Abstract
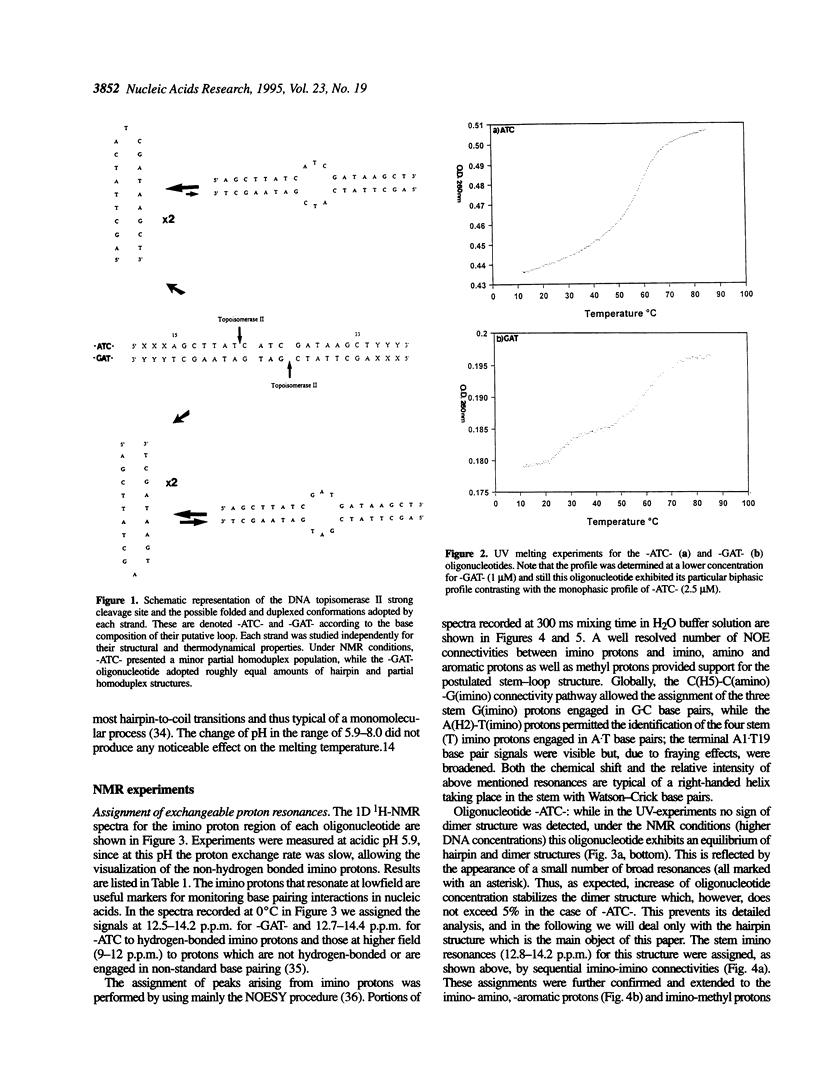
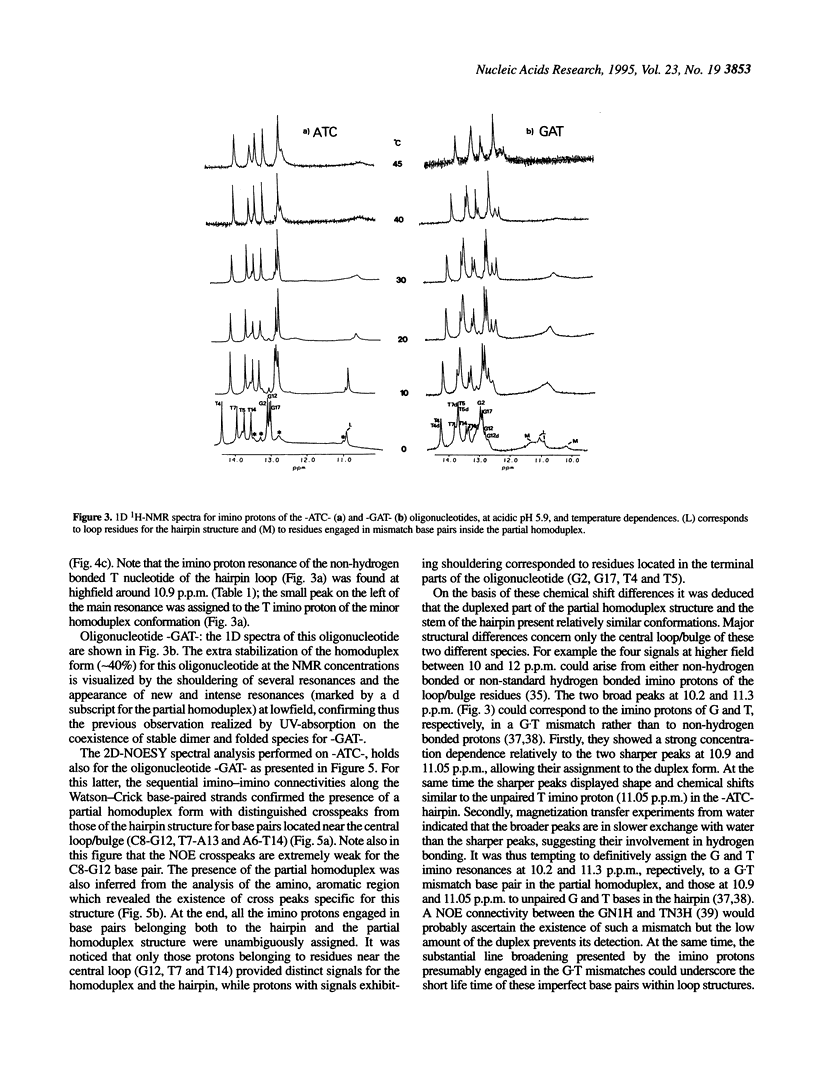
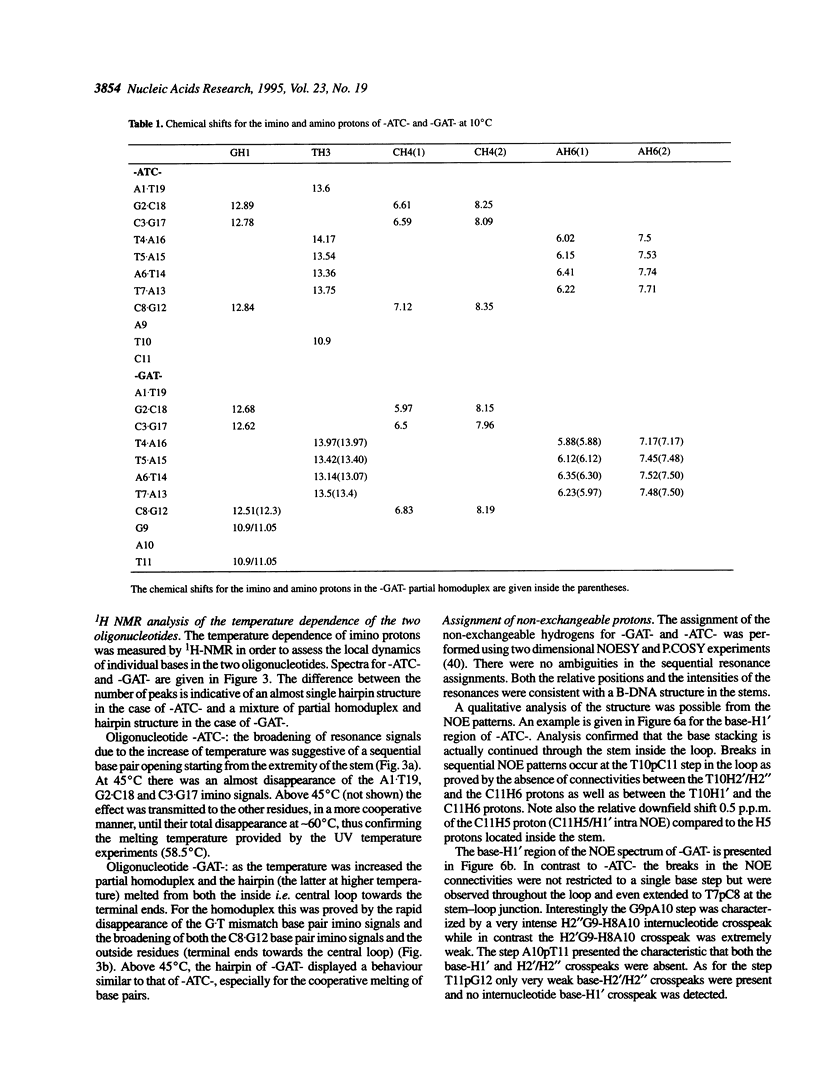
1H- and 31P-NMR and UV-absorption studies were carried out with the oligonucleotide strands d(AGCT-TATC-ATC-GATAAGCT) (-ATC-) and d(AGCTTATC-GAT-GATAAGCT) (-GAT-) contained in the strongest and salt resistant cleavage site for topoisomerase II in pBR322 DNA. We found that the two oligonucleotides were stabilized under a hairpin structure characterized by a eight base pair stem and a three base loop at low DNA and salt concentrations. In such experimental conditions, only the -GAT- oligonucleotide displayed a partial homoduplex structure in slow equilibrium with its folded structure. Temperature dependencies of imino protons showed that the partial homoduplex of -GAT- melted at a lower temperature than the hairpin structure. It was suggested that the appearance of the partial homoduplex in -GAT- is related to the formation of two stabilizing (G.T) mismatched base pairs in the central loop of this structure. Finally, it was inferred from the dispersion of chemical shifts in the 31P-NMR spectra that the distortions affecting the backbone of the hairpin loop are larger in the case of -ATC- compared with -GAT-. At the same time NOEs proved that the base stacking was stronger within the loop of the -ATC- hairpin.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aboul-ela F., Koh D., Tinoco I., Jr, Martin F. H. Base-base mismatches. Thermodynamics of double helix formation for dCA3XA3G + dCT3YT3G (X, Y = A,C,G,T). Nucleic Acids Res. 1985 Jul 11;13(13):4811–4824. doi: 10.1093/nar/13.13.4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avizonis D. Z., Kearns D. R. Kinetic and thermodynamic characterization of DNA duplex-hairpin interconversion for two DNA decamers: d(CAACGGGTTG) and d(CAACCCGTTG). Biopolymers. 1995 Feb;35(2):187–200. doi: 10.1002/bip.360350207. [DOI] [PubMed] [Google Scholar]
