Abstract
Objective
This study evaluated whether genes involved in the metabolism of steroid hormones are associated with hormone levels or menopausal symptoms.
Methods
We used a population-based prospective sample of 436 African American (AA) and European American (EA) women who were premenopausal at enrollment and were followed longitudinally through menopause. We evaluated the relationship between steroid hormone metabolism genotypes at COMT, CYP1A2, CYP1B1, CYP3A4, CYP19, SULT1A1, and SULT1E1 with hormone levels and menopausal features.
Results
In EA women, SULT1E1 variant carriers had lower levels of dehydroepiandrosterone sulfate, and SULT1A1 variant carriers had lower levels of estradiol, dehydroepiandrosterone sulfate, and testosterone compared with women who did not carry these variant alleles. In AA women, CYP1B1*3 genotypes were associated with hot flashes (odds ratio [OR], 0.62; 95% CI, 0.40–0.95). Interactions of CYP1A2 genotypes were associated with hot flashes across menopausal stage (P = 0.006). Interactions of CYP1B1*3 (P = 0.02) and CYP1B1*4 (P = 0.03) with menopausal stage were associated with depressive symptoms. In EA women, SULT1A1*3 was associated with depressive symptoms (OR, 0.53; 95% CI, 0.41–0.68) and hot flashes (OR, 2.08; 95% CI, 1.64–2.63). There were significant interactions between SULT1A1*3 and hot flashes (P < 0.001) and between SULT1A1*2 and depressive symptoms (P = 0.007) on menopausal stage, and there were race-specific effects of SULT1A1*2, SULT1A1*3, CYP1B1*3, and CYP3A4*1B on menopause.
Conclusions
Our results suggest that genotypes are associated with the occurrence of menopause-related symptoms or the timing of the menopausal transition.
Keywords: Genotype, Steroid hormones, Metabolism, Menopause, Symptoms
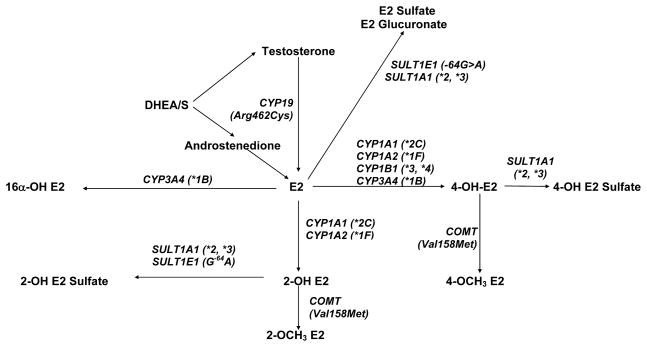
Changes in steroid hormone levels during the transition to menopause are associated with a variety of symptoms.1 The metabolism of steroid hormones may be mediated by inherited genotypes to exert clinically relevant effects. The genes involved in the disposition of endogenous steroid hormones are well known and include catechol-O-methyltransferase (COMT); the sulfotransferases SULT1A1 and SULT1E1; members of the cytochrome P450 family, including CYP19 (aromatase), CYP1B1, CYP1A2, CYP1A1, and CYP3A4; and others (Fig. 1). A number of studies have evaluated the associations between several hormone metabolism genes and endogenous hormone levels (Tables 1 and 2), symptoms associated with menopause,2–4 and timing of menopause.5–7 Most of these associations have been inconsistent or have not been replicated. The only replicated association that has been reported to date is the effect of CYP19 genotypes on estrone (E1) or estradiol (E2) in postmenopausal women.8,9
FIG. 1.
Steroid hormone metabolism pathways with genetic variants studied at these genes shown in parentheses. E2, estradiol; DHEA/S, dehydroepiandrosterone/sulfate.
TABLE 1.
Summary of candidate steroid hormone metabolism genes and serum steroid hormone levels in premenopausal women
| Hormone | COMT | CYP1A1 | CYP1A11a | CYP1A2 | CYP1B1 | CYP17 | CYP19 | ESR1 | HSD3B |
|---|---|---|---|---|---|---|---|---|---|
| Androstenedione | Null2,26,a | Null2 | Null26 | Null26 | Null2,25,a | Null2,a | |||
| DHEA | Null2,26,a | Null2 | Null26 | Null26 | Null 26, ↑2,a | Null2,a | |||
| Estrone | Null2,27 | Null27 | Null27 | Null2,27,a | Null2,a | ||||
| Estradiol | |||||||||
| Total | Null2,27,28,a | Null2,27, effect in Japanese22 | ↓ in *1F27 | Null27 | Null2,27,a | Null2 | Effect in AA23 | Null2,a | |
| Follicular | Null26 | Null26 | Null26 | Null26 | |||||
| Luteal | Null26 | Null26 | ↑ in L432V, S453N26 | Null26 | |||||
| Free | Null28 | ||||||||
| 2-OH estrone | Null27 | Null27, effect in Chinese22,a | Null27 | Null27 | Null2,27,a | ||||
| 16α-OH estrone | ↑ in V158M27 | Null26, effect in AA22,a | ↑ in 1*F27 | Null27 | Null2,27,a | Effect in AA23,a | |||
| FSH | Null28 | ||||||||
| Progesterone | |||||||||
| Total | Null 2,26,a, ↑ in V158M28 | Null2,a | Null26 | Null26 | Null2,a | Null2,26,a | Null2,a | ||
| Follicular | Null26 | Null26 | Null26 | Null26 | |||||
| Luteal | Null26 | Null26 | Null26 | Null26 | |||||
| SHBG | Null28 | Null26 | Null26 | Null26 | |||||
| Testosterone | Null2,26 | Null2,a | Null26 | Null26 | Null2,a | Null2,26,a | Effect in Japanese23,a | Null2,a | |
“Null” denotes no association was observed, using adjusted results when possible. The symbol ↑ denotes that the variant allele or genotype was associated with an increased level of a specified hormone, and ↓ denotes that the variant allele or genotype was associated with a decreased level of specified hormone. Allele names are provided when significant associations were reported. Empty cells indicate that the relationship has not been reported. COMT, catechol-O-methyltransferase; DHEA, dehydroepiandrosterone; AA, African American; FSH, follicle-stimulating hormone; SHBG, sex hormone–binding globulin.
TABLE 2.
Summary of candidate steroid hormone metabolism genes and serum steroid hormone levels in postmenopausal women
| Hormone | COMT | CYP1A1 | CYP1A11a | CYP1A2 | CYP1B1 | CYP17 | CYP19 | ESR1 | SHBG |
|---|---|---|---|---|---|---|---|---|---|
| Androstenedione | Null26 | Null8,9,26 | Null8,9 | Null9,26 | Null9 | ||||
| DHEA | Null26 | Null8,26 | Null8 | Null26 | |||||
| Estrone | Null9 | Null9 | Null8,9 | ↓ in 7r(−3)8, ↑ in 8r8, ↓ in 3′UTRt>c9 | Null9 | ||||
| Estradiol | ↓ in 7r(−3)8, ↑ in 8r(8), ↓ in 3′UTRt>c9 | ||||||||
| Total | Null9,28, ↑ in V158M7 | Null6 | Null8,9 | Null8,9 | Null9 | ||||
| Follicular | Null26 | Null26 | Null26 | ||||||
| Luteal | Null26 | ↑ in L432V, S453N26 | Null26 | ||||||
| Free | Null28 | ↑ in *46 | Null8 | Null8 | ↓ in 7r(−3)8, ↑ in 8r8 | ||||
| 2-OH estrone | Null8 | Null6 | Null8 | Null8 | |||||
| 16α-OH estrone | ↑ in V158M8 | ↑ in *46 | Null8 | Null8 | |||||
| FSH | Null28 | ||||||||
| Progesterone | |||||||||
| Total | Null9,28 | Null26 | Null9,26 | Null9 | Null26, ↓ in 3′UTRt>c9 | ||||
| Follicular | Null26 | Null26 | Null26 | ||||||
| Luteal | Null26 | Null26 | Null26 | ||||||
| SHBG | Null9,28 | ↑ in *46 | Null26 | Null8,9,26 | Null8 | Null9,26, ↑ in 7r(−3)8 | ↑ in 5 ′UTRg>a9, D356N9 | ||
| Testosterone | Null9 | Null26 | Null8,9,26 | Null8 | Null9,26 | Null9 | |||
“Null” denotes that no association was observed, using adjusted results when possible. The symbol ↑ denotes that the variant allele or genotype was associated with an increased level of specified hormone, and ↓ denotes that the variant allele or genotype was associated with a decreased level of specified hormone. Allele names are provided when significant associations were reported. Empty cells indicate that the relationship has not been reported. DHEA, dehydroepiandrosterone; FSH, follicle-stimulating hormone; SHBG, sex hormone–binding globulin.
Studies have also evaluated whether steroid hormone metabolism genotypes are associated with menopausal symptoms. One group has reported that CYP1B1 Val432Leu was associated with the occurrence and severity of hot flashes in a sample of women aged 45 to 54 years.2,3 The same group reported a borderline association with genetic variation at 3BHSD, as well as a possible joint effect of CYP1B1 and 3BHSD.2 A second group4 also reported that CYP1911r genotypes were associated with hot flashes in 174 midlife women. These two groups found no association of hot flashes with genotypes at CYP1A1, CYP17, ESR1, or COMT or for other variants at CYP19 or CYP1B1.
Relatively little information is available that evaluates the association of genotypes with the natural menopausal transition. Hefler et al5 reported that the presence of CYP1B1*4 was associated with a slightly earlier age at menopause than in women who did not carry this variant. However, other studies have reported no association of CYP1A16 or COMT7 genotypes on age at natural menopause. Importantly, studies to date have not examined the association between steroid hormone metabolism genotypes and longitudinal changes in steroid hormones and menopausal symptoms that occur with late reproductive aging.
Although there is some information about the relationship of hormone metabolism genes and hormone levels or menopausal traits and symptoms, the literature remains inconclusive about these associations. Therefore, we undertook a prospective cohort study to assess the association between candidate steroid hormone metabolism genes and hormone or menopausal traits. We hypothesized that inherited variation in hormone metabolism genes may be associated with baseline hormone levels and changes in these levels across the menopausal transition and that these genetically influenced endogenous hormonal phenotypes may further influence menopausal symptoms and timing. We assessed the association of candidate hormone metabolism genotypes on endogenous hormone levels, symptoms accompanying menopause, and timing of the menopausal transition in a population-based prospective cohort of women who were followed from premenopause through menopause.
METHODS
Study cohort
Participants in the Penn Ovarian Aging Study10 were identified by random-digit dialing to households in Philadelphia County. Recruitment of the cohort has been described in detail elsewhere1,11,12 and was stratified to enroll equal numbers of African American (AA) and European American (EA) women. Eligibility criteria for enrollment included age between 35 and 47 years, menstrual cycles in the reference range (22–35 d) for the previous 3 months, and presence of the uterus and at least one ovary. Exclusion criteria included current use of psychotropic or hormonal medications (including hormonal contraception and replacement therapies), pregnancy or lactation, serious health problems known to compromise ovarian function (eg, diabetes, liver disease, and breast or endometrial cancer), abuse of alcohol or drugs within the past year, and non–English speaking. The study was approved by the University of Pennsylvania Institutional Review Board, and all participants provided written informed consent forms. A total of 436 women (75% of those eligible) were enrolled in the cohort (218 AA and 218 EA women), of whom 413 had DNA available for this study. Two hormone measures taken at each assessment period (each year) were averaged for each participant. As an outcome variable, the average hormone value per participant in each year was used with repeated-measures analysis. At the 11th assessment period, approximately 11 years after study inception, complete data had been collected from 301 women, or 69% of the eligible women who were originally enrolled. Comparisons of baseline data between the participants in the present study and individuals who discontinued participation revealed no significant differences in demographic background variables or any of the variables in this report.
Assessments of study variables
Data were collected at approximately 9-month intervals in the first 5 years of the study and then annually with a 2-year interval between assessment periods 10 and 11. During each assessment period, two visits were scheduled between days 1 and 6 of two consecutive menstrual cycles, to obtain blood samples for hormone assays. The narrow visit window was selected to assess hormone levels in the early follicular phase, when levels are the most reliable13,14 and changes associated with ovarian aging are most pronounced.15 At each visit, a trained research interviewer conducted a standardized interview, collected blood samples for hormone assays, and measured height and weight to determine body mass index. The interview focused on overall health and included demographic background information, menstrual cycle dates, reproductive history, general health status and behaviors (including medications, smoking, alcohol and caffeine consumption, and history of depressive disorders), and common menopausal symptoms. Participants completed self-administered standard questionnaires at study enrollment and at each assessment period. All study questionnaires were completed within the first 6 days of the menstrual cycle in conjunction with the hormone assessments.
We defined five stages of the menopausal transition based on menstrual bleeding patterns and adapted from the Stages of Reproductive Aging Workshop16 to capture the early changes in the menopausal transition. The following five categories were defined in this study: (1) premenopausal—regular menstrual cycles in the 22- to 35-day range; (2) late premenopausal defined as a change in cycle length of 7 days or more either direction from the participant’s personal baseline at enrollment in the cohort and observed for at least one cycle in the study; (3) early transition defined as changes in cycle length of 7 days or more in either direction from the participant’s personal baseline at enrollment in the cohort and observed for at least two consecutive cycles in the study or 60 days of amenorrhea; (4) late transition—90 days to 11 months amenorrhea; and (5) postmenopausal—12 months or more amenorrhea excluding hysterectomy. Menopausal stage was identified at each assessment period using the menstrual dates at each study visit (visits were conducted within 6 d of bleeding) and the two previous menstrual dates obtained at each visit. Additional confirmatory data were obtained from the daily symptom diaries that participants recorded for one menstrual cycle at each assessment period, the reported number of menstrual periods between assessments, cycle length, and number of bleeding days.
In addition to menopausal stage and hormones, the following risk factors were selected based on their significance in previous studies of menopausal symptoms9–12: age, race, depressive symptoms based on the Center for Epidemiologic Studies Depression (CES-D) instrument,17 body mass index (calculated as weight in kilograms divided by the square of height in meters), and current smoking status (yes, no). The symptoms analyzed were depressive symptoms as assessed by CES-D and hot flashes. Hot flashes were included in a validated symptom list that was embedded in the structured interview at each assessment.18 The participants were asked whether hot flashes occurred in the past month, the frequency of their occurrence, and the severity rated on a 4-point scale from 0 (none) to 4 (severe). The presence or absence of hot flashes at each assessment in the 11-year interval was used in this study. Current depressive symptoms were assessed by the CES-D, a validated 20-item self-report questionnaire. The standard CES-D cutoff score of 16 or greater was used to define high depressive symptoms or depressed mood.
Laboratory measures
Nonfasting blood samples for the hormone assays were collected between days 2 and 6 of the menstrual cycle in two consecutive cycles (or at monthly intervals in noncycling women) during each assessment period. The samples were centrifuged and frozen in aliquots at −80°C. Assays were conducted in batches that included four visits per participant to reduce the within-subject variability due to assay conditions. E2, dehydroepiandrosterone sulfate (DHEAS), and testosterone levels were measured by radioimmunoassay using Coat-A-Count commercial kits (Diagnostic Products, Los Angeles, CA). Assays were performed in duplicate for all hormones and repeated if values differed by more than 15%. The interassay and intra-assay coefficients of variation calculated from the assays were less than 5%. The intra-assay and interassay coefficients of variation were less than 8% and less than 20%, respectively, for concentrations of 50 to 500 pg/mL; the analytical sensitivity was 15 pg/mL.
Genomic DNA was extracted from 300 blood samples and 88 serum samples obtained for serum hormone analyses. In a subset of 27 women, genotype analysis using serum-derived DNA was not successful, and a buccal swab was obtained by mail. Extraction of genomic DNA was performed using the QIAamp 96 DNA Buccal Swab Biorobot Kit and performed on a 9604 Biorobot (Qiagen, Inc., Valencia, CA). To limit the number of hypotheses tested, we identified the seven genes involved in the downstream metabolism of estrogen. We chose functionally relevant single nucleotide polymorphisms (SNPs) with a sufficiently high allele frequency to provide adequate power for testing first-order interactions. These were COMT Val158Met (rs4680), CYP19 Arg264Cys (rs700519), CYP1A2*1F (rs762551), CYP1B1*4 (Asn452Ser, rs1800440), CYP1B1*3 (Leu432Val, rs1056836), CYP3A4*1B (rs2740574), SULT1A1 Arg213His (*2; rs9282861), SULT1A1*3 (Met223Val, rs1801030), SULT1E1 (–64G>A Promoter Variant; rs3736599), and SULT1E1 A220G 3′UTR Variant (rs3786599). Genotypes were determined using previously described methods.19,20 Genotype coding was based on knowledge of the predicted function of the variants, as well as the frequency of genotypes of interest (Table 3).
TABLE 3.
Descriptive characteristics of study sample
| Variable | Group | African American (n = 206), n (%) | European American (n = 207), n (%) |
|---|---|---|---|
| Ever smoker | No | 116 (56) | 139 (67) |
| Yes | 90 (44) | 67 (32) | |
| Missing | – | 1 (1) | |
| Body mass index | <25 | 49 (23) | 101 (49) |
| 25–30 | 51 (25) | 54 (26) | |
| >30 | 102 (50) | 47 (23) | |
| Missing | 4 (2) | 5 (2) | |
| COMT | Met/Met | 151 (73) | 141 (68) |
| Any Val | 21 (10) | 50 (24) | |
| Missing | 34 (17) | 16 (8) | |
| CYP19 | Any 264Cys | 57 (28) | 14 (7) |
| 264 Arg/Arg | 133 (64) | 184 (89) | |
| Missing | 16 (8) | 9 (4) | |
| CYP1A2 | Any *1F | 31 (15) | 20 (10) |
| *1/*1 | 150 (73) | 170 (82) | |
| Missing | 25 (12) | 17 (8) | |
| CYP1B1*3 | Any *3 | 107 (52) | 37 (18) |
| *1/*1 | 77 (37) | 154 (74) | |
| Missing | 22 (11) | 16 (8) | |
| CYP1B1*4 | Any *4 | 163 (79) | 136 (66) |
| *1/*1 | 21 (10) | 54 (26) | |
| Missing | 22 (11) | 17 (8) | |
| CYP3A4 | Any *1B | 38 (18) | 177 (86) |
| *1/*1 | 142 (69) | 14 (6) | |
| Missing | 26 (13) | 16 (8) | |
| SULT1A1*2 | Any *2 | 98 (48) | 93 (45) |
| *1/*1 | 58 (28) | 88 (42) | |
| Missing | 50 (24) | 26 (13) | |
| SULT1A1*3 | Any *3 | 88 (42) | 183 (88) |
| *1/*1 | 68 (34) | 1 (1) | |
| Missing | 50 (24) | 23 (11) | |
| SULT1E1 5′UTR | Any A | 56 (27) | 24 (12) |
| G/G | 82 (40) | 129 (62) | |
| Missing | 68 (33) | 54 (26) | |
| SULT1E1 3′UTR | Any C | 175 (85) | 96 (47) |
| T/T | 19 (9) | 94 (45) | |
| Missing | 12 (6) | 17 (8) |
Statistical analysis
Generalized linear regression models for repeated measures21 were used to estimate the unadjusted association of genotypes and each study hormone. Repeated-measures extensions of logistic regression were also used to evaluate the association between each genotype and symptom severity, dichotomized as moderate or severe versus none or mild, to examine the level of symptom severity that is likely to be reported in a clinical setting. We then examined menopausal stage and other selected risk factors simultaneously in multivariable models to estimate the adjusted associations of genotype with hormones or menopausal symptoms.
Hormone measures were transformed to the natural log values. The two hormone values obtained in each assessment period were averaged for each participant, with the mean value and the SD around the average of the two hormone measurements for that assessment period used in the analysis. In cases where two hormone values were not obtained in an assessment period, the single value was used for the mean, and the SD was set as missing for that period. This approach was used to provide a measure of the women’s hormone fluctuations apart from the measured levels. Averaging the two hormone measures at each assessment period reduces the measurement error in the hormone values and avoids complications in the analysis of relating two correlated hormone measures with a single measure of the symptoms and other risk factors at each assessment period.
Cox proportional hazards models were used to estimate hazard ratios (HRs) for age at late premenopause, early menopausal transition, or postmenopause. Observations were censored if the participant reached the end of follow-up without having recorded the menopause stage of interest, observations of breast-feeding, or exogenous hormone use were censored at the times of their observation. Premenopausal observations were used as the reference group. Variance estimates were adjusted for the repeated observations from each participant using generalized estimates equations.14 Statistical analyses were conducted using the SAS statistical package, version 9.1 (SAS Institute, Cary, NC), and Stata version 9 (College Station, TX). All statistical tests were two tailed.
RESULTS
The participants were nearly equally distributed by race (Table 3). At the time of the last assessment, 95 (35%) women were postmenopausal, 60 (22%) were in the late menopausal transition, 103 (38%) were in the early menopausal transition, 8 (3%) were in the late premenopausal stage, and 6 (2%) were still premenopausal. The remaining 29 active participants at period 11 were either censored because of hysterectomy or hormone use or were not included in the study because of lack of DNA.
Hormone levels
Among EA women, those who carried variant alleles of SULT1E1 5′UTR had lower levels of DHEAS over time compared with women who were wildtype at this locus (P = 0.04; Table 4). In addition, those who carried SULT1A1*3/*3 genotypes had lower levels of E2, DHEAS, and testosterone over time compared with women who carried any *1 allele at this locus (P = 0.04). For the remaining genotypes in EA women and for all genotypes in AA women, there were no mean differences in hormone levels or differences in the change of hormone levels over time (ie, over the menopausal transition; Table 4).
TABLE 4.
Association of candidate genotypes with hormone levels: geometric means (95% CIs)a
| Gene | Genotype comparison | African American |
European American |
||||
|---|---|---|---|---|---|---|---|
| Estradiol | DHEAS | Testosterone | Estradiol | DHEAS | Testosterone | ||
| COMT | Met/Met | 35.2 (27.9–44.4) | 73.3 (59.1–90.9) | 6.2 (4.3–9.1) | 37.7 (33.7–42.2) | 86.1 (72.7–101.8) | 8.1 (7.2–9.1) |
| Any Val | 33.8 (30.0–38.1) | 72.5 (63.2–83.1) | 6.4 (5.1–7.9) | 40.1 (36.8–43.7) | 95.2 (87.4–103.8) | 7.6 (6.3–9.1) | |
| CYP19 | Any 264Cys | 35.2 (31.2–39.9) | 74.0 (64.2–85.2) | 6.3 (5.1–7.8) | 39.3 (36.0–42.8) | 93.0 (85.9–100.7) | 7.7 (6.9–8.6) |
| 264 Arg/Arg | 32.2 (27.3–38.1) | 69.1 (57.8–82.6) | 6.0 (4.7–7.8) | 36.3 (30.6–43.1) | 83.2 (66.9–103.3) | 6.2 (4.9–7.8) | |
| CYP1A2 | Any *1F | 34.7 (30.8–39.2) | 71.4 (62.5–81.6) | 8.2 (7.3–9.2) | 39.8 (36.6–43.2) | 93.6 (86.1–101.7) | 6.1 (4.9–7.6) |
| *1/*1 | 32.9 (27.0–40.0) | 72.9 (56.4–94.2) | 7.3 (5.5–9.6) | 37.9 (32.8–43.9) | 88.1 (66.2–117.1) | 6.0 (4.6–7.9) | |
| CYP1B1*3 | Any *3 | 34.1 (29.7–39.1) | 73.3 (61.6–87.3) | 6.4 (5.1–8.0) | 40.1 (37.1–43.4) | 90.7 (83.2–98.8) | 7.9 (7.0–8.8) |
| *1/*1 | 33.6 (29.2–38.7) | 70.7 (61.1–81.7) | 6.3 (5.0–8.0) | 36.6 (31.9–42.0) | 102.0 (89.0–116.9) | 8.0 (6.8–9.5) | |
| CYP1B1*4 | Any *4 | 34.0 (26.8–43.0) | 62.1 (43.5–88.5) | 5.8 (4.3–7.8) | 38.9 (35.0–43.2) | 86.7 (76.4–98.4) | 7.6 (6.4–9.0) |
| *1/*1 | 33.5 (29.5–38.0) | 72.6 (64.0–82.5) | 6.4 (5.1–7.9) | 39.7 (36.4–43.2) | 95.5 (87.2–104.5) | 8.0 (7.1–9.0) | |
| CYP3A4*1B | Any *1B | 34.0 (30.0–38.5) | 73.4 (63.6–84.7) | 6.1 (4.9–7.6) | 36.7 (27.9–48.4) | 76.0 (47.4–122.0) | 7.5 (5.3–10.5) |
| *1/*1 | 32.2 (27.1–38.2) | 66.1 (55.1–79.4) | 6.1 (4.5–8.3) | 39.7 (36.8–42.9) | 94.8 (87.8–102.3) | 7.9 (7.0–8.9) | |
| SULT1A1*2 | Any *2 | 34.5 (29.6–40.2) | 72.3 (61.7–84.8) | 7.0 (5.4–9.1) | 38.8 (35.3–42.6) | 88.4 (78.4–99.8) | 7.8 (6.8–8.9) |
| *1/*1 | 33.7 (29.3–38.8) | 67.6 (57.3–79.7) | 6.3 (4.9–8.0) | 40.6 (36.8–44.9) | 94.9 (86.2–104.4) | 8.1 (6.9–9.4) | |
| SULT1A1*3 | Any *3 | 35.2 (30.6–40.5) | 70.4 (57.8–85.8) | 6.4 (5.0–8.1) | 13.1 (11.4–15.2) | 9.5 (8.4–10.8) | 3.9 (3.2–4.8) |
| *1/*1 | 34.0 (29.4–39.3) | 68.4 (58.4–80.1) | 6.8 (5.3–8.8) | 39.4 (36.4–42.6) | 93.0 (85.9–100.6) | 7.9 (7.0–8.9) | |
| SULT1E1 5′UTR | Any A | 39.3 (34.1–45.3) | 73.3 (60.7–88.6) | 6.7 (5.2–8.7) | 35.0 (29.9–41.0) | 73.5 (59.3–91.2) | 8.2 (6.6–10.0) |
| G/G | 32.6 (28.4–37.4) | 76.3 (65.2–89.3) | 5.9 (4.6–7.6) | 40.6 (37.2–44.3) | 93.4 (85.3–102.2) | 7.7 (6.8–8.7) | |
| SULT1E1 3′UTR | Any C | 34.2 (27.8–42.2) | 85.0 (59.6–121.3) | 7.0 (5.1–9.6) | 40.4 (36.9–44.3) | 95.5 (87.0–105.0) | 7.8 (6.7–9.0) |
| T/T | 33.4 (29.4–37.9) | 70.7 (62.3–80.1) | 6.1 (4.9–7.5) | 38.9 (35.5–42.7) | 93.2 (83.0–104.6) | 7.8 (6.8–8.9) | |
DHEAS, dehydroepiandrosterone sulfate; BMI, body mass index.
Effect estimates were adjusted for the following variables: menopause status (late premenopause, early transition, late transition, postmenopausal), smoking (ever/never), and BMI (<25, ≥25–<30, and ≥30 kg/m2).
Symptoms
CYP1B1*3 genotypes were associated with a significantly decreased risk of hot flashes in AA women (OR, 0.62; 95% CI, 0.40–0.95; Table 5). We also observed an interaction of CYP1A2 genotypes and menopausal stage with the occurrence of hot flashes (P = 0.006) and of menopausal stage with CYP1B1*3 (P = 0.02) or CYP1B1*4 (P = 0.03) genotypes for depressive symptom outcomes in AA women. In EA women, SULT1A1*3 genotypes were associated with a significantly decreased risk of depressive symptoms (OR, 0.53; 95% CI, 0.41–0.68; Table 5) and an increased risk of hot flashes (OR, 2.08; 95% CI, 1.64–2.63). We also observed an interaction of SULT1A1*3 genotypes and menopausal stage with the occurrence of hot flashes (P < 0.001). Finally, there was a significant interaction of SULT1A1*2 genotypes across menopausal stage with depressive symptoms (P = 0.007). These significant interactions suggest that the degree to which some genotype was associated with depressive symptoms or hot flashes varied by menopausal stage.
TABLE 5.
Association of candidate genotypes with features of menopause: ORs with 95% CIsa associated with genotype main effect as well as interaction with menopause status [with interaction P value]
| Gene | Variant vs reference genotype comparison | African American |
European American |
||
|---|---|---|---|---|---|
| Depressive symptoms | Hot flashes | Depressive symptoms | Hot flashes | ||
| COMT | Met/Met vs Any Val | 0.61 (0.31–1.17) [0.88] | 1.29 (0.67–2.48) [0.76] | 1.10 (0.67–1.79) [0.98] | 0.82 (0.49–1.35) [0.09] |
| CYP19 | 264 Arg/Arg vs Any 264Cys | 0.91 (0.58–1.44) [0.07] | 0.68 (0.43–1.07) [0.62] | 0.58 (0.27–1.25) [0.41] | 0.86 (0.41–1.80) [0.75] |
| CYP1A2 | Any *1F vs *1/*1 | 0.82 (0.46–1.46) [0.62] | 1.04 (0.64–1.69) [0.006] | 0.97 (0.55–1.73) [0.11] | 0.96 (0.48–1.92) [0.54] |
| CYP1B1*3 | Any *3 vs *1/*1 | 0.74 (0.48–1.15) [0.02] | 0.62 (0.40–0.95) [0.93] | 0.61 (0.35–1.06) [0.47] | 0.94 (0.55–1.59) [0.39] |
| CYP1B1*4 | Any *4 vs *1/*1 | 0.61 (0.30–1.21) [0.03] | 0.66 (0.33–1.30) [0.54] | 0.80 (0.51–1.26) [0.37] | 1.20 (0.76–1.89) [0.44] |
| CYP3A4 | Any *1B vs *1/*1 | 0.98 (0.58–1.66) [0.13] | 1.10 (0.63–1.94) [0.50] | 1.03 (0.48–2.19) [0.69] | 0.87 (0.39–1.94) [0.12] |
| SULT1A1*2 | Any *2 vs *1/*1 | 1.28 (0.82–2.0) [0.95] | 0.77 (0.48–1.24) [0.31] | 0.77 (0.50–1.17) [0.007] | 0.75 (0.49–1.14) [0.34] |
| SULT1A1*3 | Any *3 vs *1/*1 | 1.27 (0.81–1.97) [0.58] | 0.95 (0.59–1.50) [0.88] | 0.53 (0.41–0.68) – | 2.08 (1.64–2.63) [<0.001] |
| SULT1E1 5′UTR | Any A vs G/G | 0.68 (0.41–1.11) [0.89] | 1.39 (0.86–2.23) [0.80] | 0.97 (0.53–1.78) [0.12] | 1.43 (0.77–2.63) [0.92] |
| SULT1E1 3′UTR | Any C vs T/T | 1.33 (0.67–2.66) [0.08] | 1.01 (0.49–2.10) [0.07] | 0.91 (0.59–1.38) [0.87] | 0.86 (0.57–1.31) [0.24] |
OR, odds ratio.
ORs were estimated from repeated-measures logistic regression analysis adjusted for the following variables: menopause status (late premenopause, early transition, late transition, postmenopausal).
Menopausal stage
The results of the analysis of age at late premenopause (stage 2), early menopausal transition (stage 3), or postmenopause (stage 5) are presented in Table 6. Age at stage 2 was significantly earlier among EA women who carried any SULT1A1*2 allele (HR, 1.80; 95% CI, 1.20–2.69; P = 0.004) or among AA women who carried any CYP1B1*3 allele (HR, 1.55; 95% CI, 1.03–2.34; P = 0.035). We also observed a significant interaction between race and SULT1A1*2 genotypes (interaction P = 0.003) such that EA women who carried any SULT1A1*2 allele reached the late premenopause stage at an earlier age than did EA women who were wildtype for SULT1A1*2. This is in contrast to AA women, of whom women who carried any SULT1A1*2 allele tended to reach the late premenopause stage at a later age than did AA women who were wildtype for the SNP. Mean age (95% CI) at stage 2 was 43.8 years (35.8–49.5 y) for EA women who were wildtype for SULT1A1*2 compared with 44.5 years (36.1–51.1 y) for EA women who carried any SULT1A1*2 allele. For AA women, mean age (95% CI) at stage 2 was 43.1 years (36.1–59.9 y) for those who were wildtype compared with 43.9 years (37.9–49.5 y) for those who carried any SULT1A1*2 allele.
TABLE 6.
Effect of genotype on time to menopausal stage in a prospective cohort of 413 women followed from premenopause to postmenopause
| Time to | Na | Race | Main effectsb | Na | Interaction by raceb |
|---|---|---|---|---|---|
| Late premenopause (stage 2) | 174 | EA | SULT1A1*2 (HR, 1.80; P = 0.004) | 326 | Race × SULT1A1*2 (Pint = 0.003) |
| 180 | AA | CYP1B1*3 (HR, 1.55; P = 0.035) | |||
| Early menopausal transition (stage 3) | 185 | EA | CYP3A4*1B (HR, 3.17; P = 0.001) | ||
| 175 | EA | SULT1A1*2 (HR, 1.63; P = 0.01) | 327 | Race × SULT1A1*2 (Pint = 0.026) | |
| 178 | EA | SULT1A1*3 (HR, 37.01; P = 0.001) | 330 | Race × SULT1A1*3 (Pint = 0.001) | |
| Postmenopause (stage 5) | 175 | EA | CYP3A4*1B (HR, 4.67; P = 0.017) | ||
| EA | SULT1A1*2 (HR, 0.53; P = 0.035) |
EA, European American; AA, African American; HR, hazard ratio; BMI, body mass index.
Denotes the number of women with the indicated genotype and race who had reached the indicated menopausal stage and were included in each analysis. Variation in numbers within stage reflects genotype success rates at each locus.
Adjusted for ever smoking and BMI (<25, ≥25–<30, and ≥30 kg/m2).
Age at early menopausal transition (stage 3) was significantly earlier among EA women who carried any CYP3A4*1B allele (HR, 3.17; 95% CI, 1.62–6.22), any SULT1A1*2 allele (HR, 1.77; 95% CI, 1.20–2.60; P = 0.004), or any SULT1A1*3 allele (HR, 0.01; 95% CI, 0.0005–0.13; P = 0.001). However, this last result was not considered to be interpretable because there was only one individual with the variant genotype. We also observed significant interactions between race and SULT1A1*2 genotype (interaction P = 0.026): AA women reached the menopausal transition later if they carried a SULT1A1*2 allele, whereas EA women reached the menopausal transition earlier if they carried any SULT1A1*2 allele.
Time from study enrollment to stage 5 was significantly earlier among EA women who carried any SULT1A1*2 allele (HR, 1.88; 95% CI, 1.04–3.37; P = 0.035).
DISCUSSION
We hypothesized that inherited variation in hormone metabolism genes are associated with baseline steroid hormone levels and changes in these levels across the menopausal transition and that genotypes are further associated with endogenous hormonal phenotypes to affect menopausal symptoms and timing. We report that SULT1E1 variant allele carriers had lower levels of DHEAS and carriers of SULT1A1 variants had lower levels of E2, DHEAS, and testosterone compared with women who did not carry these variant alleles. These genotype-phenotype associations have not been previously reported but are biologically plausible. The biological function of sulfotransferases and the genes that encode them is well characterized. The SULT1A1*2 genotype is associated with lower enzyme thermostability, lower enzyme activity29 and lower estrogen sulfation ability than the non-variant form.30 The SULT1A1 enzyme acts on E2. Thus, the decreased ability to sulfate endogenous E2 may explain the effect of this pathway on hormone levels.
There is only limited consistent evidence for associations of genotypes with hormone levels. The effects of genotypes on hormone levels are generally nonsignificant. This is also consistent with prior reports associating genotype with hormone levels.22,23 In a few cases, there have been two or more reports of specific genotypes being associated with hormone levels. In premenopausal women (Table 1), most associations have been null, although there have been a few reports of associations in population subsets. However, no true validations in comparable populations have been reported to date, except multiple associations consistent with the null hypothesis. In postmenopausal women (Table 2), most reports have again presented null associations, and there are few replication/validation studies of specific hormones and genetic variants. In published papers, the only gene for which some suggestion of replication exists is genetic variation in CYP19 (aromatase) and E1 or E2 levels. The genotypes evaluated are not all the same across studies, but the report of CYP19 genotypes having an effect on E1/E2 levels tends to be consistent with the predicted biological effects of these variants (when this information is known). Because CYP19 falls at a critical step in estrogen biosynthesis (Fig. 1), there is also a strong biological basis for a role of CYP19 in determining E2 or E1 levels. In our analysis, we observed no association of CYP19 genotypes with hormone levels. In part, this may reflect the fact that our study sample set entered the cohort when they were still premenopausal, and no association of CYP19 genotype with E1/E2 levels in premenopausal women has been reported (Table 1). Thus, if CYP19 does affect endogenous E1/E2 levels, this effect may be most likely to be observed in postmenopausal women.
There are a number of possible explanations for the limited consistency in the associations of genotypes and hormone levels. First, many studies may have had limited statistical power to detect small differences in hormone levels. Second, most studies have analyzed a small number of candidate SNPs in candidate genes. It is therefore possible that we have yet to capture the relevant genotype classes if a genotype-hormone relationship does exist. Haplotype-based studies may be undertaken to further explore this hypothesis. Third, it is possible that variability in hormone levels is so strongly influenced by other factors (eg, reproductive history, diet, exercise, and lifestyle) that the effect of single SNPs in candidate genes has become undetectable within the large amount of variability induced by other factors. We and others have attempted to account for measurement error and variability by undertaking appropriate assays, but the appropriate measurements may still not have been possible to obtain, particularly if the genotype effect is acting at a particular stage in life, at specific points in the menstrual cycle, and others. Similarly, if these genes and variants affect hormone levels, it is possible that they do so only in combinations (ie, as joint interaction effects), not as single SNP associations. Whereas most studies, including ours, attempt to measure confounders and other covariates of interest, many studies may not have appropriately accounted for reproductive events, menstrual cycle, or other factors that may influence the ability to detect single SNP effects on levels. Even if these factors could be accounted for, the present data are consistent with the hypothesis that single SNP variation in the genes that metabolize steroid hormones do not explain a substantial amount of interindividual variability in steroid hormone levels. However, the new evidence presented here suggests that sulfation may be associated with hormone levels. Additional studies are required to confirm these results.
We observed several race-specific associations of genotype with hot flashes. In our study, CYP1B1*3 was associated with the occurrence and severity of hot flashes in AA women, consistent with a previous study of women aged 45 to 54 years.2,3 Among EA women, SULT1A1*3 was associated with an increased risk of hot flashes. Similar to prior studies,2–4 our study did not show an association of hot flashes with genotypes at COMT, CYP19, or other CYP1B1 variants. Our results do suggest that the association between genotype and menopausal symptoms may vary by stage of reproductive aging. However, not all studies have provided consistent inferences in the relationship between menopausal symptoms and inherited genotype,24,25 so additional studies must be undertaken to confirm associations in this area.
Despite the biological plausibility of our findings, there are a number of limitations in this work. First, we evaluated the association between candidate genes and circulating hormone levels. These levels represent only one aspect of steroid hormones that may be relevant to menopause. We did not evaluate the association between candidate genotypes and metabolic clearance of hormones. Therefore, additional research that considers the complete spectrum of hormone metabolism is required to fully understand the associations identified here. Second, our sample size may have been inadequate to detect some small effects or may have involved rare genotypes, resulting in small numbers of observations in some groups. Thus, additional larger studies may be required to identify very small associations of genotypes with the hormone-related phenotypes studied here. However, it may be impractical to translate this research if genotypic effects are so small as not to be detectable here. Similarly, although we have been able to study both AA and EA women, our sample size may have been inadequate to detect statistically significant differences or interaction effects between these groups. Finally, the women in this study represent a select group in that they were pre-menopausal and could not be using hormonal contraception. This could have excluded a sizable proportion of women, and therefore, results may not be generalizable to the general population. Despite these limitations, the strengths of this study include prospectively collected longitudinal data with relatively long follow-up. These data include both endogenous hormone measures and symptom data.
We have confirmed the pattern seen in much of the literature that suggests that single SNP effects in candidate steroid hormone metabolism genes are not generally responsible for interindividual variation in steroid hormone levels or with changes in these levels across the menopausal transition. However, we do observe race-specific associations with CYP1B1, CYP1A2, and SULT1A1 on menopausal symptoms; race-specific effects of SULT1A1*2, SULT1A1*3, CYP1B1*3, and CYP3A4*1B on time to late premenopause, early menopausal transition, and menopause; and interactions of race with SULT1A1*2 and SULT1A1*3 on time to menopause.
CONCLUSIONS
If these associations are confirmed, they may provide information about the prediction of menopausal symptoms and allow clinicians to individualize and target hormone therapy in women experiencing menopausal symptoms. Because hormone exposures, genotypes involved in hormone metabolism, and the phenotypic manifestations of these factors on symptoms are all associated epidemiologically with risk of cancer and other diseases, a better understanding of the role of genotypes and intermediate phenotypes such as hormone levels may ultimately assist our understanding of steroid hormone– related disease etiology and prevention.
Acknowledgments
We thank Dr. Shiv Kapor of the University of Pennsylvania Clinical and Translational Research Center for performing the hormone assays.
Funding/support: R01-AG-12745 (E.W.F.) and RR024134 (Clinical and Translational Research Center). K12HD001265 and ACS MRSG08110-01 (HIS).
Footnotes
Financial disclosure/conflicts of interest: None reported.
References
- 1.Freeman EW, Grisso JA, Berlin J, et al. Symptom reports from a cohort of African American and white women in the late reproductive years. Menopause. 2001;8:33–42. doi: 10.1097/00042192-200101000-00007. [DOI] [PubMed] [Google Scholar]
- 2.Schilling C, Gallicchio L, Miller SR, et al. Genetic polymorphisms, hormone levels, and hot flashes in midlife women. Maturitas. 2007;57:120–131. doi: 10.1016/j.maturitas.2006.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Visvanathan K, Gallicchio L, Schilling C, et al. Cytochrome gene polymorphisms, serum estrogens, and hot flushes in midlife women. Obstet Gynecol. 2005;106:1372–1381. doi: 10.1097/01.AOG.0000187308.67021.98. [DOI] [PubMed] [Google Scholar]
- 4.Woods NF, Mitchell ES, Tao Y, et al. Polymorphisms in the estrogen synthesis and metabolism pathways and symptoms during the menopausal transition: observations from the Seattle Midlife Women’s Health Study. Menopause. 2006;13:902–910. doi: 10.1097/01.gme.0000227058.70903.9f. [DOI] [PubMed] [Google Scholar]
- 5.Hefler LA, Grimm C, Heinze G, et al. Estrogen-metabolizing gene polymorphisms and age at natural menopause in Caucasian women. Hum Reprod. 2005;20:1422–1427. doi: 10.1093/humrep/deh848. [DOI] [PubMed] [Google Scholar]
- 6.Napoli N, Villareal DT, Mumm S, et al. Effect of CYP1A1 gene polymorphisms on estrogen metabolism and bone density. J Bone Miner Res. 2005;20:232–239. doi: 10.1359/JBMR.041110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Worda C, Sator MO, Schneeberger C, et al. Influence of the catechol-O-methyltransferase (COMT) codon 158 polymorphism on estrogen levels in women. Hum Reprod. 2003;18:262–266. doi: 10.1093/humrep/deg059. [DOI] [PubMed] [Google Scholar]
- 8.Tworoger SS, Chubak J, Aiello EJ, et al. Association of CYP17, CYP19, CYP1B1, and COMT polymorphisms with serum and urinary sex hormone concentrations in postmenopausal women. Cancer Epidemiol Biomarkers Prev. 2004;13:94–101. doi: 10.1158/1055-9965.epi-03-0026. [DOI] [PubMed] [Google Scholar]
- 9.Dunning AM, Dowsett M, Healey CS, et al. Polymorphisms associated with circulating sex hormone levels in postmenopausal women. J Natl Cancer Inst. 2004;96:936–945. doi: 10.1093/jnci/djh167. [DOI] [PubMed] [Google Scholar]
- 10.Freeman EW, Sammel MD, Lin H, et al. Symptoms associated with menopausal transition and reproductive hormones in midlife women. Obstet Gynecol. 2007;110:230–240. doi: 10.1097/01.AOG.0000270153.59102.40. [DOI] [PubMed] [Google Scholar]
- 11.Freeman EW, Sammel MD, Grisso JA, et al. Hot flashes in the late reproductive years: risk factors for Africa American and Caucasian women. J Womens Health Gend Based Med. 2001;10:67–76. doi: 10.1089/152460901750067133. [DOI] [PubMed] [Google Scholar]
- 12.Hollander LE, Freeman EW, Sammel MD, et al. Sleep quality, estradiol levels, and behavioral factors in late reproductive age women. Obstet Gynecol. 2001;98:391–397. doi: 10.1016/s0029-7844(01)01485-5. [DOI] [PubMed] [Google Scholar]
- 13.Muti P, Trevisan M, Micheli A, et al. Reliability of serum hormones in premenopausal and postmenopausal women over a one-year period. Cancer Epidemiol Biomarkers Prev. 1996;5:917–922. [PubMed] [Google Scholar]
- 14.Santoro N, Adel T, Skurnick JH. Decreased inhibin tone and increased activin A secretion characterize reproductive aging in women. Fertil Steril. 1999;71:658–662. doi: 10.1016/s0015-0282(98)00529-9. [DOI] [PubMed] [Google Scholar]
- 15.Cramer DW, Barbieri RL, Xu H, et al. Determinants of basal follicle-stimulating hormone levels in premenopausal women. J Clin Endocrinol Metab. 1994;79:1105–1109. doi: 10.1210/jcem.79.4.7962282. [DOI] [PubMed] [Google Scholar]
- 16.Soules MR, Sherman S, Parrott E, et al. Stages of Reproductive Aging Workshop (STRAW) J Womens Health Gend Based Med. 2001;10:843–848. doi: 10.1089/152460901753285732. [DOI] [PubMed] [Google Scholar]
- 17.Radloff LS. The CES-D scale: a self-report depression scale for research in the general population. Appl Psychol Meas. 1977;1:385–401. [Google Scholar]
- 18.Freeman EW, Sammel MD, Liu L, Martin P. Psychometric properties of a menopausal symptom list. Menopause. 2003;2010:258–265. doi: 10.1097/00042192-200310030-00014. [DOI] [PubMed] [Google Scholar]
- 19.Rebbeck TR, Troxel AB, Wang Y, et al. Estrogen sulfation genes, hormone replacement therapy, and endometrial cancer risk. J Natl Cancer Inst. 2006;98:1311–1320. doi: 10.1093/jnci/djj360. [DOI] [PubMed] [Google Scholar]
- 20.Shatalova EG, Walther SE, Favorova OO, et al. Genetic polymorphisms in human SULT1A1 and UGT1A1 genes associate with breast tumor characteristics: a case-series study. Breast Cancer Res. 2005;7:R909–R921. doi: 10.1186/bcr1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCullagh P, Nelder J. Generalized Linear Models. London, England: Chapman & Hall; 1989. [Google Scholar]
- 22.Sowers MR, Wilson AL, Kardia SR, et al. CYP1A1 and CYP1B1 polymorphisms and their association with estradiol and estrogen metabolites in women who are premenopausal and perimenopausal. Am J Med. 2006;119:S44–S51. doi: 10.1016/j.amjmed.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 23.Sowers MR, Wilson AL, Karvonen-Gutierrez CA, et al. Sex steroid hormone pathway genes and health-related measures in women of 4 races/ethnicities: the Study of Women’s Health Across the Nation (SWAN) Am J Med. 2006;119:S103–S110. doi: 10.1016/j.amjmed.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 24.Kravitz HM, Meyer PM, Seeman TE, et al. Cognitive functioning and sex steroid hormone gene polymorphisms in women at midlife. Am J Med. 2006;119:S94–S102. doi: 10.1016/j.amjmed.2006.07.030. [DOI] [PubMed] [Google Scholar]
- 25.Kravitz HM, Janssen I, Lotrich FE, et al. Sex steroid hormone gene polymorphisms and depressive symptoms in women at midlife. Am J Med. 2006;119:S87–S93. doi: 10.1016/j.amjmed.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 26.Garcia-Closas M, Herbstman J, Schiffman M, et al. Relationship between serum hormone concentrations, reproductive history, alcohol consumption and genetic polymorphisms in pre-menopausal women. Int J Cancer. 2002;102:172–178. doi: 10.1002/ijc.10651. [DOI] [PubMed] [Google Scholar]
- 27.Lurie G, Maskarinec G, Kaaks R, et al. Association of genetic polymorphisms with serum estrogens measured multiple times during a 2-year period in premenopausal women. Cancer Epidemiol Biomarkers Prev. 2005;14:1521–1527. doi: 10.1158/1055-9965.EPI-04-0746. [DOI] [PubMed] [Google Scholar]
- 28.Hong CC, Thompson HJ, Jiang C, et al. Val158Met polymorphism in catechol-O-methyltransferase gene associated with risk factors for breast cancer. Cancer Epidemiol Biomarkers Prev. 2003;12:838–847. [PubMed] [Google Scholar]
- 29.Raftogianis RB, Wood TC, Otterness DM, Van Loon JA, Weinshilboum RM. Phenol sulfotransferase pharmacogenetics in humans: association of common SULT1A1 alleles with TS PST phenotype. Biochem Biophys Res Comm. 1997:298–304. doi: 10.1006/bbrc.1997.7466. [DOI] [PubMed] [Google Scholar]
- 30.Shatalova EG, Walther SE, Favorova OO, Rebbeck TR, Blanchard RL. Genetic polymorphisms in human SULT1A1 and UGT1A1 genes associated with breast tumor characteristics: a case series study. Breast Cancer Res. 2005;7:R909–R921. doi: 10.1186/bcr1318. [DOI] [PMC free article] [PubMed] [Google Scholar]