Abstract
Background
Membrane proteins (MPs) play key roles in signal transduction. However, understanding their function at a molecular level is mostly hampered by the lack of protein in suitable amount and quality. Despite impressive developments in the expression of prokaryotic MPs, eukaryotic MP production has lagged behind and there is a need for new expression strategies. In a pilot study, we produced a Drosophila glutamate receptor specifically in the eyes of transgenic flies, exploiting the naturally abundant membrane stacks in the photoreceptor cells (PRCs). Now we address the question whether the PRCs also process different classes of medically relevant target MPs which were so far notoriously difficult to handle with conventional expression strategies.
Principal Findings
We describe the homologous and heterologous expression of 10 different targets from the three major MP classes - G protein-coupled receptors (GPCRs), transporters and channels in Drosophila eyes. PRCs offered an extraordinary capacity to produce, fold and accommodate massive amounts of MPs. The expression of some MPs reached similar levels as the endogenous rhodopsin, indicating that the PRC membranes were almost unsaturable. Expression of endogenous rhodopsin was not affected by the target MPs and both could coexist in the membrane stacks. Heterologous expression levels reached about 270 to 500 pmol/mg total MP, resulting in 0.2–0.4 mg purified target MP from 1 g of fly heads. The metabotropic glutamate receptor and human serotonin transporter - both involved in synaptic transmission - showed native pharmacological characteristics and could be purified to homogeneity as a prerequisite for further studies.
Significance
We demonstrate expression in Drosophila PRCs as an efficient and inexpensive tool for the large scale production of functional eukaryotic MPs. The fly eye system offers a number of advantages over conventional expression systems and paves the way for in-depth analyses of eukaryotic MPs that have so far not been accessible to biochemical and biophysical studies.
Introduction
Membrane proteins (MPs) represent more than 30% of the cell proteome [1] and play key roles in signal transduction. Dysfunction often leads to major disorders or death and therefore, MPs account for more than 50% of the current drug targets [2]. However, drug discovery as well as detailed biochemical and structural studies are still hindered by a number of problems already encountered in the production of eukaryotic MPs. It is therefore not surprising that the majority of eukaryotic MPs found in the structural database (Membrane Proteins of Known 3D-Structure, http://blanco.biomol.uci.edu) are naturally abundant [3], [4] and that their structures were determined using material from wild-type organisms. Most of them are localized in specialized cells from i.e. the retina for rhodopsin, the lens for aquaporins, the sarcoplasmic reticulum for calcium ATPases and the electric organ of Torpedo for the nicotinic acetylcholine receptor pore. These cells are adapted to the massive production of MPs, which are often densely packed in their respective membrane environment.
In contrast to eukaryotic MPs, our understanding of prokaryotic MPs has tremendously increased in the past decade due to the optimization of bacterial strains and expression tools for MP production [4], as well as by the use of extremophilic organisms (e.g. Archaea) as a source for MPs of increased stability [5]. Bacteria enriched in membranes are widely used for MP expression as they seem to offer increased membrane surface as well as an optimized insertion machinery [6]. The crystal structures of close prokaryotic homologs provided relevant models for many mammalian MPs. However, some eukaryotic MPs which are of prime interest in neuropharmacology, like the sodium-dependent serotonin transporter (SERT or 5HTT), do not have close bacterial homologs [7]. Importantly, differences in the active sites have been observed e.g. in rhodopsin [8] or potassium channels [9] that distinguish the pro- and eukaryotic proteins. The precise architecture of these binding sites can be difficult to model which leads to controversies in the perception of their reaction mechanisms. For MPs regulated by allosteric mechanisms [10], focusing on the ligand binding site is not sufficient. Among G protein-coupled receptors (GPCRs), metabotropic glutamate receptors (mGluRs) are prototypes for allosteric regulation and have been subjected to random high-troughput ligand screens for drug design as well as structure-based virtual screening [11], [12]. Both, high-throughput pharmacological and structural analyses of MPs require amounts of material which are often not provided in sufficient quality and quantity by conventional expression systems.
Eukaryotic cells in culture, like insect cells and yeast are commonly used for the overexpression of eukaryotic MPs [3]. However, a major drawback is the often limited capacity of these cells for trafficking, folding and membrane insertion of the target MPs and therefore, a significant portion of immature MPs remain trapped in internal membranes [13]. In a pilot study, we engineered a transgenic fly overexpressing a recombinant Drosophila metabotropic glutamate receptor (DmGluRA) specifically in the eyes [14]. The idea was to target the receptor to the naturally abundant membrane stacks in the photoreceptor cells (PRCs), the rhabdomeres, housing the GPCR-prototype rhodopsin. Drosophila melanogaster was chosen because fly genetics offers the possibility of regulating ectopic expression in intensity, kinetics and localization using specific promoters (drivers). The DmGluRA production in fly eyes gave higher yields than the baculovirus overexpression system in Sf9 cells and the receptor was functional. In addition, the purified protein was clearly superior in homogeneity compared to protein obtained from Sf9 membranes [14] which typically suffers from the presence of immature receptors [3]. The receptor could be purified in mg amounts [14] and biochemical analysis suggested cholesterol as an allosteric regulator that switches the receptor to a high affinity state [15]. Recently, the expression protocol was improved by the use of GFP-fusion constructs [16]. However, the question remained whether overexpression in fly eyes would be also applicable to the heterologous expression for MPs like transporters and channels which are often difficult to express in conventional systems.
In this study, we show the exceptional properties of the PRCs in offering seemingly unsaturable membrane space for target MP insertion. We describe the heterologous expression of functional MPs including mammalian GPCRs, neurotransmitter transporters and the channelrhodopsin ChR2. We establish overexpression in fly eyes as a general, efficient and inexpensive method for large scale production of functional eukaryotic MPs and exemplify our findings with an in depth analysis of mGluR5 and SERT.
Results
Photoreceptor cells have a large capacity for recombinant MPs
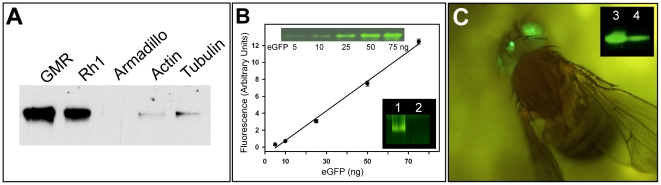
The successful expression of a functional Drosophila metabotropic glutamate receptor DmGluRA in fly eyes recommended this system for the production of eukaryotic MPs [14] (see Supporting Information: Primer of the fly eye system (Primer S1)). We now addressed the question whether overexpression in the eyes is superior to overexpression e.g. in the whole fly or other body parts. DmGluRA was expressed in transgenic flies under the control of different drivers [17] inducing specific expression in the eyes (GMR- or Rh1-GAL4) or ubiquitous expression (Tubulin-, Actin- or Armadillo-GAL4). The expression driven by eye-specific promoters was impressive compared to the insignificant levels obtained with ubiquitous promoters (Figure 1A). Using an eye-specific driver was a prerequisite for high expression.
