Abstract
Cancer genomes frequently contain somatic copy number alterations (SCNA) that can significantly perturb the expression level of affected genes and thus disrupt pathways controlling normal growth. In melanoma, many studies have focussed on the copy number and gene expression levels of the BRAF, PTEN and MITF genes, but little has been done to identify new genes using these parameters at the genome-wide scale. Using karyotyping, SNP and CGH arrays, and RNA-seq, we have identified SCNA affecting gene expression (‘SCNA-genes’) in seven human metastatic melanoma cell lines. We showed that the combination of these techniques is useful to identify candidate genes potentially involved in tumorigenesis. Since few of these alterations were recurrent across our samples, we used a protein network-guided approach to determine whether any pathways were enriched in SCNA-genes in one or more samples. From this unbiased genome-wide analysis, we identified 28 significantly enriched pathway modules. Comparison with two large, independent melanoma SCNA datasets showed less than 10% overlap at the individual gene level, but network-guided analysis revealed 66% shared pathways, including all but three of the pathways identified in our data. Frequently altered pathways included WNT, cadherin signalling, angiogenesis and melanogenesis. Additionally, our results emphasize the potential of the EPHA3 and FRS2 gene products, involved in angiogenesis and migration, as possible therapeutic targets in melanoma. Our study demonstrates the utility of network-guided approaches, for both large and small datasets, to identify pathways recurrently perturbed in cancer.
Introduction
Somatic copy number alterations (SCNA) are a recurrent characteristic of malignant cancers [1], [2], [3]. The amplification and subsequent over-expression or, conversely, deletion and loss of expression of key regulators of cell proliferation, senescence or death have been shown in many cases to contribute significantly to the progression from the normal to the malignant state [4], [5], [6], [7]. Therefore, the discovery and characterization of chromosomal regions involved in SCNA and of the genes encoded in them has been a crucial contributor to our understanding of the molecular mechanisms of carcinogenesis.
The methods used to detect and characterize SCNA have evolved significantly over the last decades. Initial cytogenetic observations have been supplemented with Southern blots and quantitative PCR. Almost twenty years ago, the availability of BAC clones delineating a tiling path through the entire human genome made it possible to detect SCNA in a genome-wide fashion, but with limited resolution [8]. More recently, oligonucleotide-based arrays have enabled comparative genome hybridizations (CGH) at high resolution, and CGH has become the method of choice to detect copy-number variations [9], [10], [11]. A recent SNP-based survey [1] of 3,131 copy-number profiles derived from over 26 different types of cancer has provided a dramatic illustration of the power of high-throughput techniques in distinguishing random alterations in the genome from those that may have a direct impact on tumorigenesis.
Genomic alterations in many tumors, especially at late stages in their development, are so extensive that the copy-number status of individual genes or chromosomal regions can vary over a very wide range of values. A mixture of chromosomal rearrangements and focal expansions can create genomic landscapes that are very difficult to analyze using standard CGH techniques. Moreover, the exact boundaries of SCNA or the expression status of the genes encoded within them are usually not known, precluding a thorough assessment of their impact on the phenotype of the cancer cells. It has recently been proposed that SNP arrays may be better suited for the determination of copy number states in tumor samples because the analysis of data derived from such arrays can make use of allelic imbalance information in addition to hybridization intensity [12].
In the present study, we analyzed the genome-wide copy-number status of seven highly aneuploid metastatic melanoma cell lines and determined the expression of their genes using a sequencing-based approach. We show that a combination of SNP-based and CGH arrays is necessary to obtain a reliable estimate of the true copy-number status of the entire genome in the face of extensive genomic instability, and that the combination of copy-number and expression status provides powerful clues as to the possible role of genes encoded within SCNA in tumorigenesis. Moreover, we show that a protein-based network-guided analysis of SCNA-affected genes with altered expression in our data and two published datasets [13], [14] identifies pathways commonly altered in melanoma.
Results
CGH and SNP arrays are required to comprehensively document somatic copy-number alterations in metastatic melanoma cell lines
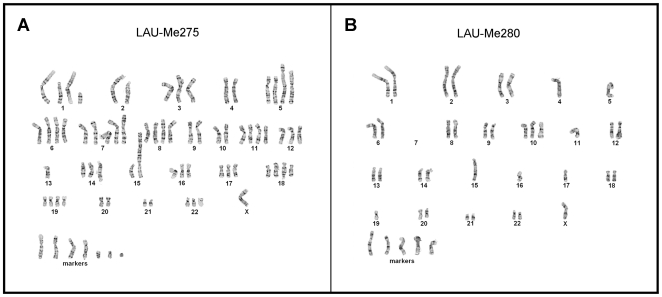
We analyzed seven low-passage melanoma cell lines that were established from metastases (see Table 1) together with matched controls from the same patients (see Materials and Methods). Karyotyping of the melanoma cell lines revealed extreme levels of aneuploidy. For example, LAU-Me280, the most extensively deleted line, had a per cell content of 34 to 42 chromosomes (median: 40), whereas LAU-Me275, one of the most amplified melanomas, harbored 68 to 81 chromosomes (median 73.5) (see Fig. 1). Additionally, the presence of many unassigned chromosomal fragments (markers) made it difficult to determine the true level of aneuploidy.
Table 1. Melanoma cell lines.
| Melanoma | Site | BRAF mutation | Number of chromosomes (karyotype) |
| LAU-Me280.R.LN | Lymph node | G593M, L597R | 34–42 |
| LAU-Me246.M1 | Skin | V600E | 45–82 |
| LAU-T618A | Skin | wt but NRAS mutation (Q61R) | 55–71 |
| LAU-T50B | Skin | V600E | 65–71 |
| LAU-T149D | Visceral | V600E | 68–81 |
| LAU-Me275 | Lymph node | V600E | 68–81 |
| LAU-Me235 | Skin | K601E | 73–103 |
Figure 1. Karyotypes of two malignant melanomas.
Representative karyotype (Giemsa stain) for LAU-Me275, one of the most hyperploid melanoma (here 76 chromosomes including 7 markers); and LAU-Me280, the most extensively deleted line (42 chromosomes including 5 markers).
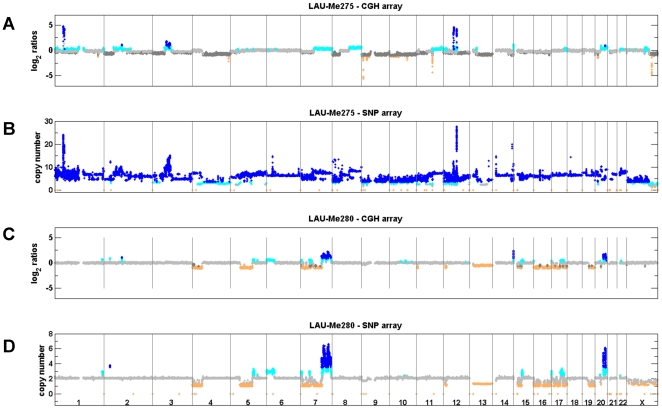
In initial CGH (Agilent 244k) experiments we observed that in LAU-Me275, and other highly hyperploid cell lines, the hybridization ratios between cancer cells and matched controls did not reflect the chromosome-wide aberrations observed in the karyotypes. For example tetraploid regions were measured as triploid or less by the CGH arrays (see Fig. S1). We considered whether this was due to the normalization protocol and subsequent segmentation analysis. Using technical replicates of LAU-Me275 DNA, we tested three independent normalization schemes, two of which were specifically developed for cancer genome analysis (see Materials and Methods) and found that the methodology proposed by Chen and colleagues [15] was the most reproducible (Spearman correlation 0.96; see Fig. S2). We then partitioned the genome into regions reflecting copy number changes and assigned copy number using two independent classification methods (see Materials and Methods). Since neither of these classification methods gave entirely satisfactory results (see Fig. S4 and Methods S1), we developed a Gaussian Mixture Model (GMM) approach that was highly reproducible based on a technical replicate analysis (Spearman correlation 0.9).
The GMM method found only 42 regions in the LAU-Me275 genome that were amplified (CN≥4; see Table 2). This number was less than expected based on the karyotype analysis, which documented a high number of arm-level chromosome amplifications (see Fig. 1). Thus, while CGH-based methods are well adapted to document differences in copy number status between the genomes of normal cells derived from different individuals, our results clearly show that they are inadequate to deal with the large-scale rearrangements and amplifications typical of hyperploid cancer cells. The most likely reason is that the total DNA content of cancer cells is too different from that of normal cells to allow a robust experimental normalization. Given this limitation, we asked whether SNP arrays might be better suited to detect chromosome-wide changes in a highly amplified genome.