- Avizonis D. Z., Kearns D. R. Structural characterization of d(CAACCCGTTG) and d(CAACGGGTTG) mini-hairpin loops by heteronuclear NMR: the effects of purines versus pyrimidines in DNA hairpins. Nucleic Acids Res. 1995 Apr 11;23(7):1260–1268. doi: 10.1093/nar/23.7.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blommers M. J., Walters J. A., Haasnoot C. A., Aelen J. M., van der Marel G. A., van Boom J. H., Hilbers C. W. Effects of base sequence on the loop folding in DNA hairpins. Biochemistry. 1989 Sep 5;28(18):7491–7498. doi: 10.1021/bi00444a049. [DOI] [PubMed] [Google Scholar]
- Fossé P., René B., Le Bret M., Paoletti C., Saucier J. M. Sequence requirements for mammalian topoisomerase II mediated DNA cleavage stimulated by an ellipticine derivative. Nucleic Acids Res. 1991 Jun 11;19(11):2861–2868. doi: 10.1093/nar/19.11.2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froelich-Ammon S. J., Gale K. C., Osheroff N. Site-specific cleavage of a DNA hairpin by topoisomerase II. DNA secondary structure as a determinant of enzyme recognition/cleavage. J Biol Chem. 1994 Mar 11;269(10):7719–7725. [PubMed] [Google Scholar]
- Gorenstein D. G., Luxon B. A. High-resolution phosphorus nuclear magnetic resonance spectra of yeast phenylalanine transfer ribonucleic acid. Melting curves and relaxation effects. Biochemistry. 1979 Aug 21;18(17):3796–3804. doi: 10.1021/bi00584a024. [DOI] [PubMed] [Google Scholar]
- James J. K., Tinoco I., Jr The solution structure of a d[C(TTCG)G] DNA hairpin and comparison to the unusually stable RNA analogue. Nucleic Acids Res. 1993 Jul 11;21(14):3287–3293. doi: 10.1093/nar/21.14.3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallick D. A., Wemmer D. E. 1H NMR of 5'CGCGTATATACGCG3', a duplex and a four-membered loop. Nucleic Acids Res. 1991 Nov 11;19(21):6041–6046. doi: 10.1093/nar/19.21.6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamhasni S., Larsen A. K., Barray M., Monnot M., Delain E., Fermandjian S. Changes of self-association, secondary structure, and biological activity properties of topoisomerase II under varying salt conditions. Biochemistry. 1995 Mar 21;34(11):3632–3639. doi: 10.1021/bi00011a018. [DOI] [PubMed] [Google Scholar]