Figure 1. Recombinant expression of Drosophila and mammalian GPCRs in fly eyes.
(A). Western blot analysis of DmGluRA expression using eye-specific (GMR, Rh1) or ubiquitous (Armadillo, Actin, Tubulin) promoter elements. DmGluRA was detected with the 7G11 antibody. β-tubulin was used as a loading control (not shown). (B). Quantification of DmGluRA-GFP expressed under the control of the GMR driver. Intrinsic fluorescence signal of DmGluRA-GFP (1) compared with the GMR driver fly (2)(lower inset). The heads of three flies were analyzed. The GFP standard curve shown as graph was obtained by fluorescence scanning of a clear-native gel (using 5, 10, 25, 50, 75 ng GFP; upper inset). Fluorescence signals were integrated with the ImageJ software. (C). Typical fluorescence image of a transgenic Drosophila expressing the human vasopressin receptor V2R-GFP under the control of the eye-specific GMR1104 driver. The inset shows the fluorescence signals of V2R-GFP (3) and Rh1-GFP (4) from three fly heads.
The green fluorescent protein (GFP) was fused to the C-terminus of all MP-targets in this study for efficient monitoring [18], e.g. to select the best expressing flies, for quantification, localization of expression and for quality control of large-scale cultures. GFP fluorescence indicates also correct folding of the N-terminally fused partner protein [19]. Flies expressing different GPCR-GFP fusion constructs under the control of GMR-GAL4 were generated (for technical details see [16]). Quantification by fluorescence-scanning of native gels (Figure 1B) showed that e.g. DmGluRA expression levels reached about 50% of endogenous rhodopsin (Rh1) present in the PRCs (Table 1). Recombinant Rh1 could be expressed at similar levels (502 pmol/mg MP or 18 ng/eye) as endogenous Rh1 (3 to 6×107 Rh1 molecules/rhabdomere, corresponding to 10 to 20 ng/eye [20], [21]) and similar to recombinant Rh1 not fused to GFP (15 ng/eye [22]) (Table 1). A number of rhodopsin-type GPCRs (Class A GPCRs [23]) were tested for heterologous expression. Among them, the mammalian vasopressin receptor (V2R) was one of the best expressing test cases (>1 µmol/mg MP; Table 1, Figure 1C). V2R is involved in the regulation of water homeostasis by the kidney and in X-linked nephrogenic diabetes insipidus [24]. The expression level of V2R in PRCs is higher than the best ones previously reported using conventional overexpression systems optimized for eukaryotic MPs [4], [25]. Human CCR5, a chemokine receptor currently serving as a major therapeutic target against HIV cell-entrance [26], was expressed at levels similar to Drosophila Rh1 (555 pmol/mg MP; Table 1). These examples suggest that heterologous expression in the fly eye can be applied to most class A GPCRs. Since fly Rh1 is the predominant MP in rhabdomere membranes [27], it is remarkable that the overexpression of recombinant MPs did not affect the amount of endogenous Rh1 as analyzed by Western blot (not shown). On the other hand, the high level of endogenous Rh1 does not seem to limit the expression of recombinant MPs. The rhabdomere membranes appear to have seemingly unsaturable capacity to accommodate MPs.
Table 1. Expression levels of target MPs.
| Target MP_ Species | Expression level [pmol/mg total MP] | |
| GPCRs (7 TMs) | ||
| Endogenous Rh1_Drosophila | rhodopsin | 272–544* |
| Rh1_Drosophila | rhodopsin | 502 |
| V2R_Human | vasopressin receptor | >1000 |
| CCR5_Human | chemokine receptor | 555 |
| DmGluRA_Drosophila | metabotropic glutamate receptor | 226 |
| mGluR5_Rat | metabotropic glutamate receptor | 192 |
| Channel (7 TMs) | ||
| ChR2 **_Clamydomonas | channelrhodopsin | 206 |
| Transporters (12TMs) | ||
| SERT_Drosophila | serotonin transporter | 493 |
| SERT_Human | serotonin transporter | 220 |
| EAAT2_Human | glutamate transporter | 173 |
| EAAT1_Drosophila | glutamate transporter | 716 |
A rhodopsin knock-down is not required for high expression levels
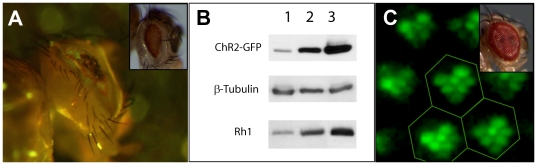
The capacity of the PRCs to host large amounts of recombinant MPs in the presence of endogenous Rh1 indicates that there is no need to down-regulate Rh1 in order to increase the expression levels. In contrary, a fly knock-out for Rh1 would alter the biogenesis of the rhabdomere membrane [28], [29]. Moreover, the expression of algal channelrhodopsin ChR2 which contains retinal as a cofactor [30] was shown to directly correlate with the levels of endogenous Rh1 (Figure 2). Chlamydomonas reinhardtii ChR2 was expressed under the control of different drivers including GMR drivers of diverse origins. Briefly, the use of a GMR driver (Bloomington #1104) [31] constructed on a gl60j genetic background missing the glass protein [32] and therefore Rh1 [33] gave a surprisingly strong eye-phenotype (Figure 2A) not seen i.e. for V2R-expressing flies (not shown), and ChR2 was barely detectable (Figure 2B, lane 1). Two other GMR drivers (Bloomington #9146 and #8605) expressing higher amounts of Rh1 (Figure 2B, lane 2 and 3, respectively) induced also a higher expression of ChR2 (Figure 2B, lanes 2 and 3, respectively). A correlation with Rh1 levels was not observed for other MPs targets e.g. the V2R (not shown). Therefore, expression of Rh1 and ChR2 are somehow linked. ChR2 expression reached 200 pmol/mg MP (Table 1). In the presence of Rh1, the channel localized in the rhabdomeres (Figure 2C) and the eye morphology was normal (Figure 2C, Inset). The observed retinal (Figure S1Aand Rh1 dependence for the proper processing of recombinant ChR2 indicated that the photoreceptor cells are specially adapted for the expression of retinal-binding membrane proteins.
Figure 2. High expression levels of Channelrhodopsin ChR2 correlate with endogenous rhodopsin (Rh1).
(A). Fluorescence image of a fly expressing ChR2-GFP under the control of the GMR1104 driver (Inset: bright light picture of the head). (B). ChR2-GFP expression driven by different GMR promoter elements (GMR1104 (1), GMR9146 (2), and GMR8605 (3)) analyzed by Western blot and compared with endogenous Rh1 levels. ChR2-GFP, Rh1 and β-tubulin were detected with GFP, Rh1 and β-tubulin antibodies, respectively. (C). Analysis of an intact head using a water-immersion objective shows rhabdomere localization of ChR2-GFP expressed under the control of GMR9146. Magnification was 10×20. Inset: the bright light picture shows normal eye morphology. For easier recognition, the facettes of the fly eye are marked by hexagons.
