP9-DL1 cocultures raised the question about their maturity. Thus, we sought to determine whether naEML cells from EML/OP9-DL1 cocultures can produce IFN-γ in response to TCR stimulation (Fig. 5G) [28,32]. Harvested naEML cells from day 17 EML/OP9-DL1 cocultures and C57BL/6 J thymocytes were stimulated with plate-bound α-CD3 and α-CD28 monoclonal antibodies for 72 hours. Presence of intracellular IFN-γ was detected by antibody staining and compared with IFN-γ production by B6 thymocytes. Notably, a significant percentage of stimulated naEML cells were found to produce IFN-γ (Fig. 5G).
Cumulatively, these results support the notion that in addition to development of DN-like cells, coculture with OP9-DL1 stroma supports differentiation of EML cell line into cells resembling DP and SP T cells, and cells that produce IFN-γ after TCR stimulation.
T-lineage differentiation of EML cell line is accompanied by expression of genes associated with T-cell development
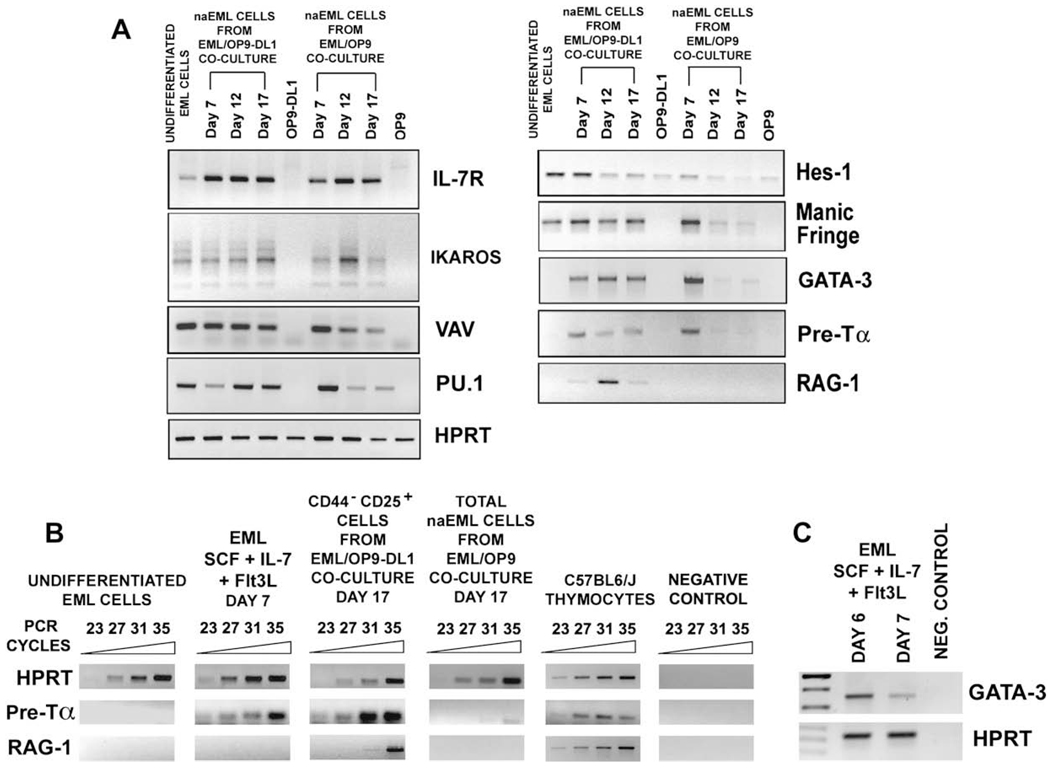
To further characterize differentiation of EML cells into T-cell lineage, we examined temporal expression pattern of genes from the Notch pathway (Hes-1, Fringe genes, Delta, and Jagged ligands), and several genes associated with T-cell development (Ikaros, IL-7R, Vav, PU.1, GATA-3, pre-Tα, and RAG-1) in 1) undifferentiated EML cells; 2) naEML cells from day 7, 12, and 17 OP9-DL1 cocultures; 3) naEML cells from day 7, 12, and 17 OP9 cocultures; and 4) OP9-DL1 and OP9 stromal cells [33–39,41,51–61].
The IL-7 receptor is a critical mediator of cell proliferation and differentiation during T-cell development, and DN thymocytes express high levels of IL-7R [44,46, 59–61]. Ikaros and Vav1 play important roles during T-cell development and in T-cell receptor signaling [51–54], whereas PU.1, which is expressed from thymic precursors to the DN3 stage, regulates transcription of IL-7Rα gene [55,56]. Transcripts for IL-7R, Ikaros, Vav1, and PU.1 were detected by RT-PCR in undifferentiated EML cells as well as in naEML cells from OP9-DL1 and OP9 cocultures (Fig. 6A).