- Liu L. F. DNA topoisomerase poisons as antitumor drugs. Annu Rev Biochem. 1989;58:351–375. doi: 10.1146/annurev.bi.58.070189.002031. [DOI] [PubMed] [Google Scholar]
- Marion D., Wüthrich K. Application of phase sensitive two-dimensional correlated spectroscopy (COSY) for measurements of 1H-1H spin-spin coupling constants in proteins. Biochem Biophys Res Commun. 1983 Jun 29;113(3):967–974. doi: 10.1016/0006-291x(83)91093-8. [DOI] [PubMed] [Google Scholar]
- Marky L. A., Blumenfeld K. S., Kozlowski S., Breslauer K. J. Salt-dependent conformational transitions in the self-complementary deoxydodecanucleotide d(CGCAATTCGCG): evidence for hairpin formation. Biopolymers. 1983 Apr;22(4):1247–1257. doi: 10.1002/bip.360220416. [DOI] [PubMed] [Google Scholar]
- McMurray C. T., Juranić N., Chandrasekaran S., Macura S., Li Y., Jones R. L., Wilson W. D. Hairpin formation within the human enkephalin enhancer region. 2. Structural studies. Biochemistry. 1994 Oct 4;33(39):11960–11970. doi: 10.1021/bi00205a035. [DOI] [PubMed] [Google Scholar]
- Moe J. G., Russu I. M. Kinetics and energetics of base-pair opening in 5'-d(CGCGAATTCGCG)-3' and a substituted dodecamer containing G.T mismatches. Biochemistry. 1992 Sep 15;31(36):8421–8428. doi: 10.1021/bi00151a005. [DOI] [PubMed] [Google Scholar]
- Monnot M., Mauffret O., Simon V., Lescot E., Psaume B., Saucier J. M., Charra M., Belehradek J., Jr, Fermandjian S. DNA-drug recognition and effects on topoisomerase II-mediated cytotoxicity. A three-mode binding model for ellipticine derivatives. J Biol Chem. 1991 Jan 25;266(3):1820–1829. [PubMed] [Google Scholar]
- Mooren M. M., Pulleyblank D. E., Wijmenga S. S., van de Ven F. J., Hilbers C. W. The solution structure of the hairpin formed by d(TCTCTC-TTT-GAGAGA). Biochemistry. 1994 Jun 14;33(23):7315–7325. doi: 10.1021/bi00189a037. [DOI] [PubMed] [Google Scholar]
