Abstract
The E. coli periplasmic proteins CusF and CusB, as part of the CusCFBA efflux system, aid in the resistance of elevated levels of copper and silver by direct metal transfer between the metallochaperone CusF and the membrane fusion protein CusB, before metal extrusion from the periplasm to the extracellular space. Although previous in vitro experiments have demonstrated highly specific interactions between CusF and CusB that are crucial for metal transfer to occur, the structural details of the interaction have not been determined. Here, the interactions between CusF and CusB are mapped through nuclear magnetic resonance (NMR) spectroscopy and chemical cross-linking coupled with high-resolution mass spectrometry to better understand how recognition and metal transfer occur between these proteins. The NMR 1H-15N correlation spectra reveal that CusB interacts with the metal-binding face of CusF. In vitro chemical cross-linking with a 7.7 Å homobifunctional amine-reactive cross-linker, BS2G, was used to capture the CusF/CusB interaction site and mass spectral data acquired on an LTQ-Orbitrap confirm the following two cross-links: CusF K31 to CusB K29 and CusF K58 to CusB K32; thus, revealing that the N-terminal region of CusB interacts with the metal-binding face of CusF. The proteins transiently interact in a metal-dependent fashion and contacts between CusF and CusB are localized to regions near their respective metal binding sites.
Excess levels of intracellular heavy metal ions, including copper and silver, have toxic effects on cells due to the metals’ redox capabilities. Metal ion concentrations must be carefully regulated in order to avoid cell damage and death. Silver is used extensively as an antibacterial agent and is rising in popularity for its implementation into commercial products (1–3). Escherichia coli, as well as many other Gram-negative bacteria, respond to elevated levels of intracellular copper or silver by up-regulation of the cusCFBA operon (4). Three of the proteins in this system, CusCBA, are believed to form a protein complex that spans the periplasm, fulfilling its role as a metal efflux pump by transporting Cu(I)/Ag(I) from the periplasm to the extracellular space (5). CusA, CusB, and CusC are thought to share similarities in structure and function with the well-characterized multidrug resistance systems, for example AcrAB-TolC (6). However, while multidrug export systems generally demonstrate broad substrate specificity, the Cus system is highly specific for Cu(I) and Ag(I) (7–9).
The periplasmic membrane fusion protein (MFP), CusB, serves an essential role in conferring copper/silver resistance in the Cus system. CusB has a substrate binding function where it binds a single Cu(I) or Ag(I) ion through an N-terminal 3-methionine coordination site (10). If this metal binding site is disrupted, a loss of metal resistance is observed, demonstrating the importance of metal binding by CusB in the efflux process (10). This metal binding site in CusB may serve as the metal entry point into the efflux complex. However, structural details about this metal binding site are still needed, as this region of CusB is missing from the recently reported crystal structure (11).
CusF is a small periplasmic metal-binding protein that is essential for maximal function of the Cus system. CusF homologs are found only in putative copper/silver transport systems, making them a unique component when compared to the multidrug resistance systems (12). CusF functions as a metallochaperone, similar to previously studied yeast cytoplasmic copper chaperone systems (13–15), though it is structurally unrelated to other characterized copper chaperones (12).
Previous isothermal titration calorimetry (ITC) experiments demonstrated highly specific interactions between CusF and CusB. Furthermore, these interactions only take place when one of the proteins is bound to metal, and no interaction is observed when the proteins are either both in the apo state or both in the metal-bound state. This result suggests that the metal sites are important for promoting the interaction between CusF and CusB. As evidence for the role of CusF as a metallochaperone for CusB, experiments have shown that CusF directly transfers metal to CusB (16). In vitro, metal transfer occurs in both directions regardless of which protein was initially bound to metal, and metal is distributed approximately equally between the two proteins when CusF and CusB are at equimolar concentrations (16). This distribution in vitro is a reflection of the similar metal binding affinities of these two proteins (8,10).
The interaction between CusF and CusB is critical for direct metal transfer to occur, though the structural details of the interaction are not yet understood. In this manuscript, results of experiments designed to probe the molecular details of the interaction between CusF and CusB are described. First, NMR spectroscopy is used to identify CusF residues affected by interactions with CusB. Resonance positions are a result of the local environment of each nucleus influenced by both covalent and non-covalent interactions; thus, CusF residues involved in interaction with CusB can be mapped through analysis of chemical shift perturbations (17). Chemical cross-linking coupled to mass spectrometry is a very powerful tool for determining protein-protein interactions and is used in the present work to identify the region of CusB involved in the CusF/CusB interaction site. Cross-linking captures proteins interacting in their native/dynamic state and can resolve interaction sites down to the peptide/amino acid level. When coupled to mass spectrometry, this technique has advantages over other structural techniques such as NMR and X-ray crystallography in that it requires very low concentrations of protein and is unlimited by protein size (18). The results of these experiments demonstrate a specific interaction between the proteins in a region that involves each protein’s metal binding site.
Materials and Methods
Cell growth and protein purification
For preparation of uniformly 15N–labeled CusF, E. coli BL21-(λDE3) containing cusF in pASK-IBA3 was grown in M9 minimal media (19) containing 1.0 g/liter 15NH4Cl (Cambridge Isotopes Laboratories) as the sole source of nitrogen. Cells were grown in 20 mL of lysogeny broth (LB) media overnight, then centrifuged and transferred to 1 L of M9 media and grown at 37 °C until they reached an O.D.600 of 0.6–1.0, then induced with 200 µg/L of anhydrotetracycline (AHT). Growth was continued at 30 °C for 8–10 hrs. For the uniform 15N, 13C and 2H-labeled sample, the procedure was slightly modified. From the glycerol stock of cells maintained at −80 °C, 3.0 mL of LB media was inoculated. Cells were grown for approximately 3 hours at 37 °C after which they were transferred to 50 mL of M9 media made in H2O and containing 0.15 g 13C glucose and 0.05 g 15NH4Cl (20), resulting in final concentrations of 3.0 g/L 13C glucose and 1.0 g/L 15NH4Cl. The cells were grown for another 7 hours to an O.D.600 of approximately 1.5 then pelleted by centrifugation and resuspended in 200 mL of M9 media made in 2H2O and containing 13C glucose and 15NH4Cl at the above stated concentrations. Cell growth was continued for approximately 20 minutes before transferring to 800 mL of M9 made with 13C-glucose (3.0 g/L), 15NH4Cl (1.0 g/L) and 2H2O at 37 °C. At an O.D.600 of 0.2, cells were induced with AHT (200 µg/L) and the temperature was reduced to 28 °C. Cells were grown for another 12 hours before being harvested by centrifugation. The purification of CusF was performed as described previously (12). Cell growth and purification of CusB were performed as previously described (10).
NMR sample preparation, data collection and analysis
For NMR samples, the [15N]-CusF, [U-15N,13C,2H]-CusF, and CusB were dialyzed against 50 mM cacodylate pH 7.0. AgNO3 was added to either CusB or CusF at the described stoichiometry. The proteins were then re-dialyzed against 50 mM cacodylate pH 7.0 to remove unbound Ag(I). CusF and CusB were separately concentrated using Amicon concentrators with 5 kDa molecular weight cut-offs. Protein concentrations were determined using the BCA assay (Pierce Biotechnology).