Figure 6.
Temporal expression pattern of genes associated with T-cell development in nonadherent EML (naEML) cells from EML/OP9 stromal cells expressing Notch ligand Delta-like 1 (OP9-DL1) or EML/OP9 cocultures. (A) Reverse transcription polymerase chain reaction (RT-PCR) analysis of the temporal expression pattern of genes from the Notch pathway (HES-1, Fringe genes), transcription factors (Ikaros, PU.1, GATA-3), and other genes (IL-7Rα, Vav1, Pre-Tα, and RAG-1) associated with the T-cell development, in (1) undifferentiated EML cells; (2) naEML cells from day 7, 12, and 17 EML/OP9-DL1 or EML/OP9 cocultures; and (3) OP9-DL1 and OP9 stromal cells. Hypoxanthine phosphoribosyltransferase (HPRT) was amplified from all cell samples as an internal positive control. (B) Semi-quantitative RT-PCR analysis of Pre-Tα RAG-1 and HPRT gene expression in 1) undifferentiated EML cells, 2) EML cells cultured for 7 days in the presence of cytokines (SCF, IL-7, and Flt3L) without OP9 stromal cells, 3) purified CD44−CD25+ DN3 naEML cells sorted from day 17 EML/OP9-DL1 cocultures, 4) total naEML cells from day 17 control EML/OP9 cocultures, and 5) thymocytes from C57BL/6 J mice. Semi-quantitative RT-PCR reactions were performed for 23, 27, 31, and 35 cycles. Data shown are inverted images of ethidium bromide–stained gels. (C) RT-PCR analysis of GATA-3 expression in EML cells that were cultured for 6 and 7 days with cytokines (SCF, IL-7, and Flt3L), but in the absence of OP9 or OP9-DL1 stromal cells.
Hes-1, a downstream target of Notch signaling that is expressed at high levels in DN cells and in thymic stromal cells [24,34,35], was already transcribed in undifferentiated EML cells, and its expression was maintained in cocultured EML cells (Fig. 6A). On the other hand, expression of Delta and Jagged ligands was not detected in EML cells (data not shown). Interaction of Notch1 with Delta or Jagged ligands is influenced by Lunatic, Manic, and Radical Fringe proteins [36–38]. Undifferentiated EML and naEML cells from both cocultures transcribed the Manic Fringe (Fig. 6A), but not the Lunatic and Radical Fringe (data not shown). In agreement with the previous report, OP9 and OP9-DL1 stromal cells transcribe Hes-1, but do not express any of the Fringe genes (Fig. 6A and data not shown) [62].
GATA-3 is essential for the earliest stages of T-cell development, and is indispensable for SP CD4 cell development [57,58]. The Pre-Tα pairs with the TCR-β chains expressed in CD44−CD25+ DN3 cells and forms the pre – T-cell receptor complex, which mediates survival, proliferation, and differentiation of immature thymocytes [41,42,44,47,48]. The RAG-1 is necessary for TCR gene rearrangement, which begins at the DN3 stage and proceeds through the development of DP cells [39].
Notably, transcription of GATA-3 and Pre-Tα was induced in naEML cells from both OP9-DL1 and OP9 cocultures on day 7 (Fig. 6A). Interestingly however, while expression of GATA-3 and Pre-Tα was maintained in naEML cells throughout EML/OP9-DL1 coculture, it was barely detectable in naEML cells from day 12 and 17 OP9 cocultures (Fig. 6A). Most importantly, the RAG-1 expression was detected only in naEML cells from EML/OP9-DL1 cocultures (Fig. 6A).
These observations were confirmed by semi-quantitative RT-PCR analysis of Pre-Tα and RAG-1 expression in: 1) undifferentiated EML cells; 2) EML cells cultured for 7 days in the presence of cytokines (SCF, IL-7, and Flt3L) alone (without OP9 or OP9-DL1 stroma); 3) purified CD44−CD25+ DN3 naEML cells, sorted from day 17 EML/OP9-DL1 cocultures; 4) total naEML cells from day 17 control EML/OP9 cocultures, and 5) thymocytes from C57BL6/J mice (Fig. 6B).
The Pre-Tα transcript was expressed at high level in purified CD44−CD25+ DN3 naEML cells from day 17 EML/OP9-DL1 cocultures and in thymocytes, but was again barely detectable in naEML cells from day 17 EML/OP9 cocultures and undetectable in undifferentiated EML cells (Fig. 6B). Importantly, both the Pre-Tα and GATA-3 were also transcribed in EML cells cultured for 7 days with cytokines only (SCF, IL-7, and Flt3L), indicating that cytokines alone can induce GATA-3 and Pre-Tα expression in the absence of OP9-DL1 stromal cells (Fig. 6B and C). Expression of RAG-1, on the other hand, was detected only in the purified CD44−CD25+ DN3 naEML cells from EML/OP9-DL1 cocultures, and thymocytes from C57BL6/J mice (Fig. 6B).
Discussion
The phenotypic, molecular, and functional analysis of EML cells from OP9 coculture system has revealed that in EML/OP9-DL1 cocultures with cytokines (SCF, IL-7, and Flt3L), but not in control EML/OP9 cocultures, EML cell line undergoes T-cell lineage commitment and differentiation. During coculture with OP9-DL1 cells EML cell line differentiated into cells that 1) exhibited DN, DP, and SP cell-surface phenotype; 2) expressed CD3-TCR and TCR-Vβ chains; 3) initiated and maintained expression of GATA-3, Pre-Tα, and RAG-1 genes, associated with T-cell lineage commitment and differentiation; and 4) responded to TCR stimulation by IFN-γ production. Overall, the kinetics of EML cell differentiation into distinct stages of T-cell development was remarkably similar to although not as robust as previously described T-cell development from mouse fetal HPCs and adult HSC in OP9-DL1 cultures [27,29–32]. In addition, the T-cell differentiation of EML cell line was very similar to recently described T-lineage differentiation of adult DN1 and DN2 thymocytes, Lin−c-kit+ progenitors from fetal liver and Lin−Sca-1+c-kit+ Thy-1.1neg BM progenitor cells in OP9-DL1 cocultures [46,63,64].
Similar to the previous report, CD4+ CD8− cells that likely represent developmental intermediates between DN and DP cells, appeared early in EML/OP9-DL1 cocultures [27,32]. Also, development of CD8+ SP cells in EML/OP9-DL1 cocultures was more robust than development of CD4+ SP cells, likely due to the fact that OP9 cells express major histocompatibility complex class I but not class II, and the absence of the proper positive selection [27,32].
Exclusive development of cells with the DN, DP, and SP phenotypes in EML/OP9-DL1 cocultures with cytokines, together with the observation that in the absence of DL1 (EML/OP9 cocultures or EML cultured in the presence of cytokines alone) EML cells differentiate poorly into DN3 cells, indicates that continuous Notch1-DL1 interactions are required for T-lineage commitment and differentiation of EML cells [32,65].
Finding that coculture of EML cells with OP9-DL1 stroma without IL-7 and Flt3L does not support significant development of DN3 cells suggests that Notch1 – DL1 interaction and cytokine signaling are both necessary for inducing and sustaining differentiation of EML cells into T-cell lineage. In fact, several studies have shown that IL-7, Flt3L, and SCF are required for Notch-induced T lymphopoiesis in vitro, thus underlining a synergistic effect of Notch1 and cytokine signaling in inducing and supporting T-cell development [28,60,64,66,67].
Importantly, in contrast to undifferentiated EML cells, the naEML cells from EML/OP9-DL1 cocultures initiated and continued to express CD3, TCR, GATA-3, Pre-Tα, and RAG-1 genes. On the other hand, the naEML cells from control EML/OP9 cocultures did not express RAG-1, and have initiated but did not maintain the transcription of GATA-3 and Pre-Tα genes in the absence of DL1. Notably, the induction of GATA-3 and Pre-Tα expression was also observed in EML cells cultured in the presence of cytokines alone.
The latter observation supports the notion that expression of GATA-3 and Pre-Tα in EML cells is induced by IL-7 and Flt3L, and that the presence of OP9-DL1 is not required for that to occur. However, the continued expression of GATA-3 and Pre-Tα in naEML cells requires the presence of DL1 ligand. Thus, our results are consistent with earlier findings that suggested that GATA-3 and Pre-Tα expression can be induced by cytokines, and although being a part of the T-lineage specification process, it is not sufficient per se for differentiation into later stages of T-cell development [45,46].
More importantly, the induction of RAG-1 expression only in naEML cells from EML/OP9-DL1 cocultures indicates that interaction of Notch1 with DL1 is necessary for initiation and maintenance of RAG-1 expression in cocultured EML cells. The OP9 stromal cells support robust myelopoiesis and erythropoiesis, as well as B and natural killer lymphopoiesis from mouse embryonic stem cells, and fetal and adult HSC and MPPs [15,16]. The original article that described derivation of EML cell line has reported that in coculture with W20 stromal cell line and in the presence of IL-7, EML cells give rise to pro-B cells that express RAG-1 and undergo D-J rearrangements [1]. Because the coculture of EML cells on OP9 stroma did not result in development of cells expressing RAG-1, we examined the expression of B220 and CD19 B cell markers on naEML cells from multiple EML/OP9-DL1 and EML/OP9 cocultures. Although 20% to 30% of undifferentiated EML cells express B220 antigen, none of them express CD19 (Suppl. Fig. 1 and data not shown). Similarly, a portion of naEML cells from OP9-DL1 and OP9 cocultures continued to express B220 marker, but none expressed CD19 (Suppl. Fig. 1 and data not shown). However, while the percentage of B220+ EML cells in EML/OP9 cocultures remained relatively unchanged for the first 17 days, in EML/OP9-DL1 cocultures the percentage of B220+ EML cells steadily decreased (Suppl. Fig. 1).
Thus, it seems that conditions used to coculture EML cells on OP9 stroma in this study are not sufficient to support differentiation of EML cells into a B-cell lineage, thus explaining the lack of RAG-1 expressing naEML cells in EML/OP9 cocultures. Because the B220+ EML cells were maintained in EML/OP9 cocultures, it is possible that modified OP9 coculture conditions could induce and support B cell differentiation of EML cell line.
Overall, the observed induction and temporal kinetics of T-lineage gene expression in EML cells from OP9-DL1 cocultures was similar to gene expression pattern and kinetics during T-cell development of mouse embryonic stem cells and fetal liver Lin−c-kit+ cells in OP9-DL1 cultures [27,29–32,63].
The coexpression of Notch1 and Manic Fringe could be contributing to the capacity of EML cells to undergo T-cell development in OP9-DL1 cultures. Manic and Lunatic Fringe proteins regulate Notch signaling by increasing the sensitivity of Notch1 to Delta-like ligands, and decreasing Notch sensitivity to Serrate-like ligands and [36–38]. Thus, an interaction of DL1 with Notch1 receptor on EML cells and their T-cell development could be positively regulated by Manic Fringe. Interestingly, multipotent and committed hematopoietic progenitors express high levels of Manic Fringe, and it was suggested that Manic protein could regulate the activation of Notch receptors during hematopoietic progenitor cell fate decisions [17].
Taken together, these results support the conclusion that EML cell line has the capacity for T-cell differentiation, and confirm the notion that EML cell line is a unique multipotent hematopoietic cell line with the capacity to differentiate into all major blood cell lineages (Fig. 1) [1–3]. Additional flow cytometry analysis of IL-7 receptor α-chain (CD127), CD44 and CD25 expression on Sca-1+ c-kithi undifferentiated EML cells has revealed that EML cell line contains a subpopulation of Sca-1+ c-kithi IL-7R−/low CD44+ CD25− cells, which phenotypically resemble early thymic progenitors (Suppl. Fig. 2) [64,68,69]. Thus, it is conceivable that the OP9-DL1 coculture of purified Sca-1+ c-kithi IL-7R−/low CD44+ CD25− EML cells, or coculture with OP9 cells coexpressing DL1 and DL4 ligands could lead to even more robust T-cell development of EML cell line [64,68–71].
Despite noted limitations of the OP9-DL1 system [32], the EML/OP9-DL1 coculture could be very useful for studying various molecular and cellular aspects of T-cell differentiation [63–65,67]. For example, the EML/OP9-DL1 coculture could be a valuable experimental system to study the impact of overexpression and short hairpin RNA-mediated knockdown of particular genes on T-cell lineage specification and differentiation [3,8,11–14].
Supplementary Material
Acknowledgments
Cell analysis and sorting were done at the UM Sylvester Cancer Center Flow Cytometry Core Lab. We are indebted to Dr. Zúñiga-Pflücker for sharing with us the OP9-DL1 and OP9 stromal cell lines, and for critical advice and very helpful comments. We also thank our colleagues Drs. Adkins, Strbo, and Malek for helpful comments and for sharing some of the reagents. This work was supported by the National Institutes of Health RO1 RR15242 grant (Bethesda, MD, USA) (R.J.) and the University of Miami Pilot Study Grant (Miami, FL, USA) (R.J.).
Footnotes
Conflict of Interest
No financial interest/relationships with financial interest relating to the topic of this article have been declared.
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.exphem.2009.05.002
References
- 1.Tsai S, Bartelmez S, Sitnicka E, Collins S. Lymphohematopoietic progenitors immortalized by a retroviral vector harboring a dominant-negative retinoic acid receptor can recapitulate lymphoid, myeloid, and erythroid development. Genes Dev. 1994;8:2831–2841. doi: 10.1101/gad.8.23.2831. [DOI] [PubMed] [Google Scholar]
- 2.Lawson ND, Krause DS, Berliner N. Normal neutrophil differentiation and secondary granule gene expression in the EML and MPRO cell lines. Exp Hematol. 1998;26:1178–1185. [PubMed] [Google Scholar]
- 3.Zayas J, Spassov DS, Nachtman RG, Jurecic R. Hematopoietic stem cells and multipotent progenitors express new truncated intracellular form of c-kit receptor. Stem Cells Dev. 2008;17:343–353. doi: 10.1089/scd.2007.0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christensen JL, Weissman IL. Flk-2 is a marker in hematopoietic stem cell differentiation: a simple method to isolate long-term stem cells. Proc Natl Acad Sci U S A. 2001;98:14541–14546. doi: 10.1073/pnas.261562798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Terskikh AV, Miyamoto T, Chang C, Diatchenko L, Weissman IL. Gene expression analysis of purified hematopoietic stem cells and committed progenitors. Blood. 2003;102:94–101. doi: 10.1182/blood-2002-08-2509. [DOI] [PubMed] [Google Scholar]
- 6.Park IK, He Y, Lin F, et al. Differential gene expression profiling of adult murine hematopoietic stem cells. Blood. 2002;99:488–498. doi: 10.1182/blood.v99.2.488. [DOI] [PubMed] [Google Scholar]
- 7.Pineault N, Helgason CD, Lawrence HJ, Humphries RK. Differential expression of Hox, Meis1, and Pbx1 genes in primitive cells throughout murine hematopoietic ontogeny. Exp Hematol. 2002;30:49–57. doi: 10.1016/s0301-472x(01)00757-3. [DOI] [PubMed] [Google Scholar]
- 8.Chiang MY, Monroe JG. BSAP/Pax5A expression blocks survival and expansion of early myeloid cells implicating its involvement in maintaining commitment to the B-lymphocyte lineage. Blood. 1999;94:3621–3632. [PubMed] [Google Scholar]
- 9.Bourette RP, De Sepulveda P, Arnaud S, Dubreuil P, Rottapel R, Mouchiroud G. Suppressor of cytokine signaling 1 interacts with the macrophage colony-stimulating factor receptor and negatively regulates its proliferation signal. J Biol Chem. 2001;276:22133–22139. doi: 10.1074/jbc.M101878200. [DOI] [PubMed] [Google Scholar]
- 10.Du Y, Campbell JL, Nalbant D, et al. Mapping gene expression patterns during myeloid differentiation using the EML hematopoietic progenitor cell line. Exp Hematol. 2002;30:649–658. doi: 10.1016/s0301-472x(02)00817-2. [DOI] [PubMed] [Google Scholar]
- 11.Si J, Collins SJ. IL-3-induced enhancement of retinoic acid receptor activity ismediated through Stat5, which physically associates with retinoic acid receptors in an IL-3–dependent manner. Blood. 2003;100:4401–4409. doi: 10.1182/blood-2001-12-0374. [DOI] [PubMed] [Google Scholar]
- 12.Kirito K, Fox N, Kaushansky K. Thrombopoietin stimulates Hoxb4 expression: an explanation for the favorable effects of TPO on hematopoietic stem cells. Blood. 2003;102:3172–3178. doi: 10.1182/blood-2003-03-0944. [DOI] [PubMed] [Google Scholar]
- 13.Jing X, Infante J, Nachtman RG, Jurecic R. E3 ligase FLRF (Rnf41) regulates differentiation of hematopoietic progenitors by governing steady-state levels of cytokine and retinoic acid receptors. Exp Hematol. 2008;36:1110–1120. doi: 10.1016/j.exphem.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang HH, Hemberg M, Barahona M, Ingber DE, Huang S. Transcriptome-wide noise controls lineage choice in mammalian progenitor cells. Nature. 2008;453:544–547. doi: 10.1038/nature06965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maillard I, Adler SH, Pear WS. Notch and the immune system. Immunity. 2003;19:781–791. doi: 10.1016/s1074-7613(03)00325-x. [DOI] [PubMed] [Google Scholar]
- 16.Baron M. An overview of the Notch signaling pathway. Semin Cell Dev Biol. 2003;14:113–119. doi: 10.1016/s1084-9521(02)00179-9. [DOI] [PubMed] [Google Scholar]
- 17.Singh N, Phillips RA, Iscove NN, Egan SE. Expression of notch receptors, notch ligands, and fringe genes in hematopoiesis. Exp Hematol. 2000;28:527–534. doi: 10.1016/s0301-472x(00)00146-6. [DOI] [PubMed] [Google Scholar]