Table 2. Number of genes affected by SCNA in seven melanoma cell lines.
| CGH arrays | LAU-Me280 | LAU-Me246 | LAU-T618A | LAU-T50B | LAU-T149D | LAU-Me275 | LAU-Me235 | Unique gene count |
| Deletion | 3668 | 4281 | 986 | 3656 | 108 | 122 | 1059 | 10711 |
| Arm-level amplification | 222 | 0 | 549 | 99 | 998 | 42 | 0 | 1884 |
| Focal amplification | 0 | 0 | 0 | 26 | 379 | 0 | 4 | 409 |
| SNP arrays | LAU-Me280 | LAU-Me246 | LAU-T618A | LAU-T50B | LAU-T149D | LAU-Me275 | LAU-Me235 | Unique gene count |
| Deletion | 2294 | 3157 | 2 | 113 | 70 | 2 | 39 | 5544 |
| Arm-level amplification | 0 | 0 | 16584 | 1033 | 3477 | 16398 | 10384 | 19496 |
| Focal amplification | 213 | 0 | 978 | 438 | 894 | 1853 | 161 | 4055 |
Number of genes affected by somatic deletions, arm-level amplifications (≥4 copies but <1 copy above the chromosome arm baseline) and focal amplifications (≥4 copies and ≥1 copy above the chromosome arm baseline), as measured using SNP or CGH arrays.
We hybridized DNA from LAU-Me275 to Illumina 1M SNP arrays and analyzed the signals using the OverUnder algorithm [16], which uses minor allele frequencies in heterozygous loci to improve copy number estimation. These results correlated well (Spearman correlation 0.77) with a technical replicate analyzed on the Affymetrix SNP platform (see Fig. S5C and S5D), and indicated that 18,251 genes in the LAU-Me275 genome had a copy number of at least four (see Table 2). Within this group, 132 genes had undergone focal amplifications of at least 10-fold. These SNP-based results were more consistent with the karyotype observations. For example, CGH had predicted two copies of chromosome 7p (Chr7p) and three copies of Chr7q (see Fig. 2), while the SNP results indicated three and five copies, respectively (see Fig. 2 and 3), which was more consistent with the cytogenetic data (see Fig. 1).
Figure 2. Copy number analysis using CGH and SNP arrays.
A. and B. shows the analysis of LAU-Me275 on CGH and SNP arrays. C. and D. shows results for LAU-Me280. Probe/SNP are plotted as a function of their genomic position on the X axis. Y axis for CGH arrays corresponds to hybridization ratios. Y axis for SNP arrays corresponds to the predicted copy number. Colors indicate a copy number state (orange<2 copies; gray = 2 copies; cyan = 3 copies; dark blue>3 copies). Dark gray in the CGH panels indicates regions identified as diploid in the analysis, but where the karyotype analysis indicated copy neutral or deleted states, possibly due to cell heterogeneity.
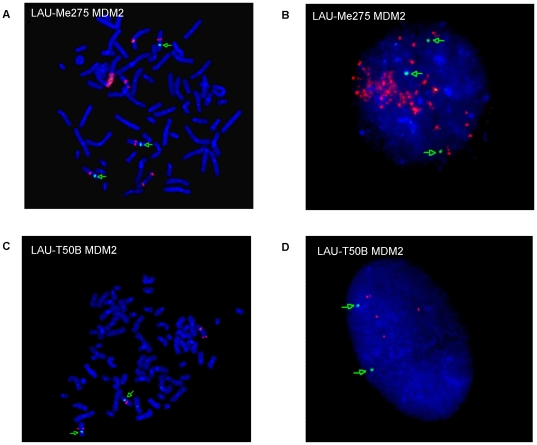
Figure 3. Determination of MDM2 copy number by FISH.
The MDM2 gene was assayed in two melanoma samples (LAU-Me275 and LAU-T50B) derived from the same patient. Panels A and C show a metaphase and B and D an interphase. MDM2 probe is in red; centromere-specific probe is in green. FISH shows amplification for both LAU-Me275 (more than eight copies) and LAU-T50B (four copies). Metaphase-FISH helps to identify homogeneously staining regions and Interphase-FISH to estimate the copy number.
Therefore, based on our findings with LAU-Me275, we determined the SCNA in the six other melanoma cell lines using both CGH (Agilent 244k) and SNP (Illumina 1M) array platforms. Identification of genes within amplifications or deletions was determined as described in Materials and Methods, and the number of SCNA for each cell line is given in Table 2. In all cases CGH predicted more deletions than did SNP arrays, agreeing with our initial observations using LAU-Me275. Also, with the exception of the LAU-Me280 cell line, amplifications were better predicted by SNP arrays. This bias is evident in a graphical representation of the intersection between CGH and SNP predictions (see Fig. S7) and with CGH/SNP genome-wide copy number profiles (see Fig. S8). These results confirmed our conclusion that CGH is more suitable for detecting deletions while SNP arrays are better for identifying amplifications.
Few SCNA-genes are recurrent in different melanoma cell lines
A potential problem with SCNA studies performed in isolation is that they cannot assess the expression status of the genes contained within the altered genomic regions. Amplified genes are not necessarily highly expressed, and the exact boundaries of deletions may or may not encompass a gene of interest. We reasoned that the combination of precise copy number determination and gene expression measurement would allow us to highlight with much higher confidence those genes whose expression is affected by SCNA (SCNA-genes) in the melanoma cell lines. We therefore analyzed gene expression in each of the melanoma cell lines by RNA-seq using the Roche/454 pyrosequencing method. Additionally, we performed RNA-seq on a pool of epidermal melanocytes to determine a reference level of expression for each gene in normal melanocytic cells (see Materials and Methods).
We first looked for genes within focal amplifications with at least two-fold over-expression relative to the reference melanocytes (see Table S1). Only KIAA0090, a protein coding gene of unknown function not previously associated with cancer, was affected in three melanomas (see Table S2). A further 56 genes were altered in two melanomas, but the only known cancer-related gene was MDM2, an oncogene previously demonstrated to be amplified in sarcoma, glioma, colorectal and other cancers including melanoma [5], [17]. In LAU-Me275, MDM2 was 3.9-fold over-expressed relative to melanocytes and had a copy number greater than ten (as predicted by SNP array). By contrast, in LAU-T50B, MDM2 was predicted by SNP array to be diploid (CN = 2), and by CGH to be duplicated (CN = 3). This potential difference between these cell lines is intriguing because they were derived from metastases surgically removed from the same patient at a 12 year interval. We therefore determined the copy number status of the MDM2 gene in these two samples using fluorescent in situ hybridization (FISH). In LAU-Me275, the fluorescence signal indicated that at least 8 MDM2 copies were present at the locus on Chr12 in addition to a homogeneously staining region on Chr5 (see Fig. 3A and 3B) which is in agreement with results from the SNP arrays. In LAU-T50B, FISH revealed a total of four MDM2 copies, two on Chr12 and two located on an unidentified chromosome (see Fig. 3C and 3D), which is higher than the copy number estimated by CGH and SNP arrays. Re-investigation of the raw SNP data for LAU-T50B showed that there was indeed a small amplification signal at MDM2, but this had not been detected using our optimization parameters (see Methods S1). This highlights the challenge of determining optimal parameters that are usable on a genome-wide scale for all samples in a study.
We next derived a list of genes within deletions detected by CGH that were expressed in melanocytes but not in the melanoma cell lines (see Table S1). We reasoned that such genes are likely to be enriched for melanocyte functions that have been lost during tumorigenesis. The vast majority of such genes (554) were private to a single melanoma sample; seventy genes were shared by two samples; and only ten genes were shared by three melanomas: ADAMTSL1, ARMC4, DLL1, HSD17B3, LOC441177, OSTCL, PARK2, PLXDC2, SLC24A2 and ULBP3 (see Table S2).
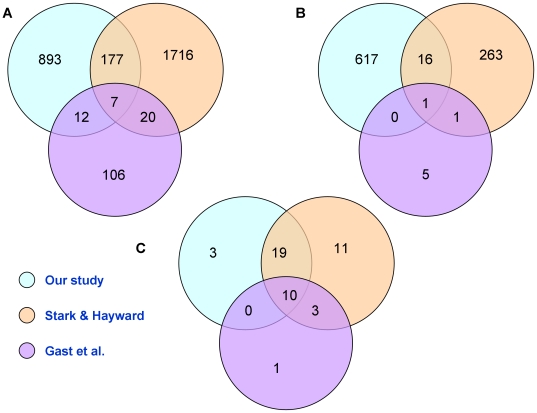
Altogether, we identified a total of 1,710 SCNA-genes affected by amplification or deletion (summarized in Table S2; complete dataset in Table S3). To determine the relevance to melanoma of this set of altered genes from our small sample set, we compared it to gene lists in two published studies that used larger melanoma collections [13], [14]. These two studies provided a list of genes recurrently affected by amplification or deletion in 76 (primary and metastatic) and 60 (metastatic) melanoma samples, respectively. Only 196 of our 1,089 amplified genes (p-value<0.001, see Fig. 4A), and 17 of our 634 genes within deletions (p-value<0.007, see Fig. 4B) were present in the Stark and Hayward or Gast et al. datasets. Surprisingly, the number of genes common to the two published gene sets was also small (27 amplified genes and 2 genes within deletions; p-values<0.005), and demonstrates the difficulty to identify commonly affected genes relevant to tumor progression even within larger melanoma collections.
Figure 4. Intersection between our dataset and two published datasets of SCNA-genes and derived pathways.
A. Intersection between amplified genes in published melanoma datasets (Stark and Hayward 2007; Gast et al., 2010) and our list of over-expressed genes within focal amplifications B. Intersection between genes within homozygous deletions from the Stark and Hayward and Gast et al. datasets and our list of non-expressed genes within deletions C. Intersection between pathways found significantly affected by SCNAs from our analysis of the three datasets.
Correlation between mRNA and protein levels
A potential problem with assessing gene expression by mRNA quantitation is that this may not reflect cellular protein levels. To check whether SCNA-genes were also expressed at the protein level in the melanoma cell lines, we performed a SILAC proteomic analysis (stable isotope labeling with amino acids in cell culture [18], [19]). This technique is used to quantify relative protein amounts as measured by mass spectrometry in extracts from cells grown in unlabeled (light) medium and cells grown in (heavy) medium containing non-radioactively-labeled amino acids. Since the normal human melanocytes did not grow well in the labeling media, heavy isotopes were incorporated into the Lau-Me275 cell line, and protein quantification in all cases was relative to this sample. After stringent data filtering (see Materials and Methods), we investigated whether transcript levels reflected protein levels as detected by SILAC. As shown in Fig. S11 there was a moderate but significant correlation between global mRNA and protein levels in all samples (mean Spearman correlation 0.62).
Pathways significantly enriched in SCNA-genes are recurrent in melanoma
As described above, we found that very few of the SCNA-genes were altered in more than one melanoma cell line, which is not unexpected given the small number of samples in our dataset. An idea popular in the current literature is that signaling pathways, rather than individual genes, are recurrently perturbed in cancer [20]. To determine whether the SCNA-genes from different melanoma cell lines shared membership of one or more cellular pathways, we investigated whether the proteins encoded by the SCNA-genes were connected in known human protein interaction networks (see Materials and Methods). Out of a total of 1,563 proteins analyzed, 377 (24%) were connected within the network, and clustering of the proteins based on the topology of the sub-networks identified 14 protein networks, or ‘clusters’, containing at least five significantly connected members. For each cluster, we identified genes belonging to known signaling and metabolic pathways including nine clusters that significantly overlapped known pathways (FDR≤0.05; listed in Table S4). Following detailed manual annotation, the resulting pathways were ranked according to the number of contributing melanomas and to the number of SCNA-genes involved in the pathway. The pathways common to at least four melanoma samples are shown in Table 3 (for the complete list see Table S4). Interestingly, the vast majority of the recurrent pathways we identified involve signal transduction and have been implicated in one or more cancer types. In addition, that we identified ten pathways common to at least five of the melanoma samples confirms the idea that protein network-guided analysis is a good method for detecting recurrently affected pathways in small datasets.
Table 3. Pathways identified by network-guided analysis.
| Pathway | #Melanomas | Genes | #genes |
| G protein signaling | 6 | ADORA1, ADRA2A, CHRM1, CHRM5, DRD2, GNAO1, GNB3, GNG4, HTR1F, OPRL1, PLCB2, RGS10, RGS11, RGS14, RGS19 | 15 |
| WNT signaling (includes Apoptosis and Hedgehog signaling) | 6 | CDH19, CDH2, CDH4, DVL1, FRAT1, FZD8, PCDH17, PCDH9, SFRP1, WNT11, WNT16, WNT2B, WNT4, WNT5B | 14 |
| Cadherin signaling | 6 | ACTG2, CDH19, CDH2, CDH4, FZD8, PCDH17, PCDH9, WNT11, WNT16, WNT2B, WNT4, WNT5B | 12 |
| Melanogenesis | 6 | CAMK2A, CAMK2G, DVL1, FZD8, NRAS, WNT11, WNT16, WNT2B, WNT4, WNT5B | 10 |
| Angiogenesis | 5 | BRAF, DVL1, EFNB2, EPHA3, EPHB2, FGF1, FRS2, NRAS, PIK3R3, PRKCZ, SFRP1, WNT2B, WNT5B | 13 |
| Axon guidance (migration and adhesion) | 5 | CDK5, EFNB2, EPHA3, EPHB2, EPHB6, FES, NRAS | 7 |
| MAPK signaling | 5 | DUSP1, DUSP12, DUSP2, FGF1, FGF14, FGFR4, MAPK9 | 7 |
| TGF beta signaling | 5 | ACVRL1, AMHR2, FOXH1, LEFTY1, SMAD9, TGFB1, TLL2 | 7 |
| Alzheimer disease | 5 | CHRM1, CHRM5, PKN3, PRKCZ | 4 |
| FGF signaling | 5 | FGF1, FGF14, FGFR4, FRS2 | 4 |
| Calcium signalling | 4 | CAMK2A, CAMK2G, CHRM1, CHRM5, GNAO1, GRIN2C, PRKCZ, RGS10, RGS11, RGS14, RGS19 | 11 |
| Huntington_disease (vesicle-mediated transport) | 4 | ACTG2, CLTB, GRIN2B, GRIN2C, GRIN3A, KALRN | 6 |
| Neuroreceptor (Muscarinic, Metabotropic) | 4 | GRIN2B, GRIN2C, GRIN3A, KCNQ2, PKN3, PRKCZ | 6 |
| Cell cycle (G1 progression) | 4 | CCNA1, CDC20, CDC26, CDKN2B, HDAC1 | 5 |
In our search for genes recurrently affected by SCNA, we found only ∼10% overlap between our list of SCNA-genes and those derived from studies with much larger sample sizes [13], [14]. To determine if this was also true at the level of pathways, we performed a protein network-guided analysis as described above on each of these datasets using the published gene lists (neither study originally presented this type of analysis). Detailed annotation and comparison of the results for each dataset is given in Table S4. In contrast to what we had found at the gene level, all but three of our pathway modules were also present among those identified from one or both of the published gene datasets (see Fig. 4C). Ten pathway modules (angiogenesis, EGF, ERBB, integrin signaling, long term potentiation, MAPK, natural killer cell mediated toxicity, PDGF, regulation of actin cytoskeleton and VEGF) were common to all three datasets, and the combined overlap with our pathway dataset was 66%. Thus, the majority of pathways defined by SCNA affected genes in our melanoma samples were recurrent in the three datasets, whereas the individual genes were not.
An additional benefit of the protein network-guided approach is that it generates a list of genes affected by SCNA that contributed significantly to a given pathway (see Table S4). Although two-thirds of the pathways were common between our dataset and the published datasets, only two genes, NRAS and BRAF, were present in all three (see Table S5). Of the genes shared by two datasets, four were components of the angiogenesis pathway, including EPHA3 and FRS2. We noted also that several members of the WNT (WNT3A, 4, 5B, 7A, 9A, 11, 16) or cadherin (CDH2, 4, 9, 12, 17, 18, 19) gene families were affected by SCNA in only one dataset, further reinforcing the idea that different genes can potentially alter the same pathway (WNT or cadherin) in different melanoma samples.
Discussion
Our goal to identify somatic copy number aberrations in metastatic melanoma cell lines revealed extreme levels of aneuploidy characteristic of this cancer type [21], [22], and complicated the application of standard CGH array protocols [9], [10], [11]. Nevertheless, using our GMM method we were able to demonstrate that although CGH arrays fail to identify all large-scale amplifications, they are able to detect deletions very efficiently, including genes having lost expression compared to melanocytes (see Table 2 and Fig. S7). Conversely, SNP arrays, which measure hybridization intensities for both alleles at heterozygous loci, allow the consideration of an additional parameter (the so-called B-allele frequency) and greatly improve the measurement of DNA copies beyond the normal diploid complement (as implemented in the OverUnder algorithm, Attiyeh et al., 2009; see Fig. S6). We did notice, however, that this algorithm systematically detected deletions located in sub-telomeric regions for both tumors and controls, which indicates a systematic bias and suggests that the algorithm is optimized to detect duplications and amplifications but not deletions. Therefore, it can be argued that CGH and SNP techniques should be combined to obtain a reliable assessment of all copy number states from deletion to high-level focal amplification.
To enrich for genes that might be involved in the oncogenic process, we focused on two groups: focally amplified genes that were over-expressed relative to melanocytes; and deleted genes with no expression in the melanoma cell lines, but that were expressed in normal melanocytes. In the first group, MDM2 [5], [17] was the only cancer gene amplified and over-expressed in more than one melanoma sample. Comparison of genes amplified in our samples with published gene lists from two large melanoma studies (Stark and Hayward 2007; Gast et al., 2010) while revealing very little overlap (see Fig. 4) did identify BRAF, MDM2, and NRAS, genes known to be important in melanoma [5], [17], [23], [24], [25], [26], [27], [28], [29]. In the second group, ten genes were deleted in three of the melanoma samples (see Table S2). These genes are located on Chr6q25, Chr6q27, Chr9 or Chr10p, consistent with previous observations that both arms of chromosomes 9 and 10 and Chr6q frequently undergo hemizygous deletion or copy neutral LOH in melanoma [14]. Of the ten genes, the Parkinson's disease-associated gene PARK2 has been recently described as a tumor suppressor gene in glioblastoma and other malignancies [30], while DLL1, HSD17B3 and ULBP have been reported to be associated with cancer, although not as tumor suppressors [31], [32], [33], [34], [35], [36], [37]. Experimental investigation will be required to determine if any of these ten genes performs an anti-oncogenic function in melanoma cells. The only deleted gene common to our study and those of Stark and Hayward and Gast et al. was PTEN, a tumor suppressor gene already known to be deleted in melanoma [7], [38].