All spectra were acquired at 298 K on a 600 MHz Varian Inova instrument equipped with a four-channel pulsed-field gradient triple-resonance cold probe, using pulse sequences from Varian Biopack. 1H-15N Heteronuclear Single Quantum Coherence (HSQC) (21) spectra with the TROSY option (22) were collected on four different samples: (i) 150 µM apo-[U-15N,13C,2H]-CusF; (ii) 150 µM Ag(I)-[U-15N,13C,2H]-CusF; (iii) 170 µM Ag(I)-[U-15N,13C,2H]-CusF mixed 1:1 with apo-CusB; and (iv) 160 µM apo-[U-15N,13C,2H]-CusF mixed 1:1 with Ag(I)-CusB. All samples contained 10% 2H2O and 0.02% NaN3. 256 increments of 2048 complex data points were collected for each experiment. Spectral widths of 7.2 kHz and 1.82 kHz were used in the 1H and 15N dimensions respectively. Spectra were processed with NMRPIPE (23) and analyzed with NMRView (24).
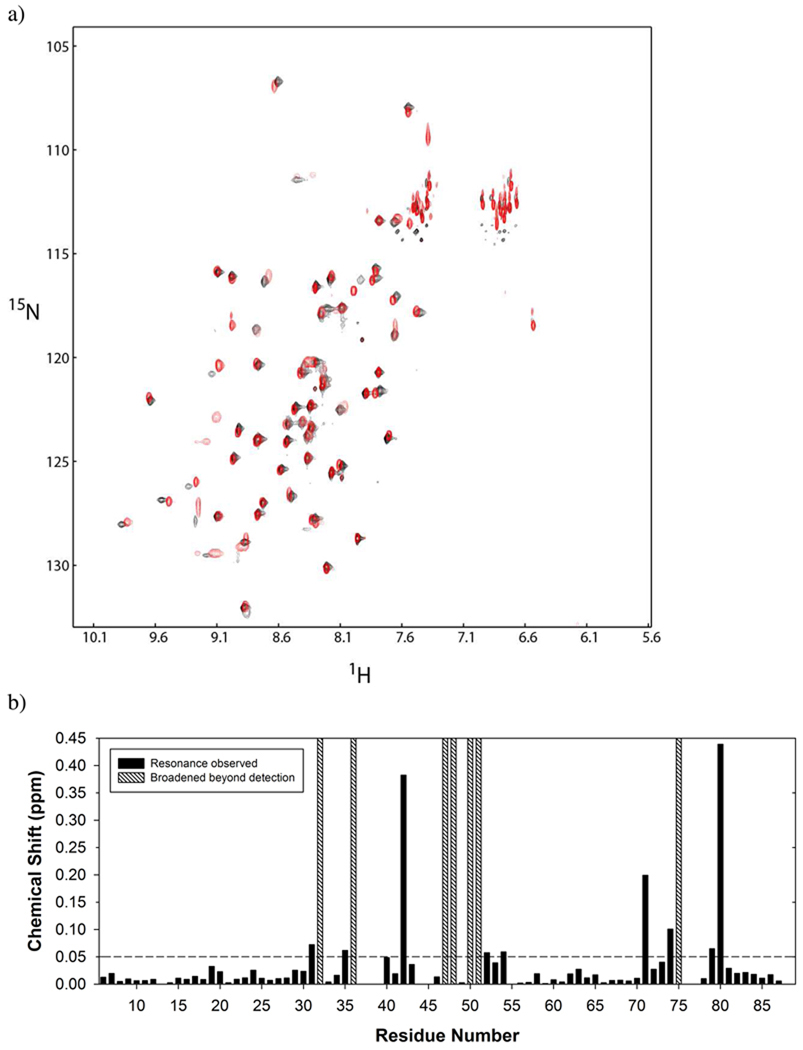
To determine the 1HN, 15N and 13Cα backbone assignments of CusF in the presence of CusB and Ag(I), a three-dimensional HNCA (25) spectrum was acquired on 170 µM Ag(I)-[U-15N,13C,2H]-CusF mixed with apo-CusB in a ratio of 1:1. The resonances were assigned by following the sequential connectivities in the HNCA spectrum. Of the 88 residues in CusF, the four proline residues and five N-terminal residues have not been assigned, in addition to I39, G76 and N77, which are not observed in either the Ag(I)-bound (8) or apo forms of CusF (12). Of the remaining 76 residues, 69 1HN-15N backbone resonances have been determined (Supplementary Table 1). The unassigned residues include I32, H36, M47, T48, R50, F51, and Q75, which are broadened beyond detection. Changes in chemical shifts upon addition of apo-CusB to Ag(I)-[U-15N,13C,2H]-CusF were calculated as the weighted average of the changes in the peak positions in the 1HN and 15N dimensions as compared to the CusF spectra containing a 0.5 molar ratio of Ag(I) using Δav= [(ΔδNH2 + ΔδN2/25)/2]1/2, where Δav is the combined chemical shift and ΔδNH and ΔδN are the chemical shift changes in the 1HN and 15N dimensions respectively.
To determine the fraction of CusF and CusB in association under NMR conditions, the spin-lattice (T1) and spin-spin (T2) relaxation values for Ag(I)-[15N]-CusF and Ag(I)-[15N]-CusF in the presence of apo-CusB were acquired using the standard 15N HSQC pulse sequence from Varian Biopack. 15N T1 values were measured from the spectra recorded with 7 different durations of delay T: T= 0, 30, 60, 120, 240, 480 and 750 msec. Similarly, 15N T2 values were measured from the spectra recorded with 8 different durations of delay T: T= 10, 30, 50, 70, 90, 130, 170, and 190 msec. The standard NMRView module was used to calculate the T1 and T2 values using the 66 resonances observable in both samples.
An approximation for the fraction of protein bound under NMR conditions and the protein-protein dissociation constant was obtained through the following procedure. From experimental average rotational correlation times (τm), adjusted for thermal and viscosity effects, from proteins ranging from 35 to 82 kDa (26–32), an estimate for the average rotational correlation time per kDa of protein was calculated as 0.55 nsec/kDa at 25 °C. The τm curve as a function of molecular weight is roughly linear in this range with near-zero intercept. Based on this value, the τm for a 54 kDa protein (CusF is ~10 kDa and CusB is ~44 kDa) is roughly 30 nsec. Using this value and an order parameter of 0.86, a 1/T1 value of 0.41 sec−1 and a 1/T2 value of 35.6 sec−1 on a 600 MHz spectrometer was obtained. The assumptions in this simplistic approach are: (i) fast exchange between the free and bound states for the observable resonances, (ii) isotropic molecular rotational diffusion, and (iii) similar order parameters in the free and bound proteins and negligible contributions to relaxation from terms related to fast internal motion correlation times. The fraction-bound was determined by utilizing the equation (33, 34):
where f is the fraction bound, (1/T)obs is the observed relaxation rate for Ag(I)-[15N]-CusF in the presence of CusB, (1/T)free is the observed rate for Ag(I)-[15N]-CusF alone, (1/T)bound is the estimated rate for a 54 kDa protein, and T is either T1 or T2. The dissociation constant (kD) of the complex was estimated using a single binding site model (34). The reported fraction bound value is the average from a set of residues whose extracted fraction bound values from T1 and T2 agree within 20% of each other. This criterion insures only simultaneous changes in both T1 and T2 due to the same molecular weight increase are included in the results, removing complications from other contributing factors.
Chemical cross-linking materials, reaction, and digestion
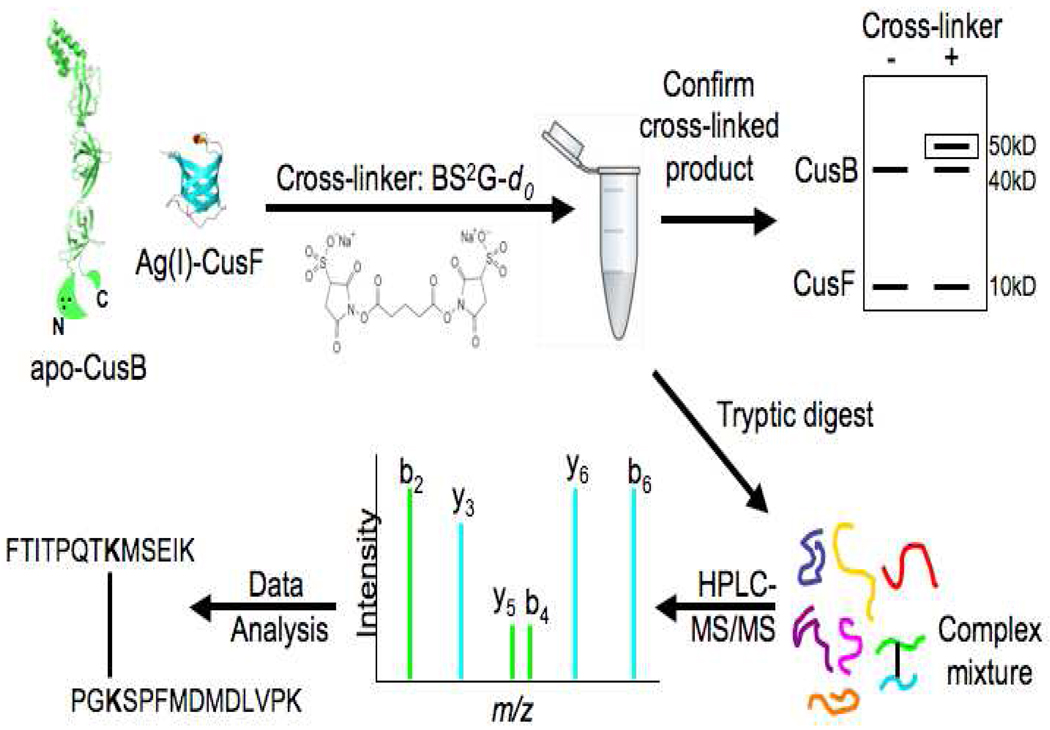
An overview of the cross-linking strategy is provided in Figure 1. Cross-linking reagent BS2G-d0 (bis(sulfosuccinimidyl)glutarate-d0) was purchased from Pierce (Rockford, IL). Sequencing-grade trypsin and all other chemicals were purchased from Sigma-Aldrich unless otherwise stated. Micro Bio-Spin Chromatography Columns were purchased from Bio-Rad (Hercules, CA). UltraMicro Spin C18 cartridges were purchased from the Nest Group (Southborough, MA).
Figure 1.
Schematic illustrating cross-linking strategy: Ag(I)-CusF and apo-CusB are mixed in 1:1 molar ratios with 50-fold excess cross-linker (BS2G-d0). Cross-linked product is confirmed using one-dimensional SDS-PAGE prior to in-solution tryptic digest. The complex peptide mixture is separated with nano-HPLC and analyzed by an LTQ-Orbitrap. LC-MS/MS data are searched with Phenyx and Popitam to identify cross-links. (CusF pdb code 1ZEQ (12), CusB pdb code 3H9I (11), modified with a cartoon depiction of the missing N and C terminal regions. The three black circles within the N-terminus represent the three-methionine metal binding site, M21, M36, and M38.)
For the cross-linking reactions, Ag(I)-CusF and apo-CusB were mixed in a 1:1 molar ratio (10 µM final concentration each) in 40 mM MOPS buffer, pH 7.0. The homobifunctional cross-linking reagent BS2G-d0 was prepared as a stock solution (50 mM) in DMSO shortly prior to addition. BS2G-d0 was added immediately upon protein mixing with a final concentration of 500 µM (50-fold molar excess of protein concentration) with a final reaction volume of 100 µL. The reaction was carried out at room temperature for 30 minutes and was quenched upon addition of NH4HCO3 (20 mM final concentration). A small portion (12 µL) of the reaction mixture was saved for cross-linked product verification with one-dimensional SDS-PAGE (4–20% gradient gel) and silver staining. The remaining reaction mixture was buffer exchanged into 25 mM NH4HCO3 and dried to 1 mg/mL protein concentration in a Speed Vac. The cross-linked sample was denatured with 6 M urea, diluted, and digested with trypsin (50:1 ratio of protein:trypsin) overnight at 37 °C. Prior to MS analysis, the cross-linked sample was desalted using a mini C18 column.
Nano-HPLC, mass spectrometry and data analysis
The digested peptide sample was analyzed on a hybrid linear ion trap-Orbitrap instrument, the LTQ-Orbitrap (Thermo Fisher, San Jose, CA). Peptides were ionized by electrospray ionization in positive ion mode. The LTQ-Orbitrap mass spectrometer was coupled to a nanoflow HPLC system (NanoAcquity; Waters Corporation, Milford, MA) equipped with a 100 µm i.d. × 18 mm long precolumn packed with 200 Å C18 stationary phase (5 µm, C18AQ; Michrom) for trapping peptides and a 75 µm i.d. × 150 mm long analytical column packed with 100 Å C18 stationary phase (5 µm, C18AQ; Michrom) for separation of peptides prior to MS analysis.
About 0.5 µg of the peptide digest was injected onto the precolumn at a flow of 4 µL/min in water/acetonitrile (95/5) with 0.1% (v/v) formic acid and eluted with an acetonitrile gradient with a flow of 250 nL/min. The mobile phase consisted of: (A) water/acetonitrile (95/5), 0.1% formic acid, (B) acetonitrile, 0.1% formic acid. A linear HPLC gradient was applied, as previously described (35).
MS survey scans were acquired in the Orbitrap over a m/z range of 400–2000, a resolution of 60000 (m/z 400) and an ion population of 5 × 105. For tandem MS scans in the Orbitrap, the precursor isolation width was set to 4 m/z units, resolution to 7500 and an ion population of 2 × 105. Collision Induced Dissociation (CID) was performed in the LTQ with collision energy of 40%. The five most abundant peaks in an MS survey scan were selected for tandem MS using the data dependent scanning mode. The data-dependent selection was set up to reject all un-assigned, singly-, doubly- and triply-charged precursors (as cross-linked peptides often have a charge state of 4 or higher, e.g. two amino termini and two basic C-terminal residues from tryptic cleavage). Dynamic exclusion was used to minimize data redundancy by excluding previously selected precursor ions (−0.1/+1.1 Da) for 45 sec after they have been fragmented once.
Tandem mass spectra were converted into peaklists (.dta files), deconvoluted, and searched by the open-modification pipeline as described previously (35). Briefly, Phenyx was used to identify and filter out the spectra originating from linear peptides and the unidentified spectra were searched with the open-modification search engine, Popitam. Popitam aids in the identification of cross-links by identifying all peptides with modifications of any mass on any amino acid and associates these modified peptides with their corresponding tandem mass spectra.
Results
CusB and CusF interact only in the presence of metal
CusF and CusB interact specifically in a metal dependent manner and metal transfer can occur between the two proteins (16). To gather further information on the interactions of these proteins, NMR 1H-15N correlation (HSQC) spectra were collected on samples of isotopically labeled CusF in the presence of unlabeled CusB (Supplementary Figures 1a–d). In agreement with the previously published results from isothermal titration calorimetry data (16), the NMR spectra showed chemical shift changes indicative of an interaction only when either one protein or the other is bound to metal (Supplementary Figure 1a–b) but not in the absence of metal (Supplementary Figure 1c) or when both proteins are in the Ag(I)-bound state (Supplementary Figure 1d). Furthermore, similar spectra of CusF are obtained regardless of which protein was initially bound to metal (Supplementary Figure 1e), supporting the previous finding that metal transfer can occur from either protein to the other and that an approximately equal distribution of metal occurs when the proteins are in equimolar amounts (16). Control spectra of CusF taken in the presence of bovine serum albumin or the E. coli periplasmic multicopper oxidase CueO in the presence of Ag(I) show no chemical shift changes indicative of a specific interaction (data not shown).
The CusB interaction face on CusF
The residues of CusF that interact with CusB were identified from NMR experiments. 1H-15N correlation spectra were collected of Ag(I)-2H/15N/13C-CusF mixed with apo-CusB at an equimolar concentration, hereafter referred to as the CusF/CusB/Ag(I) sample, though note that only CusF is isotopically labeled. A comparison of the spectra acquired for CusF/CusB/Ag(I) and either apo-CusF or Ag(I)-CusF shows that significant changes have occurred. Because of metal redistribution between the two proteins, the CusF/CusB/Ag(I) spectrum reflects a spectrum in which half of the CusF proteins are bound to metal, plus additional spectral effects from the interaction of CusF with CusB. In order to interpret the chemical shift effects from CusB, the resonances in the CusF/CusB/Ag(I) spectrum were assigned as described in the methods and then compared to the spectrum of CusF with a half stoichiometric equivalent of Ag(I) (Figure 2a–b). From this comparison, the CusF residues that were most significantly affected by the addition of CusB are L80, V42, N71, Q74, K31, S79, H35, T54, and T52, in decreasing order of effect. The resonances of seven residues: I32, H36, M47, T48, R50, F51 and Q75 were not observed due to line broadening, which also indicates an effect at these residues though the magnitude of the effect cannot be determined. The CusF-binding site of CusB could not be obtained by NMR experiments, since CusB spectra of sufficient quality could not be acquired. Therefore, chemical cross-linking coupled with mass spectrometry was used to obtain further information regarding the interaction.
Figure 2.
a) Overlay of 1H-15N correlation (HSQC) spectra of Ag(I)-15N-CusF (0.5:1) (red) and Ag(I)-15N-CusF (1:1) mixed equimolar with unlabeled CusB (black). b) Combined 1HN and 15N chemical shift changes between the spectra shown in a) are plotted for each residue in CusF. Residues whose chemical shifts were broadened beyond detection are indicated with a hatched bar.
The interaction site of CusF and CusB
The general strategy for determining the interaction site of CusF and CusB by chemical cross-linking mass spectrometry is illustrated in Figure 1. A 7.7 Å homobifunctional amine-reactive cross-linker, BS2G, was used to capture the metal-dependent interaction. No cross-linking between CusF and CusB was observed in the presence of cross-linker when both proteins were in their apo form. This is consistent with the NMR data above that show no evidence of an interaction in the absence of metal. However, when either CusF or CusB was metal-bound and the other protein was in the apo state, cross-linked products were observed on silver-stained SDS-PAGE gels at a position consistent with the sum of molecular weights of CusF and CusB (Supplementary Figure 2).
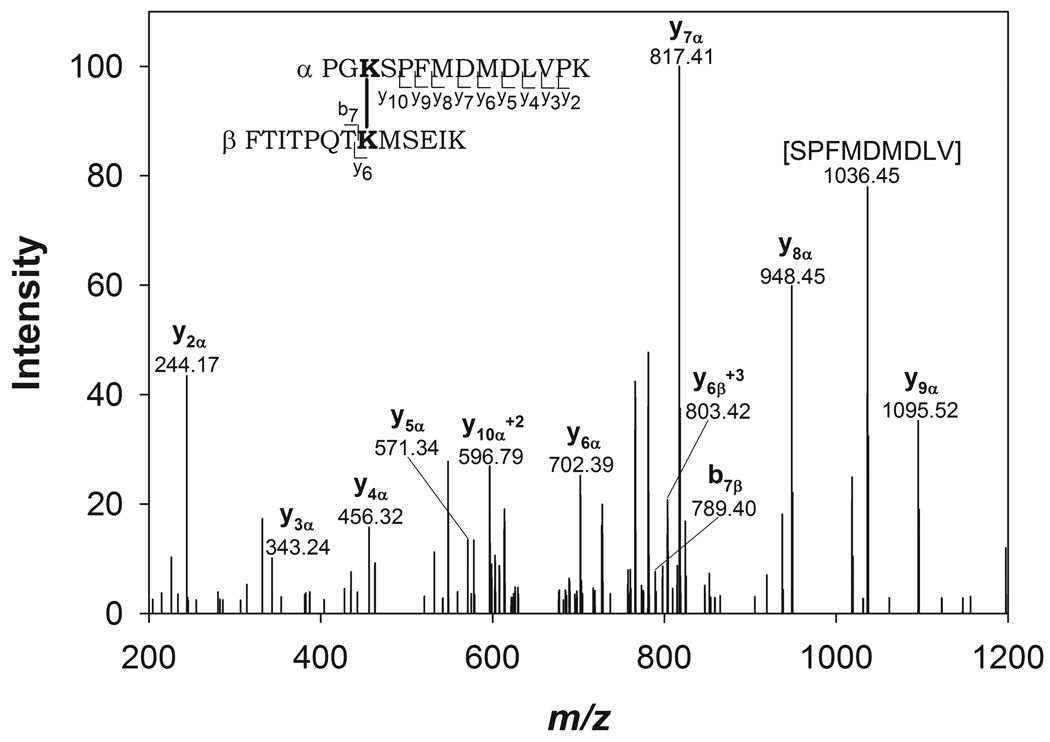
Following tryptic digestion of the cross-linked sample and LC-MS/MS data acquisition on an LTQ-Orbitrap, two CusF/CusB cross-linked products were identified using the open-modification pipeline. The mass to charge ratios of both cross-linked product precursor ions were within 20 ppm of the theoretical masses (Table 1). The first cross-link that was identified consisted of peptides with residues 51–63 of CusF and residues 30–43 of CusB. The second cross-linked product consisted of residues 31–50 of CusF and residues 27–55 of CusB. To further confirm the identity of the cross-linked products, fragment ions were assigned in the MS/MS spectra of the precursor ions (shown for the first cross-linked product in Figure 3 and the second cross-linked product in Supplementary Figure 3) using General Protein Mass Analysis for Windows (GPMAW) (Lighthouse Data, Odense, Denmark). Based on the MS/MS fragmentation, the cross-linking positions were identified as CusF K58 and CusB K32 in the first product and CusF K31 and CusB K29 in the second product.
Table 1.
List of amino acid sequences and residue numbers involved in CusF/CusB cross-linked products. The measured precursor masses are included, along with the error of the measured precursor ion compared to the theoretical mass.
| CusF peptide Residues (cross-linked residue) |
CusB peptide Residues (cross-linked residue) |
Measured precursor (amu) |
Error (ppm) |
|---|---|---|---|
| FTITPQTKMSEIK 51–63 (K58) | PGKSPFMDMDLVPK 30–43 (K32) | 3195.66 | 18.8 |
| KITIHHDPIAAVNWPEMTMR 31–50 (K31) | FDKPGKSPFMDMDLVPKYADEESSASGVR 27–55 (K29) | 5673.73 | 2.1 |
Figure 3.
MS/MS spectrum of the cross-linked precursor: 3195.66 amu. CusB30–43 and CusF51–63 are labeled as α and β respectively. Fragmentation resulting in b and y ions is marked within the peptide sequence and labeled in the spectrum.
Overlapping regions of CusB were identified in the two cross-linked products, while adjacent peptides of CusF were identified. In each cross-linked product, distinct lysine residues from both CusF and CusB peptides were involved in forming the cross-links. The peptides of CusB belong to the N-terminal region of the protein, which was previously identified as the metal-binding region (10). One of the peptides of CusF contains the previously determined metal binding residues (H36, M47, M49) (9, 12), and the other peptide is adjacent in sequence to this region. Based on the crystal structure of CusF (12), lysine residues CusF K31 and CusF K58, which participate in the two cross-linked products, are 13.79 Å apart (distance measured between alpha-carbons).
The interaction of CusF and CusB is transient
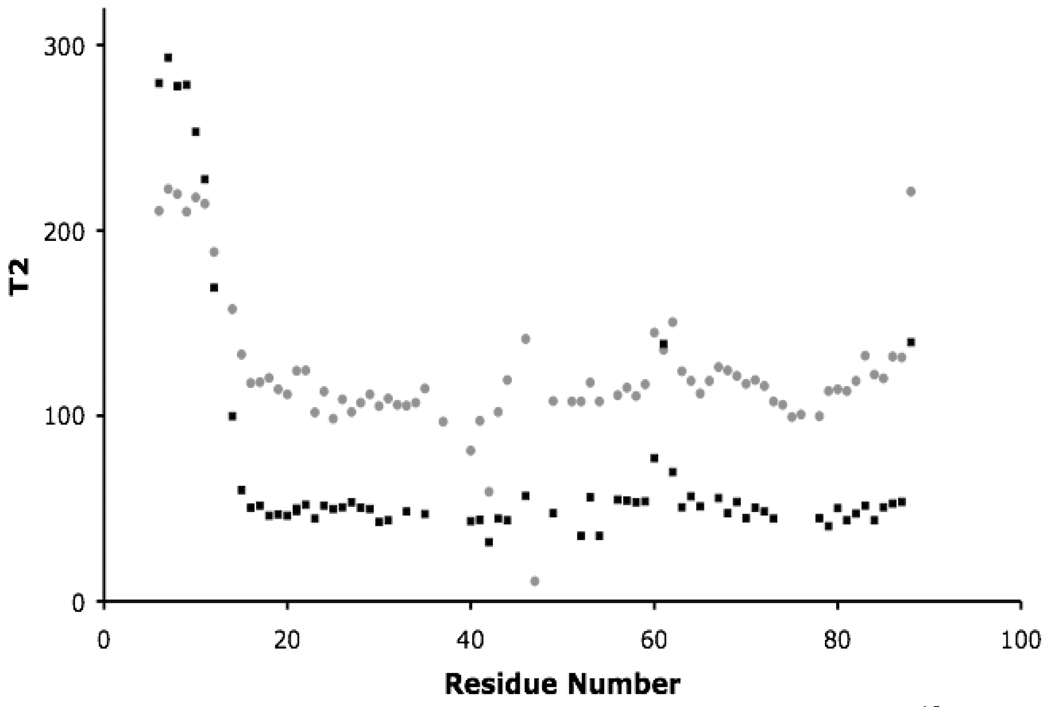
The HSQC NMR spectra of CusF do not show appreciable uniform linebroadening effects when CusB is added (Supplementary Figures 1a–b), suggesting that CusF and CusB form a transient complex. If a stable complex were formed, broadening of the linewidths in the spectra of CusF/CusB (~50 kD) would be expected compared to the spectrum of CusF alone (~10 kD). However, the effects of the CusF/CusB interaction are apparent from the NMR relaxation data for CusF/CusB/Ag(I) when compared to Ag(I)-CusF alone. The 15N relaxation times (T1 and T2) of Ag(I)-CusF in the presence of CusB are significantly changed as compared to that of Ag(I)-CusF alone (Figure 4 and Supplementary Figure 4). The average T1 value of CusF/CusB/Ag(I) is 568.5 msec compared to 457.1 msec for Ag(I)-CusF. The average T2 value of CusF/CusB/Ag(I) is 50 msec, while the average T2 value for Ag(I)-CusF alone is 110 msec.
Figure 4.
Plot of CusF residue number and corresponding T2 for Ag(I)-15N-CusF (gray circles) and 15N-CusF/CusB/Ag(I) (black squares).
An estimate of the fraction of CusF/CusB complex that is formed under these conditions was obtained from the difference between the observed relaxation times for the CusF/CusB/Ag(I) sample and that for a 54 kD molecule, as described in the methods. These calculations, using both the T1 and T2 data, suggest that approximately 38% of the CusF in the sample is in complex with CusB, corresponding to a dissociation constant of approximately 160 µM.