Heterologous and homologous expression of glutamate receptors give similar amounts
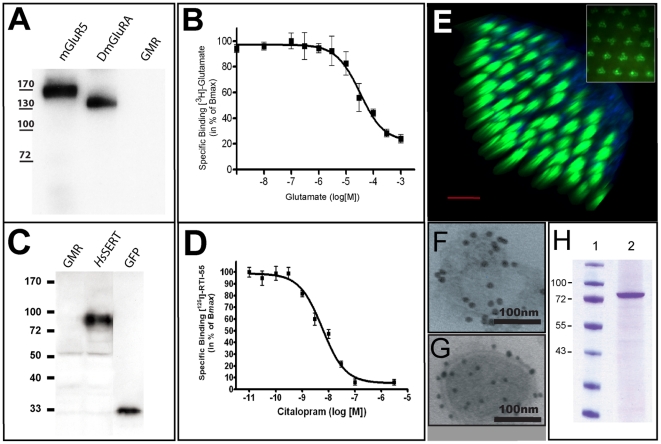
We have shown that GPCRs can be expressed in high amounts in the fly eyes. In order to compare heterologous and homologous expression we choose mGluRs. Mammalian mGluR5 is involved in antipsychotic medication and subject of intensive pharmacological and structural characterization [34], [35]. Expression of mGluR5 gave strong eye fluorescence (not shown) with expression levels similar to DmGluRA according to Western blot and fluorescence-scanning analyses (Figure 3A; Table 1). For functional tests fly heads were collected as previously described [16] and membranes were prepared for radioactive glutamate binding assays. mGluR5 had an affinity for glutamate (31±2 µM) (Figure 3B) in the same range as reported previously for DmGluRA (54 µM) [15] suggesting proper folding of the heterologously expressed receptor. The results showed that heterologous expression of functional GPCRs was efficient and reached similar levels as homologous expression.
Figure 3. Heterologous expression of functional GPCRs and transporters in fly eyes.
(A). Western blot analysis of Drosophila (DmGluRA-GFP) or mammalian (mGluR5-GFP) metabotropic glutamate receptors expressed in fly eyes using a GFP antibody. The GMR driver fly is shown as a negative control. (B). Homologous competitive binding experiment with glutamate [15] on membranes from flies expressing mGluR5-GFP. (C). Western blot analysis of the human serotonin transporter HsSERT-GFP expressed in fly eyes using a GFP antibody. The GMR driver fly and GFP standard are shown as negative and positive controls, respectively. (D). Competition binding experiment on membranes from flies expressing HsSERT-GFP. Binding of [125I]-RTI55 was competed with racemic citalopram (Ki 4.5±2.7 nM). (E). Three-dimensional reconstruction from laser scanning confocal microscopy images of a fly eye expressing HsSERT-GFP. GFP fluorescent signal, showing the presence of HsSERT in all rhabdomeres of all ommatidia (in green); natural autofluorescence delimits the surface of the eyes (in blue). The scale bar represents 40 µm. Inset: Analysis of an intact head using a water-immersion objective shows rhabdomere localization of HsSERT-GFP expressed under the control of GMR1104. Magnification was 10×20. (F–G). EM-double immunogold labeling of recombinant HsSERT-GFP and endogenous Rh1 with the GFP (10 nm gold) and the Rh1 (15 nm gold) antibodies, respectively, using purified rhabdomere membranes. (F). Typical Rh1-positive domain. (G). Typical HsSERT-labeled domain. (H). Coomassie-stained SDS-PAGE of HsSERT-GFP purified [52] in one step using a nickel column (lane 2). MW standards are shown in lane 1.
Heterologous expression of neurotransmitter transporters
Encouraged by the success with heterologous expression of GPCRs and channelrhodopsin ChR2, we set out to test the fly eye system also for membrane transporters. For eukaryotic neurotransmitter transporters low level expression and heterogeneity have been reported from classical overexpression systems. The serotonin transporter (SERT) seems to require rather sophisticated overexpression systems i.e. with engineered chaperones [36]. We tested serotonin transporters from human (HsSERT) and Drosophila (DmSERT). Strong expression was detected by epifluorescence microscopy and by Western blot analysis for HsSERT (Figure 3C). DmSERT and HsSERT expression quantified by fluorescence scanning of native gels was 493 and 220 pmol/mg MP, respectively (corresponding to 43 and 20 ng SERT/eye, respectively; Table 1). These expression levels are in range with endogenous rhodopsin. Proper folding of HsSERT is indicated by binding the inhibitors R,S-citalopram (nanomolar affinity; Figure 3D) and cocaine (309±38 nM, not shown) with similar affinities as reported previously [37], [38]. Similarly, the glutamate transporters DmEAAT1 and HsEAAT2 both expressed well in fly eyes (Table 1). These data show that the fly eye system is suitable for heterologous and homologous expression of functional neurotransmitter transporters.
SERT and Rh1 localize in distinct domains in the rhabdomere membrane
We have shown that despite the high quantities of endogenous Rh1, SERT is expressed in similarly high amounts. In order to test whether HsSERT and Rh1 co-localize in the PRCs, HsSERT localization was analyzed by 3D-laser-scanning confocal microscopy (LSCM) of the intact fly head (Figure 3E) as well as by epifluorescence microscopy using water-immersion objectives [39] (Figure 3E,inset). HsSERT is expressed in all 7 rhabdomeres of each compound eye called ommatidia. The distribution of HsSERT (Figure 3E) and DmSERT (not shown) in the rhabdomeres is similar to rhodopsins [29], [40]. However, it was possible to separate domains containing endogenous Rh1 from those containing DmSERT using an Ultra-Turrax for membrane disruption and subsequent ultracentrifugation in a density gradient (Figure S3). This suggests that Rh1 and DmSERT are accommodated in separate membrane areas of the rhabdomeres. To further investigate this, electron microscopy with double gold-immunolabeling using anti-Rh1- and anti-GFP antibodies was performed on rhabdomere membranes (Figure 3F–G). 56% and 42% of the membrane structures were either positive for Rh1 (Figure 3F) or SERT-GFP (Figure 3G) respectively, and only 2% contained a Rh1/SERT-GFP mixture. Therefore, Rh1 and DmSERT indeed localize in separate membrane domains. This shows the feasibility of further analysis of the supramolecular organization of SERT and probably other recombinant proteins in rhabdomere membranes by biophysical methods like cryo-electron microscopy [41] or atomic force microscopy [42], [43].
Large-scale purification of MP targets
Some overexpression systems like Pichia pastoris display often impressive levels of MP production at a small scale but expression at a larger scale is tricky and requires sophisticated devices [44]. In order to test the scalability of the fly eye system, the fly cultures were expanded (Figure S2) and HsSERT was subjected to large scale purification. Fly heads were collected for membrane preparation [16]. A volume of 4 ml (2 g) frozen fly heads gave typically 45 mg of total MP with 0.5 mg HsSERT (1 mg DmSERT) purified routinely using an affinity column (Figure 3H). The transporters and receptors are now used for detergent optimization and crystallization trials. Taken together, the amounts obtained with the fly eye system in combination with the superior homogeneity of the protein provide the basis for further biochemical, pharmacological and structural analyses.
Discussion
We show that the expression of eukaryotic membrane proteins in the eye of transgenic Drosophila is a powerful tool for the production of functional GPCRs, neurotransmitter transporters and channels. For SERT we demonstrate that the fly eye system can be scaled up to the amounts needed for routine crystallization studies and biochemical characterization. The expression levels of a number of test cases come close to that of endogenous rhodopsin. Using a GFP tag for monitoring allows for easy in vivo and in vitro MP analysis and quality control of the fly cultures.
Specific properties of the fly eye system offer major advantages compared to conventional expression systems. These include accessibility, low cost and superior quality of the expressed proteins [14], [45]. The PRCs maintain a high turnover of rhodopsin in their specialized membrane stacks [21], [46] which relies on high-throughput MP production, folding and targeting. Being specialized and polarized cells, PRCs [47], [48] harbor the rhabdomeres as an ideal storage compartment for MPs. PRC targeting of MPs that are often toxic for the host cell might benefit from the absence of endogenous ligand or from having only minor effects on local metabolism. We observed that the capacity of the PRCs to host MPs seems almost unsaturable, as in addition to endogenous rhodopsin equivalent amounts of recombinant MP can be accommodated. Heterologous expression can reach a similar level as homologous expression as shown for the mammalian mGluRs and SERT. The fly eye system is therefore particularly suited for heterologous expression.
In conventional eukaryotic expression systems ER retention of recombinant GPCRs and transporters can indicate improper folding and is often a problem e.g. for expression in yeast. In the fly eye system the majority of the target proteins were localized entirely in rhabdomere membranes. This also demonstrates that MPs with various intrinsic signal sequences are targeted to the rhabdomeres. The expression of the channelrhodopsin ChR2 was dependent on the endogenous Rh1 levels, suggesting a co-transport to the rhabdomeres. Also, there is indication that ChR2 expressed in PRCs binds its cofactor retinal, necessary for folding and activity. In addition to the classical post-translational modifications like glycosylation [49], the PRCs can efficiently produce retinal-binding proteins, while classical eukaryotic cell cultures or cell-free expression systems would require an exogenous supply of cofactor [50], [51].
Expression of MPs in the fly eye system is also a cheap alternative to expensive eukaryotic cell cultures and their requirement to work sterile. The costs for making a transgenic fly (e.g. through collaboration or using a Drosophila injection service) and maintaining even large scale cultures is negligible (Note: the food being made of cornmeal, moult, yeast and sugar is inexpensive with only around 10$/40 large vials). In addition, making a fly can be faster than producing baculovirus stocks for overexpression in insect cells. Due to the short life cycle of the flies, about one month is sufficient starting from the DNA-construct of the target MP to the first expression test with the transgenic fly. While an overall comparison of different expression systems is straightforward concerning the costs, the comparison of yields, workload and most importantly the protein quality requires more attention. Compared with expression systems that require liters of sterile medium, the continuous fly cultures and the handling of small volumes (125 ml of flies corresponding to 25 kflies or ≥1 mg of pure target MP) provide important advantages. When the workload of membrane preparation and the quality of the purified MPs are compared with conventional expression systems, the fly eye system is superior.
Taken together, we developed a fly eye system for the heterologous and homologous expression of different classes of eukaryotic membrane proteins. It offers a number of advantages compared to conventional expression systems and is more easily accessible than one would probably imagine. The fly eye system opens the door for studying eukaryotic membrane proteins that have so far not been accessible to biochemical and biophysical studies.
Materials and Methods
Cloning strategy
MP targets: the rat mGluR5 (mGluR5), human sodium-dependent serotonin transporter (HsSERT), glutamate transporters (EAATs) and channelrhodopsin (ChR2) constructs were generous gifts from J.-P. Pin (Montpellier, France), R. D. Blakely (Nashville, USA), S. Birman (Marseille, France) and P. Hegemann (Berlin, Germany), respectively. The Drosophila melanogaster SERT (DmSERT) cDNA from the Berkeley Drosophila Genome Project was provided by BioCat/Open Biosystems (Heidelberg, Germany).
The general protocol for cloning of target MPs has been described previously [16]. Typically, the gene coding for the target MP was amplified using EcoRI and a XhoI restriction sites and cloned in frame with eGFP (GFP) into the Drosophila pUAST vector [17]. GFP was flanked at the N-terminus by a Leu-Glu linker encoded by the XhoI site followed by the TEV-cleavage site ENLYFQG and at the C-terminus by a 6-his tag (TEV-GFP-6his). The construct in pUAST was sequenced and tested for expression in Schneider S2 cells as described [16].
Transgenic fly generation
The MP-GFP construct cloned in the pUAST vector was used for classical P-element-mediated transformation of embryos [53] of the Drosophila host line w1118 (carried out by Vanedis (Oslo, Norway) or BestGene (Chino Hills, U.S.A.)). Most of the driver lines were provided by the Bloomington center. The various driver lines used in this study were eye-specific using either the minimal rhodopsin promoter for the Rh1-GAL4 line or a glass-binding enhancer element GMR (Glass Multiple Reporter or Glass Minimum Response) for the GMR-GAL4 lines [33], [54]. The GMR driver lines used a pentameric arrangement of an enhancer region of the Rh1 promoter (glass binding site). The GMR8506 driver (Bloomington #8506) has a longer pentameric repeat (38 bp, “long GMR” driver) [55] than the GMR1104 driver (29 bp, “short GMR” driver) [31]. An advantage of the GMR8506 driver is that the longer enhancer site sequence confers a strict PRC specificity [31]. The ELAV-GAL4 driver (Bloomington #8765) was chosen for its predominant induction of expression in neurons [56].
Flies were reared at room temperature on standard fly food (yeast, corn syrup and agar) in a 12 hours light/12 hours dark cycle and stocks were kept at 18°C and 60% humidity. For scaling-up the fly cultures, we opted for a continuous culture in vials at room temperature instead of large cages that are difficult to handle for fly harvesting (Figure S2). For retinal depletion experiments, flies were reared for minimum two generations on carotenoid-free medium (10% yeast, 10% sucrose, 0.02% cholesterol, 2% agar) [57]. Replenishment with retinal was performed by adding 80 µg all trans-retinal on the surface of the carotenoid-free medium [49].
Fluorescence microscopy on fly heads
For selection and sorting according to GFP fluorescence, flies were kept anaesthetized under CO2 on a glass filter (Neolab) and observed using a MZ 12-5 Leica stereomicroscope mounted with a 10× objective and equipped with an epifluorescence device (illumination path: BP 480/40 nm, dichroic mirror/reflector: 505 nm, observation path: LP 510 nm).
For rhabdomere localization experiments, flies were put asleep in CO2 and over-anaesthetized for 10 min in diethylether vapors, mounted on a needle and observed under water using a water-immersion objective [39] (HCX APO, L 20×/0.5 W or L 40×/0.80 W U-V-I, Leica, Germany) on a DM LFS microscope (Leica, light source: ebq 100 dc-1 [100 W], Jena GmbH, Germany; I3 filter set (illumination path: BP 450–490 nm, dichroic mirror/reflector: 510 nm, observation path: LP 515 nm)). The fluorescence was documented with a digital camera (DC200, Leica, Germany). Confocal laser scanning microscopy was performed on intact heads mounted in PBS between two coverslips spaced by clay on the stage of a Nikon TE2000-E inverted fluorescence microscope. Heads were subjected to series scan (300 z-stacks) with a 488 nm laser over half a mm depth to build a 3D-image of a whole eye.
Harvesting of fly heads
10 ml frozen flies in liquid nitrogen were gently shaken in a 50 ml-Falcon tube together with 5 ml of glass beads (diameter 4 mm) as described [16]. Briefly, the flies and beads were transferred on a set of sieves with decreasing meshes (Neolab #6-2380 (the three smaller meshes)) pre-cooled in liquid nitrogen. After shaking, the heads were collected from the middle compartment and stored at −80°C.
Membrane preparation
Frozen fly heads were homogenized in sucrose buffer (TRIS-HCl 50 mM, NaCl 150 mM, MgCl2 2 mM, EGTA 1 mM, Sucrose 250 mM, pH 7.4) and membranes were prepared as described [16]. It is noteworthy that fly eye tissue is much easier to homogenize than cells in culture.
Western blot analysis and quantification by fluorescence
For Western Blot, 12 µl of a sample containing 5 fly heads homogenized in 30 µl of a classical loading buffer were analyzed and detection was performed by classical enhanced chemiluminescence (ECL™, GE Healthcare) using an antibody against GFP (Biovision, Mountain View), Rh1 (4C5 ascites, DSHB, Iowa), β-tubulin (E7, DSHB, Iowa) or the Drosophila glutamate receptor (monoclonal 7G11 [45]).
Quantification of the fluorescent recombinant proteins was done in native gradient (4–10%)-polyacrylamide gel electrophoresis in the presence of n-Dodecyl-β-D-maltoside (DDM) or digitonin 0.1% in the gel [58]. Six fly heads were homogenized in 8 µl sucrose buffer complemented with the protease inhibitors (see membrane preparation). DDM or digitonin was added (final concentration 1%) for solubilization and left on ice for two hours. The samples were ultracentrifuged at 4°C for 10 min and 3 µl supernatant was mixed with 3 µl native loading buffer (TRIS-HCl 100 mM, glycerol 20%, Bromophenol blue). The samples were loaded in parallel with a GFP standard curve (eGFP, Biovision, Mountain View) and run at 180 V in the dark for about three hours. The gel was analyzed using the Ettan DIGE imager (GE Healthcare). Image J software was used to integrate the pixel values.
Ligand binding
2.5 µg Drosophila head membranes [14] from flies expressing HsSERT were incubated in 100 µl sodium phosphate buffer 50 mM, NaCl 100 mM, BSA 0.2% (pH 7.2) with [125I]-RTI-55 (Perkin Elmer) and increasing concentrations of racemic citalopram (Sigma) or cocaine (Sigma). Bound and free were separated by rapid filtration on a GF/B glass filter saturated with BSA 1% and polyethylene imine 0.5% using a Brandel M-48 harvester. GraphPad Prism 4.0 software was used for curve fitting and data analysis.
Preparation of rhabdomere membranes
The eyes from 50 flies expressing HsSERT were dissected and retina membranes were released using a reciprocating shaker (Mini-Bead-Beater, GlenMills, New Jersey) in the presence of 0.1 mm zirconia/silica beads (50 mg) in 125 µl ice-cold Optiprep 10%, HEPES-NaOH 10 mM, NaCl 120 mM, KCl 4 mM, sucrose 32 mM, pH 7,4 buffer. The resulting membranes were collected in the 35% Optiprep-fraction of an Optiprep-gradient (10 to 55%) after centrifugation 2.5 h at 20,000 g, 20°C. The presence of both rhodopsin and HsSERT in this fraction was confirmed by Western Blot using the monoclonal 4C5 and the GFP antibody, respectively (not shown). Alternatively, the use of an ULTRA-TURRAX disperser instead of the Mini-Bead-Beater produced smaller membranes containing separated Rh1- and HsSERT rhabdomere membrane sub-populations that could be recovered on a 40% and 20% Optiprep-gradient fraction, respectively (Figure S3).
EM double immunogold labelling
Membranes resuspended in Ringer buffer at 0.1 mg/ml were adsorbed on 300-mesh carbon-coated EM grids (EM Sciences, Munich, Germany) for 2 min at RT. For immunogold labeling of GFP fusion proteins, unspecific labeling was blocked by incubating the grids on blocking solution (0.8% bovine serum albumin, 0.1% fish skin gelatin in PBS) for 10 min at RT. The samples were then double-labeled according to Slot et al. [59] except that the antibody and protein A incubation times were reduced to 15 and 10 min, respectively. The antibody against GFP (Molecular Probes, dil. 1/200) was used first followed by rhodopsin antibody (4C5, DSHB Iowa, dil. 1/1000) and rabbit anti-mouse (DaKoCytomation, Denmark). After the last incubation with protein A coupled to gold (University of Utrecht, the Netherlands), the grids were washed 5 times in PBS, 5 times in water and the samples were embedded by looping out the grids in a mixture of 8 parts methyl cellulose (Sigma, 25 centipoise; 2%) and 2 parts uranyl acetate (Fluka, Heidelberg, Germany, 3% in water) and removing excess liquid on a filter. Grids were analyzed with a Zeiss electron microscope EM10 and images taken with a Gatan MultiScan™ camera and Digital Micrograph™ software and further processed using Adobe Photoshop CS3.
Additional methods
Additional information on large scale fly cultures is available in Figure S2.
Supporting Information
ChR2 expression depends on retinal. Like rhodopsin, ChR2 is a retinal-binding protein1. Transgenic flies expressing ChR2-GFP grown on carotenoid-depleted food2, which prevents retinal synthesis, showed a clear drop in ChR2 expression (lane 2) compared to flies grown on normal medium (lane 1). ChR2 expression was recovered by replenishing the food with synthetic all-trans retinal (lane 3), indicating that the observed effect is specific for retinal. Lane 4 shows a driver fly as a control. A Western blot using a GFP antibody with two fly heads is shown. The same blot was analyzed with antibodies against β-tubulin as a control of protein load and against Rh1, respectively. The well-known dependence on retinal is observed for endogenous Rh1 expression3. The requirement of the chromophore for ChR2 expression could be a prerequisite for folding or could indicate that it follows the endogenous Rh1 levels.
(DOC)
Large scale Drosophila cultures. 1. Initial cultures: 12 crosses (in 12 vials) were made between the UAS-MP-GFP fly line and the driver line in small 2,5 cm-diameter vials (10 ml fly food). Alternatively, a stable expressing line GMR-GAL4;UAS-MP-GFP can be used (described in4). 2. Egg-laying Flies: the offspring was collected into larger vials (35 ml fly food) i.e. flies from 4 small vials transferred in one large vial with 5 cm diameter. Those flies of the first generation were used to lay eggs in large vials and were passed every fourth day in new large 5 cm-diameter vials. 3. Harvesting Tour: the vials emptied of flies and full of larvae were used for the fly harvesting. The whole culture consisted of 12 small vials (first generation flies), around twelve larger vials used for laying eggs (first generation flies) and three racks each containing 40 large harvesting-vials (third and fourth generation flies). The time required to scale-up the culture for MP purification in milligram amounts is about one month and the culture is kept running continuously. Harvesting by flushing CO2 into the 3×40 vials to anaesthetize the flies and freeze them in liquid nitrogen, takes about 40 min. The harvested flies were stored at −80°C. Note: for fly harvesting vials were better than the large cages utilized for larvae collection5.
(DOC)
Endogenous Rh1 and recombinant Hs SERT localize in separate rhabdomere domains. The heads of 50 flies expressing HsSERT under the control of the GMR1104 driver were dispersed with an Ultra Turrax in 300 µl of a buffer containing NaCl 120 mM, KCl 4 mM, sucrose 30 mM, Hepes-NaOH 10 mM pH 7.4, 8% Optiprep® and protease inhibitors (Complete®). The resulting membranes were loaded on the top of an Optiprep gradient (10 to 55%) in the same buffer, centrifuged 2.5 h at 20,000 g, 20°C and the fractions (1 to 8 from top to bottom, respectively) were analyzed by Western blot with an antibody against GFP or Rh1, respectively. The results indicate that HsSERT, which localizes in rhabdomeres (Figure 3E), accumulates in different membrane areas than endogenous Rh1. HsSERT-containing membranes were less dense than Rh1 domains. This difference is most likely due to the density of the membrane proteins packed in these areas.
(DOC)
Primer for MP expression in fly eyes.
(DOC)
Acknowledgments
The authors thank Silke Adrian and Lisa Jödicke for excellent technical help as well as the Bloomington Drosophila Stock Center (Indiana University, US) for some driver fly lines. We thank Ulrike Engel from the Nikon Imaging Center (Heidelberg University) for initial training and access to confocal microscopy. We thank Claude Desplan (New York, US) for helpful discussions, Gunter Merdes (ZMBH, Heidelberg) and Armin Huber (Hohenheim University) for advise and some fly lines, and Dimitris Liakopoulos (BZH) for support in confocal microscopy.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by the European Community Specific Targeted Research Project grant IMPS (FP6-2003-LifeSciHealth 513770). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Wallin E, von Heijne G. Genome-wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms. Protein Sci. 1998;7:1029–1038. doi: 10.1002/pro.5560070420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nat Rev Drug Discov. 2006;5:993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 3.Midgett CR, Madden DR. Breaking the bottleneck: eukaryotic membrane protein expression for high-resolution structural studies. J Struct Biol. 2007;160:265–274. doi: 10.1016/j.jsb.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 4.Junge F, Schneider B, Reckel S, Schwarz D, Dotsch V, et al. Large-scale production of functional membrane proteins. Cell Mol Life Sci. 2008;65:1729–1755. doi: 10.1007/s00018-008-8067-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berezovsky IN, Shakhnovich EI. Physics and evolution of thermophilic adaptation. Proc Natl Acad Sci U S A. 2005;102:12742–12747. doi: 10.1073/pnas.0503890102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miroux B, Walker JE. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J Mol Biol. 1996;260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- 7.Henry LK, Meiler J, Blakely RD. Bound to be different: neurotransmitter transporters meet their bacterial cousins. Mol Interv. 2007;7:306–309. doi: 10.1124/mi.7.6.4. [DOI] [PubMed] [Google Scholar]
- 8.Okada T, Palczewski K. Crystal structure of rhodopsin: implications for vision and beyond. Curr Opin Struct Biol. 2001;11:420–426. doi: 10.1016/s0959-440x(00)00227-x. [DOI] [PubMed] [Google Scholar]
- 9.Armstrong CM. Voltage-gated K channels. Sci STKE. 2003;2003:re10. doi: 10.1126/stke.2003.188.re10. [DOI] [PubMed] [Google Scholar]
- 10.Conn PJ, Christopoulos A, Lindsley CW. Allosteric modulators of GPCRs: a novel approach for the treatment of CNS disorders. Nat Rev Drug Discov. 2009;8:41–54. doi: 10.1038/nrd2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Graaf C, Rognan D. Customizing G Protein-coupled receptor models for structure-based virtual screening. Curr Pharm Des. 2009;15:4026–4048. doi: 10.2174/138161209789824786. [DOI] [PubMed] [Google Scholar]
- 12.Fraley ME. Positive allosteric modulators of the metabotropic glutamate receptor 2 for the treatment of schizophrenia. Expert Opin Ther Pat. 2009;19:1259–1275. doi: 10.1517/13543770903045009. [DOI] [PubMed] [Google Scholar]
- 13.Griffith DA, Delipala C, Leadsham J, Jarvis SM, Oesterhelt D. A novel yeast expression system for the overproduction of quality-controlled membrane proteins. FEBS Lett. 2003;553:45–50. doi: 10.1016/s0014-5793(03)00952-9. [DOI] [PubMed] [Google Scholar]
- 14.Eroglu C, Cronet P, Panneels V, Beaufils P, Sinning I. Functional reconstitution of purified metabotropic glutamate receptor expressed in the fly eye. EMBO Rep. 2002;3:491–496. doi: 10.1093/embo-reports/kvf088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eroglu C, Brugger B, Wieland F, Sinning I. Glutamate-binding affinity of Drosophila metabotropic glutamate receptor is modulated by association with lipid rafts. Proc Natl Acad Sci U S A. 2003;100:10219–10224. doi: 10.1073/pnas.1737042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Panneels V, Sinning I. Membrane protein expression in the eyes of transgenic flies. Methods Mol Biol. 2010;601:135–147. doi: 10.1007/978-1-60761-344-2_9. [DOI] [PubMed] [Google Scholar]
- 17.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 18.Drew D, Newstead S, Sonoda Y, Kim H, von Heijne G, et al. GFP-based optimization scheme for the overexpression and purification of eukaryotic membrane proteins in Saccharomyces cerevisiae. Nat Protoc. 2008;3:784–798. doi: 10.1038/nprot.2008.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Waldo GS, Standish BM, Berendzen J, Terwilliger TC. Rapid protein-folding assay using green fluorescent protein. Nat Biotechnol. 1999;17:691–695. doi: 10.1038/10904. [DOI] [PubMed] [Google Scholar]
- 20.Zuker CS. The biology of vision of Drosophila. Proc Natl Acad Sci U S A. 1996;93:571–576. doi: 10.1073/pnas.93.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hardie RC. Phototransduction in Drosophila melanogaster. J Exp Biol. 2001;204:3403–3409. doi: 10.1242/jeb.204.20.3403. [DOI] [PubMed] [Google Scholar]
- 22.Vought BW, Salcedo E, Chadwell LV, Britt SG, Birge RR, et al. Characterization of the primary photointermediates of Drosophila rhodopsin. Biochemistry. 2000;39:14128–14137. doi: 10.1021/bi001135k. [DOI] [PubMed] [Google Scholar]
- 23.Foord SM, Bonner TI, Neubig RR, Rosser EM, Pin JP, et al. International Union of Pharmacology. XLVI. G protein-coupled receptor list. Pharmacol Rev. 2005;57:279–288. doi: 10.1124/pr.57.2.5. [DOI] [PubMed] [Google Scholar]
- 24.Oksche A, Rosenthal W. The molecular basis of nephrogenic diabetes insipidus. J Mol Med. 1998;76:326–337. doi: 10.1007/s001090050224. [DOI] [PubMed] [Google Scholar]
- 25.Grisshammer R, Tate CG. Overexpression of integral membrane proteins for structural studies. Q Rev Biophys. 1995;28:315–422. doi: 10.1017/s0033583500003504. [DOI] [PubMed] [Google Scholar]
- 26.Nazari R, Joshi S. CCR5 as target for HIV-1 gene therapy. Curr Gene Ther. 2008;8:264–272. doi: 10.2174/156652308785160674. [DOI] [PubMed] [Google Scholar]
- 27.Paulsen R, Schwemer J. Biogenesis of blowfly photoreceptor membranes is regulated by 11-cis-retinal. Eur J Biochem. 1983;137:609–614. doi: 10.1111/j.1432-1033.1983.tb07869.x. [DOI] [PubMed] [Google Scholar]
- 28.Koenig J, Merriam J. Autosomal ERG mutants. Dros Inf Serv. 1977;42:50–51. [Google Scholar]
- 29.Kumar JP, Ready DF. Rhodopsin plays an essential structural role in Drosophila photoreceptor development. Development. 1995;121:4359–4370. doi: 10.1242/dev.121.12.4359. [DOI] [PubMed] [Google Scholar]
- 30.Kateriya S, Nagel G, Bamberg E, Hegemann P. “Vision” in single-celled algae. News Physiol Sci. 2004;19:133–137. doi: 10.1152/nips.01517.2004. [DOI] [PubMed] [Google Scholar]
- 31.Ellis MC, O'Neill EM, Rubin GM. Expression of Drosophila glass protein and evidence for negative regulation of its activity in non-neuronal cells by another DNA-binding protein. Development. 1993;119:855–865. doi: 10.1242/dev.119.3.855. [DOI] [PubMed] [Google Scholar]
- 32.Moses K, Ellis MC, Rubin GM. The glass gene encodes a zinc-finger protein required by Drosophila photoreceptor cells. Nature. 1989;340:531–536. doi: 10.1038/340531a0. [DOI] [PubMed] [Google Scholar]
- 33.Moses K, Rubin GM. Glass encodes a site-specific DNA-binding protein that is regulated in response to positional signals in the developing Drosophila eye. Genes Dev. 1991;5:583–593. doi: 10.1101/gad.5.4.583. [DOI] [PubMed] [Google Scholar]
- 34.Gasparini F, Kuhn R, Pin JP. Allosteric modulators of group I metabotropic glutamate receptors: novel subtype-selective ligands and therapeutic perspectives. Curr Opin Pharmacol. 2002;2:43–49. doi: 10.1016/s1471-4892(01)00119-9. [DOI] [PubMed] [Google Scholar]
- 35.Niswender CM, Conn PJ. Metabotropic glutamate receptors: physiology, pharmacology, and disease. Annu Rev Pharmacol Toxicol. 50:295–322. doi: 10.1146/annurev.pharmtox.011008.145533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tate CG, Haase J, Baker C, Boorsma M, Magnani F, et al. Comparison of seven different heterologous protein expression systems for the production of the serotonin transporter. Biochim Biophys Acta. 2003;1610:141–153. doi: 10.1016/s0005-2736(02)00719-8. [DOI] [PubMed] [Google Scholar]
- 37.Barker EL, Perlman MA, Adkins EM, Houlihan WJ, Pristupa ZB, et al. High affinity recognition of serotonin transporter antagonists defined by species-scanning mutagenesis. An aromatic residue in transmembrane domain I dictates species-selective recognition of citalopram and mazindol. J Biol Chem. 1998;273:19459–19468. doi: 10.1074/jbc.273.31.19459. [DOI] [PubMed] [Google Scholar]
- 38.Ramamoorthy S, Bauman AL, Moore KR, Han H, Yang-Feng T, et al. Antidepressant- and cocaine-sensitive human serotonin transporter: molecular cloning, expression, and chromosomal localization. Proc Natl Acad Sci U S A. 1993;90:2542–2546. doi: 10.1073/pnas.90.6.2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pichaud F, Desplan C. A new visualization approach for identifying mutations that affect differentiation and organization of the Drosophila ommatidia. Development. 2001;128:815–826. doi: 10.1242/dev.128.6.815. [DOI] [PubMed] [Google Scholar]
- 40.Harris WA, Ready DF, Lipson ED, Hudspeth AJ, Stark WS. Vitamin A deprivation and Drosophila photopigments. Nature. 1977;266:648–650. doi: 10.1038/266648a0. [DOI] [PubMed] [Google Scholar]
- 41.Bartesaghi A, Subramaniam S. Membrane protein structure determination using cryo-electron tomography and 3D image averaging. Curr Opin Struct Biol. 2009;19:402–407. doi: 10.1016/j.sbi.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muller DJ, Wu N, Palczewski K. Vertebrate membrane proteins: structure, function, and insights from biophysical approaches. Pharmacol Rev. 2008;60:43–78. doi: 10.1124/pr.107.07111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scheuring S, Sturgis JN. Atomic force microscopy of the bacterial photosynthetic apparatus: plain pictures of an elaborate machinery. Photosynth Res. 2009 doi: 10.1007/s11120-009-9413-7. [DOI] [PubMed] [Google Scholar]
- 44.Macauley-Patrick S, Fazenda ML, McNeil B, Harvey LM. Heterologous protein production using the Pichia pastoris expression system. Yeast. 2005;22:249–270. doi: 10.1002/yea.1208. [DOI] [PubMed] [Google Scholar]
- 45.Panneels V, Eroglu C, Cronet P, Sinning I. Pharmacological characterization and immunoaffinity purification of metabotropic glutamate receptor from Drosophila overexpressed in Sf9 cells. Protein Expr Purif. 2003;30:275–282. doi: 10.1016/s1046-5928(03)00100-1. [DOI] [PubMed] [Google Scholar]
