Abstract
Enzyme inhibition via allostery, in which the ligand binds remotely from the active site, is a poorly understood phenomenon, and represents a significant challenge to structure-based drug design. Dipicolinic acid (DPA), a major component of Bacillus spores, is shown to inhibit glutamate racemase from Bacillus anthracis, a monosubstrate/monoproduct enzyme, in a novel allosteric fashion. Glutamate racemase has long been considered an important drug target for its integral role in bacterial cell wall synthesis. The DPA binding mode was predicted via multiple docking studies and validated via site-directed mutagenesis at the binding locus, while the mechanism of inhibition was elucidated with a combination of Blue Native PAGE, molecular dynamics simulations, free energy and pKa calculations. Inhibition by DPA not only reveals a novel cryptic binding site, but also represents a form of allosteric regulation that exploits the interplay between enzyme conformational changes, fluctuations in the pKa values of buried residues and catalysis. The potential for future drug development is discussed.
Keywords: Glutamate racemase, dipicolinic acid (DPA), B. anthracis, enzyme flexibility, allostery
1. Introduction
D-glutamate (D-glu) has been shown to be an essential feature of the peptidoglycan layer of bacterial cell walls in a number of pathogenic organisms, strongly suggesting that blocking its biosynthesis would be an attractive mode of action for antimicrobial drug discovery1. The enzyme glutamate racemase (GR) is responsible for the biosynthesis of D-glu in bacteria, employing a cofactor independent 1,1 proton transfer to invert the stereochemistry of the L-glutamate substrate. GR is a highly pursued antimicrobial drug target, and has been the subject of numerous ligand discovery studies2–9.
Extensive inhibitor development against GR has been directed toward the CDC’s Category A agents 5,10–12. B. anthracis, the causative agent of inhalational anthrax and a Category A agent, is unique in possessing two functional GR isozymes (RacE1 and RacE2)5. These particular enzymes have been the subject of extensive structural and kinetic characterization5,11. Active site differences between the two isozymes, specifically a key valine residue that bridges the active site and an adjacent hydrophobic pocket at the entrance of RacE2 (but not RacE1), have hampered efforts to develop a common and potent inhibitor for both B. anthracis GR isozymes5.
Accurately modeling enzyme (or receptor) flexibility has long been recognized as one of the grand challenges in structure-based drug discovery. A diverse set of crystallographic, computational and screening studies have pointed to the extensive flexibility of the GR enzyme6–8,11,13, thus creating challenges in elucidating both its catalytic mechanism, as well as the functional pharmacophore for effective structure-based drug discovery campaigns. An important corollary to receptor flexibility is the possibility for allosteric regulation by an effector molecule, with previous successes seen across several classes of enzymes14. According to the Monod-Wyman-Changeus (MWC) model15, a flexible (unliganded) enzyme samples a range of conformational states, which may be captured or recognized by a ligand or set of ligands. If significant distortion of a target enzyme is populated in these pre-binding equilibria, then capture of an inactive conformation represents a reasonable strategy for structure-based drug design. Accordingly, an alternative approach to finding inhibitors for GRs would be to identify noncompetitive (or uncompetitive) inhibitors, which act remotely from the active site. This strategy was utilized by Lundqvist and coworkers in the discovery of a pyrazolopyrimidinedione analogue (“Compound A”) inhibitor against H. pylori GR4. Compound A was identified in HTS against AstraZeneca’s compound library as an inhibitor of racemization in the D→L direction. Lundqvist et al. elegantly demonstrated that Compound A was an extremely rare uncompetitive inhibitor of a monosubstrate-monoproduct enzyme in the D→L direction4. Interestingly, this inhibitory approach exploits the fact that reduction of the kcat/KM of a racemase in one direction must equal its reduction in the reverse direction (vide infra), which results in a net decrease in the production of D-glu in vivo as indicated by the accumulation of peptidoglycan precursors in the cytoplasmic extract of H. pylori cells treated with Compound A4. This inability to produce peptidoglycan is lethal for H. pylori cells, as demonstrated by a minimal inhibitor concentration (MIC) of 8 µg/mL for Compound A4. However, the natural effector molecule acting at the Compound A binding pocket remains unknown.
In the current study we have discovered a novel inhibitory property for a natural product, dipicolinic acid (DPA), which is acting as an allosteric inhibitor of both B. anthracis GR isozymes, and has a distinct binding pocket from Compound A of Lundqvist et al.4. Furthermore, we employ experimental and computational studies to elucidate this remote binding locus as well as propose an atomistic rationale for DPA inhibition supported by MD simulations, free energy and pKa calculations of the dynamic GR ensemble. Interestingly, DPA is a natural product occurring at high concentrations within the B. anthracis spore and has been implicated to play a key role in that organism's life cycle. The implications for B. anthracis sporulation and drug discovery are discussed.
2. Computational and Experimental Details
2.1. Site-Directed Mutagenesis
Mutants racE2_K106A and racE2_S207A were prepared using a QuikChange II XL Site-Directed Mutagenesis Kit (Stratagene, Santa Clara, CA) and primers obtained from Eurofins MWG Operon (Huntsville, AL). Previously prepared and recently isolated pET15b (Novagen, San Diego, CA) containing the gene of interest was used as the template DNA. A BioRad MJ Mini Personal Thermal Cycler (BioRad, Hercules, CA) was used for all PCR reactions. Mutagenesis was confirmed via in-house DNA sequencing using an ABI 3730XL capillary sequencer. Primer sequences are detailed in Supplementary Table 4.
2.2. Protein Expression and Purification
Proteins were expressed using the BL21 strain (Novagen, San Diego, CA) of E. coli and pET15b expression vector (Novagen, San Diego, CA). Purification was composed of three key components: Cobalt-affinity chromatography, ATP incubation to remove chaperone contaminants, and anion-exchange chromatography. Expression and purification are described in full detail in Supplementary Information.
2.3. Enzyme Kinetics – Circular Dichroism
Stereoisomerization of D-glutamate by GR was assayed by measuring angle of rotation (mdeg) at 220–225nm using a J-720 CD spectropolarimeter (JASCO, Oklahoma City, OK). Data was fit to Michaelis-Menten equations as well as nonlinear regression curves for noncompetitive inhibition using GraphPad Prism v5.0 (GraphPad Software Inc., La Jolla, CA). Complete details of kinetic assay conditions can be found in Supplementary Information.
2.4. Colloidal Aggregation Control
Inhibitors were analyzed for the possibility of colloidal aggregation using a detergent-based assay previously established by Feng and Shoichet and successfully applied to this particular system in Whalen et al.7,16. Further details can be found in Supplementary Information.
2.5. Blue Native – Polyacrylamide Gel Electrophoresis
Protein samples were incubated in loading buffer (100 mM Tris-Cl, 40% glycerol, 0.01% Coomassie Brilliant Blue G, pH 8.0) and the compound of interest (1 mM or 45 mM) at room temperature for 15 minutes. Blue-native (BN) polyacrylamide gels were prepared at a resolving concentration of 10% (30:0.8 total acrylamide:bis ratio) with a 5% stacking gel. The compound of interest was incorporated into both the gel and running buffer (100 mM Tris-Histidine, pH 8.0) at the desired concentration. Gels were run at constant voltage (100 V) and constant amperage (15 mA) for approximately 3.5 h at 4 °C. Gels were stained using Imperial Protein Stain and destained in water. Respective intensities of protein bands were analyzed by pixel quantification of a gel scan using Adobe Photoshop CS4 Extended v11.0 (Adobe Systems Inc., San Jose, CA).