In an alternative approach to detect recurrent events in these samples, we used a protein network-guided analysis [20], [39], [40], [41], [42], [43] to identify pathways affected by SCNA-genes in the seven melanoma cell lines. In contrast to the low level of recurrence in these melanoma samples at the individual gene level, we found that six pathways were shared by five of the samples, and four pathways (G protein, WNT, cadherin signaling and melanogenesis) were common to six (see Table 3). Several of these pathways are highly relevant to melanoma (e.g. MAPK, cadherin and FGF signaling) and have also emerged from cDNA expression studies [44], lending support to our results. G proteins transduce signals from G protein-coupled receptors (GPCRs), the largest family of membrane receptors involved in signal transduction, and whose over-expression in tumors can contribute to tumor progression, angiogenesis and metastasis [45]. Alteration of G proteins could impact the activities of GPCRs key to melanocytic cells, such as MC1R (melanocortin receptor), chemokine (e.g. CXCR2), and endothelin receptors. [46]. The recent identification of activating mutations in two G protein alpha subunits, GNAQ and GNA11, in a large proportion of uveal melanomas [47], [48], further underscores the relevance of this class of proteins to melanoma.
Although annotated as distinct pathways, WNT, cadherin signaling and melanogenesis shared six SCNA-genes in common (FZD8 and several members of the WNT family). This may reflect interactions between these pathways, an interplay between the WNT and cadherin pathways is known to exist [49], or may be a consequence of poor pathway annotation. The cadherin pathway controls cell-adhesion and plays a role in invasion and metastasis [50]. WNT (and Hedgehog) control development and growth in the embryo; aberrant activation of their transcriptional components ultimately affects cell fate, proliferation, and migration [51], [52], [53]. The only common non-signaling pathway was melanogenesis. Melanoma develops from melanocytes, cells highly specialized in the synthesis of melanin pigment, a process that requires a complex enzymatic machinery and unique organelle structures [54]. Our pathway analysis predicted that melanoma SCNA affect melanogenesis. Loss of pigmentation in metastases compared to primary tumors is commonly observed in cutaneous melanoma, and although not completely understood, it can be brought about by different mechanisms, such as premature degradation of melanogenic proteins [55] or downregulation of MITF transcription program [56]. Our study suggests that SCNA may also contribute to these alterations. An unexpected pathway that emerged from our analysis, and perhaps merits further exploration, is neurotransmission. These results suggest an involvement of neuronal pathways in melanoma, possibly related to the neural crest origin of melanocytes. Lending support to this hypothesis, the metabotropic glutamate receptor GRM1 has recently been implicated in the development of spontaneous melanoma in a mouse model, and an autocrine glutamate/GRM1 loop has been described in human melanoma [57].
Comparison of the pathways generated from SCNA-genes in our data and genes affected by copy number changes in two published datasets (Stark and Hayward [14] and Gast et al. [13]) revealed a high level of overlap, much higher than we expected based on the number of commonly affected genes (see Fig. 4). An explanation for this outcome is that different genes within the same pathway are affected in different datasets, and the commonalities are apparent only at the pathway level. The number of affected genes in a given pathway would be expected to increase with increasing sample size, and this is largely the case between our data and those of Stark and Hayward, but not in the Gast et al dataset (see Table S4). The reason for the low number of SCNA affected genes and corresponding pathways in the latter case may be the high stringency criteria employed in their analysis [13].
The angiogenesis pathway was one of ten common to all three datasets. Its up-regulation is a well-known hallmark of cancer [58], and it has long been proposed as a target for therapeutic treatment [59], [60]. Activation signals for angiogenesis include vascular endothelial growth factor (VEGF) and acidic fibroblast growth factor (FGF), and both were in our list of significantly affected pathways (see Table 3) and within our analysis of the Stark and Hayward (VEGF and FGF) and Gast et al (VEGF) datasets (see Table S4). Two genes in this pathway, EPHA3 and FRS2, were designated SCNA-genes in both our dataset and in Stark and Hayward, and were annotated as amplified, in skin-derived tumors, in the Cancer Genome Project dataset [5], [61].
In our analysis EPHA3, an ephrin tyrosine kinase receptor, was both focally amplified and over-expressed only in LAU-Me275. However, EPHA3 was highly over-expressed in LAU-T149D and LAU-Me246 (see Table S3) and amplified in LAU-T618A (CN = 6.4), LAU-Me235 (CN = 4) and LAU-T50B (CN = 4.2). EPHA3 is recurrently mutated in adenocarcinoma [62], [63] and has been implicated in renal carcinoma, glioblastoma, colorectal, breast and lung cancer [63], [64], [65], [66], [67]. Mutations in EPHA3 have been detected in melanoma [68], and several ephrin-derived peptide antigens (from EPHA2, EPHA3 and EPHB6) can be recognized by cancer-specific cytotoxic T-cells [69], . In addition, the feasibility of specific EPHA3 targeting has been reported [73]. These observations indicate that EPHA3 might be a promising target for therapeutic treatment in melanoma and other cancers.
FRS2, fibroblast growth factor receptor substrate 2, is an adaptor that acts downstream of a limited number of receptor tyrosine kinases, in particular FGF and neurotrophin receptors, RET and ALK, and plays a major role in tumorigenesis [74]. Dey and coworkers [75] recently targeted the FGF receptors (FGFR) using tyrosine kinase inhibitors to decrease the activity of AKT and ERK kinases, inducing apoptosis in breast cancer cell lines. FGFR inhibition is highly relevant to melanoma, where autocrine stimulation via FGF2/FGFR1 constitutes a pivotal role in proliferation and survival [76]. FRS2 has been suggested as a therapeutic target in cancer [77] and because of its downstream activities to FGFR and other receptors, it might offer new insights in melanoma treatment. In our data FRS2 was both focally amplified and over expressed in two melanoma samples (LAU-T149D and LAU-Me275) and amplified (CN = 4) in two additional melanomas (LAU-T618A and LAU-Me235). Inspection of its amplification status in larger melanoma collections would be useful to confirm its potential role as a target of interest in melanoma.
Using SILAC, we demonstrated a global correlation between mRNA and protein levels across all samples. However, we are aware that the levels of individual proteins may not always reflect mRNA levels, and that the activity of certain proteins, and therefore the activation state of cellular pathways, can be further modulated by post-translational modifications. Further investigations, beyond the scope of the current study, will be required to address these possibilities. Such studies should include a functional assessment of the implicated pathways using various manipulations to activate or inhibit them (e.g. with siRNA or other inhibitors) to determine the role of the pathways in these melanoma cell lines.
In conclusion, we have identified SCNA-genes and pathways potentially altered in our metastatic melanoma samples and two published datasets (Stark and Hayward 2007; Gast et al., 2010) which should be investigated by screening larger tumor collections and in functional studies. Two SCNA-genes, EPHA3 and FRS2, emerged from our analysis as potential therapeutic targets. These genes were replicated in our analysis of the two published melanoma collections, have been extensively studied in other cancer types, and thus might offer new insights in the treatment of malignant melanoma.
Materials and Methods
Melanoma samples, DNA and RNA extraction
Melanoma cell lines were established from metastases from patients with cutaneous melanoma and were used at low passage (<10). Donor matched cells were either peripheral blood lymphocytes (PBL) or Epstein-Barr virus transformed lymphoblastoid (EBV) cell lines. EBV cell lines were karyotyped to ensure genome stability and diploidy. Approval to use these samples for this project was given by the CHUV (Centre Hospitalier Universitaire Vaudois) ethical committee for clinical research. Melanoma cell lines were cultured conditions in RPMI-1640 medium supplemented with 10% fetal calf serum (FCS), and no antibiotics. Human foreskin melanocytes were grown in HAM-F10 medium supplemented with 2% FCS, 5% MelanoMax supplement (Gentaur, Belgium) and 6 mM HEPES. EBV cell lines were cultured in IMDM/10% FCS medium. All cultures were without mycoplasma. DNA (Gentra kit, Qiagen) and RNA (guanidinium/cesium chloride gradient) isolation and karyotype preparations were performed from parallel cultures.
Cytogenetic and FISH analysis
Cytogenetic (GTG-banding) and fluorescence in situ hybridization (FISH) metaphase analyses of melanoma cell lines were performed using standard protocols. We performed 15 spreads for each melanoma cell line, except for Lau-Me246 [14] and Lau-Me275 [21]. Dual color FISH was done using a commercially available set consisting of a locus-specific MDM2 combined with a chromosome 12 centromeric probe (Kreatech Poseidon FISH probe) to distinguish aneuploidy of chromosome 12 and specific locus loss or gain. Chromosomes with homogeneously staining region (HSR) were identified with the analysis of FISH metaphases in inverted digital images. Copy number estimation of HSR was done using FISH interphases.
Comparative genomic arrays (CGH)
CGH arrays were processed according to the manufacturer's protocol (Agilent Technologies, Inc.) and as described in Martinet et al. [78].
The normalization and detection of copy number aberration is detailed in Methods S1. In brief, signal intensities were normalized using three independent normalization schemes: Loess [79]; PopLowess [80]; and the statistical framework from Chen et al. [15].
Then probe-level data were segmented using Circular Binary Segmentation [81], [82] and attributed a discrete copy number to segments using three independent methods: 1) a naive scoring-based approach, where outliers relative to the chromosomal baseline are detected using a non-parametric score; 2) the MergeLevels method [83] and 3) our own classification algorithm based on Gaussian Mixture Model which models the observed distribution of intensity ratios as a combination of Gaussian distributions that can be subsequently classified into deletion (CN<2), copy neutral event (CN = 2), duplication and amplification (CN = 3 and CN≥4) (see Fig. S3).
Single Nucleotide polymorphism arrays (SNP)
Illumina 1M SNP arrays
Genomic DNA from each of the 7 melanoma and their matched normal cells (either EBV cell line or PBL); as well as two control melanocytes were genotyped on the Illumina Infinium Human1M-Duo arrays. Aliquots of DNA (30 µl at 50 ng/µl) for each sample were processed according to the manufacturer's protocol (Infinium HD Gemini protocol). Subsequently we used the OverUnder algorithm [16] to correct the hybridization log ratios for polyploidy and to attribute a continuous copy number value to each SNP. We estimated that the window size parameter set to 201 SNPs, gave the highest reproducibility between technical replicates (see Fig. S5A and S5B).
Affymetrix 6.0 arrays
As part of the technical replicate design, we analyzed LAU-Me275 on Affymetrix 6.0 SNP arrays. The experiment was performed in accordance with the manufacturer's instructions. Normalization and copy number prediction were done using the PICNIC algorithm [84].
Transcriptome sequencing
The transcriptome from all seven melanoma as well as a pool of two melanocytes was sequenced using the Roche 454 Titanium technology. mRNA isolation and cDNA preparation were performed following the protocol used by Bainbridge et al. [85], with some modifications (See Methods S1). 3–5 µg of cDNA were used for 454 libraries preparation, according to manufacturer's protocol. All experiments produced about 1M single end reads, with a median length of 367 nucleotides (interquartile range 265–436). We derived transcript tag counts using our own published methodology [86] (see also Methods S1). Using tag counts from the pool of melanocytes, we were able to derive a ratio of expression for each melanoma with respect to these control melanocytes.
Detection of somatic copy number alterations with altered expression (SCNA-genes)
We computed the median copy number at each Refseq gene. To overcome density limitations, we included SNPs that were within 2 kb of the gene boundaries. For CGH arrays, we included probes within 3 kb of the gene boundaries. We defined SCNA-genes as follows. A gene was flagged as within a focal amplification when its CN, as computed from SNP arrays, was ≥4, the difference in CN relative to the chromosomal arm was ≥1, the gene was diploid (CN = 2) in the matched control cell line, and the expression in the melanoma cell line was at least 2-fold greater than that in the control melanocytes. For deletions, a gene needed to have CN<2, as detected by CGH, without expression in the melanoma cell line and CN = 2 with detected expression in the melanocytes.
Protein network-guided analysis of SCNA
A non-redundant human protein interaction network was generated by combining iRefseq [87] and Pathway Commons [88] protein interaction databases with functional interactions from Panther pathways [89]. The resulting network has 21,876 nodes and 376,528 edges and combines interaction data from 15 primary protein interaction databases (BIND, BioGRID, CORUM, DIP, HPRD, IntAct, MINT, MPact, MPPI, OPHID, Reactome, HumanCyc, Cancer Cell Map, IMID and NCI/Nature pathway interaction database). Mapping between gene names and their protein Uniprot IDs was downloaded from the HGNC website (www.genenames.org). Among the 1710 (unique) SCNA-genes, 104 did not have a UniProt ID and 43 could not be mapped onto the network. Using a walk trap community algorithm and permutation approaches (with n = 1000), we were able to extract clusters of proteins from the network (Details are available in Methods S1).
Pathway analysis
Significance of overlap between the modules and pathways from Panther, Kegg and MSigDB [89], [90], [91] was calculated with an hypergeometric test, P-values were corrected for multiple comparisons by calculating the false discovery rate using the Benjamini and Hochberg procedure [92].
To reduce the redundancy present in pathway database annotation, we first combined pathways that contained the same SCNA-genes from the same melanoma cell lines, then we reviewed the pathway list to either remove redundancies or un-merge unrelated pathways affected by similar genes. We also excluded KEGG “cancer” annotated pathways.
Proteomic analysis
SILAC
Cell growth conditions and labeling are detailed in Methods S1. LC-MS/MS data were analyzed using Mascot 2.2 (Matrix Science, London, UK) for database search and MaxQuant 1.0.13.13 for peak list export from raw data and quantitative analysis [93]. Database search was carried out against the IPI human database (version 3.52, [94]) including decoy sequences and a list of common contaminants. A maximum false discovery rate of 1% was used for protein identification.
A total of 5522 proteins were detected by the SILAC experiment. We excluded 55 proteins that matched the list of common contaminants. To check whether culture condition (with the heavy isotope) could influence the protein quantification, we performed a self-self experiment with LAU-Me275. Although no significant bias was found (see Fig. S9), we excluded 79 proteins that were outliers in this experiment. Next, we checked the number of peptides detected per protein (see Fig. S10). Since protein quantification is not accurate when only few peptides are observed, we excluded 2,134 proteins that had less than three peptides and 42 with missing values in all seven experiments. This led to a cleaned dataset of 3,212 proteins; 2,922 of them could be remapped to Refseq genes using their Uniprot ID and were used for subsequent comparison with RNA-seq data.
Comparison with RNA-seq data
For comparison, we normalized the RNA seq tag count in the same manner as the SILAC data (i.e. we expressed the transcript tag count from LAU-Me275 with respect to all other samples, including the control melanocytes). Next, we computed the Spearman correlation coefficient between the expression and protein log2 ratios (see Fig. S11). P values to test for significantly positive correlation were estimated using large-sample approximations (as implemented by the ‘corr’ function in MATLAB).
Accession number
Microarray and sequencing data were deposited in NCBI GEO and are available under accession number GSE23056.
Supporting Information
CGH hybridization ratio in a tetraploid region in LAU-Me275. Each plot shows the hybridization log2 ratio at each CGH probe (in gray) obtained using three normalization methods. Ridge refers to the framework from Chen et al. Red segments were obtained using Circular Binary Segmentation. Karyotype analysis of LAU-Me275 revealed 11q amplification (CN≥4), so the expected CGH log ratio would be two. Here the ratios obtained from three different normalizations failed to reflect the amplification (both Loess and Ridge were close to 0; PopLowess was close to 0.6 indicating 3 copies).
(DOC)
Correlation between replicates using different normalization schemes. The scatter plots illustrate the correlation at each CGH probe between two replicates. The heatmaps show the correlation for each pairs of replicates. The normalization method is indicated in each plot title. RIDGE refers to the framework from Chen et al.
(DOC)
Gaussian Mixtures identified in four replicates from LAU-Me275. Each histogram shows the distribution of CBS segment log2 ratios, colors highlight the Gaussian components. The number of components identified and the Bayes Information Criterion are indicated in each figure title.
(DOC)
Comparison of CNV detection algorithm on CGH data. Panels are, from left to right: a melanoma with large deletions (LAU-Me280); a melanoma with large amplifications (LAU-Me275); and a control EBV cell line (male) hybridized using a pool of female references. From top to bottom: CNV classification (following CBS segmentation) using 1) Gaussian Mixture Model (GMM), 2) MergeLevels, 3) the scoring-based approach. Each dot corresponds to a CGH probe with its genomic position on the X axis and its log2 ratio of hybridization on the Y axis. Colors indicate the copy number state: orange< = 1 copy gray = 2 copies, cyan = 3 copies and dark blue more than 3 copies. For the scoring approach distinction is made between 1 copy (orange) and 0 copy (red).
(DOC)
Optimization of Illumina analysis and comparison with Affymetrix prediction in LAU-Me275. A. Pearson correlation between SNP CN, as a function of OverUnder window size. B. Copy number concordance at each SNP for different window sizes of OverUnder. Colors indicate window size parameters, the bar height indicates the total number of SNPs (in log10 scale) found in one replicate. The gray bar indicates the intersection between two technical replicates. The percentage of concordance (number of SNPs found with the same copy number bin in both replicates/total number of SNPs from this given copy number bin in the first replicate) is shown on top of each bar. C. Copy number prediction on chromosome 1 using OverUnder with a window size of 201 SNPs. D. Copy number prediction on chromosome 1 using an Affymetrix 6.0 array (with the PICNIC algorithm).
(DOC)
Copy number analysis using Illumina SNP arrays. DNA from LAU-Me275 was hybridized to Illumina SNP arrays, and the data were analyzed using the method of Attiyeh et al. The top panel shows genome-wide copy number: dark blue indicates more than three copies; cyan:three copies; gray:copy neutral; orange: deletion. Subsequent panels show chromosome 7 with, from top to bottom: Hybridization log2 ratio; B allele frequency; and copy number prediction.
(DOC)
Intersection between CGH and SNP predictions. A. Intersection between CGH and SNP predictions for genes with more than 4 copies. B. Intersection for genes within deletions. C. Intersection for genes within deletions for which expression was not detected.
(DOC)
Copy number prediction from CGH and SNP arrays, LOH prediction from SNP arrays.
(PDF)
Boxplots of SILAC heavy/light normalized log2 ratios. In all experiments, LAU-Me275 was labeled with the heavy isotope; the unlabeled sample is indicated in the boxplot label on the X axis. ‘NHM’ refers to the pool of normal melanocytes and ‘self-self’ to a control experiment using only LAU-Me275 to check for any bias due to the label/no label culture conditions (no significant bias was detected).
(DOC)
Histogram of unique peptides identified per protein in the SILAC data.
(DOC)
Correlation between mRNA expression and protein levels. Both SILAC and RNA seq log2 ratios are expressed for LAU-Me275 with respect to the sample indicated in each plot title. Spearman Rho correlation coefficient is also indicated in the title. In all experiments, the correlation is significantly positive (p<0.001).
(DOC)
Count of genes affected by SCNA.
(DOC)
Processed list of SCNA genes in all seven melanoma cell lines.
(XLS)
Genomic and transcriptomic data for SCNA-genes in all seven melanoma cell lines.
(XLS)
List of pathways significantly enriched in SCNA, and pathway comparison between three melanoma datasets.
(XLS)
List of genes contributing to pathway enrichment in three melanoma datasets.
(XLS)
(DOC)
Acknowledgments
We thank Keith Harshman and Johann Weber (Genomic Technologies Facility, Lausanne) for helpful discussions; Anguraj Sadanandam (EPFL-ISREC, Lausanne) and Bob Strausberg (Ludwig Institute for Cancer Research, New York) for useful comments on the manuscript; Ghanem Ghanem (Institut Bordet, Belgium) for his generous gift of melanocytes; Patrick Descombes and the Genomics Platform of the NCCR “Frontiers in Genetics” for performing the SNP experiments; Sven Bergmann (Department of Medical Genetics, Lausanne) for access to his computing resources; and Bastian Peter for system administration. Part of the computation was performed on the cluster at the Vital-IT computing center, Lausanne. We are very grateful to Katja Muehlethaler (Ludwig Institute for Cancer Research, Lausanne); Alexandra Potts (UNIL Protein Analysis Facility) and Marianne Wicht, Nathalie Besuchet-Schmutz, and Anne-Claude Magnin (CHUV cytogenetic laboratory) for excellent technical assistance.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by the Ludwig Institute for Cancer Research Ltd. (www.licr.org). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baudis M. Genomic imbalances in 5918 malignant epithelial tumors: an explorative meta-analysis of chromosomal CGH data. BMC Cancer. 2007;7:226. doi: 10.1186/1471-2407-7-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mitelman F, Johansson B, Mertens F. Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer 2010 [Google Scholar]
- 4.Lockwood WW, Chari R, Coe BP, Girard L, Macaulay C, et al. DNA amplification is a ubiquitous mechanism of oncogene activation in lung and other cancers. Oncogene. 2008;27:4615–4624. doi: 10.1038/onc.2008.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Futreal PA, Coin L, Marshall M, Down T, Hubbard T, et al. A census of human cancer genes. Nat Rev Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bignell GR, Santarius T, Pole JC, Butler AP, Perry J, et al. Architectures of somatic genomic rearrangement in human cancer amplicons at sequence-level resolution. Genome Res. 2007;17:1296–1303. doi: 10.1101/gr.6522707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stahl JM, Cheung M, Sharma A, Trivedi NR, Shanmugam S, et al. Loss of PTEN promotes tumor development in malignant melanoma. Cancer Res. 2003;63:2881–2890. [PubMed] [Google Scholar]
- 8.Kallioniemi A, Kallioniemi OP, Sudar D, Rutovitz D, Gray JW, et al. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science. 1992;258:818–821. doi: 10.1126/science.1359641. [DOI] [PubMed] [Google Scholar]
- 9.Bignell GR, Huang J, Greshock J, Watt S, Butler A, et al. High-resolution analysis of DNA copy number using oligonucleotide microarrays. Genome Res. 2004;14:287–295. doi: 10.1101/gr.2012304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pinkel D, Albertson DG. Array comparative genomic hybridization and its applications in cancer. Nat Genet. 2005;37(Suppl):S11–17. doi: 10.1038/ng1569. [DOI] [PubMed] [Google Scholar]
- 11.Kallioniemi A. CGH microarrays and cancer. Curr Opin Biotechnol. 2008;19:36–40. doi: 10.1016/j.copbio.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 12.LaFramboise T, Weir BA, Zhao X, Beroukhim R, Li C, et al. Allele-specific amplification in cancer revealed by SNP array analysis. PLoS Comput Biol. 2005;1:e65. doi: 10.1371/journal.pcbi.0010065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gast A, Scherer D, Chen B, Bloethner S, Melchert S, et al. Somatic alterations in the melanoma genome: a high-resolution array-based comparative genomic hybridization study. Genes Chromosomes Cancer. 2010;49:733–745. doi: 10.1002/gcc.20785. [DOI] [PubMed] [Google Scholar]
- 14.Stark M, Hayward N. Genome-wide loss of heterozygosity and copy number analysis in melanoma using high-density single-nucleotide polymorphism arrays. Cancer Res. 2007;67:2632–2642. doi: 10.1158/0008-5472.CAN-06-4152. [DOI] [PubMed] [Google Scholar]
- 15.Chen HI, Hsu FH, Jiang Y, Tsai MH, Yang PC, et al. A probe-density-based analysis method for array CGH data: simulation, normalization and centralization. Bioinformatics. 2008;24:1749–1756. doi: 10.1093/bioinformatics/btn321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Attiyeh EF, Diskin SJ, Attiyeh MA, Mosse YP, Hou C, et al. Genomic copy number determination in cancer cells from single nucleotide polymorphism microarrays based on quantitative genotyping corrected for aneuploidy. Genome Res. 2009;19:276–283. doi: 10.1101/gr.075671.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oliner JD, Kinzler KW, Meltzer PS, George DL, Vogelstein B. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature. 1992;358:80–83. doi: 10.1038/358080a0. [DOI] [PubMed] [Google Scholar]
- 18.Ong S-E, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, et al. Stable Isotope Labeling by Amino Acids in Cell Culture, SILAC, as a Simple and Accurate Approach to Expression Proteomics. Molecular & Cellular Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 19.Colzani M, Schutz F, Potts A, Waridel P, Quadroni M. Relative protein quantification by isobaric SILAC with immonium ion splitting (ISIS). Mol Cell Proteomics. 2008;7:927–937. doi: 10.1074/mcp.M700440-MCP200. [DOI] [PubMed] [Google Scholar]
- 20.Cerami E, Demir E, Schultz N, Taylor BS, Sander C. Automated network analysis identifies core pathways in glioblastoma. PLoS One. 2010;5:e8918. doi: 10.1371/journal.pone.0008918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ozisik YY, Meloni AM, Altungoz O, Peier A, Karakousis C, et al. Cytogenetic findings in 21 malignant melanomas. Cancer Genet Cytogenet. 1994;77:69–73. doi: 10.1016/0165-4608(94)90152-x. [DOI] [PubMed] [Google Scholar]
- 22.Becher R, Gibas Z, Karakousis C, Sandberg AA. Nonrandom chromosome changes in malignant melanoma. Cancer Res. 1983;43:5010–5016. [PubMed] [Google Scholar]
- 23.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 24.Miller AJ, Mihm MC., Jr Melanoma. N Engl J Med. 2006;355:51–65. doi: 10.1056/NEJMra052166. [DOI] [PubMed] [Google Scholar]
- 25.Bloethner S, Chen B, Hemminki K, Muller-Berghaus J, Ugurel S, et al. Effect of common B-RAF and N-RAS mutations on global gene expression in melanoma cell lines. Carcinogenesis. 2005;26:1224–1232. doi: 10.1093/carcin/bgi066. [DOI] [PubMed] [Google Scholar]
- 26.Wellbrock C, Ogilvie L, Hedley D, Karasarides M, Martin J, et al. V599EB-RAF is an oncogene in melanocytes. Cancer Res. 2004;64:2338–2342. doi: 10.1158/0008-5472.can-03-3433. [DOI] [PubMed] [Google Scholar]
- 27.Karasarides M, Chiloeches A, Hayward R, Niculescu-Duvaz D, Scanlon I, et al. B-RAF is a therapeutic target in melanoma. Oncogene. 2004;23:6292–6298. doi: 10.1038/sj.onc.1207785. [DOI] [PubMed] [Google Scholar]
- 28.Wellbrock C, Rana S, Paterson H, Pickersgill H, Brummelkamp T, et al. Oncogenic BRAF regulates melanoma proliferation through the lineage specific factor MITF. PLoS One. 2008;3:e2734. doi: 10.1371/journal.pone.0002734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Omholt K, Platz A, Kanter L, Ringborg U, Hansson J. NRAS and BRAF mutations arise early during melanoma pathogenesis and are preserved throughout tumor progression. Clin Cancer Res. 2003;9:6483–6488. [PubMed] [Google Scholar]
- 30.Veeriah S, Taylor BS, Meng S, Fang F, Yilmaz E, et al. Somatic mutations of the Parkinson's disease-associated gene PARK2 in glioblastoma and other human malignancies. Nat Genet. 2010;42:77–82. doi: 10.1038/ng.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ayyanan A, Civenni G, Ciarloni L, Morel C, Mueller N, et al. Increased Wnt signaling triggers oncogenic conversion of human breast epithelial cells by a Notch-dependent mechanism. Proc Natl Acad Sci U S A. 2006;103:3799–3804. doi: 10.1073/pnas.0600065103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma D, Dai J, Zhu X, Yan S, Zhao P, et al. Aberrant expression of Notch signaling molecules in patients with immune thrombocytopenic purpura. Ann Hematol. 2010;89:155–161. doi: 10.1007/s00277-009-0790-y. [DOI] [PubMed] [Google Scholar]