- 46.Hardie RC, Raghu P, Moore S, Juusola M, Baines RA, et al. Calcium influx via TRP channels is required to maintain PIP2 levels in Drosophila photoreceptors. Neuron. 2001;30:149–159. doi: 10.1016/s0896-6273(01)00269-0. [DOI] [PubMed] [Google Scholar]
- 47.Ready DF. A multifaceted approach to neural development. Trends Neurosci. 1989;12:102–110. doi: 10.1016/0166-2236(89)90166-5. [DOI] [PubMed] [Google Scholar]
- 48.Knust E, Bossinger O. Composition and formation of intercellular junctions in epithelial cells. Science. 2002;298:1955–1959. doi: 10.1126/science.1072161. [DOI] [PubMed] [Google Scholar]
- 49.Ahmad ST, Natochin M, Barren B, Artemyev NO, O'Tousa JE. Heterologous expression of bovine rhodopsin in Drosophila photoreceptor cells. Invest Ophthalmol Vis Sci. 2006;47:3722–3728. doi: 10.1167/iovs.06-0281. [DOI] [PubMed] [Google Scholar]
- 50.Klaassen CH, Bovee-Geurts PH, Decaluwe GL, DeGrip WJ. Large-scale production and purification of functional recombinant bovine rhodopsin with the use of the baculovirus expression system. Biochem J. 1999;342(Pt 2):293–300. [PMC free article] [PubMed] [Google Scholar]
- 51.Cappuccio JA, Blanchette CD, Sulchek TA, Arroyo ES, Kralj JM, et al. Cell-free co-expression of functional membrane proteins and apolipoprotein, forming soluble nanolipoprotein particles. Mol Cell Proteomics. 2008;7:2246–2253. doi: 10.1074/mcp.M800191-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamashita A, Singh SK, Kawate T, Jin Y, Gouaux E. Crystal structure of a bacterial homologue of Na+/Cl–dependent neurotransmitter transporters. Nature. 2005;437:215–223. doi: 10.1038/nature03978. [DOI] [PubMed] [Google Scholar]
- 53.Voie AM, Cohen S. Press SDA, editor. Germ-line transformation of Drosophila melanogaster. In cell biology: a laboratory handbook. 1998. pp. 510–517.
- 54.Mismer D, Rubin GM. Analysis of the promoter of the ninaE opsin gene in Drosophila melanogaster. Genetics. 1987;116:565–578. doi: 10.1093/genetics/116.4.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wernet MF, Labhart T, Baumann F, Mazzoni EO, Pichaud F, et al. Homothorax switches function of Drosophila photoreceptors from color to polarized light sensors. Cell. 2003;115:267–279. doi: 10.1016/s0092-8674(03)00848-1. [DOI] [PubMed] [Google Scholar]
- 56.Yao KM, White K. Neural specificity of elav expression: defining a Drosophila promoter for directing expression to the nervous system. J Neurochem. 1994;63:41–51. doi: 10.1046/j.1471-4159.1994.63010041.x. [DOI] [PubMed] [Google Scholar]
- 57.Nichols R, Pak WL. Characterization of Drosophila melanogaster rhodopsin. J Biol Chem. 1985;260:12670–12674. [PubMed] [Google Scholar]
- 58.Wittig I, Karas M, Schagger H. High resolution clear native electrophoresis for in-gel functional assays and fluorescence studies of membrane protein complexes. Mol Cell Proteomics. 2007;6:1215–1225. doi: 10.1074/mcp.M700076-MCP200. [DOI] [PubMed] [Google Scholar]
- 59.Slot JW, Geuze HJ, Gigengack S, Lienhard GE, James DE. Immuno-localization of the insulin regulatable glucose transporter in brown adipose tissue of the rat. J Cell Biol. 1991;113:123–135. doi: 10.1083/jcb.113.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ChR2 expression depends on retinal. Like rhodopsin, ChR2 is a retinal-binding protein1. Transgenic flies expressing ChR2-GFP grown on carotenoid-depleted food2, which prevents retinal synthesis, showed a clear drop in ChR2 expression (lane 2) compared to flies grown on normal medium (lane 1). ChR2 expression was recovered by replenishing the food with synthetic all-trans retinal (lane 3), indicating that the observed effect is specific for retinal. Lane 4 shows a driver fly as a control. A Western blot using a GFP antibody with two fly heads is shown. The same blot was analyzed with antibodies against β-tubulin as a control of protein load and against Rh1, respectively. The well-known dependence on retinal is observed for endogenous Rh1 expression3. The requirement of the chromophore for ChR2 expression could be a prerequisite for folding or could indicate that it follows the endogenous Rh1 levels.
(DOC)
Large scale Drosophila cultures. 1. Initial cultures: 12 crosses (in 12 vials) were made between the UAS-MP-GFP fly line and the driver line in small 2,5 cm-diameter vials (10 ml fly food). Alternatively, a stable expressing line GMR-GAL4;UAS-MP-GFP can be used (described in4). 2. Egg-laying Flies: the offspring was collected into larger vials (35 ml fly food) i.e. flies from 4 small vials transferred in one large vial with 5 cm diameter. Those flies of the first generation were used to lay eggs in large vials and were passed every fourth day in new large 5 cm-diameter vials. 3. Harvesting Tour: the vials emptied of flies and full of larvae were used for the fly harvesting. The whole culture consisted of 12 small vials (first generation flies), around twelve larger vials used for laying eggs (first generation flies) and three racks each containing 40 large harvesting-vials (third and fourth generation flies). The time required to scale-up the culture for MP purification in milligram amounts is about one month and the culture is kept running continuously. Harvesting by flushing CO2 into the 3×40 vials to anaesthetize the flies and freeze them in liquid nitrogen, takes about 40 min. The harvested flies were stored at −80°C. Note: for fly harvesting vials were better than the large cages utilized for larvae collection5.
(DOC)
Endogenous Rh1 and recombinant Hs SERT localize in separate rhabdomere domains. The heads of 50 flies expressing HsSERT under the control of the GMR1104 driver were dispersed with an Ultra Turrax in 300 µl of a buffer containing NaCl 120 mM, KCl 4 mM, sucrose 30 mM, Hepes-NaOH 10 mM pH 7.4, 8% Optiprep® and protease inhibitors (Complete®). The resulting membranes were loaded on the top of an Optiprep gradient (10 to 55%) in the same buffer, centrifuged 2.5 h at 20,000 g, 20°C and the fractions (1 to 8 from top to bottom, respectively) were analyzed by Western blot with an antibody against GFP or Rh1, respectively. The results indicate that HsSERT, which localizes in rhabdomeres (Figure 3E), accumulates in different membrane areas than endogenous Rh1. HsSERT-containing membranes were less dense than Rh1 domains. This difference is most likely due to the density of the membrane proteins packed in these areas.
(DOC)
Primer for MP expression in fly eyes.
(DOC)