2.6. Docking of DPA to GR
Protein Receptor Preparation: The PDB file 2GZM was downloaded from RCSB for use as the protein receptor. One dimer was deleted from the crystal cell as only one dimer would be utilized in docking. The remaining dimer was submitted to steepest descent minimization and 50 picoseconds of molecular dynamics simulation with YASARA v9.11.9 (further details below)17. All water molecules and ligands were removed from the finals structure as the last step of preparation. DPA in its deprotonated state was used as the ligand for docking.
AutoDock (through YASARA): A simulation cell was created 5 Å from all atoms with dimensions: 102 Å × 72.8 Å × 71.3 Å. The following general docking parameters were used: 25 independent docking runs, each with a total of 2.5 × 106 energy evaluations, a torsional degrees of freedom value of 8, grid point spacing was left at the default of 0.375 Å, and the force field selected was AMBER03. Specific to the genetic algorithm, the following parameters were used: a population size of 150, 2.7 × 104 generations, an elitism value of 1, a mutation rate of 0.02, and a crossover rate of 0.8. Final poses were considered distinct if they varied by > 5 Å RMSD.
GOLD: The genetic algorithm parameters were defined as such: number of islands set to 5, population size of 100, selection pressure value of 1.1, maximum number of operations set to 125,000, and a niche size of 2. Additionally, both crossover and mutation frequency were set to 95, while the migration frequency was set to 10. GoldScore was used as the sole scoring function (without the optional fifth component, internal hydrogen bond energy). DPA was docked a total of 30 times and all solutions were retained.
FRED: A box ~200,000 Å2 was drawn around the entire structure, and a high quality shape potential was created. In enumerating all possible ligand poses, a translational step size of 1 Å and a rotational step size of 1.5 Å were used. Inner and outer contour filtering was enabled and no additional user-defined constraint filters were used. Ensemble poses resulting from exhaustive docking are scored using Chemgauss3 and the top 100 poses are retained. DPA was docked a total of 32 times.
2.7. Molecular Dynamics Simulations
The procedures and parameters for molecular dynamics simulations of GR as well as free energy calculations are described in full in the Supplementary Information.
2.8. Selection of Non-Redundant Structures from MD Simulation
Structure snapshots taken every 150 picoseconds of the molecular dynamics simulations were converted to PDB format with all water molecules removed. The collection of structures (133 structures per complex) were then imported to the Multiseq extension of Visual Molecular Dynamics v1.8.6 for Windows (VMD, Univ. of Illinois – Urbana-Champaign) and submitted to STAMP alignment (npass =1, scanscore =1, and scanslide =2). Multiseq was further used to create a phylogenetic tree based on RMSD of the Cα atoms. Finally, non-redundant structures were extracted from the phylogenetic tree using a QH cutoff value of 0.90.
2.9. H++ pKa Calculations
Structures were submitted in pdb format to the H++ server. All water, ligand and inhibitor molecules were deleted prior to pKa calculation. Structure “clean-up” is described in full by Gordon et al.18 and the method of electrostatic calculation of pKa values for all titratable groups is described in Bashford and Gerwert19. pKa values presented in the text were taken from the pK1/2 output of H++.
3. Results
3.1. Inhibition of RacE1 and RacE2 by DPA
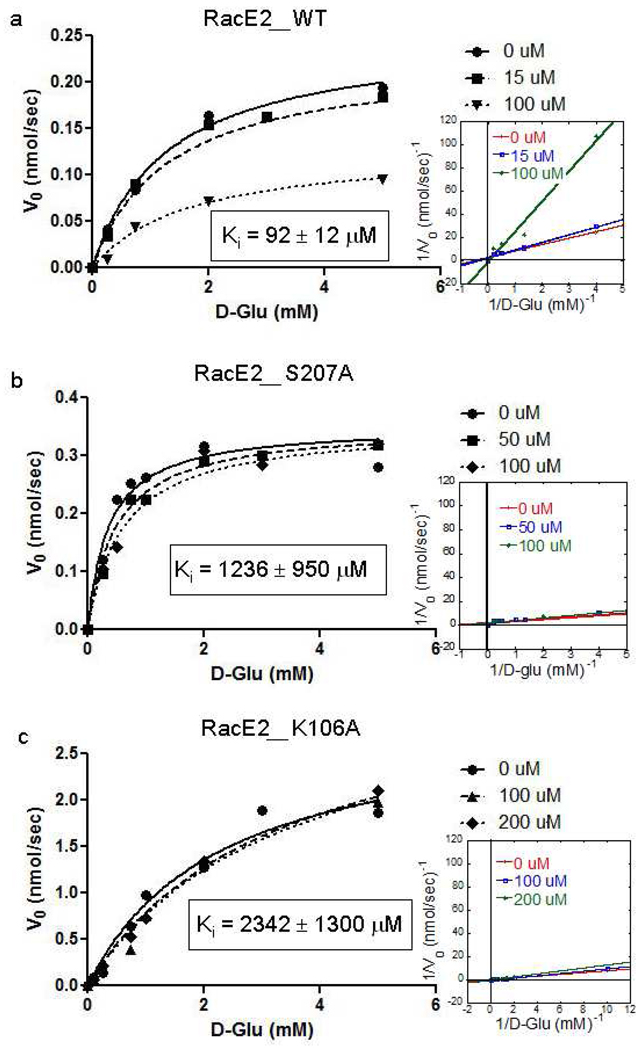
DPA was originally found in a virtual screening campaign for B. subtilis RacE but exhibited only low mM inhibition6. When tested on the two glutamate racemase isozymes from B. anthracis, RacE1 and RacE2, DPA exhibited low µM inhibition (Ki = 75 ± 16 µM and 92 ± 12 µM, respectively) with clear noncompetitive behavior, as confirmed via the F test when data is fit to varying models of inhibition (Fig. 1a and S1, Table S1 and S2). To our knowledge, this is the first noncompetitive inhibitor against a glutamate racemase. In general, it is quite rare to find noncompetitive or uncompetitive inhibitors for any monosubstrate-monoproduct (i.e. Uni-Uni) enzymes20,21. The immediate tendency is to be wary of noncompetitive inhibitors discovered from screening campaigns, as they are often revealed to be colloidal aggregators that inhibit enzymes in a non-drug-like manner22. Colloidal aggregators can be exposed using a simple detergent-based kinetic assay16. Inhibition by DPA does not exhibit the characteristic alleviation in the presence of detergent, thus eliminating the possibility of colloidal-aggregation-based inhibition (Fig. S2). Previously, our group successfully employed this detergent-based assay to identify a colloidal aggregator in a screening campaign against GR from B. subtilis that exhibited apparent noncompetitive inhibition in the low-µM range7. RacE2 was the main isozyme studied in the following work since B. anthracis genetic knockout studies identified the absence of RacE2 as resulting in the more severe growth defect, relative to the RacE1 isozyme10. Furthermore, the inhibitor studies described here focus on the D→L direction, as in the studies of Lundqvist et al., on GR inhibition by Compound A4. However, it should be noted that the Haldane relationship dictates that any reduction of kcat/KM for a racemase in one direction will result in an equivalent reduction in kcat/KM in the opposite direction20,23,24. This is because the equilibrium constant for a racemase is unity, and leads to the following constraint:
| Eq. 1 |
Figure 1.
Experimental results for noncompetitive inhibition of RacE2_WT (a), RacE2_S207A (b) and RacE2_K106A (c) by DPA. Kinetic data was attained via circular dichroism and three independent Michaelis-Menten curves were globally fit to a noncompetitive inhibition model to produce the presented Ki values (further details in Supporting Information).
3.2. Identification of a Novel Allosteric Pocket by Docking; Location of the DPA Binding Site
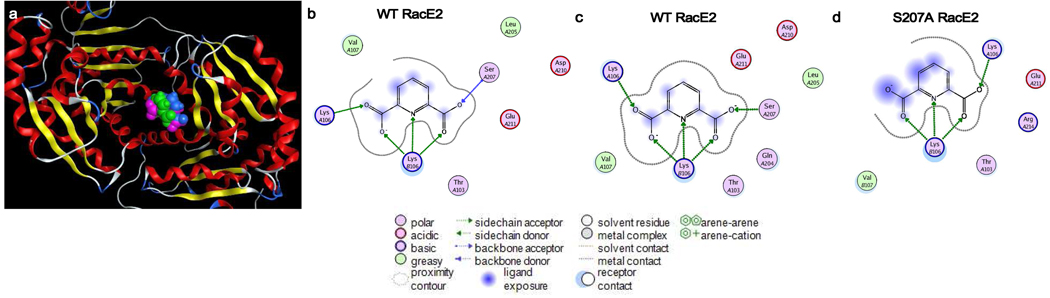
A blind docking campaign targeting the RacE2 dimer was carried out in order to identify the binding site of DPA. GOLD v4.1, FRED v2.2.5 and AutoDock 4 (via Yasara v9.11.9) were each utilized in this campaign and represent three different types of docking and scoring methods17,25,26. Despite using very different methods of docking and scoring (these differences are expounded upon in the Computational Procedures section of the Supplementary Information), all three programs came to the same conclusion with regards to the location of the DPA binding site (Fig. 2a). Given a RacE2 dimer from the original crystal structure (2GZM) and no user-defined specifications for the binding location, DPA was consistently positioned with highest rank in all three docking programs in a small pocket at the dimer interface, making direct contacts with Lys106 and Ser207 of one monomer and the Lys106 of the second monomer. Specifically, the Ser207 backbone amide acts as a H-bond donor to one carboxyl substituent of DPA while the Lys106 side chain of the same monomer is a donor for the second carboxyl substituent of DPA (Fig. 2b). The Lys106 side chain of the second monomer coordinates both carboxylates as well as the pyridine nitrogen of DPA. Interestingly, energy minimization results in the formation of an H-bond between the carboxylate of DPA and the beta-hydroxyl of Ser207 while the Lys contacts remain unchanged relative to the original docking pose (Fig. 2c). An MD simulation using S207A RacE2 with DPA bound was conducted to further elucidate the role of Ser207 in DPA binding. MD simulation shows that the mutagenesis of Ser207 to alanine results in a complete loss of contact with the backbone in that region, resulting in a highly solvent-exposed, and presumably unfavorable, binding position for DPA (Fig. 2d). These results point to the presence of Ser207 as being essential for organization in this region of the enzyme and accordingly, formation of the DPA binding pocket. With all three docking programs in agreement, the predicted site was probed via site-directed mutagenesis (vide infra). Importantly, these models predicting that DPA binds at a site distal to the active site are consistent with the steady state kinetic data suggesting non-competitive inhibition.
Figure 2.
Superpose of top-docked positions of DPA (space-filling) to the RacE2 dimer (ribbon, 2GZM) as determined by GOLD v4.1 (magenta), Autodock v4 (blue) and FRED v2.2.5 (green) (a). The binding pocket is located at the dimer interface and is composed of residues from both monomers, as detailed by the interaction map (b). After minimization, the backbone contact of Ser207 is swapped for a contact with the beta-hydroxyl group (c). After MD simulation of the top docked complex with Ser207 replaced by Ala, the binding site lacks any contact with the region previously containing Ser207 (d). Letters immediately preceding the residue numbers indicate the monomer, A or B. Ligand interaction maps were constructed using the LigX function of MOE v2009.10.
3.3. Validation of DPA Binding Site via Mutagenesis
In order to experimentally confirm the predicted DPA binding mode, the two residues predicted to interact directly with the inhibitor, Lys106 and Ser207, were mutated to alanine independently and the purified mutant enzyme was assayed for inhibition via DPA. Enzyme activity was unaltered for both mutants relative to wild-type protein, as indicated by their kcat/KM values (Table S3). First, RacE2_S207A was constructed and predicted to abolish a single H-bond between enzyme and ligand. The observed Ki (Ki = 1236 ± 950 µM) increased 13× compared to wildtype RacE2, which is consistent with the loss of a single H-bond (loss of 0.68–1.88 kcal/mol, strength of a normal hydrogen bond within a protein ranges from 1 to 3 kcal/mol27, Fig. 1b). Second, RacE2_K106A was constructed, abolishing four hydrogen bonds (according to the docking model). The observed Ki (Ki = 2342 ± 1300 µM) increased 24× compared to wildtype (loss of 1.43–2.17 kcal/mol, slightly less than predicted; Fig. 1c). The attenuation of DPA inhibition in both the mutant constructs agrees well with the predicted site being the true DPA binding site. This is a testament to the precision of GOLD, FRED and AutoDock at correctly positioning a small molecule in a blind docking situation.
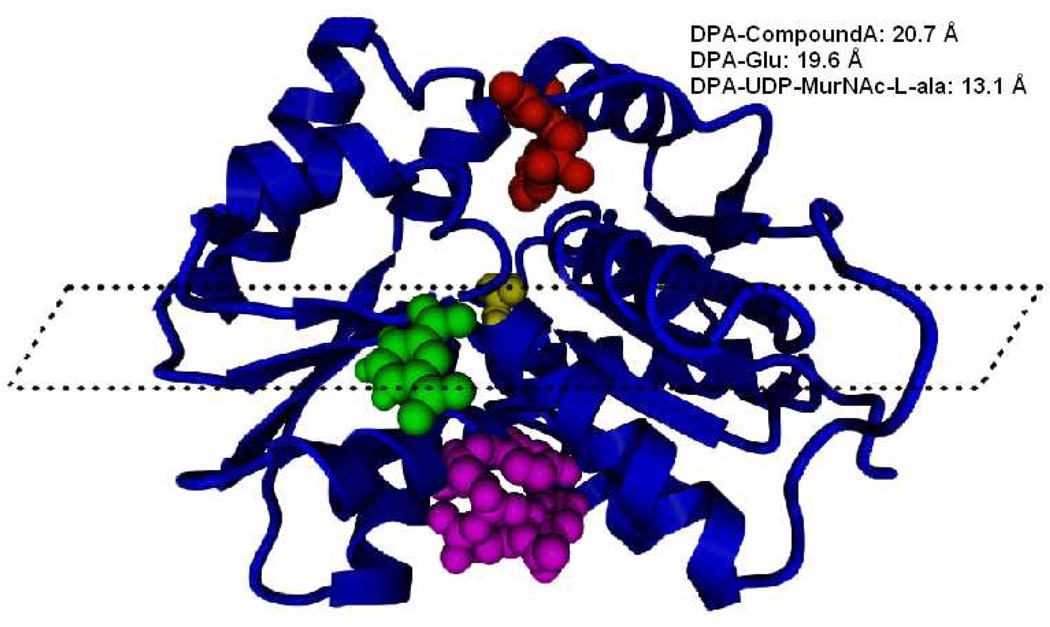
Cryptic binding sites are not entirely unprecedented for glutamate racemases. In 1994, Doublet and coworkers characterized the activation of GR from E. coli by UDP-MurNAc-L-Ala binding to a cryptic back pocket on the monomer (later co-crystallized by Lundqvist et al.)4,28. As previously mentioned, a team at AstraZeneca discovered Compound A, a novel uncompetitive inhibitor in a high-throughput screening against GR from H. pylori and determined this compound to bind to a pocket located on the opposite face from the active site4. Thus far, these two examples have been the only homologues of GR with established allosteric regulation. Crystal structures of the activator bound to E. coli GR (2JFN) and Compound A bound to H. pylori GR (2JFZ) were superposed with the top-docked conformation of DPA into RacE2 in order to see whether DPA was exploiting either of these pre-established binding sites (Fig. 3). When the dimer interface is revealed by hiding one monomer, one can clearly see that the DPA binding site is distinct from those of the activator and Compound A. With this perspective, one can envision D-glutamate bound in the active site and DPA bound to the back side as being on a shared horizontal plane. The uncompetitive inhibitor from Lundqvist et al. is bound above this plane and UDP-MurNAc-L-Ala is bound below4. Thus, the DPA binding site is unlike any established cryptic sites of GR. With the addition of DPA, three distinct cryptic sites exist within the GR receptor class, each exhibiting a different mode of regulation.