- 18.Walker L, Carlson A, Tan-Pertel HT, Weinmaster G, Gasson J. The notch receptor and its ligands are selectively expressed during hematopoietic development in the mouse. Stem Cells. 2001;19:543–552. doi: 10.1634/stemcells.19-6-543. [DOI] [PubMed] [Google Scholar]
- 19.Radtke F, Wilson A, Mancini SJC, MacDonald HR. Notch regulation of lymphocyte development and function. Nat Immunol. 2004;5:247–253. doi: 10.1038/ni1045. [DOI] [PubMed] [Google Scholar]
- 20.Radtke F, Wilson A, Stark G, et al. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity. 1999;10:547–558. doi: 10.1016/s1074-7613(00)80054-0. [DOI] [PubMed] [Google Scholar]
- 21.MacDonald HR, Wilson A, Radtke F. Notch1 and T-cell development: insights from conditional knockout mice. Trends Immunol. 2001;22:155–160. doi: 10.1016/s1471-4906(00)01828-7. [DOI] [PubMed] [Google Scholar]
- 22.Wilson A, MacDonald HR, Radtke F. Notch1-deficient common lymphoid precursors adopt a B cell fate in the thymus. J Exp Med. 2001;194:1003–1012. doi: 10.1084/jem.194.7.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harman BC, Jenkinson EJ, Anderson G. Entry into the thymic microenvironment triggers Notch activation in the earliest migrant T cell progenitors. J Immunol. 2003;170:1299–1303. doi: 10.4049/jimmunol.170.3.1299. [DOI] [PubMed] [Google Scholar]
- 24.Pear WS, Radtke F. Notch signaling in lymphopoiesis. Semin Immunol. 2003;15:69–79. doi: 10.1016/s1044-5323(03)00003-4. [DOI] [PubMed] [Google Scholar]
- 25.Hernandez-Hoyos G, Alberoila-Ila J. A Notch so simple influence on T cell development. Semin Cell Dev Biol. 2003;14:121–125. doi: 10.1016/s1084-9521(02)00180-5. [DOI] [PubMed] [Google Scholar]
- 26.Jaleco AC, Neves H, Hooijberg E, et al. Differential effects of Notch ligands Delta-1 and Jagged-1 in human lymphoid differentiation. J Exp Med. 2001;194:991–1002. doi: 10.1084/jem.194.7.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmitt TM, Zúñiga-Pflücker JC. Induction of T cell development from hematopoietic progenitor cells by delta-like-1 in vitro. Immunity. 2002;17:749–756. doi: 10.1016/s1074-7613(02)00474-0. [DOI] [PubMed] [Google Scholar]
- 28.Hozumi K, Abe N, Chiba S, Hirai H, Habu S. Active form of notch members can enforce T lymphopoiesis on lymphoid progenitors in the monolayer culture specific for B cell development. J Immunol. 2003;170:4973–4979. doi: 10.4049/jimmunol.170.10.4973. [DOI] [PubMed] [Google Scholar]
- 29.Schmitt TM, de Pooter RF, Gronski MA, Cho SK, Ohashi PS, Zúñiga-Pflücker JC. Induction of T cell development and establishment of T cell competence from embryonic stem cells differentiated in vitro. Nat Immunol. 2004;5:410–417. doi: 10.1038/ni1055. [DOI] [PubMed] [Google Scholar]
- 30.La Motte-Mohs RN, Herer E, Zúñiga-Pflücker JC. Induction of T cell development from human cord blood hematopoietic stem cells by Delta-like 1 in vitro. Blood. 2005;105:1431–1439. doi: 10.1182/blood-2004-04-1293. [DOI] [PubMed] [Google Scholar]
- 31.Zúñiga-Pflücker JC. T-cell development made simple. Nat Rev Immunol. 2004;4:67–72. doi: 10.1038/nri1257. [DOI] [PubMed] [Google Scholar]
- 32.de Pooter R, Zúñiga-Pflücker JC. T-cell potential and development in vitro: the OP9-DL1 approach. Curr Opin Immunol. 2007;19:163–168. doi: 10.1016/j.coi.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 33.Bellavia D, Campese AF, Vacca A, Gulino A, Screpanti I. Notch3, another Notch in T cell development. Semin Immunol. 2003;15:107–112. doi: 10.1016/s1044-5323(03)00007-1. [DOI] [PubMed] [Google Scholar]
- 34.Tomita K, Hattori M, Nakamura E, Nakanishi S, Minato N, Kageyama R. The bHLH gene Hes1 is essential for expansion of early T cell precursors. Genes Dev. 1999;13:1203–1210. doi: 10.1101/gad.13.9.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaneta M, Osawa M, Sudo K, Nakauchi H, Farr AG, Takahama Y. A role for pref-1 and HES-1 in thymocyte development. J Immunol. 2000;164:256–264. doi: 10.4049/jimmunol.164.1.256. [DOI] [PubMed] [Google Scholar]
- 36.Shimizu K, Chiba S, Saito T, Kumano K, Takahashi T, Hirai H. Manic fringe and lunatic fringe modify different sites of the notch2 extracellular region, resulting in different signaling modulation. J Biol Chem. 2002;276:25753–25758. doi: 10.1074/jbc.M103473200. [DOI] [PubMed] [Google Scholar]
- 37.Hicks C, Johnston SH, diSibio G, Collazo A, Vogt TF, Weinmaster G. Fringe differentially modulates Jagged1 and Delta1 signalling through Notch1 and Notch2. Nat Cell Biol. 2000;2:515–520. doi: 10.1038/35019553. [DOI] [PubMed] [Google Scholar]
- 38.Koch U, Lacombe TA, Holland D, et al. Subversion of the T/B lineage decision in the thymus by lunatic fringe-mediated inhibition of Notch-1. Immunity. 2001;15:225–236. doi: 10.1016/s1074-7613(01)00189-3. [DOI] [PubMed] [Google Scholar]
- 39.Mombaerts P, Iacomini J, Johnson RS, Herrup K, Tonegawa S, Papaioannou VE. RAG-1 deficient mice have no mature B and T lymphocytes. Cell. 1992;68:869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- 40.Pannetier C, Cochet M, Darche S, Casrouge A, Zoller M, Kourilsky P. The sizes of the CDR3 hypervariable regions of the murine T-cell receptor beta chains vary as a function of the recombined germ-line segments. Proc Natl Acad Sci U S A. 1993;90:4319–4323. doi: 10.1073/pnas.90.9.4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shortman K, Wu L. Early T lymphocyte progenitors. Annu Rev Immunol. 1996;14:29–47. doi: 10.1146/annurev.immunol.14.1.29. [DOI] [PubMed] [Google Scholar]
- 42.Godfrey DI, Kennedy J, Suda T, Zlotnik A. A developmental pathway involving four phenotypically and functionally distinct subsets of CD3-CD4-CD8- triple-negative adult mouse thymocytes defined by CD44 and CD25 expression. J Immunol. 1993;150:4244–4252. [PubMed] [Google Scholar]
- 43.Hough MR, Takei F, Humphries RK, Kay R. Defective development of thymocytes overexpressing the costimulatory molecule, heat-stable antigen. J Exp Med. 1994;179:177–184. doi: 10.1084/jem.179.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Crompton T, Outram SV, Buckland J, Owen MJ. Distinct roles of the interleukin-7 receptor alpha chain in fetal and adult thymocyte development revealed by analysis of interleukin-7 receptor alpha-deficient mice. Eur J Immunol. 1998;28:1859–1866. doi: 10.1002/(SICI)1521-4141(199806)28:06<1859::AID-IMMU1859>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 45.Rothenberg EV. T-lineage specification and commitment: a gene regulation perspective. Semin Immunol. 2002;14:431–440. doi: 10.1016/s1044532302000787. [DOI] [PubMed] [Google Scholar]
- 46.Balciunaite G, Ceredig R, Fehling HJ, Zúñiga-Pflücker JC, Rolink AG. The role of Notch and IL-7 signaling in early thymocyte proliferation and differentiation. Eur J Immunol. 2005;35:1292–1300. doi: 10.1002/eji.200425822. [DOI] [PubMed] [Google Scholar]
- 47.von Boehmer H, Aifantis I, Feinberg J, et al. Pleiotropic changes controlled by the pre-T-cell receptor. Curr Opin Immunol. 1999;11:135–142. doi: 10.1016/s0952-7915(99)80024-7. [DOI] [PubMed] [Google Scholar]
- 48.Kruisbeek AM, Haks MC, Carleton M, Michie AM, Zúñiga-Pflücker JC, Wiest DL. Branching out to gain control: how the pre-TCR is linked to multiple functions. Immunol Today. 2000;21:637–644. doi: 10.1016/s0167-5699(00)01744-8. [DOI] [PubMed] [Google Scholar]
- 49.Hayday AC, Barber DF, Douglas N, Hoffman ES. Signals involved in gamma/delta T cell versus alpha/beta T cell lineage commitment. Semin Immunol. 1999;11:239–249. doi: 10.1006/smim.1999.0180. [DOI] [PubMed] [Google Scholar]
- 50.MacDonald HR, Radtke F, Wilson A. T cell fate specification and alphabeta/gammadelta lineage commitment. Curr Opin Immunol. 2001;13:219–224. doi: 10.1016/s0952-7915(00)00207-7. [DOI] [PubMed] [Google Scholar]
- 51.Georgopoulos K, Bigby M, Wang JH, et al. The Ikaros gene is required for the development of all lymphoid lineages. Cell. 1994;79:143–156. doi: 10.1016/0092-8674(94)90407-3. [DOI] [PubMed] [Google Scholar]
- 52.Yoshida T, Ng SY, Zúñiga-Pflücker JC, Georgopoulos K. Early hematopoietic lineage restrictions directed by Ikaros. Nat Immunol. 2006;7:382–391. doi: 10.1038/ni1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Harker N, Naito T, Cortes M, et al. The CD8alpha gene locus is regulated by the Ikaros family of proteins. Mol Cell. 2002;10:1403–1415. doi: 10.1016/s1097-2765(02)00711-6. [DOI] [PubMed] [Google Scholar]
- 54.Reynolds LF, Smyth LA, Norton T, et al. Vav1 transduces T cell receptor signals to the activation of phospholipase C-gamma1 via phosphoinositide 3-kinase-dependent and -independent pathways. J Exp Med. 2002;195:1103–1114. doi: 10.1084/jem.20011663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rothenberg EV, Dionne CJ. Lineage plasticity and commitment in T-cell development. Immunol Rev. 2002;187:96–115. doi: 10.1034/j.1600-065x.2002.18709.x. [DOI] [PubMed] [Google Scholar]
- 56.DeKoter RP, Lee HJ, Singh H. PU.1 regulates expression of the interleukin-7 receptor in lymphoid progenitors. Immunity. 2002;16:297–309. doi: 10.1016/s1074-7613(02)00269-8. [DOI] [PubMed] [Google Scholar]
- 57.Ting CN, Olson MC, Barton KP, Leiden JM. Transcription factor GATA-3 is required for development of the T-cell lineage. Nature. 1996;384:474–478. doi: 10.1038/384474a0. [DOI] [PubMed] [Google Scholar]
- 58.Pai SY, Truitt ML, Ting CN, Leiden JM, Glimcher LH, Ho IC. Critical roles for transcription factor GATA-3 in thymocyte development. Immunity. 2003;19:863–875. doi: 10.1016/s1074-7613(03)00328-5. [DOI] [PubMed] [Google Scholar]
- 59.Yu Q, Erman B, Bhandoola A, Sharrow SO, Singer A. In vitro evidence that cytokine receptor signals are required for differentiation of double positive thymocytes into functionally mature CD8+ T cells. J Exp Med. 2003;197:475–487. doi: 10.1084/jem.20021765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Munitic I, Williams JA, Yang Y, et al. Dynamic regulation of IL-7 receptor expression is required for normal thymopoiesis. Blood. 2004;104:4165–4172. doi: 10.1182/blood-2004-06-2484. [DOI] [PubMed] [Google Scholar]
- 61.Purohit SJ, Stephan RP, Kim HG, Herrin BR, Gartland L, Klug CA. Determination of lymphoid cell fate is dependent on the expression status of the IL-7 receptor. EMBO J. 2003;15:5511–5521. doi: 10.1093/emboj/cdg522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hoflinger S, Kesavan K, Fuxa M, et al. Analysis of Notch1 function by in vitro T cell differentiation of Pax5 mutant lymphoid progenitors. J Immunol. 2004;173:3935–3944. doi: 10.4049/jimmunol.173.6.3935. [DOI] [PubMed] [Google Scholar]
- 63.Taghon TN, David ES, Zúñiga-Pflücker JC, Rothenberg EV. Delayed, asynchronous, and reversible T-lineage specification induced by Notch/Delta signaling. Genes Dev. 2005;19:965–978. doi: 10.1101/gad.1298305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang H, Pierce LJ, Spangrude GJ. Lymphoid potential of primitive bone marrow progenitors evaluated in vitro. Ann N Y Acad Sci. 2005;1044:210–219. doi: 10.1196/annals.1349.026. [DOI] [PubMed] [Google Scholar]
- 65.Schmitt TM, Ciofani M, Petrie HT, Zúñiga-Pflücker JC. Maintenance of T cell specification and differentiation requires recurrent notch receptor-ligand interactions. J Exp Med. 2004;200:469–479. doi: 10.1084/jem.20040394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Varnum-Finney B, Brashem-Stein C, Bernstein ID. Combined effects of Notch signaling and cytokines induce a multiple log increase in precursors with lymphoid and myeloid reconstituting ability. Blood. 2003;101:1784–1789. doi: 10.1182/blood-2002-06-1862. [DOI] [PubMed] [Google Scholar]
- 67.Massa S, Balciunaite G, Ceredig R, Rolink AG. Critical role for c-kit (CD117) in T cell lineage commitment and early thymocyte development in vitro. Eur J Immunol. 2006;36:526–532. doi: 10.1002/eji.200535760. [DOI] [PubMed] [Google Scholar]
- 68.Porritt HE, Rumfelt LL, Tabrizifard S, Schmitt TM, Zúñiga-Pflücker JC, Petrie HT. Heterogeneity among DN1 prothymocytes reveals multiple progenitors with different capacities to generate T cell and non-T cell lineages. Immunity. 2004;20:735–745. doi: 10.1016/j.immuni.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 69.Allman DA, Sambandam SK, Miller JP, et al. Thymopoiesis independent of common lymphoid progenitors. Nat Immunol. 2003;4:168–174. doi: 10.1038/ni878. [DOI] [PubMed] [Google Scholar]
- 70.Bhandoola A, von Boehmer H, Petrie HT, Zúñiga-Pflücker JC. Commitment and developmental potential of extrathymic and intrathymic T cell precursors: plenty to choose from. Immunity. 2007;26:678–689. doi: 10.1016/j.immuni.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 71.Koch U, Fiorini E, Benedito R, et al. Delta-like 4 is the essential, nonredundant ligand for Notch1 during thymic T cell lineage commitment. J Exp Med. 2008;205:2515–2523. doi: 10.1084/jem.20080829. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.