Abstract
We report TERS imaging of individual 50-nm, biotin-labeled gold nanoparticles bound to a streptavidin-derivatized glass slide. Individual gold nanoparticles detected by a nanoparticle TERS tip generate Raman enhancements in both the biotin and streptavidin signals. These results indicate that nanoparticles are capable of investigating nanoscale spatial and chemical environments with non-resonant Raman enhancements.
Nanoparticles are commonly used as labels for imaging in biological systems. The electric fields established, from the collective oscillation of conduction band electrons, known as the localized surface plasmon resonances (LSPR), in silver and gold nanostructures give rise to the electromagnetic enhancement involved in surface-enhanced Raman spectroscopy (SERS).1 Studies have shown that the enhanced field generates substantial Raman enhancements from molecules with electronic transitions that overlap with the LSPR, a technique commonly known as surface-enhanced resonance Raman spectroscopy (SERRS). The utility of functionalized-nanoparticles is well known. The narrow spectral bands characteristic of Raman provide opportunities for multiplex labeling. While SERRS tags are proving to be useful,2–5 it would be advantageous to use the Raman enhancements of nanoparticles to investigate the intrinsic molecules present in a sample.6 Tip-enhanced Raman scattering (TERS) merges SERS and atomic force microscopy (AFM) to obtain chemical, structural and spatial information simultaneously.7, 8 A distinct advantage of TERS is the enhanced Raman scattering in the tip’s immediate vicinity, which is generally a much smaller area than a diffraction limited laser focus; however, individual nanostructures often do not generate sufficient enhancements to distinguish samples with large far-field backgrounds. Since the largest SERS enhancements arise from nanoparticle junctions, combining nanoparticle labels with a nanoparticle TERS tip appears beneficial for amplifying the Raman signal associated with individual nanoparticles. Thus, we have combined nanoparticle labels with TERS imaging methodology to enhance the intrinsic Raman scattering of a protein – ligand complex with spatial resolution of approximately 50 nm.
We observe Raman scattering from a single biotin-labeled nanoparticle on a streptavidin-derivatized glass slide. Samples were prepared by pipetting 20 μL of 0.05% solution of monodisperse 50-nm gold nanoparticles functionalized with biotin (Nanocs, Inc) onto the streptavidin-functionalized slides and allowing the sample to air dry. Similar to previous experiments,9 the TERS microscope places an AFM (Nanonics MV4000) in the sample position of an upright microscope. A 35-mW, 633-nm HeNe laser irradiates the sample at normal incidence. Using a glass pipette with a 100-nm gold nanoparticle at the apex for our TERS tip, we simultaneously collect an AFM topographical map and generate an enhanced Raman signal at the tip position. The tip is irradiated with 0.5 mW of laser power. To avoid polarization artifacts associated with linear light in a backscattering geometry,10 radial polarization was achieved using a liquid crystalline mode converter (ArcOptix). Focused radial polarization has been shown to induce a longitudinal mode at the focus that results in increased enhancement normal to sample, improving coupling to the AFM tip.10, 11 The Raman backscattering signal is epi-collected through the objective and sent to a spectrograph (Horiba Jobin Yvon iHR320), equipped with a 600 grooves/mm grating to disperse the signal onto a CCD camera (Jobin Yvon, Synapse). Raman images were collected with 1-second spectral acquistions spaced every 20 nm during an AFM scan.
We performed an AFM scan to find isolated nanoparticles for TERS analysis. During the TERS scan, the electromagnetic field surrounding the nanoparticle AFM tip generates an enhanced Raman signal as shown in Figure 1. As the tip is scanned across the streptavidin surface, scattering corresponding to the vibrational modes of the protein are observed. There is a large area of streptavidin that produces a weak signal seen in the upper right corner of Fig 1d as evident in the corresponding spectrum (Fig 1a, blue). Vibrational bands are observed at 960, 1160(shoulder), and 1500–1600 cm−1 associated with the, Trp/Val, C-N/Trp/Val/Lys, and Trp modes of the protein, respectively. Enhancement was verified by repeating the scan with the nanoparticle-AFM tip retracted from the surface (Fig 1c) where no Raman scattering was observed above the noise. The strongest Raman spectra comes from the small area that also contains the biotin-labeled nanoparticle (Fig. 1a, red). In the spectrum of the nanoparticle, the features at 560, 870,1030, 1160,1270, and 1345 cm−1 are a combination of the biotin and streptavidin signals; however, the bands at 962, , 1400–1600 cm−1 correspond only to streptavidin. Tryptophan residues comprise a majority of the observed streptavidin spectrum, while features between 1160–1280 cm−1 are associated with other amino acid residues.12
Figure 1.
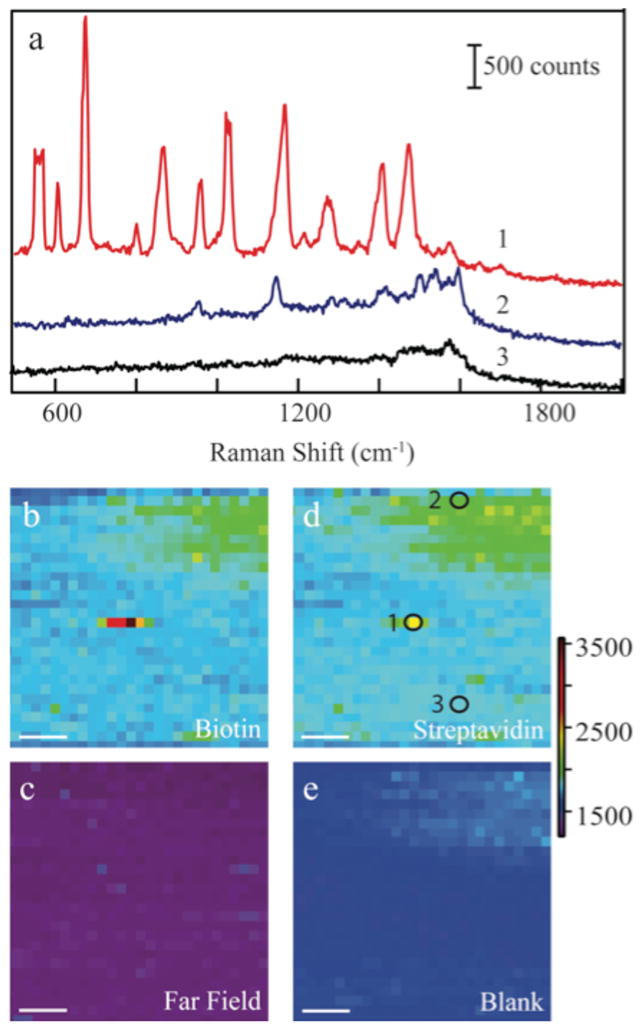
The Raman spectra (a) from selected points of the TERS image are shown. Peak assignments are provided in the electronic supplemental information. The Raman maps are displayed corresponding to the 680 cm−1 band of biotin (b) with and (c) without the TERS tip, the Streptavidin 962 cm−1 feature (d) with the TERS tip, and of (e) a blank region at 1800 cm−1. The circles and numbers in Fig. 1d correspond to the numbered spectra in panel a. The scale bar in all images (b–e) is 100 nm.
When the nanoparticle tip comes into proximity of a biotin-functionalized nanoparticle bound to the streptavidin protein, a dramatic increase in signal is observed. The increased signal originates from two effects associated with coupled nanostructures. It has been demonstrated that the maximum SERS enhancements occur in the gap or crevices between nanostructures.13 By bringing the nanoparticle tip into proximity of the functionalized nanoparticle, molecules residing in the junction will exhibit increased scattering. As shown in Figure 1a, Raman spectra taken at various points during the scan, show the difference between an area with streptavidin only and an area where the streptavidin has bound a biotin-labeled nanoparticle. In previous studies, the biotin contributes only a small component to the Raman spectrum of the biotin-avidin complex, which was attributed to either a much weaker biotin Raman cross-section12, 14 or a shielding effect by the protein, as the biotin is bound within the avidin.15 Instead, the biotin band at 680 cm−1 is the most prominent feature present in the spectrum of the nanoparticle – tip dimer. It is reasonable to expect that biotin, unbound to streptavidin, is present on the top of the nanoparticle and resides in the junction formed with the nanoparticle tip.
The second effect is attributed to a shift in the plasmon frequency associated with coupled nanostructures. We measured the plasmon resonance frequency of our 50 nm at 530 nm. As a second gold nanoparticle is brought close, the individual plasmons interact and shift the observed plasmon resonance to longer wavelenghts. A dimer of gold nanoparticles red-shifts the plasmon band maximum above 600 nm. 16–18 This shift moves the plasmon resonance closer to the 633 nm laser excitation wavelength and increases the enhancement of the Raman signal from molecules surrounding the nanoparticle dimer. This enhanced Raman signal increases the Raman scattering from the streptavidin protein bound to the biotin molecules on the nanoparticle giving rise to the spectrum shown in Figure 1a.
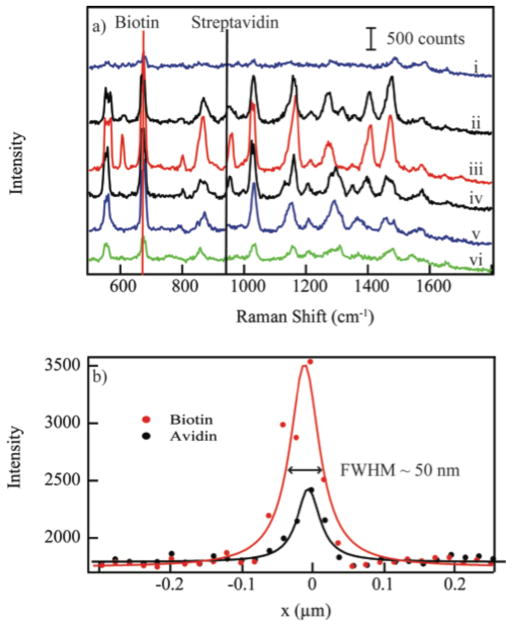
Previous reports have examined the interaction between metallic TERS tips and metal surfaces. Pettinger and coworkers showed that an Au-STM tip coming into contact with single crystal Au surface exhibited a signal increase as the gap distance was decreased.19 The enhancements observed in these experiments are sufficient to detect picomole quantities of nucleotides.20 Recently, Novotny and colleagues showed that a nanoparticle tip interacting with dye-functionalized nanoparticles exhibited a similar dependence.21 The Raman intensity vs. the tip-nanoparticle separation distance follows a Lorentz line shape in agreement with these previous reports. In Figure 2a, the Raman intensity associated with the 680 cm−1 band of biotin exhibits a larger magnitude of enhancement consistent with the larger fields expected within the junctions. The 962 cm−1 band of streptavidin also shows an increase indicating significant enhancement exists surrounding the exterior of the nanoparticle-complex.
Figure 2.
The upper graph shows Raman spectra in line with the biotin-labeled nanoparticle at (i) 40 nm, (ii) 20 nm, (iii) 0 nm, (iv) −20 nm, (v) −40 nm, and (vi) −60 nm from the centroid of the nanooparticle. The lower graph depicts the intensity profile associated with the 680 cm−1 and 962 cm−1 band of biotin and streptavidin, respectively. The FWHM of both traces is 50 nm. The points are experimental data, the solid line is a fit to a Lorentz line.
The full width at half maximum (FWHM) of the signal increase (Fig. 2b) is 50 nm, the size of the biotin-labeled nanoparticle. The coupling of the LSPRs of the two nanoparticles is a short range effect, generating the greatest Raman enhancements when the AFM tip is in close proximity to the biotin-functionalized nanoparticle. The observed FWHM suggests that the distance dependence of the red shift in the LSPR upon dimer formation is controlled by the smaller nanoparticle.
The biotin-streptavidin complex is often utilized due to it’s stability and high binding affinity with a dissociation constant of 10−15 M.22 Numerous studies have characterized the binding site in an effort to understand the extremely tight binding. This binding results from a non-covalent interaction where streptavidin forms a classic β-barrel structure with one end containing the binding site which is shaped similar to a biotin molecule. The streptavidin signals detected provide direct insight into the chemical nature of streptavidin binding complex. The streptavidin signals at, 962, 1030 (shoulder), 1170 (shoulder), 1340–1365, 1576 cm−1 that are observed to be strongly enhanced by the nanoparticle-tip complex are assigned to bands associated with the amino acid tryptophan. A small band shift between free and complexed streptadvidin show these tryptophan residues are located within the binding region of the protein.12 Since the SERS enhancement is known to rapidly decrease as you move away from the particle, it is expected that the binding pocket residues should show the greatest enhancement. Moscovits and coworkers similarly reported SERS enhancements of proteins from amino acid residues near a thiol-linker that bound to nanoparticles to form a dimer-complex.23 There is enhancement of other protein modes in comparison to the far-field spectrum, but the greatest signal enhancements are associated with residues directly involved in the binding interaction.
The critical aspect for the detection of these non-resonant molecules is the formation of the dimer-complex. Utilizing the AFM tip to form the dimer, we gain both signal enhancement and spatial resolution. As evident in Figure 2, the spatial resolution of our spectroscopic imaging experiment is 50 nm, as determined by the onset of enhancement to the maximum in the Raman signal. This value matches the size of our functionalized nanoparticle and suggests that gains may be possible utilizing smaller nanoparticle probes.
The ability to image and directly detect the residues involved in protein – ligand binding opens new avenues to exploring receptor function of proteins in cellular environments. The molecules detected here do not rely on an electronic enhancement enabling the study of any molecule – receptor interaction using nanoparticles as probes of cellular membranes.
Supplementary Material
Acknowledgments
This research was supported by the National Institutes of Health Award R00RR024367 and the University of Notre Dame.
Footnotes
Electronic Supplementary Information (ESI) available: Peak assignments for the spectra in Figure 1 and additional examples of single particles and images of aggregated particles are provided. See DOI: 10.1039/b000000x/
Notes and references
- 1.Stiles PL, Dieringer JA, Shah NC, Van Duyne RR. Annu Rev Anal Chem. 2008;1:601–626. doi: 10.1146/annurev.anchem.1.031207.112814. [DOI] [PubMed] [Google Scholar]
- 2.Graham D, Faulds K. Chemical Society Reviews. 2008;37:1042–1051. doi: 10.1039/b707941a. [DOI] [PubMed] [Google Scholar]
- 3.Golightly RS, Doering WE, Natan MJ. Acs Nano. 2009;3:2859–2869. doi: 10.1021/nn9013593. [DOI] [PubMed] [Google Scholar]
- 4.Zavaleta CL, Smith BR, Walton I, Doering W, Davis G, Shojaei B, Natan MJ, Gambhir SS. P Natl Acad Sci USA. 2009;106:13511–13516. doi: 10.1073/pnas.0813327106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Larmour IA, Faulds K, Graham D. Chemical Science. 2010;151–160 [Google Scholar]
- 6.Li JF, Huang YF, Ding Y, Yang ZL, Li SB, Zhou XS, Fan FR, Zhang W, Zhou ZY, WuDe Y, Ren B, Wang ZL, Tian ZQ. Nature. 2010;464:392–395. doi: 10.1038/nature08907. [DOI] [PubMed] [Google Scholar]
- 7.Verma P, Inouye Y, Kawata S. Top Appl Phys. 2006;103:241–260. [Google Scholar]
- 8.Yeo BS, Stadler J, Schmid T, Zenobi R, Zhang WH. Chem Phys Lett. 2009;472:1–13. [Google Scholar]
- 9.Schultz ZD, Stranick SJ, Levin IW. Appl Spectrosc. 2008;62:1173–1179. doi: 10.1366/000370208786401635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schultz ZD, Stranick SJ, Levin IW. Analytical Chemistry. 2009;81:9657–9663. doi: 10.1021/ac901789w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stanciu C, Sackrow M, Meixner AJ. Journal of Microscopy. 2008;229:247–253. doi: 10.1111/j.1365-2818.2008.01894.x. [DOI] [PubMed] [Google Scholar]
- 12.Fagnano C, Torreggiani A, Fini G. Biospectroscopy. 1996;2:225–232. [Google Scholar]
- 13.Wustholz KL, Henry AI, McMahon JM, Freeman RG, Valley N, Piotti ME, Natan MJ, Schatz GC, Duyne RPV. J Am Chem Soc. 2010;132:10903–10910. doi: 10.1021/ja104174m. [DOI] [PubMed] [Google Scholar]
- 14.Fagnano C, Fini G, Torreggiani A. J Raman Spectrosc. 1995;26:991–995. [Google Scholar]
- 15.Livnah O, Bayer EA, Wilchek M, Sussman JL. P Natl Acad Sci USA. 1993;90:5076–5080. doi: 10.1073/pnas.90.11.5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Braun G, Lee SJ, Dante M, Nguyen TQ, Moskovits M, Reich N. J Am Chem Soc. 2007;129:6378. doi: 10.1021/ja070514z. [DOI] [PubMed] [Google Scholar]
- 17.Link S, El-Sayed MA. J Phys Chem B. 1999;103:4212–4217. [Google Scholar]
- 18.Sonnichsen C, Reinhard BM, Liphardt J, Alivisatos AP. Nat Biotechnol. 2005;23:741–745. doi: 10.1038/nbt1100. [DOI] [PubMed] [Google Scholar]
- 19.Pettinger B, Domke KF, Zhang D, Schuster R, Ertl G. Phys Rev B. 2007;76:4. [Google Scholar]
- 20.Domke KF, Zhang D, Pettinger B. J Am Chem Soc. 2007;129:6708–6709. doi: 10.1021/ja071107q. [DOI] [PubMed] [Google Scholar]
- 21.Bharadwaj P, Beams R, Novotny L. Chemical Science. 2011 [Google Scholar]
- 22.Green NM. Method Enzymol. 1990;184:51–67. doi: 10.1016/0076-6879(90)84259-j. [DOI] [PubMed] [Google Scholar]
- 23.Pavel I, McCarney E, Elkhaled A, Morrill A, Plaxco K, Moskovits M. The Journal of Physical Chemistry C. 2008;112:4880–4883. doi: 10.1021/jp710261y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.