Figure 3.
Superpose of a variety of GR structures in order to highlight the diversity of known allosteric positions. Superpose of DPA bound to B. a. RacE2, UDP-MurNAc-L-Ala bound to E. c. GR (2JFN), and pyrazolopyrimidinedione analogue (aka Compound A) bound to H. p. GR (2JFZ). Only the trace of one monomer is shown for clarity and the perspective is looking at the enzyme face directly opposite of the entrance to the active site. D-glutamate (yellow) is seen in the background bound to the active site, while DPA (green), UDP-MurNAc-L-Ala (magenta), and Compound A (red) are seen in the foreground. All three cryptic binding sites are distinct. Indicated are center of mass distances between molecules. There is no evidence that any one GR structure possesses all of these allosteric pockets. Rather, the figure is meant to illustrate the distinctive positions and identities of these three different effectors relative to the glutamate binding pocket.
3.4. Quaternary Organization and Inhibition by DPA
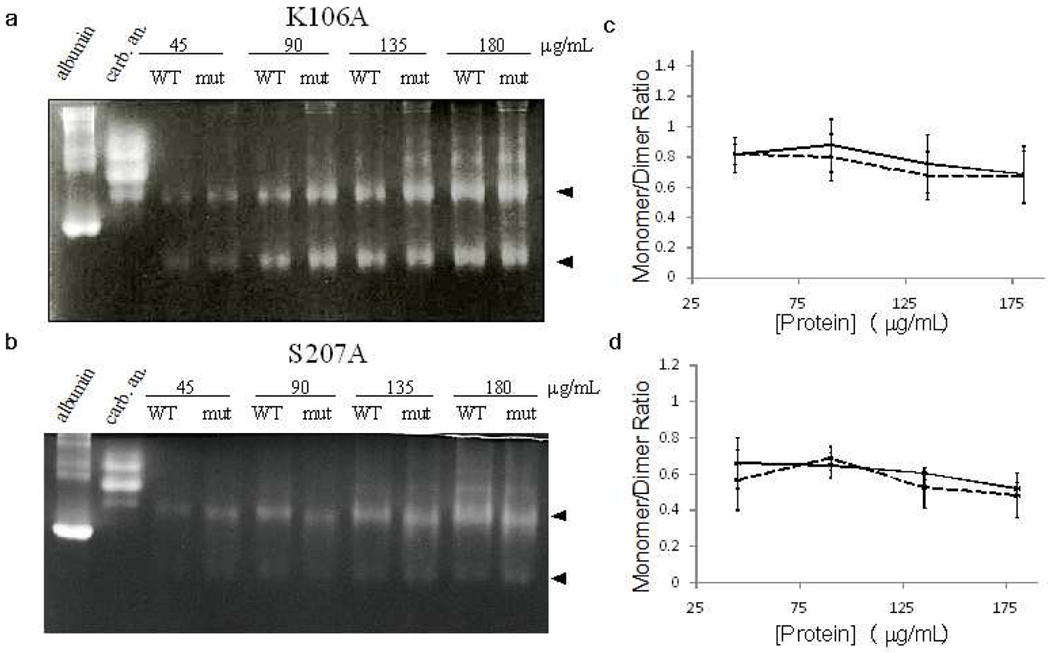
One possible mechanism of GR inhibition is a DPA-induced shift in the oligomeric equilibrium. For instance, Johnson and coworkers have hypothesized that the RacE2 monomer experiences increased conformational flexibility and thus higher rates of catalysis29. In order to investigate any DPA-induced changes of the oligomeric equilibrium of GR, RacE2 dimerization was analyzed using Blue Native PAGE (BN-PAGE) gels supplemented with DPA. Gels supplemented with the DPA-analogue, 3,4-pyridinedicarboxylic acid, were used as a control. This analogue shows no inhibition against RacE2 nor RacE1, and thus is predicted not to bind the enzyme; but its presence provides a control for nonspecific interactions that may affect the BN-PAGE results. Both compounds were present in the gel, running buffer and loading buffer (respectively) at a concentration well above the Ki calculated for DPA. DPA does not induce any significant change in the monomer-dimer equilibrium of RacE2 relative to the control (Fig. S4). Although BN-PAGE rules out the occurrence of a DPA-induced shift in the oligomeric equilibrium, the possibility remains that allostery is the source of inhibition by DPA.
As recently reported by Johnson and coworkers, certain mutations in residues located at the dimer interface result in an altered RacE2 monomer-dimer equilibrium29. Thus, both mutant constructs were analyzed via Blue Native PAGE to assess the monomer-dimer equilibrium and rule out this phenomenon as having any affect on DPA binding or overall RacE2 catalysis. Both RacE2_S207A (Fig. 4a) and RacE2_K106A (Fig. 4b) showed no significant difference in monomer-dimer equilibrium compared to wild type protein. For RacE2_K106A, these results are contrary to what was reported by Johnson and coworkers, as they observed only the monomer state for this mutant. Differences could be attributed to the method (Blue Native PAGE versus size-exclusion chromatography) or protein concentration (45 µg/mL versus 5 mg/mL); particularly since we observed that the monomer-dimer equilibrium exhibited a dependence on protein concentration in all gels. Specifically, as protein concentration increased, the ratio of monomer to dimer decreased (Fig. 4 and S4). We chose to examine a protein concentration in the range of those used for kinetic assays. Furthermore, these GR concentrations may be more physiologically relevant than the much higher concentrations used in the study by Johnson and coworkers29.
Figure 4.
BN-PAGE to determine the oligomerization of wild-type and mutant RacE2. Wildtype RacE2 and RacE2_S207A (a) or RacE2_K106A (b) were run side-by-side at concentrations varying from 45 to 180 µg/mL. Albumin and carbonic anhydrase were included as running controls. Arrowheads indicate bands representing the dimer and monomer. Band intensity was quantified via pixel counting and the ratio of monomer to dimer was plotted against protein concentration for RacE2_S207A (solid line = WT, dotted line = mutant; c) and RacE2_K106A (d). Data represents an average of three or more independent trials with standard error shown. Data was additionally fitted to the expression for M/C ratio as a function of total protein concentration and the monomer:dimer equilibrium constant (see Supplementary Methods for derivation of this expression and model fitting parameters).The results indicate that the two mutants do not have any significant effect on the oligomeric equilibrium. Additionally, see Figure S3 for BN-PAGE of RacE2 and running controls with NativeMark ladder.
3.5. Effect of DPA on Free Energy of Binding of D-Glu to RacE2
Molecular dynamics simulations were performed for both RacE2 dimer with D-glu bound to both active sites (abbreviated as E2·D-glu2), as well as the top-docked complex of RacE2 and DPA with glutamate bound to both active sites (abbreviated as E2·D-glu2·DPA). Both simulations were carried out for 20 nanoseconds (see Materials and Methods for a complete description of the simulation parameters). Separately, a collection of snapshots taken every 150 picoseconds (totally 133 structures) from each simulation were submitted to STAMP structural alignment30 and used to compose a structural phylogenetic tree based on RMSD differences between α-carbon atoms. A structural phylogenetic tree provides a graphical representation of the structure-based relationship between different simulation snapshots. Non-redundant structures were then selected from the collection to represent the most unique conformations using the widely-accepted technique of QR factorization, thus more efficiently examining phase space31,32. A structural homology (QH) value of 0.90 was used as the cutoff for structural redundancy (where QH represents the fraction of Cα atoms that superimpose, total overlap = 1 and no overlap = 0)33, resulting in 14 unique structures from E2·D-glu2 and 2 unique structures from E2·D-glu2·DPA. The QR factorization results immediately indicate that the presence of bound-DPA results in limited conformational diversity for RacE2 within the respective equilibrated time series. An estimate of the binding free energy was calculated for glutamate bound to either monomer using an “endpoint” approach (recently reviewed by Steinbrecher and Labahn)34. The method employed here, MM-BEMSA, is a variation on the popular MM-PBSA method, with electrostatic potentials calculated with the Boundary Element Method (BEM), instead of numerically solving the Poison-Boltzmann (PB) equation. The details of this approach are described in the Computational Methods section of the Supplementary Information. Briefly, the BEM technique for calculating electrostatic potentials has some advantages in dealing with highly curved surfaces of macromolecules, such as clefts and buried pockets, which made it ideal for the GR receptor35,36.
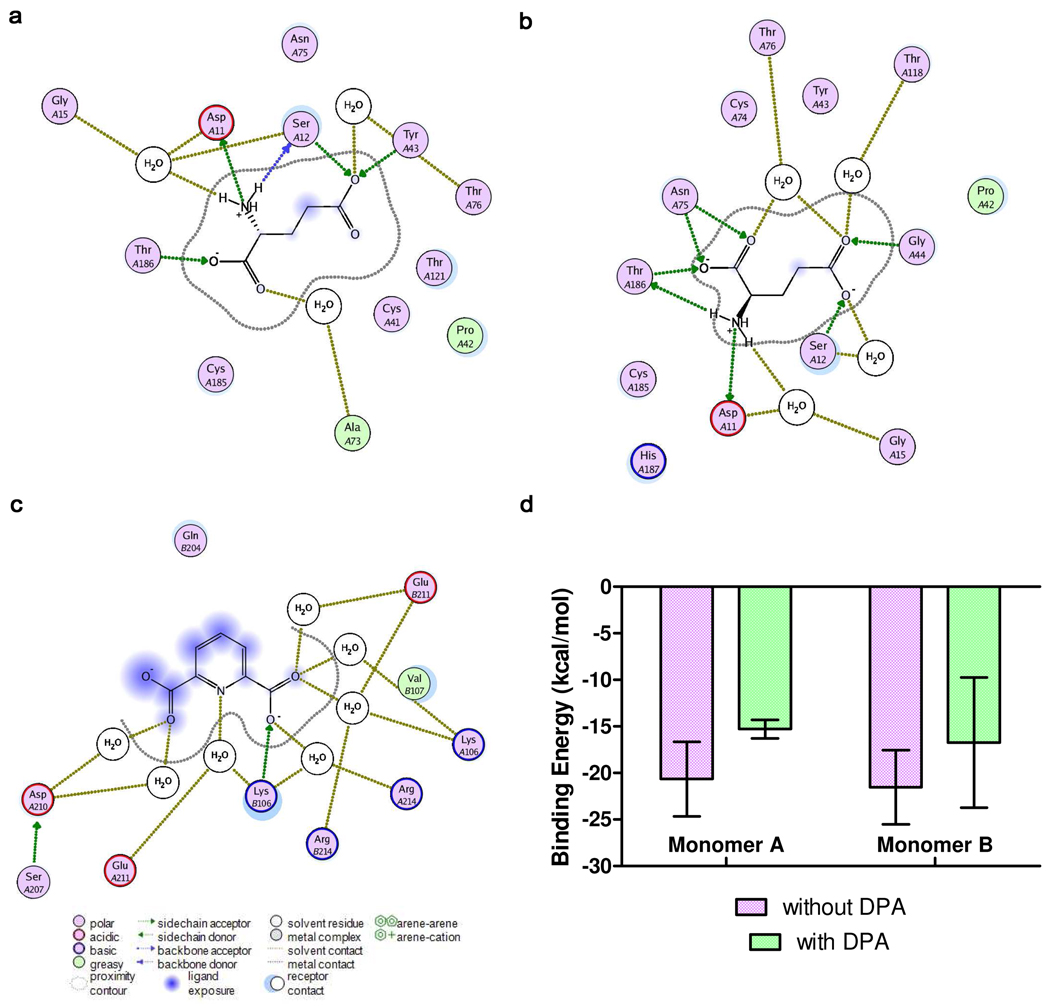
Instead of doing an endpoint free energy calculation of D-glu binding at every time point of an MD simulation, we sought a more efficient use of computational resources. The predicted binding energy was averaged for all non-redundant structures from each respective trajectory. In the past, McCammon and coworkers applied this technique to an MD simulation of Kinetoplastid RNA editing ligase 1 (KREL1), where non-redundant representatives were shown to possess as much binding energy information as the entire set of structures resulting from the simulation31. Thus, the non-redundant set of MD structures, obtained from QR factorization was used in the end point free energy calculations of D-glu to the RacE2 dimer, with and without DPA bound (i.e. E2·D-glu2 and E2·D-glu2·DPA). For glutamate bound to monomer B, no significant difference in free energy of binding exists between E2·D-glu2·DPA and E2·D-glu2 (−17 ± 5.0 kcal/mol versus −22 ± 2.0 kcal/mol, respectively). On the contrary, for glutamate bound to the A monomer, a less negative free energy of binding (i.e. weaker complexation) is seen in E2·D-glu2·DPA compared to E2·D-glu2 (−15 ± 1.0 kcal/mol versus −21 ± 2.0 kcal/mol, respectively, Fig. 5d). End point free energy calculations (such as MM-PBSA) do not yield accurate absolute free energies37, but have been shown to be highly accurate in terms of relative free energy values38, and may be used as powerful predictor of trends in relative binding affinities.