- Orbons L. P., van der Marel G. A., van Boom J. H., Altona C. Hairpin and duplex formation of the DNA octamer d(m5C-G-m5C-G-T-G-m5C-G) in solution. An NMR study. Nucleic Acids Res. 1986 May 27;14(10):4187–4196. doi: 10.1093/nar/14.10.4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel D. J., Kozlowski S. A., Ikuta S., Itakura K. Deoxyguanosine-deoxyadenosine pairing in the d(C-G-A-G-A-A-T-T-C-G-C-G) duplex: conformation and dynamics at and adjacent to the dG X dA mismatch site. Biochemistry. 1984 Jul 3;23(14):3207–3217. doi: 10.1021/bi00309a015. [DOI] [PubMed] [Google Scholar]
- Pognan F., Paoletti C. Does cruciform DNA provide a recognition signal for DNA-topoisomerase II? Biochimie. 1992 Nov;74(11):1019–1023. doi: 10.1016/0300-9084(92)90022-7. [DOI] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Schroth G. P., Ho P. S. Occurrence of potential cruciform and H-DNA forming sequences in genomic DNA. Nucleic Acids Res. 1995 Jun 11;23(11):1977–1983. doi: 10.1093/nar/23.11.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle M. S., Williams D. H. On the stability of nucleic acid structures in solution: enthalpy-entropy compensations, internal rotations and reversibility. Nucleic Acids Res. 1993 May 11;21(9):2051–2056. doi: 10.1093/nar/21.9.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senior M. M., Jones R. A., Breslauer K. J. Influence of loop residues on the relative stabilities of DNA hairpin structures. Proc Natl Acad Sci U S A. 1988 Sep;85(17):6242–6246. doi: 10.1073/pnas.85.17.6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers M. F., Byrd R. A., Gallo K. A., Samson C. J., Zon G., Egan W. Nuclear magnetic resonance and circular dichroism studies of a duplex--single-stranded hairpin loop equilibrium for the oligodeoxyribonucleotide sequence d(CGCGATTCGCG). Nucleic Acids Res. 1985 Sep 11;13(17):6375–6386. doi: 10.1093/nar/13.17.6375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells R. D., Collier D. A., Hanvey J. C., Shimizu M., Wohlrab F. The chemistry and biology of unusual DNA structures adopted by oligopurine.oligopyrimidine sequences. FASEB J. 1988 Nov;2(14):2939–2949. [PubMed] [Google Scholar]
- