Abstract
For over 3 decades, the rate of replacement mutations has been assumed to be equal to, and estimated from, the rate of “strictly” neutral sequence divergence in noncoding regions and in silent-codon positions where mutations do not alter the amino acid encoded. This assumption is fundamental to estimating the fraction of harmful protein mutations and to identifying adaptive evolution at individual codons and proteins. We show that the assumption is not justifiable because a much larger fraction of codon positions is involved in hypermutable CpG dinucleotides as compared with the introns, leading to a higher expected replacement mutation rate per site in a vast majority of the genes. Consideration of this difference reveals a higher intensity of purifying natural selection than previously inferred in human genes. We also show that a much smaller number of genes are expected to be evolving with positive selection than that predicted using sequence divergence at intron and silent positions in the human genome. These patterns indicate the need for using new approaches for estimating rates of amino acid–altering mutations in order to find positively selected genes and codons in genomes that contain hypermutable CpG’s.
Keywords: adaptive evolution, mutation rate, test of selection, comparative genomics
Introduction
Following the lead of Kimura (1977), biologists have routinely assumed that non-synonymous (replacement) ‘mutations’ in codons occur at the same rate as silent mutations in molecular evolutionary analyses (Miyata and Yasunaga 1980; Nei and Kumar 2000; Bustamante et al. 2005; Nielsen et al. 2005). This assumption is made because replacement mutation rates often cannot be measured experimentally due to technical limitations or by comparative sequence analysis because natural selection prevents the fixation of a majority of replacement mutations between species. The equality assumption is at the heart of most widely used methods aimed at testing for selection on genes, identifying codons with adaptive evolutionary changes, estimating the number of deleterious mutations, and even finding coding regions in genomic sequences (Nei and Kumar 2000; Yang and Bielawski 2000; Nekrutenko et al. 2003; Yampolsky et al. 2005). Recently, the comparison of the chimpanzee genome with the human genome has established that there has been extensive selection against suboptimal synonymous codons in the human genome (Hellmann et al. 2003; Urrutia and Hurst 2003; Chamary and Hurst 2005; Lu and Wu 2005; Mikkelsen et al. 2005; Parmley et al. 2006). This fact has prompted the use of sequence divergence in introns and intergenic regions (excluding the regulatory) for establishing mutation rates at replacement positions in codons (Hellmann et al. 2003; Lu and Wu 2005; Mikkelsen et al. 2005; Parmley et al. 2006).
Should the actual mutation rate at replacement positions (called intrinsic mutation rate) be expected to be the same as the mutation rate inferred from sequence divergence at silent sites and in noncoding regions (called extrinsic mutation rate)? The context dependence of mutation rates suggests that it is unlikely. For example, positions involved in CpG dinucleotides are known to be hypermutable when methylated and are estimated to accumulate divergence at 5–20 times higher rate than other positions (Krawczak et al. 1998; Bird 1999; Hellmann et al. 2003; Subramanian and Kumar 2003). This higher mutation rate accounts for 25–50% of the interspecific variation between human and chimpanzee (Subramanian and Kumar 2003; Mikkelsen et al. 2005), single-nucleotide polymorphism diversity (Halushka et al. 1999), and replacement mutations associated with human diseases. Although this context dependence has been known for over 2 decades, severity of its effect on the disturbance of the assumed equality of mutation rates in different codon positions, introns, and intergenic regions remains to be seriously examined, particularly on the tests of selection in molecular evolutionary analyses.
The proportion of replacement positions in the CpG configuration is a function of the frequency of the codons that contain C’s and G’s, which is highly correlated with the frequency of amino acids in a given protein. By contrast, the fraction of CpG positions in introns is determined by the mutation-decay balance in their genomic milieu (Hwang and Green 2004; Fryxell and Moon 2005). These two fractions are rarely identical, as is evident from an analysis of all available functional human genes with at least one intron (10,196 genes; fig 1a). The distributions of the fraction of replacement and intronic positions involved in CpG dinucleotide configurations (CpG contents) are distinctly different in shapes and central tendencies. The CpG contents of replacement positions show a larger dispersion than introns, and the replacement positions of a vast majority of the genes have a higher CpG content compared with their introns. Over all genes, replacement positions are involved in CpG configurations 2 times more often than intron positions (P < 0.0001). This CpG content difference is statistically significant for 49% of the genes (Fisher’s exact test; P < 0.05), with 92% of the proteins showing an excess of replacement positions involved in CpG dinucleotides as compared with the introns.
FIG. 1.
(a) The differential distribution of the fraction of intronic (filled bars) and replacement positions (open bars) involved in CpG dinucleotide configuration from 10,196 functional human genes containing at least one intron. On average, the replacement positions involved in CpG configurations are 2 times higher than those of intronic positions (6.2% and 2.9%, respectively). The dispersion indices (the ratio of variance to mean) of the distributions of intron and replacement positions are 0.015 and 0.024, respectively. (b) Overestimation of the coefficient of selection obtained using intronic divergence (ωI = DR/DI) as compared with the replacement mutation rate (ωR = DR/MR) is shown for 6,435 human genes that are devoid of CpG islands, have at least one intron, and show at least one replacement difference between human and chimpanzee (see Methods). The average CpG contents (fraction of replacement positions involved in CpG dinucleotides) shown on the x axis were estimated from the genes with CpG contents of (numbers of genes in parenthesis) 0–0.01 (56), 0.01–0.02 (461), 0.02–0.03 (892), 0.03–0.04 (842), 0.04–0.05 (741), 0.05–0.06 (649), 0.06–0.07 (567), 0.07–0.08 (494), 0.08–0.09 (431), 0.09–0.10 (333), 0.10–0.11 (268), 0.11–0.12 (188), 0.12–0.13 (143), 0.13–0.14 (101), 0.14–0.15 (71), and >0.15 (198). The average ωI and ωR were computed for each category. The overestimations of ωR plotted on the y axis were computed as (ωI − ωR)/ωR × 100. The error bars show the standard error of the mean. The relationship between the CpG content and the overestimation of ωR is highly significant (R2 = 0.99, P < 0.01).
What effect does this larger proportion of hypermutable replacement positions have on the estimates of natural selection? This question can be examined by devising a formula that predicts the intrinsic mutational divergence per replacement position (MR) by accounting for the elevated mutation rate in the CpG dinucleotide positions. It is given by MR = [fR + (1 − fR) k](μA + μB) × t, where fR is the fraction of replacement positions not involved in the CpG dinucleotides, μA and μB are the baseline mutation rates at non-CpG positions in species A and B, respectively, t is the time of species divergence, and k is the factor by which the per site CpG sequence divergence exceeds the baseline mutation rate. This equation can be further modified in such a way that the estimation of the expected replacement divergence between species (MR) does not require the knowledge about the baseline mutation rates and the species divergence time (see Methods).
The fraction of replacement mutations eliminated by purifying selection is given by one minus the coefficient of selection (ω), where ω is equal to the observed replacement sequence divergence per site (DR) divided by the expected number of replacement mutations per site (MR). On the genomic scale, the average coefficient of selection (ωI = DR/DI) estimated using the sequence divergences at intron positions (DI) as a proxy for replacement mutation rate is significantly higher than the estimates obtained by using the intrinsic replacement mutation rates (ωR = DR/MR) (0.24 and 0.20, respectively; P < 10−20). Although the magnitude of the difference between the average estimates appears small (20%) on a genomic scale, the overestimation of the coefficient of selection (Δω) varies tremendously, with many genes showing >60% overestimation (fig. 1b). The coefficient of selection estimated using the intrinsic replacement mutation rate is higher for most of the human genes (94%) than that estimated using the extrinsic rate of their corresponding introns due to the greater fraction of replacement positions involved in CpG dinucleotides. The opposite pattern is true for a small fraction (~6%) of the genes, where ω is underestimated.
Significant overestimation of the coefficient of selection (ωS = DR/DS) results when the sequence divergence in silent positions is used as a proxy for mutation rate at replacement sites (P < 10−32). However, the combined effect of differences in the fraction of hypermutable sites in silent and replacement positions and the varying degrees of purifying selection on interspecies divergence in silent positions leads to an unpredictable relationship between the estimates of ω based on intrinsic and extrinsic mutation rates. Still, the impact of using silent substitution rates on genes evolving with positive selection is striking when we examine genes for which ωS > 1, which is traditionally used as an indicator of positive selection.
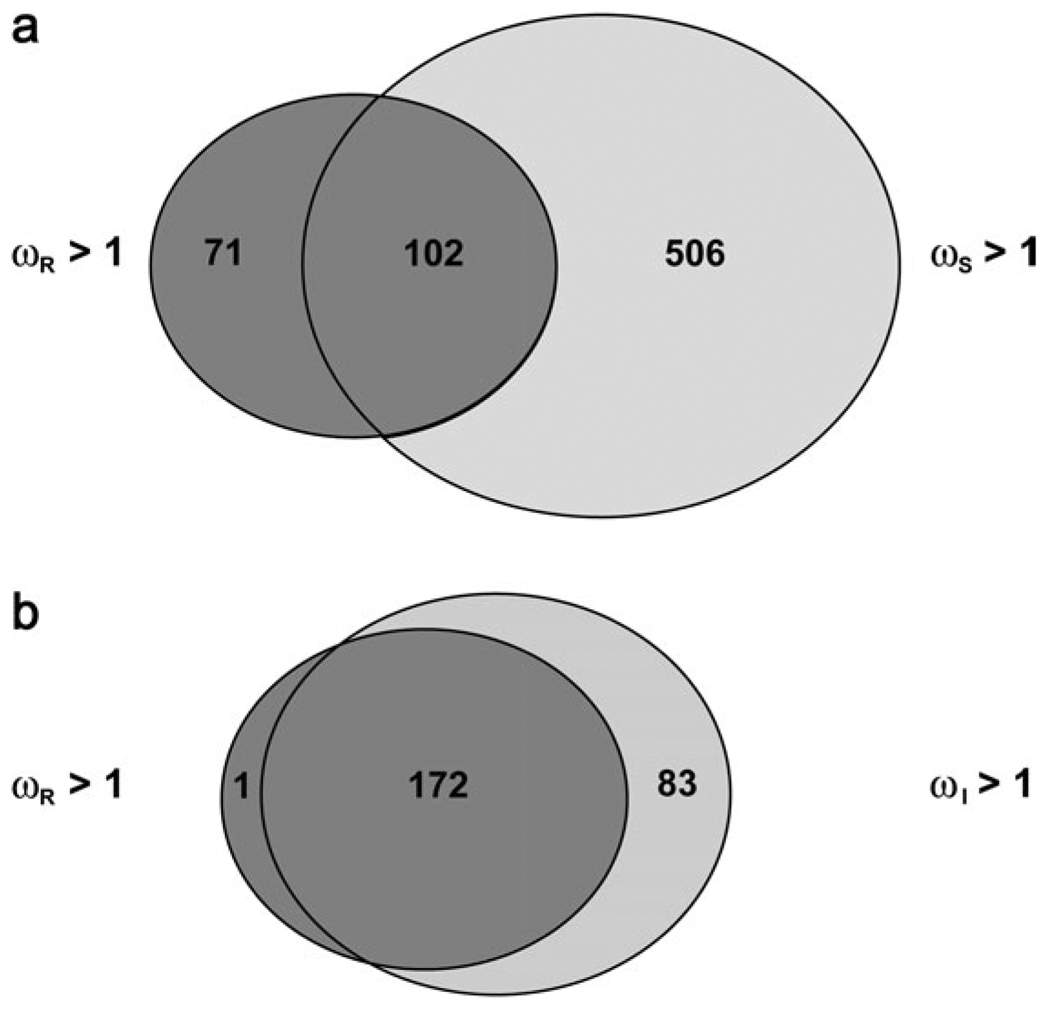
The use of ωS predicts that amino acid substitutions in 608 genes are positively selected, as compared with the use of ωR that identifies a much smaller number of genes (173). Only 102 genes are shared by the predictions based on intrinsic and extrinsic rates, and a large fraction of the genes (83%) predicted by the use of silent divergence may fall into false-positive category (fig. 2a). At the same time, ωS is too conservative for many other genes, as it fails to detect tracks of positive selection in 41% of the genes (71 out of 173). An examination of the biological processes performed by the positively selected genes indicates that a greater proportion of the genes identified using intrinsic mutation rates at replacement positions are involved in immunity and signal transduction (table 1). The use of silent divergence fails to identify over 50% of these genes, but identifies a large number of genes including those performing functions such as DNA replication and metabolism. The latter are unlikely candidates for positive selection as they are often involved in the housekeeping. Although the use of sequence divergence in introns predicts almost all the positively evolving genes that are identified by the intrinsic replacement mutation rate (fig. 2b), still 1 out of 3 predictions may be false positives. These false positives do not seem to be abundant in any specific functional category.
FIG. 2.
Venn diagrams showing the numbers of human genes under adaptive evolution (ω > 1.0) predicted by 3 different methods that use silent divergence (ωS = DR/DS), intron divergence (ωI = DR/DI), and replacement mutation rate (ωR = DR/MR) to estimate coefficient of selection. The use of intrinsic replacement mutation rate was compared with the use of (a) silent divergence and (b) intron divergence in identifying the positively evolving genes. The numbersin the overlapping area indicate the genes that were identified by both the methods, and the numbers in the individual circles represent the genes that were predicted exclusively by one of the methods.
Table 1.
Numbers of human genes identified to be positively selected (ω > 1) by using predicted replacement mutation rates, intron sequence divergences, and silent sequence divergences
| Using Extrinsic Mutation Rate (false negativesb, false positivesc) |
|||
|---|---|---|---|
| Biological Processa | Using Intrinsic Mutation Rate |
Intron divergence |
Silent divergence |
| Metabolism | 20 | 25 (1, 6) | 95 (10, 85) |
| Transport | 9 | 14 (0, 5) | 43 (2, 36) |
| Defense/immunity | 71 | 89 (0, 18) | 136 (34, 99) |
| Development | 4 | 12 (0, 8) | 20 (2, 18) |
| Signal transduction | 32 | 50 (0, 18) | 89 (17, 74) |
| Cellular process | 23 | 39 (0, 16) | 81 (6, 64) |
| Replication/transcription | 10 | 11 (0, 1) | 59 (4, 53) |
| Unknown biological process | 80 | 119 (0, 39) | 268 (31, 219) |
Genes belonging to more than one biological process are included in multiple categories, so the column sums may not equal those in figure 2.
False negative refers to genes showing ωR > 1 by using intrinsic mutation rate from replacement positions, but missed when using intron or silent divergence (ωI < 1 or ωS < 1, respectively).
False positive refers to genes that are identified by using intron or silent divergence to be positively selected (ωI > 1 or ωS > 1, respectively), but not by the use of predicted mutation rate from replacement positions (ωR < 1).
Our results demonstrate the presence of significantly more purifying selection on coding regions than previously thought because the replacement mutation rate is much higher than the observed substitution rate in introns and silent positions. These results indicate that the numbers of adaptive replacement changes in the genome of species with hypermutable CpG contents (e.g., primates) have been previously overestimated because a higher mutation rate is now implicated in producing the same observed replacement divergence between species (human and chimpanzee in the current case). These results are concerned with the strength of selection on the entire polypeptide and do not dispute existence of adaptive evolution on one or a few codons, which may occur even in highly conserved housekeeping genes (Yang and Nielsen 2002). In conclusion, however, it is clear that the estimates of mutation rates at replacement positions need to be based on the neighboring context of the replacement positions in analyses aimed at determining the proportion of replacement mutations eliminated by selection, predicting the fraction of disease-causing replacement mutations, and scanning the genome to find positively selected genes and codons.
Methods
Protein coding and intron sequences of the human genome (Build 34) were obtained from GenBank (ftp://ftp.ncbi.nih.gov/genomes/). Because we compared mutations in introns and silent positions, only the functional genes with at least one intron were included in the analysis, which resulted in 10,196 genes. CpG content of replacement positions of a gene was estimated as the number of 0-fold degenerate positions (i.e., positions in which any mutation changes the amino acid coded by the codon) that are involved in the CpG configuration divided by the total number of 0-fold degenerate positions in the gene. Similarly, the CpG content of intron is the proportion of intron positions involved in CpG dinucleotide configuration (Krawczak et al. 1998; Bird 1999; Hellmann et al. 2003; Subramanian and Kumar 2003). The biological processes of human genes were obtained from Protein Analysis Through Evolutionary Relationships classification system (http://www.pantherdb.org/). Genes belonging to more than one biological process were included in multiple categories. For simplicity, we combined the biological processes that are known to be related and created 8 categories (table 1). For instance, cell structure, motility, proliferation, and adhesion were grouped into one category called cellular process (table 1). Similarly the biological processes such as DNA replication, repair, recombination, mRNA transcription, splicing, and pre–mRNA processing were combined to form the replication/transcription category.
The intrinsic mutability of the replacement positions is largely determined by the presence of CpG dinucleotides that mutate much faster than non-CpG nucleotides. Therefore, the mutation rate at replacement sites is the sum of the rates at CpG and at non-CpG positions. If fR is the fraction of non-CpG positions and μA and μB are the baseline mutation rates at non-CpG nucleotides in lineages A and B, then the expected mutational divergence per replacement site (MR) between the species A and B is given by
| (1) |
where t is the time of species divergence, k is the ratio of CpG to non-CpG mutation rate, and fR is assumed to be equal in both the species compared (as the species considered are extremely closely related).
The equation (1) can be simplified as
| (2) |
The factor (μA + μB) × t can actually be estimated from the sequence divergence observed in pairwise orthologous sequence comparison of intronic (or genomic noncoding) positions. If fI is the fraction of non-CpG positions in the introns, then the intronic sequence divergence per site (DI) between the same 2 species is given by
| (3) |
Therefore, equation (2) can be modified as
| (4) |
Using equation (4), the expected mutational divergence at the replacement positions can be estimated without the knowledge of species divergence times as well as the baseline mutation rate in species A and B. For this purpose, we used the intronic sequence divergence (DI) between human and chimpanzee for each gene as presented in Mikkelsen et al. (2005). The use of intronic divergence from human and chimpanzee will also incorporate the effects of other determinants of instantaneous mutation rate, such as the recombination rate and region-specific G + C content (Hardison et al. 2003; Hellmann et al. 2005; Mikkelsen et al. 2005). In these calculations, the CpG positions were, at an average, assumed to accumulate divergence at 10 times the rate at other positions (k = 10) (Krawczak et al. 1998; Bird 1999; Hellmann et al. 2003; Subramanian and Kumar 2003).
It is well known that the CpG dinucleotides present in CpG islands are not hypermutable (as they are not methylated), and these islands extend to the first few exons (and introns) (Bird 1999; Rollins et al. 2006; Saxonov et al. 2006). Therefore, the methylation status of the CpG dinucleotides is ambiguous in the genes containing CpG islands. For this reason, we identified the CpG islands following the method described by Duret and Galtier (2000) and excluded the genes that contain those islands from further analysis.
For the estimation of the coefficients of selection, 6,435 human genes that are devoid of CpG islands and have at least one intron and at least one observed replacement difference between human and chimpanzee were used. The replacement (DR), silent (DS), and intronic (DI) sequence divergences between human and chimpanzee were obtained from the previous study conducted by chimpanzee sequencing and analysis consortium (Mikkelsen et al. 2005). The coefficients of selection were estimated using the intron divergence (ωI = DR/DI), silent divergence (ωS = DR/DS), and replacement mutation rate (ωR = DR/MR).
Acknowledgments
We thank Drs Brian Verrelli, Philip Hedrick, George Zhang, and Yuseob Kim for providing insightful remarks on a preliminary version of this article. We also thank an anonymous reviewer for constructive comments. This work was supported by a grant from National Institutes of Health to S.K.
Literature Cited
- Bird A. DNA methylation de novo. Science. 1999;286:2287–2288. doi: 10.1126/science.286.5448.2287. [DOI] [PubMed] [Google Scholar]
- Bustamante CD, Fledel-Alon A, Williamson S, et al. Natural selection on protein-coding genes in the human genome. Nature. 2005;437:1153–1157. doi: 10.1038/nature04240. (14 coauthors) [DOI] [PubMed] [Google Scholar]
- Chamary JV, Hurst LD. Evidence for selection on synonymous mutations affecting stability of mRNA secondary structure in mammals. Genome Biol. 2005;6:R75. doi: 10.1186/gb-2005-6-9-r75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L, Galtier N. The covariation between TpA deficiency, CpG deficiency, and G + C content of human isochores is due to a mathematical artifact. Mol Biol Evol. 2000;17:1620–1625. doi: 10.1093/oxfordjournals.molbev.a026261. [DOI] [PubMed] [Google Scholar]
- Fryxell KJ, Moon WJ. CpG mutation rates in the human genome are highly dependent on local GC content. Mol Biol Evol. 2005;22:650–658. doi: 10.1093/molbev/msi043. [DOI] [PubMed] [Google Scholar]
- Halushka MK, Fan JB, Bentley K, Hsie L, Shen N, Weder A, Cooper R, Lipshutz R, Chakravarti A. Patterns of single-nucleotide polymorphisms in candidate genes for blood-pressure homeostasis. Nat Genet. 1999;22:239–247. doi: 10.1038/10297. [DOI] [PubMed] [Google Scholar]
- Hardison RC, Roskin KM, Yang S, et al. Covariation in frequencies of substitution, deletion, transposition, and recombination during eutherian evolution. Genome Res. 2003;13:13–26. doi: 10.1101/gr.844103. (18 co-authors) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmann I, Prufer K, Ji H, Zody MC, Paabo S, Ptak SE. Why do human diversity levels vary at a megabase scale? Genome Res. 2005;15:1222–1231. doi: 10.1101/gr.3461105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmann I, Zollner S, Enard W, Ebersberger I, Nickel B, Paabo S. Selection on human genes as revealed by comparisons to chimpanzee cDNA. Genome Res. 2003;13:831–837. doi: 10.1101/gr.944903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang DG, Green P. Bayesian Markov chain Monte Carlo sequence analysis reveals varying neutral substitution patterns in mammalian evolution. Proc Natl Acad Sci USA. 2004;101:13994–14001. doi: 10.1073/pnas.0404142101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. Preponderance of synonymous changes as evidence for the neutral theory of molecular evolution. Nature. 1977;267:275–276. doi: 10.1038/267275a0. [DOI] [PubMed] [Google Scholar]
- Krawczak M, Ball EV, Cooper DN. Neighboring-nucleotide effects on the rates of germ-line single-base-pair substitution in human genes. Am J Hum Genet. 1998;63:474–488. doi: 10.1086/301965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Wu CI. Weak selection revealed by the whole-genome comparison of the X chromosome and autosomes of human and chimpanzee. Proc Natl Acad Sci USA. 2005;102:4063–4067. doi: 10.1073/pnas.0500436102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Hillier LD, Eichler EE, et al. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. (11 co-authors) [DOI] [PubMed] [Google Scholar]
- Miyata T, Yasunaga T. Molecular evolution of mRNA: a method for estimating evolutionary rates of synonymous and amino acid substitutions from homologous nucleotide sequences and its application. J Mol Evol. 1980;16:23–36. doi: 10.1007/BF01732067. [DOI] [PubMed] [Google Scholar]
- Nei M, Kumar S. Molecular evolution and phylogenetics. New York: Oxford University Press; 2000. [Google Scholar]
- Nekrutenko A, Chung WY, Li WH. An evolutionary approach reveals a high protein-coding capacity of the human genome. Trends Genet. 2003;19:306–310. doi: 10.1016/S0168-9525(03)00114-8. [DOI] [PubMed] [Google Scholar]
- Nielsen R, Bustamante C, Clark AG, et al. A scan for positively selected genes in the genomes of humans and chimpanzees. PLoS Biol. 2005;3:e170. doi: 10.1371/journal.pbio.0030170. (13 co-authors) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmley JL, Chamary JV, Hurst LD. Evidence for purifying selection against synonymous mutations in mammalian exonic splicing enhancers. Mol Biol Evol. 2006;23:301–309. doi: 10.1093/molbev/msj035. [DOI] [PubMed] [Google Scholar]
- Rollins RA, Haghighi F, Edwards JR, Das R, Zhang MQ, Ju J, Bestor TH. Large-scale structure of genomic methylation patterns. Genome Res. 2006;16:157–163. doi: 10.1101/gr.4362006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci USA. 2006;103:1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian S, Kumar S. Neutral substitutions occur at a faster rate in exons than in noncoding DNA in primate genomes. Genome Res. 2003;13:838–844. doi: 10.1101/gr.1152803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urrutia AO, Hurst LD. The signature of selection mediated by expression on human genes. Genome Res. 2003;13:2260–2264. doi: 10.1101/gr.641103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yampolsky LY, Kondrashov FA, Kondrashov AS. Distribution of the strength of selection against amino acid replacements in human proteins. Hum Mol Genet. 2005;14:3191–3201. doi: 10.1093/hmg/ddi350. [DOI] [PubMed] [Google Scholar]
- Yang Z, Bielawski JP. Statistical methods for detecting molecular adaptation. Trends Ecol Evol. 2000;15:496–503. doi: 10.1016/S0169-5347(00)01994-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Nielsen R. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol Biol Evol. 2002;19:908–917. doi: 10.1093/oxfordjournals.molbev.a004148. [DOI] [PubMed] [Google Scholar]