Figure 5.
Ligand interaction maps for glutamate bound to monomer A of the DPA-lacking RacE2 complex (a) and DPA-bound RacE2 complex (b), as well as DPA bound to the cryptic binding site located at the RacE2 dimer interface (c). Maps were generated from the final structures of 20-nanosecond MD simulations using the LigX function of MOE v2009.10. Predicted binding energy of glutamate was averaged over the set of representative structures extracted from MD simulations of the binary (red, n=15) and ternary complex (blue, n=4, d), error bars = SEM. The details of the binding energy calculations are outlined in the Computational Procedures section of Material and Methods.
Ligand interaction mapping of equilibrated E2·D-glu2·DPA after the 20 nanosecond simulation show two water bridges formed between DPA and Asp210 and Glu211 of the A monomer of RacE2 (Fig. 5c). Asp 210 goes on to form a hydrogen bond with the side-chain hydroxyl of Ser207 (previously implicated in DPA binding by initial ligand interaction mapping of the top-docked complex prior to MD). Ser207 also forms a water bridge with Glu211. This complex network of direct hydrogen bonds and water bridges between DPA and the A monomer occurs twenty residues downstream of the catalytic residues, separated by a short α-helix and β-sheet. The interaction maps of glutamate bound to the active site of the A monomer of equilibrated complexes were compared. Binding of DPA corresponds with altered hydrogen-bonding within the active site as compared to E2·D-glu2. Briefly, in E2·D-glu2·DPA, glutamate forms seven hydrogen bonds with active site residues as opposed to five hydrogen bonds in E2·D-glu2 (Fig. 5a–b). Also, glutamate of E2·D-glu2·DPA is involved in five water bridge contacts while glutamate of E2·D-glu2 involves only three. These variations in enzyme-ligand interactions point to a global conformational change translated from the dimer interface to the active site. Specific details regarding hydrogen bonding and enzyme-ligand distances for the two complexes are described in the Supplementary Information.
3.6. Effect of DPA Binding on pKa of Catalytic Cysteine 74
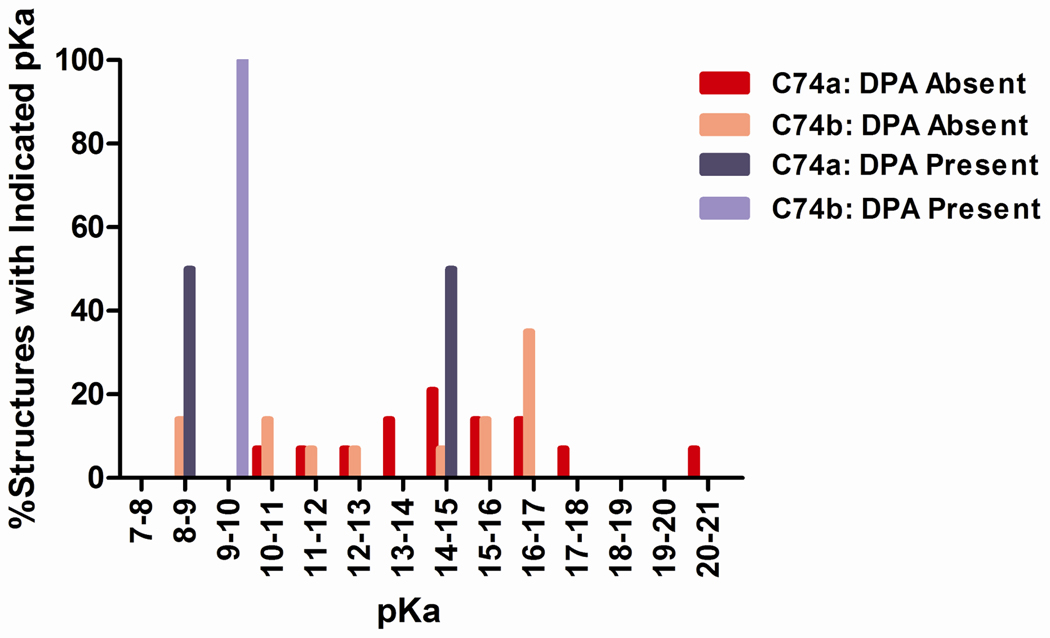
While differences in free energy of binding of D-glu may contribute partially to inhibition by DPA, they cannot explain if and why the enzyme-substrate-inhibitor (ESI) complex is enzymatically unproductive, particularly since the binding pose of D-glu does not vary widely between E2·D-glu2 and E2·D-glu2·DPA. Considering the importance of the basicity of the general acid/base for glutamate proton abstraction9,39 and the clear active site rearrangements that accompany DPA binding8, we chose to examine changes in pKa of the key catalytic residue, Cys74, for D→L racemization. Each structure selected from QR factorization of the MD simulations of both ensembles was used to calculate pKa values with the widely employed MEAD algorithm (implemented in the H++ utility18,40). The distribution of pKa values shows a significant downward shift for Cys74 of E2·D-glu2·DPA compared to Cys74 of E2·D-glu2 (Fig. 6). Overall, the E2·D-glu2·DPA structures possessed pKa values in a much more limited range and with reduced values (less basic) compared to E2·D-glu2 structures, which possess pKa values ranging from as low as 8 to as high as 20 (Fig. 6). The possibility that the loss of extreme basicity of Cys74 is responsible for a dysfunctional ESI complex is discussed below.
Figure 6.
Distribution of pKa values calculated for Cys74 (the catalytic base of D → L racemization) for E2·D-glu2 and E2·D-glu2·DPA. Each structure, selected via QR factorization of a collection of simulation snapshots, was each used in pKa calculation using the MEAD algorithm from the H++ program18. Details of parameters employed in these pKa calculations are located in the Computational Procedures section of the Materials and Methods.
4. Discussion
Here we have identified a natural compound, DPA (linked to the B. anthracis life cycle), which exhibits noncompetitive inhibition of GR from B. anthracis via binding to a remote and heretofore undiscovered allosteric pocket. A blind docking study, using three leading algorithms has determined the consensus pharmacophore for DPA, which lies at the dimer interface. This consensus pocket was experimentally validated via a multiple site-directed mutagenesis study. The loss of racemization activity due to DPA has no affect on the oligomeric equilibrium of GR, suggesting that inhibitor action is allosteric in nature.
A parallel computational study to understand the source of the DPA-induced allostery was undertaken. DPA-induced changes in the phase space of B. anthracis RacE2 showed a significant dampening of the conformational flexibility of GR. Interestingly, the large conformational fluctuations of E2·D-glu2 manifest large changes in the magnitude of the calculated pKa of the catalytic base, Cys74. The large pKa perturbations in the E2·D-glu2 are due to a variety of interactions between active site residues, particularly the interaction between the catalytic base and Asp11. Unsurprisingly, Asp11 was previously identified as being located in an area of high homology and subsequently probed via mutagenesis, resulting in a 1000-fold reduction in the observed kcat of GR39. Remarkably, the very high basicity of some enzyme catalytic bases has been shown to be critical to catalytic efficiency even when the protonation state of this species (the so called "reverse protonation" state41) is not the dominant form at a given pH. The concept of the role of the reverse protonation state has been fully developed in the case of enolase and alanine racemase42,43. Thus, what is seen here in the case of GR is a manifestation of the phenomenon exhibited in enolase and alanine racemase, in which the reverse protonation state may be the primary driver of catalytic power (i.e. a very high pKa thiolate). However, in GR there is the additional complexity of a quite flexible ensemble of protein conformations, with large fluctuations in the values of the pKa of the catalytic base. It is fascinating that the ensemble with the allosteric inhibitor (DPA) results in a conformational "freezing out" of the high pKa (i.e. catalytically reactive) forms of GR. We hypothesize that this is the source of allosteric inhibition by DPA, and refer to this mode of inhibition as Inhibition by pKa Trapping (IPKAT). We propose that this is a general phenomenon in GR that can be exploited in the DPA pocket by other small molecules, and that in principle it is possible to construct an IPKAT pharmacophore, in which GR inhibition can be predicted by the distribution of calculated pKa values of the catalytic Cys residues.
The presence of DPA in Bacillus spores has been well-established as DPA constitutes approximately 10% of the dry weight of dormant spores, but its exact role in sporulation and germination is only loosely understood44. DPA within spores is found primarily in its calcium-chelated form, Ca2+-DPA. In the developing endospore, DPA concentrations remain low until after cortex formation, which requires peptidoglycan synthesis. In the dormant spore phase of B. anthracis, Ca2+-DPA concentrations are very high and there is no detectable metabolic activity. Upon activation of spores by external factors, channel formation occurs allowing a large efflux of cations and Ca2+-DPA out of the spore core45. This event occurs concomitantly with an adjustment of the pH from 6.5 to 7.7 and increased hydration45. All of these events are required to restart metabolism. Accordingly, both the reduced DPA concentration and elevated pH are conducive with increased GR activity (pH optimum of RacE1 and RacE2 is ~8.011). As the spore core grows nearly 4–5× its original size, the demand for peptidoglycan synthesis increases and thusly the demand for D-glutamate. Additionally, D-glutamate is required for the poly-D-glutamic acid capsule surrounding the mature vegetative cell. Taking all of this into consideration, one may postulate that the fluctuation of DPA concentration over the lifetime of a differentiating B. anthracis cell coincides with the varying necessity of D-glutamate (Fig. 7). Thus, it is appealing to imagine the inhibition of GR by DPA as more than serendipitous. Of course, future in vivo studies are required to further investigate the interactions of GR and DPA in spores and vegetative cells. One point to investigate is whether the efflux of DPA actually results in a cellular DPA concentration less than the Kd, approximately 90 µM, capable of entirely alleviating GR inhibition. Compartmental distributions of DPA may allow such a concentration to be reached even if the net cellular concentration of DPA is still high.
Figure 7.
Schematic of life cycle of differentiating B. anthracis cell, with Ca2+-DPA levels and consequent GR activity indicated.
Importantly, the ligand efficiency (free energy of binding divided by the number of non-hydrogen atoms in the ligand) of DPA is quite high (−0.458 kcal mol−1 per non-H atom) compared to other known allosteric ligands such as Compound A (−0.242 kcal mol−1 per non-H atom) and the activator of GR from E. coli, UPD-MurNAc-L-Ala (−0.150 kcal mol−1 per non-H atom). Despite binding with higher affinity, the large MW of Compound A from Lundqvist et al. significantly lowers its ligand efficiency4. Ligand efficiency is one of the principal factors of lead optimization46. The high ligand efficiency of DPA means that during ligand optimization, a binding affinity in the low nM can be achieved with far fewer heavy atom additions and thus a lower MW compared to compounds of lesser efficiency. Additionally, the possible chemical modifications of DPA are manifold, providing a large potential library of small molecules to screen.
Allosteric modulators represent an emerging drug class with which some pharmaceutical companies are having notable successes such as the HIV inhibitor maraviroc (Pfizer) and the hyperparathyroidism drug cinacalcet (Amgen)47,48. There are several advantages to using allosteric binding sites as drug targets over the native substrate binding site. Primarily, the allosteric sites of GR can accommodate a chemically diverse set of compounds compared to the active site, i.e. the final drug need not be a substrate analogue. A non-amino-acid-like inhibitor will increase the selectivity for GR, while lowering the chance of undesirable interactions with enzymes containing analogous binding site motifs, such as enzymes involved in amino acid synthesis or the glutamate receptors that play an important role in neural chemistry. Also, any mutations that could circumvent inhibition at this particular allosteric site would occur within the sensitive dimer interface and are thus more likely to affect dimerization and monomer cross-talk, potentially very key components of GR catalysis. The major obstacle to virtual screening campaigns targeting allostery is pharmacophore elucidation. Once this hurdle has been overcome (i.e. a defined structure activity relationship and/or an atomistic understanding of the binding modes that lead to enzymatic inhibition), then reasonable metrics may be applied to assess binding modes and predict novel allosteric inhibitors. Such is the case now for B. anthracis RacE2, particularly in light of the IPKAT hypothesis outlined above, which permits a physical metric for inhibitory allostery. It is now possible to begin rational allostery-based virtual screening campaigns against the GR target class, potentially leading to new inhibitory small molecule anti-anthrax therapeutics.
Supplementary Material
Acknowledgements
This work was supported by NIH AI076830 (M. A. S.) and NIH AI057156 (S. R. B.). Special thanks to Dr. Keith Westcott for his contribution to the Keith R. Westcott Graduate Student Education Fund (K. L. W.). Additionally, thank you to Dr. Dylan Dodd for helpful discussions.
Abbreviations
- DPA
dipicolinic acid
- GR
glutamate racemase
- SMD
Steered Molecule Dynamics
- BN-PAGE
Blue Native Polyacrylamide Gel Electrophoresis
- IPKAT
Inhibition by pKa Trapping
Footnotes
Supporting Information Available: Additional results (including aggregation controls, statistical analysis of global fitting, BN-PAGE controls, and RMSD plots from MD simulations) as well as extensive experimental methods and computational methods may be found in the Supporting Information. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Rogers HJ, Perkins HR, Ward JB. Microbial cell walls and membranes. London, United Kingdom: Chapman and Hall; 1980. [Google Scholar]
- 2.de Dios A, Prieto L, Martin JA, Rubio A, Ezquerra J, Tebbe M, Lopez de Uralde B, Martin J, Sanchez A, LeTourneau DL, McGee JE, Boylan C, Parr TR, Jr, Smith MC. J Med Chem. 2002;45:4559. doi: 10.1021/jm020901d. [DOI] [PubMed] [Google Scholar]
- 3.Glavas S, Tanner ME. Bioorganic and Medicinal Chemistry Letters. 1997;7:2265. [Google Scholar]
- 4.Lundqvist T, Fisher SL, Kern G, Folmer RH, Xue Y, Newton DT, Keating TA, Alm RA, de Jonge BL. Nature. 2007;447:817. doi: 10.1038/nature05689. [DOI] [PubMed] [Google Scholar]
- 5.May M, Mehboob S, Mulhearn DC, Wang Z, Yu H, Thatcher GRJ, Santarsiero BD, Johnson ME, Mesecar AD. Journal of Molecular Biology. 2007;371:1219. doi: 10.1016/j.jmb.2007.05.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spies MA, Reese JG, Dodd D, Pankow KL, Blanke SR, Baudry J. J Am Chem Soc. 2009;131:5274. doi: 10.1021/ja809660g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whalen KL, Pankow KL, Blanke SR, Spies MA. ACS Medicinal Chemistry Letters. 2010;8:9. doi: 10.1021/ml900005b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruzheinikov SN, Taal MA, Sedelnikova SE, Baker PJ, Rice DW. Structure. 2005;13:1707. doi: 10.1016/j.str.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 9.Puig E, Garcia-Viloca M, Gonzalez-Lafont A, Lluch JM, Field MJ. Journal of Physical Chemistry B. 2007;111:2385. doi: 10.1021/jp066350a. [DOI] [PubMed] [Google Scholar]
- 10.Shatalin KY, Neyfakh AA. FEMS Microbiology Letters. 2005;245:315. doi: 10.1016/j.femsle.2005.03.029. [DOI] [PubMed] [Google Scholar]
- 11.Dodd D, Reese JG, Louer CR, Ballard JD, Spies MA, Blanke SR. J Bacteriol. 2007;189:5265. doi: 10.1128/JB.00352-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.LoVullo ED, Molins-Schneekloth CR, Schweizer HP, Pavelka MS., Jr Microbiology. 2009;155:1152. doi: 10.1099/mic.0.022491-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gallo KA, Tanner ME, Knowles JR. Biochemistry. 1993;32:3991. doi: 10.1021/bi00066a020. [DOI] [PubMed] [Google Scholar]
- 14.Hardy JA, Lam J, Nguyen JT, O'Brien T, Wells JA. Proc Natl Acad Sci U S A. 2004;101:12461. doi: 10.1073/pnas.0404781101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Monod J, Wyman J, Changeux JP. J Mol Biol. 1965;12:88. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 16.Feng BY, Shoichet BK. National Protocols. 2006;1:550. doi: 10.1038/nprot.2006.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.YASARA 9.11.9 ed. Vienna, Austria: YASARA Biosciences GmbH; 2010. [Google Scholar]
- 18.Gordon JC, Myers JB, Folta T, Shoja V, Heath LS, Onufriev A. Nucleic Acids Res. 2005;33:W368. doi: 10.1093/nar/gki464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bashford D, Gerwert K. Journal of Molecular Biology. 1991;224:473. doi: 10.1016/0022-2836(92)91009-e. [DOI] [PubMed] [Google Scholar]
- 20.Cook PF, Cleland WW. Enzyme kinetics and mechanism. New York City, New York: Garland Science; 2007. [Google Scholar]
- 21.Cornish-Bowden A. Fundamentals of Enzyme Kinetics. Vol. 3. Portland Press; 2004. [Google Scholar]
- 22.Coan KED, Maltby DA, Burlingame AL, Shoichet BK. Journal of Medicinal Chemistry. 2009;52:2067. doi: 10.1021/jm801605r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spies MA, Toney MD, Hynes JR, Klinman JP, Limbach HH, Schown RL. Hydrogen-Transfer Reactions. Vol. 3. Weinheim: Wiley-VCH; 2007. p. 1139. [Google Scholar]
- 24.Cleland WW. Methods in Enzymology. Vol. 87. Elsevier Inc.; 1982. An analysis of haldane relationships; p. 366. [DOI] [PubMed] [Google Scholar]
- 25.FRED; 2.2.5 ed. Sante Fe, NM, USA: OpenEye Scientific Software, Inc.; 2010. [Google Scholar]
- 26.GOLD; 4.1 ed. Cambridge, CB2 1EZ, UK: The Cambridge Crystallographic Data Centre; 2010. [Google Scholar]
- 27.Frey PA, Hegeman AD. Enzymatic reaction mechanisms. New York, New York: Oxford University Press; 2007. p. 10016. [Google Scholar]
- 28.Doublet P, van Heijenoort J, Mengin-Lecreulx D. Biochemistry. 1994;33:5285. doi: 10.1021/bi00183a035. [DOI] [PubMed] [Google Scholar]
- 29.Mehboob S, Guo L, Fu W, Mittal A, Yau T, Truong K, Johlfs M, Long F, Fung LW-M, Johnson ME. Biochemistry. 2009;48:7045. doi: 10.1021/bi9005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Russell RB, Barton GJ. Proteins: Structure, Function, and Genetics. 1992;14:309. doi: 10.1002/prot.340140216. [DOI] [PubMed] [Google Scholar]
- 31.Amaro RE, Baron R, McCammon JA. J Comput Aided Mol Des. 2008;22:693. doi: 10.1007/s10822-007-9159-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Donoghue P, Luthey-Schulten Z. J Mol Biol. 2005;346:875. doi: 10.1016/j.jmb.2004.11.053. [DOI] [PubMed] [Google Scholar]
- 33.Eastwood MP, Hardin C, Luthey-Schulten Z, Wolynes PG. IBM Journal of Research and Development. 2001;45:475. [Google Scholar]
- 34.Steinbrecher T, Andreas L. current Medicinal Chemistry. 2010;17:767. doi: 10.2174/092986710790514453. [DOI] [PubMed] [Google Scholar]
- 35.Shaw PB. Phys. Rev. A. 1985;32:2476. doi: 10.1103/physreva.32.2476. [DOI] [PubMed] [Google Scholar]
- 36.Warwicker J, Watson HC. Journal of Molecular Biology. 1982;157:671. doi: 10.1016/0022-2836(82)90505-8. [DOI] [PubMed] [Google Scholar]
- 37.Merz KR. Drug Design: Structure and Ligand-Based Approaches. Cambridge, UK: Cambridge University Press; 2010. [Google Scholar]
- 38.Brown SP, Muchmore SW. J Chem Inf Model. 2006;46:999. doi: 10.1021/ci050488t. [DOI] [PubMed] [Google Scholar]
- 39.Glavas S, Tanner ME. Biochemistry. 2001;40:6199. doi: 10.1021/bi002703z. [DOI] [PubMed] [Google Scholar]
- 40.Anandakrishnan R, Onufriev A. Journal of Computational Biology. 2008;15:165. doi: 10.1089/cmb.2007.0144. [DOI] [PubMed] [Google Scholar]
- 41.Mock WL. Bioorganic Chemistry. 1992;20:377. [Google Scholar]
- 42.Spies MA, Toney MD. Journal of the American Chemical Society. 2007;129:10678. doi: 10.1021/ja067643k. [DOI] [PubMed] [Google Scholar]
- 43.Sims PA, Larsen TM, Poyner RR, Cleland WW, Reed GH. Biochemistry. 2003;42:8298. doi: 10.1021/bi0346345. [DOI] [PubMed] [Google Scholar]
- 44.Slieman TA, Nicholson WL. Applied and Environmental Microbiology. 2001;67:1274. doi: 10.1128/AEM.67.3.1274-1279.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Setlow P. Current Opinion in Microbiology. 2003;6:550. doi: 10.1016/j.mib.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 46.Hopkins AL, Groom CR, Alex A. Drug Discovery Today. 2004;9:430. doi: 10.1016/S1359-6446(04)03069-7. [DOI] [PubMed] [Google Scholar]
- 47.Dorr P, Westby M, Dobbs S, Griffin P, Irvine B, Macartney M, Mori J, Rickett G, Smith-Burchnell C, Napier C, Webster R, Armour D, Price D, Stammen B, Wood A, Perros M. Antimicrob Agents Chemother. 2005;49:4721. doi: 10.1128/AAC.49.11.4721-4732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Iqbal J, Zaidi M, Schneider AE. IDrugs. 2003;6:587. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.