- 33.Bauer S, Groh V, Wu J, Steinle A, Phillips JH, et al. Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science. 1999;285:727–729. doi: 10.1126/science.285.5428.727. [DOI] [PubMed] [Google Scholar]
- 34.Waldhauer I, Steinle A. NK cells and cancer immunosurveillance. Oncogene. 2008;27:5932–5943. doi: 10.1038/onc.2008.267. [DOI] [PubMed] [Google Scholar]
- 35.Pende D, Rivera P, Marcenaro S, Chang CC, Biassoni R, et al. Major histocompatibility complex class I-related chain A and UL16-binding protein expression on tumor cell lines of different histotypes: analysis of tumor susceptibility to NKG2D-dependent natural killer cell cytotoxicity. Cancer Res. 2002;62:6178–6186. [PubMed] [Google Scholar]
- 36.Montgomery RB, Mostaghel EA, Vessella R, Hess DL, Kalhorn TF, et al. Maintenance of intratumoral androgens in metastatic prostate cancer: a mechanism for castration-resistant tumor growth. Cancer Res. 2008;68:4447–4454. doi: 10.1158/0008-5472.CAN-08-0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yin D, Ogawa S, Kawamata N, Tunici P, Finocchiaro G, et al. High-resolution genomic copy number profiling of glioblastoma multiforme by single nucleotide polymorphism DNA microarray. Mol Cancer Res. 2009;7:665–677. doi: 10.1158/1541-7786.MCR-08-0270. [DOI] [PubMed] [Google Scholar]
- 38.Wu H, Goel V, Haluska FG. PTEN signaling pathways in melanoma. Oncogene. 2003;22:3113–3122. doi: 10.1038/sj.onc.1206451. [DOI] [PubMed] [Google Scholar]
- 39.Klijn C, Bot J, Adams DJ, Reinders M, Wessels L, et al. Identification of networks of co-occurring, tumor-related DNA copy number changes using a genome-wide scoring approach. PLoS Comput Biol. 2010;6:e1000631. doi: 10.1371/journal.pcbi.1000631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Menashe I, Maeder D, Garcia-Closas M, Figueroa JD, Bhattacharjee S, et al. Pathway analysis of breast cancer genome-wide association study highlights three pathways and one canonical signaling cascade. Cancer Res. 2010;70:4453–4459. doi: 10.1158/0008-5472.CAN-09-4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murohashi M, Hinohara K, Kuroda M, Isagawa T, Tsuji S, et al. Gene set enrichment analysis provides insight into novel signalling pathways in breast cancer stem cells. Br J Cancer. 2010;102:206–212. doi: 10.1038/sj.bjc.6605468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heiser LM, Wang NJ, Talcott CL, Laderoute KR, Knapp M, et al. Integrated analysis of breast cancer cell lines reveals unique signaling pathways. Genome Biol. 2009;10:R31. doi: 10.1186/gb-2009-10-3-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoek K, Rimm DL, Williams KR, Zhao H, Ariyan S, et al. Expression profiling reveals novel pathways in the transformation of melanocytes to melanomas. Cancer Res. 2004;64:5270–5282. doi: 10.1158/0008-5472.CAN-04-0731. [DOI] [PubMed] [Google Scholar]
- 45.Dorsam RT, Gutkind JS. G-protein-coupled receptors and cancer. Nat Rev Cancer. 2007;7:79–94. doi: 10.1038/nrc2069. [DOI] [PubMed] [Google Scholar]
- 46.Lee HJ, Wall B, Chen S. G-protein-coupled receptors and melanoma. Pigment Cell Melanoma Res. 2008;21:415–428. doi: 10.1111/j.1755-148X.2008.00478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van Raamsdonk CD, Griewank KG, Crosby MB, Garrido MC, Vemula S, et al. Mutations in GNA11 in Uveal Melanoma. N Engl J Med. 2010 doi: 10.1056/NEJMoa1000584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van Raamsdonk CD, Bezrookove V, Green G, Bauer J, Gaugler L, et al. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature. 2009;457:599–602. doi: 10.1038/nature07586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heuberger J, Birchmeier W. Interplay of cadherin-mediated cell adhesion and canonical Wnt signaling. Cold Spring Harb Perspect Biol. 2010;2:a002915. doi: 10.1101/cshperspect.a002915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cavallaro U, Christofori G. Cell adhesion and signalling by cadherins and Ig-CAMs in cancer. Nat Rev Cancer. 2004;4:118–132. doi: 10.1038/nrc1276. [DOI] [PubMed] [Google Scholar]
- 51.Taipale J, Beachy PA. The Hedgehog and Wnt signalling pathways in cancer. Nature. 2001;411:349–354. doi: 10.1038/35077219. [DOI] [PubMed] [Google Scholar]
- 52.Lucero OM, Dawson DW, Moon RT, Chien AJ. A re-evaluation of the “oncogenic” nature of Wnt/beta-catenin signaling in melanoma and other cancers. Curr Oncol Rep. 2010;12:314–318. doi: 10.1007/s11912-010-0114-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Klaus A, Birchmeier W. Wnt signalling and its impact on development and cancer. Nat Rev Cancer. 2008;8:387–398. doi: 10.1038/nrc2389. [DOI] [PubMed] [Google Scholar]
- 54.Raposo G, Marks MS. Melanosomes–dark organelles enlighten endosomal membrane transport. Nat Rev Mol Cell Biol. 2007;8:786–797. doi: 10.1038/nrm2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Halaban R, Cheng E, Zhang Y, Moellmann G, Hanlon D, et al. Aberrant retention of tyrosinase in the endoplasmic reticulum mediates accelerated degradation of the enzyme and contributes to the dedifferentiated phenotype of amelanotic melanoma cells. Proc Natl Acad Sci U S A. 1997;94:6210–6215. doi: 10.1073/pnas.94.12.6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hoek KS, Goding CR. Cancer stem cells versus phenotype-switching in melanoma. Pigment Cell Melanoma Res. 2010;23:746–759. doi: 10.1111/j.1755-148X.2010.00757.x. [DOI] [PubMed] [Google Scholar]
- 57.Namkoong J, Shin SS, Lee HJ, Marin YE, Wall BA, et al. Metabotropic glutamate receptor 1 and glutamate signaling in human melanoma. Cancer Res. 2007;67:2298–2305. doi: 10.1158/0008-5472.CAN-06-3665. [DOI] [PubMed] [Google Scholar]
- 58.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 59.Folkman J. Tumor angiogenesis: therapeutic implications. N Engl J Med. 1971;285:1182–1186. doi: 10.1056/NEJM197111182852108. [DOI] [PubMed] [Google Scholar]
- 60.Kerbel RS. Tumor angiogenesis. N Engl J Med. 2008;358:2039–2049. doi: 10.1056/NEJMra0706596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bignell GR, Greenman CD, Davies H, Butler AP, Edkins S, et al. Signatures of mutation and selection in the cancer genome. Nature. 2010;463:893–898. doi: 10.1038/nature08768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Davies H, Hunter C, Smith R, Stephens P, Greenman C, et al. Somatic mutations of the protein kinase gene family in human lung cancer. Cancer Res. 2005;65:7591–7595. doi: 10.1158/0008-5472.CAN-05-1855. [DOI] [PubMed] [Google Scholar]
- 64.Pasquale EB. Eph receptors and ephrins in cancer: bidirectional signalling and beyond. Nat Rev Cancer. 2010;10:165–180. doi: 10.1038/nrc2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dalgliesh GL, Furge K, Greenman C, Chen L, Bignell G, et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463:360–363. doi: 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 67.Stephens P, Edkins S, Davies H, Greenman C, Cox C, et al. A screen of the complete protein kinase gene family identifies diverse patterns of somatic mutations in human breast cancer. Nat Genet. 2005;37:590–592. doi: 10.1038/ng1571. [DOI] [PubMed] [Google Scholar]
- 68.Balakrishnan A, Bleeker FE, Lamba S, Rodolfo M, Daniotti M, et al. Novel somatic and germline mutations in cancer candidate genes in glioblastoma, melanoma, and pancreatic carcinoma. Cancer Res. 2007;67:3545–3550. doi: 10.1158/0008-5472.CAN-07-0065. [DOI] [PubMed] [Google Scholar]
- 69.Chiari R, Hames G, Stroobant V, Texier C, Maillere B, et al. Identification of a tumor-specific shared antigen derived from an Eph receptor and presented to CD4 T cells on HLA class II molecules. Cancer Res. 2000;60:4855–4863. [PubMed] [Google Scholar]
- 70.Tatsumi T, Herrem CJ, Olson WC, Finke JH, Bukowski RM, et al. Disease stage variation in CD4+ and CD8+ T-cell reactivity to the receptor tyrosine kinase EphA2 in patients with renal cell carcinoma. Cancer Res. 2003;63:4481–4489. [PubMed] [Google Scholar]
- 71.Alves PM, Faure O, Graff-Dubois S, Gross DA, Cornet S, et al. EphA2 as target of anticancer immunotherapy: identification of HLA-A*0201-restricted epitopes. Cancer Res. 2003;63:8476–8480. [PubMed] [Google Scholar]
- 72.Jin M, Komohara Y, Shichijo S, Yamanaka R, Nikawa J, et al. Erythropoietin-producing hepatocyte B6 variant-derived peptides with the ability to induce glioma-reactive cytotoxic T lymphocytes in human leukocyte antigen-A2+ glioma patients. Cancer Sci. 2008;99:1656–1662. doi: 10.1111/j.1349-7006.2008.00866.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vearing C, Lee FT, Wimmer-Kleikamp S, Spirkoska V, To C, et al. Concurrent binding of anti-EphA3 antibody and ephrin-A5 amplifies EphA3 signaling and downstream responses: potential as EphA3-specific tumor-targeting reagents. Cancer Res. 2005;65:6745–6754. doi: 10.1158/0008-5472.CAN-05-0758. [DOI] [PubMed] [Google Scholar]
- 74.Gotoh N. Regulation of growth factor signaling by FRS2 family docking/scaffold adaptor proteins. Cancer Sci. 2008;99:1319–1325. doi: 10.1111/j.1349-7006.2008.00840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dey JH, Bianchi F, Voshol J, Bonenfant D, Oakeley EJ, et al. Targeting fibroblast growth factor receptors blocks PI3K/AKT signaling, induces apoptosis, and impairs mammary tumor outgrowth and metastasis. Cancer Res. 2010;70:4151–4162. doi: 10.1158/0008-5472.CAN-09-4479. [DOI] [PubMed] [Google Scholar]
- 76.Wang Y, Becker D. Antisense targeting of basic fibroblast growth factor and fibroblast growth factor receptor-1 in human melanomas blocks intratumoral angiogenesis and tumor growth. Nat Med. 1997;3:887–893. doi: 10.1038/nm0897-887. [DOI] [PubMed] [Google Scholar]
- 77.Sato T, Gotoh N. The FRS2 family of docking/scaffolding adaptor proteins as therapeutic targets of cancer treatment. Expert Opin Ther Targets. 2009;13:689–700. doi: 10.1517/14728220902942330. [DOI] [PubMed] [Google Scholar]
- 78.Martinet D, Filges I, Besuchet Schmutz N, Morris MA, Gaide AC, et al. Subtelomeric 6p deletion: clinical and array-CGH characterization in two patients. Am J Med Genet A. 2008;146A:2094–2102. doi: 10.1002/ajmg.a.32414. [DOI] [PubMed] [Google Scholar]
- 79.Smyth GK, Speed T. Normalization of cDNA microarray data. Methods. 2003;31:265–273. doi: 10.1016/s1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- 80.Staaf J, Jonsson G, Ringner M, Vallon-Christersson J. Normalization of array-CGH data: influence of copy number imbalances. BMC Genomics. 2007;8:382. doi: 10.1186/1471-2164-8-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 82.Venkatraman ES, Olshen AB. A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics. 2007;23:657–663. doi: 10.1093/bioinformatics/btl646. [DOI] [PubMed] [Google Scholar]
- 83.Willenbrock H, Fridlyand J. A comparison study: applying segmentation to array CGH data for downstream analyses. Bioinformatics. 2005;21:4084–4091. doi: 10.1093/bioinformatics/bti677. [DOI] [PubMed] [Google Scholar]
- 84.Greenman CD, Bignell G, Butler A, Edkins S, Hinton J, et al. PICNIC: an algorithm to predict absolute allelic copy number variation with microarray cancer data. Biostatistics. 2010;11:164–175. doi: 10.1093/biostatistics/kxp045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bainbridge MN, Warren RL, Hirst M, Romanuik T, Zeng T, et al. Analysis of the prostate cancer cell line LNCaP transcriptome using a sequencing-by-synthesis approach. BMC Genomics. 2006;7:246. doi: 10.1186/1471-2164-7-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jongeneel CV, Iseli C, Stevenson BJ, Riggins GJ, Lal A, et al. Comprehensive sampling of gene expression in human cell lines with massively parallel signature sequencing. Proc Natl Acad Sci U S A. 2003;100:4702–4705. doi: 10.1073/pnas.0831040100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Razick S, Magklaras G, Donaldson IM. iRefIndex: a consolidated protein interaction database with provenance. BMC Bioinformatics. 2008;9:405. doi: 10.1186/1471-2105-9-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pathway Commons website (Bader G, Cerami E, Demir E, Gross B, Sander C). Available: http://www.pathwaycommons.org. Accessed 2010 April 15. [DOI] [PMC free article] [PubMed]
- 89.Mi H, Guo N, Kejariwal A, Thomas PD. PANTHER version 6: protein sequence and function evolution data with expanded representation of biological pathways. Nucleic Acids Res. 2007;35:D247–252. doi: 10.1093/nar/gkl869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B-Methodological. 1995;57:289–300. [Google Scholar]
- 93.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 94.Kersey PJ, Duarte J, Williams A, Karavidopoulou Y, Birney E, et al. The International Protein Index: an integrated database for proteomics experiments. Proteomics. 2004;4:1985–1988. doi: 10.1002/pmic.200300721. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
CGH hybridization ratio in a tetraploid region in LAU-Me275. Each plot shows the hybridization log2 ratio at each CGH probe (in gray) obtained using three normalization methods. Ridge refers to the framework from Chen et al. Red segments were obtained using Circular Binary Segmentation. Karyotype analysis of LAU-Me275 revealed 11q amplification (CN≥4), so the expected CGH log ratio would be two. Here the ratios obtained from three different normalizations failed to reflect the amplification (both Loess and Ridge were close to 0; PopLowess was close to 0.6 indicating 3 copies).
(DOC)
Correlation between replicates using different normalization schemes. The scatter plots illustrate the correlation at each CGH probe between two replicates. The heatmaps show the correlation for each pairs of replicates. The normalization method is indicated in each plot title. RIDGE refers to the framework from Chen et al.
(DOC)
Gaussian Mixtures identified in four replicates from LAU-Me275. Each histogram shows the distribution of CBS segment log2 ratios, colors highlight the Gaussian components. The number of components identified and the Bayes Information Criterion are indicated in each figure title.
(DOC)
Comparison of CNV detection algorithm on CGH data. Panels are, from left to right: a melanoma with large deletions (LAU-Me280); a melanoma with large amplifications (LAU-Me275); and a control EBV cell line (male) hybridized using a pool of female references. From top to bottom: CNV classification (following CBS segmentation) using 1) Gaussian Mixture Model (GMM), 2) MergeLevels, 3) the scoring-based approach. Each dot corresponds to a CGH probe with its genomic position on the X axis and its log2 ratio of hybridization on the Y axis. Colors indicate the copy number state: orange< = 1 copy gray = 2 copies, cyan = 3 copies and dark blue more than 3 copies. For the scoring approach distinction is made between 1 copy (orange) and 0 copy (red).
(DOC)
Optimization of Illumina analysis and comparison with Affymetrix prediction in LAU-Me275. A. Pearson correlation between SNP CN, as a function of OverUnder window size. B. Copy number concordance at each SNP for different window sizes of OverUnder. Colors indicate window size parameters, the bar height indicates the total number of SNPs (in log10 scale) found in one replicate. The gray bar indicates the intersection between two technical replicates. The percentage of concordance (number of SNPs found with the same copy number bin in both replicates/total number of SNPs from this given copy number bin in the first replicate) is shown on top of each bar. C. Copy number prediction on chromosome 1 using OverUnder with a window size of 201 SNPs. D. Copy number prediction on chromosome 1 using an Affymetrix 6.0 array (with the PICNIC algorithm).
(DOC)
Copy number analysis using Illumina SNP arrays. DNA from LAU-Me275 was hybridized to Illumina SNP arrays, and the data were analyzed using the method of Attiyeh et al. The top panel shows genome-wide copy number: dark blue indicates more than three copies; cyan:three copies; gray:copy neutral; orange: deletion. Subsequent panels show chromosome 7 with, from top to bottom: Hybridization log2 ratio; B allele frequency; and copy number prediction.
(DOC)
Intersection between CGH and SNP predictions. A. Intersection between CGH and SNP predictions for genes with more than 4 copies. B. Intersection for genes within deletions. C. Intersection for genes within deletions for which expression was not detected.
(DOC)
Copy number prediction from CGH and SNP arrays, LOH prediction from SNP arrays.
(PDF)
Boxplots of SILAC heavy/light normalized log2 ratios. In all experiments, LAU-Me275 was labeled with the heavy isotope; the unlabeled sample is indicated in the boxplot label on the X axis. ‘NHM’ refers to the pool of normal melanocytes and ‘self-self’ to a control experiment using only LAU-Me275 to check for any bias due to the label/no label culture conditions (no significant bias was detected).
(DOC)
Histogram of unique peptides identified per protein in the SILAC data.
(DOC)
Correlation between mRNA expression and protein levels. Both SILAC and RNA seq log2 ratios are expressed for LAU-Me275 with respect to the sample indicated in each plot title. Spearman Rho correlation coefficient is also indicated in the title. In all experiments, the correlation is significantly positive (p<0.001).
(DOC)
Count of genes affected by SCNA.
(DOC)
Processed list of SCNA genes in all seven melanoma cell lines.
(XLS)
Genomic and transcriptomic data for SCNA-genes in all seven melanoma cell lines.
(XLS)
List of pathways significantly enriched in SCNA, and pathway comparison between three melanoma datasets.
(XLS)
List of genes contributing to pathway enrichment in three melanoma datasets.
(XLS)
(DOC)