Abstract
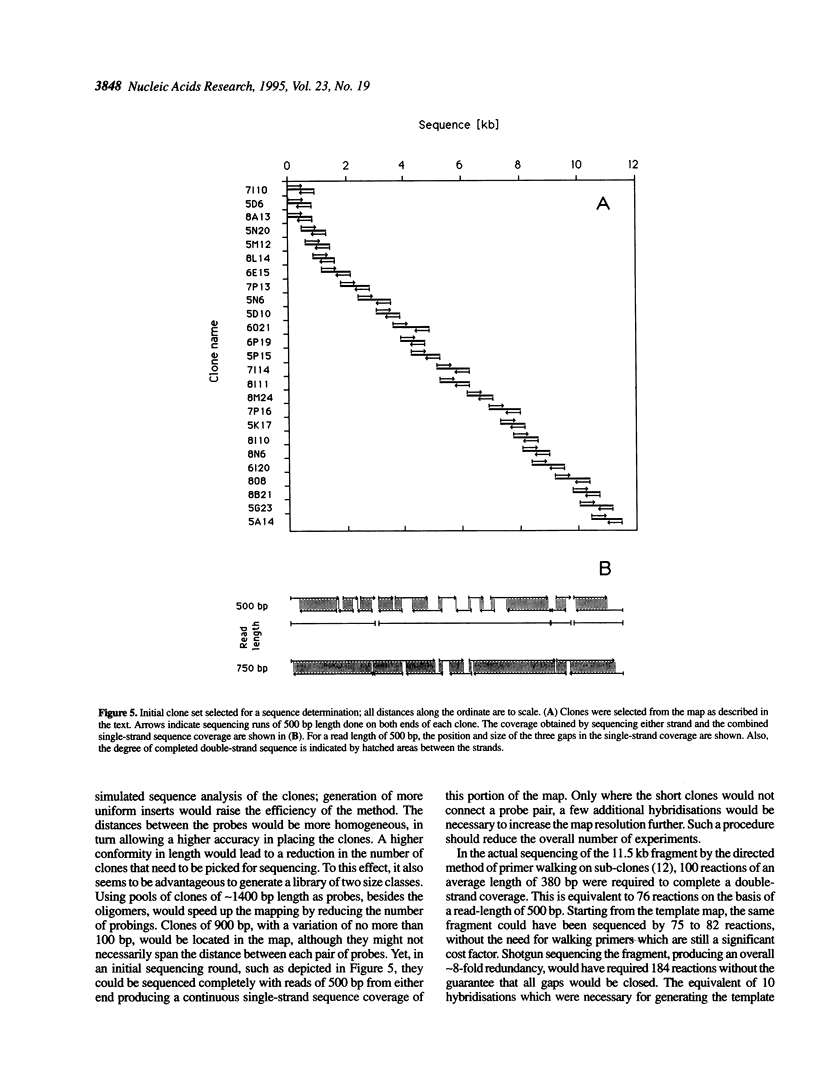
To test the effectiveness of ordering shotgun DNA-templates prior to sequence analysis, the 450 kb left arm of yeast chromosome XII was randomly subcloned into a phagemid vector. Clones were ordered by hybridisation to an average map density of one new insert every 125 bp and are currently used for sequencing the chromosomal fragment. An 11.5 kb overlap between the template map and a DNA fragment that had been sequenced earlier allowed an independent evaluation of the strategy's effectiveness. To this end, clones were selected from the map and tag-sequenced from either end, thus comparing the map position with the actual location within the 11.5 kb. Of 65 selected clones, taken mostly at random from a total of 423, 58 mapped on average about a quarter of a clone length around their predicted position, with the other seven being between 0.6 and 1.5 clone length off. 75-86 sequencing reactions on clones selected from the map would have been sufficient for completely sequencing both strands of the 11.5 kb fragment. The results demonstrate the efficacy of such template sorting, considerably assisting sequencing at relatively little cost on the mapping level.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Deininger P. L. Random subcloning of sonicated DNA: application to shotgun DNA sequence analysis. Anal Biochem. 1983 Feb 15;129(1):216–223. doi: 10.1016/0003-2697(83)90072-6. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Hoheisel J. D. Application of hybridization techniques to genome mapping and sequencing. Trends Genet. 1994 Mar;10(3):79–83. doi: 10.1016/0168-9525(94)90229-1. [DOI] [PubMed] [Google Scholar]
- Hoheisel J. D., Lennon G. G., Zehetner G., Lehrach H. Use of high coverage reference libraries of Drosophila melanogaster for relational data analysis. A step towards mapping and sequencing of the genome. J Mol Biol. 1991 Aug 20;220(4):903–914. doi: 10.1016/0022-2836(91)90362-a. [DOI] [PubMed] [Google Scholar]
- Hoheisel J. D., Maier E., Mott R., McCarthy L., Grigoriev A. V., Schalkwyk L. C., Nizetic D., Francis F., Lehrach H. High resolution cosmid and P1 maps spanning the 14 Mb genome of the fission yeast S. pombe. Cell. 1993 Apr 9;73(1):109–120. doi: 10.1016/0092-8674(93)90164-l. [DOI] [PubMed] [Google Scholar]
- Koop B. F., Rowan L., Chen W. Q., Deshpande P., Lee H., Hood L. Sequence length and error analysis of Sequenase and automated Taq cycle sequencing methods. Biotechniques. 1993 Mar;14(3):442–447. [PubMed] [Google Scholar]
- Maier E., Roest Crollius H., Lehrach H. Hybridisation techniques on gridded high density DNA and in situ colony filters based on fluorescence detection. Nucleic Acids Res. 1994 Aug 25;22(16):3423–3424. doi: 10.1093/nar/22.16.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead D. A., Szczesna-Skorupa E., Kemper B. Single-stranded DNA 'blue' T7 promoter plasmids: a versatile tandem promoter system for cloning and protein engineering. Protein Eng. 1986 Oct-Nov;1(1):67–74. doi: 10.1093/protein/1.1.67. [DOI] [PubMed] [Google Scholar]
- Meier-Ewert S., Maier E., Ahmadi A., Curtis J., Lehrach H. An automated approach to generating expressed sequence catalogues. Nature. 1993 Jan 28;361(6410):375–376. doi: 10.1038/361375a0. [DOI] [PubMed] [Google Scholar]
- Mott R., Grigoriev A., Maier E., Hoheisel J., Lehrach H. Algorithms and software tools for ordering clone libraries: application to the mapping of the genome of Schizosaccharomyces pombe. Nucleic Acids Res. 1993 Apr 25;21(8):1965–1974. doi: 10.1093/nar/21.8.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl F. M., Thomae R., Karst A. Temperature dependence of the activity of DNA-modifying enzymes: endonucleases and DNA ligase. Eur J Biochem. 1982 Mar;123(1):141–152. doi: 10.1111/j.1432-1033.1982.tb06510.x. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholler P., Schwarz S., Hoheisel J. D. High-resolution cosmid mapping of the left arm of Saccharomyces cerevisiae chromosome XII; a first step towards an ordered sequencing approach. Yeast. 1995 Jun 15;11(7):659–666. doi: 10.1002/yea.320110706. [DOI] [PubMed] [Google Scholar]