Discussion
CusF/CusB interaction is metal dependent and transient
Results from both NMR and chemical crosslinking coupled with mass spectrometry demonstrate that the interaction between CusF and CusB is metal-dependent, consistent with previous ITC and EXAFS results (16). The HSQC spectrum of apo-CusF mixed 1:1 with apo-CusB showed no change in the chemical shifts when compared with the spectrum of apo-CusF alone, indicating no interaction in the absence of metal. The HSQC spectra for apo-CusF mixed 1:1 with Ag(I)-CusB and apo-CusB mixed 1:1 with Ag(I)-CusF showed significant changes compared to the spectrum of apo-CusF alone, indicating that the two proteins interact in the presence of metal. The similarity of these spectra clearly points to an identical interaction between the two proteins regardless of which one contained metal before mixing with the apo-form of the other.
NMR spectra also demonstrate that the interaction between CusF and CusB is transient. The NMR relaxation results for CusF/CusB/Ag(I) indicate that approximately 38% of CusF is involved in a complex with CusB corresponding to a dissociation constant of approximately 160 µM. In the cell, the Cus system is expected to work by transferring metal first from CusF to CusB, and it has been recently suggested that CusB may transfer metal to CusA (36). Through transfer of metal from CusB to CusA, metal transfer from CusB to CusF (the reverse direction) may be limited, thus allowing the system to function effectively as an export system (16). The relatively weak binding measured here between CusF and CusB is likely sufficient in the organism to allow transfer of metal between proteins, while limiting metal transfer in the reverse direction. Weak association of metallochaperones and their targets are likely a general feature among these systems (37–39).
CusB interacts with CusF in the metal binding region
The CusB interaction face on CusF, as identified by the chemical shift perturbations listed above, is in the same region of the protein that contains the metal binding site. This region consists primarily of one end of the β-barrel and residues extending down the side (Figure 5). It is intuitive that CusB would interact with CusF in this region because metal is necessary for the interaction. Other proteins that share the same fold as CusF, the OB fold, primarily use the face of the β-barrel for interactions rather than the end (40). A close look at the residues in CusF affected by CusB binding reveals that three-fourths of these residues are either basic, aromatic or polar uncharged. It is thus plausible that CusF interacts with CusB via electrostatic/hydrophilic interactions. This interaction is highly specific, as evidenced by the failure of SilF, a close homolog of CusF (51% sequence identity), to interact with CusB (16). To analyze the nature of this specificity, the sequences of SilF and CusF were examined for differences in the CusB binding residues. Of the CusF residues most affected by CusB interactions, the corresponding residues in SilF are conserved in all but three positions, 35, 54, and 71 (using CusF numbering). At position 71, the residues of CusF and SilF still have somewhat similar properties, with a serine in SilF and an asparagine in CusF. However, at the other two positions, there are significant differences between the two proteins in the properties of the residues. SilF has a serine in place of H35 and a valine in place of T54. These two residues (H35 and T54) are most likely involved in specific electrostatic/hydrophilic interactions with CusB and their differences in charge or polarity with the corresponding SilF residues could explain why SilF fails to interact with CusB. Electrostatic interactions have been previously shown to be the prime recognition feature in other copper chaperone/acceptor systems like Atx1-Ccc2 (41, 42) and Cox17-Sco1 (43).
Figure 5.
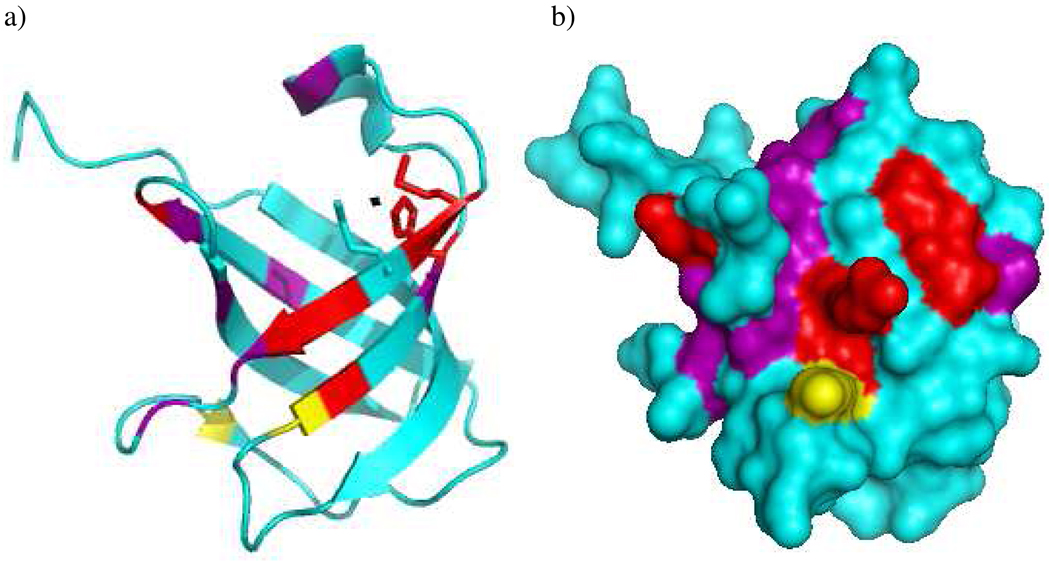
a) Ribbon diagram and b) surface representation of CusF (pdb code 2QCP (9)) indicating the residues whose resonances show significant chemical shift effects (purple), broaden beyond detection (red), and lysines K31 and K58 (yellow) that cross-link to CusB. H36, M47, and M49, which form the metal binding site, are shown in stick representation in the ribbon diagram with the Ag ion shown as a small black sphere. Figure prepared with Pymol (Delano Scientific, 2002).
Interaction site of CusF and CusB
Chemical cross-linking coupled with mass spectrometry was used to identify the interaction site of CusF/CusB during metal transfer in vitro. These results are consistent with the NMR results demonstrating that CusB interacts with the metal-binding face of CusF; however, NMR was unable to determine the region of CusB involved in the protein-protein interaction. Chemical cross-linking coupled with mass spectrometry was able to further resolve the interaction site between CusF and CusB by determining that peptides from the N-terminal region of CusB crosslink with CusF. Previous results from our lab have determined that CusB binds metal in a three-coordinate system with conserved residues M21, M36 and M38 (10). Two of these residues (M36 and M38) are included in both peptides (27–55 and 30–43) involved in cross-links with CusF. The third methionine (M21) is adjacent to this region. These cross-linked peptides identify the N-terminal metal binding region of CusB as interacting with CusF. This N-terminal region of CusB (residues 1–61, using the numbering without the leader sequence) is missing from the previously determined crystal structure (11), and this region is predicted to be mostly disordered. Though alternative CusB metal binding sites outside of the N-terminal region have been proposed (11), the residues composing these sites are not conserved and mutagenesis of the individual methionines in these proposed sites does not affect functionality (Kim, Rensing and McEvoy, unpublished results). The cross-linking data reported here further support the identification of the N-terminal M21, M36, and M38 site as the physiologically relevant metal binding site of CusB that interacts with CusF. This region of CusB could potentially prove to be the entry point of metal into the CusCBA efflux system.
The two peptides of CusF found cross-linked to CusB are adjacent to one another (31–50 and 51–63). CusF also uses a three-coordinate system for metal binding, residues H36, M47 and M49 (12). All three of these residues are present in one of the cross-linked peptides, CusF31–50. The other CusF peptide involved in cross-linking is adjacent to the first, with the lysine residues involved in both cross-links in close proximity to each other (~14 Å). Of the 16 CusF residues from the NMR results that experienced significant chemical shift changes or were broadened beyond detection, 11 are included in the CusF peptides involved in cross-linking with CusB.
Together, these results demonstrate that the interaction between the periplasmic proteins, CusF and CusB, is a transient, metal-dependent interaction that occurs at the metal binding regions of both proteins. This interaction may mark the initial entry point for metals into the Cus complex, ultimately driving the selectivity for Cu(I)/Ag(I) ions.
Supplementary Material
Acknowledgments
We gratefully acknowledge support from the National Institutes of Health GM079192 to M.M.M. and C.R., the National Institute for General Medical Sciences GM051387 to V.H.W., and the National Institutes of Health U54 AI05741 to D.R.G.
Abbreviations
- AHT
anhydrotetracycline
- BS2G
bis(sulfosuccinimidyl)glutarate
- CID
collision induced dissociation
- EXAFS
extended X-ray absorption fine structure
- GPMAW
general protein mass analysis for Windows
- HSQC
heteronuclear single quantum coherence
- ITC
isothermal titration calorimetry
- LB
lysogeny broth
- LC-MS/MS
liquid chromatography tandem mass spectrometry
- LTQ
linear trap quadrupole
- MFP
membrane fusion protein
- NMR
nuclear magnetic resonance
Footnotes
Supplemental Material
Additional information in support of this work is provided online. Figures S1a–S1e show NMR 1H-15N correlation (HSQC) spectra of 15N-labeled CusF mixed with unlabeled CusB and/or with Ag(I). Figure S2 is a silver-stained polyacrylamide gel showing the extent of cross-linking of CusF and CusB. Figure S3 shows the tandem MS spectrum of the cross-linked product. Figure S4 shows the T1 NMR data for Ag(I)-15N-CusF and 15N-CusF/CusB/Ag(I). Table S1 gives the backbone NMR resonance assignments from an HNCA experiment of Ag(I)-2H/15N/13C-CusF mixed 1:1 with unlabeled apo-CusB. This material may be accessed free of charge online at http://pubs.acs.org
References
- 1.Silver S. Bacterial silver resistance: Molecular biology and uses and misuses of silver compounds. FEMS Microbiol. Rev. 2003;27:341–353. doi: 10.1016/S0168-6445(03)00047-0. [DOI] [PubMed] [Google Scholar]
- 2.Silver S, Phung LT, Silver G. Silver as biocides in burn and wound dressings and bacterial resistance to silver compounds. J. Ind. Microbiol. Biotechnol. 2006;33:627–634. doi: 10.1007/s10295-006-0139-7. [DOI] [PubMed] [Google Scholar]
- 3.Lok CN, Ho CM, Chen R, He QY, Yu WY, Sun H, Tam PK, Chiu JF, Che CM. Silver nanoparticles: partial oxidation and antibacterial activities. J. Biol. Inorg. Chem. 2007;12:527–534. doi: 10.1007/s00775-007-0208-z. [DOI] [PubMed] [Google Scholar]
- 4.Franke S, Grass G, Nies DH. The product of the ybdE gene of the Escherichia coli chromosome is involved in detoxification of silver ions. Microbiology-UK. 2001;147:965–972. doi: 10.1099/00221287-147-4-965. [DOI] [PubMed] [Google Scholar]
- 5.Franke S, Grass G, Rensing C, Nies DH. Molecular analysis of the copper-transporting efflux system CusCFBA of Escherichia coli. J. Bacteriol. 2003;185:3804–3812. doi: 10.1128/JB.185.13.3804-3812.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nikaido H, Zgurskaya HI. AcrAB and related multidrug efflux pumps of Escherichia coli. J. Mol. Microbiol. Biotechnol. 2001;3:215–218. [PubMed] [Google Scholar]
- 7.Conroy O, Kim EH, McEvoy MM, Rensing C. Differing ability to transport nonmetal substrates by two RND-type metal exporters. FEMS Microbiol. Lett. 2010;308:115–122. doi: 10.1111/j.1574-6968.2010.02006.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kittleson JT, Loftin IR, Hausrath AC, Engelhardt KP, Rensing C, McEvoy MM. Periplasmic metal-resistance protein CusF exhibits high affinity and specificity for both Cu-I and Ag-I. Biochemistry. 2006;45:11096–11102. doi: 10.1021/bi0612622. [DOI] [PubMed] [Google Scholar]
- 9.Loftin IR, Franke S, Blackburn NJ, McEvoy MM. Unusual Cu(I)/Ag(I) coordination of Escherichia coli CusF as revealed by atomic resolution crystallography and X-ray absorption spectroscopy. Protein Sci. 2007;16:2287–2293. doi: 10.1110/ps.073021307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bagai I, Liu W, Rensing C, Blackburn NJ, McEvoy MM. Substrate-linked conformational change in the periplasmic component of a Cu(I)/Ag(I) efflux system. J. Biol. Chem. 2007;282:35695–35702. doi: 10.1074/jbc.M703937200. [DOI] [PubMed] [Google Scholar]
- 11.Su CC, Yang F, Long F, Reyon D, Routh MD, Kuo DW, Mokhtari AK, Ornam JDV, Rabe KL, Hoy JA, Lee YJ, Rajashankar KR, Yu EW. Crystal Structure of the Membrane Fusion Protein CusB from Escherichia coli. J. Mol. Biol. 2009;393:342–355. doi: 10.1016/j.jmb.2009.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loftin IR, Franke S, Roberts SA, Weichsel A, Heroux A, Montfort WR, Rensing C, McEvoy MM. A novel copper-binding fold for the periplasmic copper resistance protein CusF. Biochemistry. 2005;44:10533–10540. doi: 10.1021/bi050827b. [DOI] [PubMed] [Google Scholar]
- 13.Culotta VC, Lin SJ, Schmidt P, Klomp LW, Casareno RL, Gitlin J. Intracellular pathways of copper trafficking in yeast and humans. Adv. Exp. Med. Biol. 1999;448:247–254. doi: 10.1007/978-1-4615-4859-1_22. [DOI] [PubMed] [Google Scholar]
- 14.O'Halloran TV, Culotta VC. Metallochaperones, an intracellular shuttle service for metal ions. J. Biol. Chem. 2000;275:25057–25060. doi: 10.1074/jbc.R000006200. [DOI] [PubMed] [Google Scholar]
- 15.Rosenzweig AC. Copper delivery by metallochaperone proteins. Acc. Chem. Res. 2001;34:119–128. doi: 10.1021/ar000012p. [DOI] [PubMed] [Google Scholar]
- 16.Bagai I, Rensing C, Blackburn NJ, McEvoy MM. Direct metal transfer between periplasmic proteins identifies a bacterial copper chaperone. Biochemistry. 2008;47:11408–11414. doi: 10.1021/bi801638m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wuthrich K. Protein recognition by NMR. Nat. Struct. Biol. 2000;7:188–189. doi: 10.1038/73278. [DOI] [PubMed] [Google Scholar]
- 18.Sinz A. Chemical cross-linking and mass spectrometry to map three-dimensional protein structures and protein-protein interactions. Mass Spectrom. Rev. 2006;25:663–682. doi: 10.1002/mas.20082. [DOI] [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Vol. 1. New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 20.Gardner KH, Kay LE. The use of 2H, 13C, 15N multidimensional NMR to study the structure and dynamics of proteins. Annu. Rev. Biophys. Biomol. Struct. 1998;27:357–406. doi: 10.1146/annurev.biophys.27.1.357. [DOI] [PubMed] [Google Scholar]
- 21.Bodenhausen G, Ruben DJ. Natural Abundance N-15 Nmr by Enhanced Heteronuclear Spectroscopy. Chem. Phys. Lett. 1980;69:185–189. [Google Scholar]
- 22.Riek R, Pervushin K, Wuthrich K. TROSY and CRINEPT: NMR with large molecular and supramolecular structures in solution. Trends Biochem. Sci. 2000;25:462–468. doi: 10.1016/s0968-0004(00)01665-0. [DOI] [PubMed] [Google Scholar]
- 23.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 24.Johnson BA, Blevins RA. NMRView:a computer program for the visualization and analysis of NMR data. J. Biomol. NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 25.Bax A, Ikura M. An efficient 3D NMR technique for correlating the proton and 15N backbone amide resonances with the alpha-carbon of the preceding residue in uniformly 15N/13C enriched proteins. J. Biomol. NMR. 1991;1:99–104. doi: 10.1007/BF01874573. [DOI] [PubMed] [Google Scholar]
- 26.Liu A, Yao L, Li Y, Yan H. TROSY of side-chain amides in large proteins. J. Magn. Reson. 2007;186:319–326. doi: 10.1016/j.jmr.2007.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou H, Shatz W, Purdy MM, Fera N, Dahlquist FW, Reich NO. Long-range structural and dynamical changes induced by cofactor binding in DNA methyltransferase M.HhaI. Biochemistry. 2007;46:7261–7268. doi: 10.1021/bi602662e. [DOI] [PubMed] [Google Scholar]
- 28.Yang DW, Kay LE. TROSY triple-resonance four-dimensional NMR spectroscopy of a 46 ns tumbling protein. J. Am. Chem. Soc. 1999;121:2571–2575. [Google Scholar]
- 29.Kempf JG, Jung JY, Sampson NS, Loria JP. Off-resonance TROSY (R-1 rho-R-1) for quantitation of fast exchange processes in large proteins. J. Am. Chem. Soc. 2003;125:12064–12065. doi: 10.1021/ja037101s. [DOI] [PubMed] [Google Scholar]
- 30.Tugarinov V, Muhandiram R, Ayed A, Kay LE. Four-dimensional NMR spectroscopy of a 723-residue protein: Chemical shift assignments and secondary structure of malate synthase G. J. Am. Chem. Soc. 2002;124:10025–10035. doi: 10.1021/ja0205636. [DOI] [PubMed] [Google Scholar]
- 31.Eletsky A, Kienhofer A, Hilvert D, Pervushin K. Investigation of ligand binding and protein dynamics in Bacillus subtilis chorismate mutase by transverse relaxation optimized spectroscopy-nuclear magnetic resonance. Biochemistry. 2005;44:6788–6799. doi: 10.1021/bi0474259. [DOI] [PubMed] [Google Scholar]
- 32.Shan X, Gardner KH, Muhandiram DR, Rao NS, Arrowsmith CH, Kay LE. Assignment of N-15, C-13(alpha), C-13(beta), and HN resonances in an N-15, C-13, H-2 labeled 64 kDa trp repressor-operator complex using triple-resonance NMR spectroscopy and H-2-decoupling. J. Am. Chem. Soc. 1996;118:6570–6579. [Google Scholar]
- 33.Fischer JJ, Jardetzk O. Nuclear Magnetic Relaxation Study of Intermolecular Complexes. Mechanism of Penicillin Binding to Serum Albumin. J. Am. Chem. Soc. 1965;87:3237–3244. doi: 10.1021/ja01092a040. [DOI] [PubMed] [Google Scholar]
- 34.Fielding L. NMR methods for the determination of protein-ligand dissociation constants. Prog. Nucl. Magn. Reson. Spectrosc. 2007;51:219–242. [Google Scholar]
- 35.Singh P, Shaffer SA, Scherl A, Holman C, Pfuetzer RA, Larson Freeman TJ, Miller SI, Hernandez P, Appel RD, Goodlett DR. Characterization of Protein Cross-Links via Mass Spectrometry and an Open-Modification Search Strategy. Anal. Chem. 2008;80:8799–8806. doi: 10.1021/ac801646f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim EH, Rensing C, McEvoy MM. Chaperone-mediated copper handling in the periplasm. Nat. Prod. Rep. 2010;27:711–719. doi: 10.1039/b906681k. [DOI] [PubMed] [Google Scholar]
- 37.Banci L, Bertini I, Cantini F, Felli IC, Gonnelli L, Hadjiliadis N, Pierattelli R, Rosato A, Voulgaris P. The Atx1-Ccc2 complex is a metal-mediated protein-protein interaction. Nat. Chem. Biol. 2006;2:367–368. doi: 10.1038/nchembio797. [DOI] [PubMed] [Google Scholar]
- 38.Banci L, Bertini I, Cantini F, Rosenzweig AC, Yatsunyk LA. Metal binding domains 3 and 4 of the Wilson disease protein: solution structure and interaction with the copper(I) chaperone HAH1. Biochemistry. 2008;47:7423–7429. doi: 10.1021/bi8004736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Achila D, Banci L, Bertini I, Bunce J, Ciofi-Baffoni S, Huffman DL. Structure of human Wilson protein domains 5 and 6 and their interplay with domain 4 and the copper chaperone HAH1 in copper uptake. Proc. Natl. Acad. Sci. U.S.A. 2006;103:5729–5734. doi: 10.1073/pnas.0504472103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Agrawal V, Kishan KV. OB-fold: growing bigger with functional consistency. Curr. Protein Pept. Sci. 2003;4:195–206. doi: 10.2174/1389203033487207. [DOI] [PubMed] [Google Scholar]
- 41.Arnesano F, Banci L, Bertini I, Cantini F, Ciofi-Baffoni S, Huffman DL, O'Halloran TV. Characterization of the binding interface between the copper chaperone Atx1 and the first cytosolic domain of Ccc2 ATPase. J. Biol. Chem. 2001;276:41365–41376. doi: 10.1074/jbc.M104807200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.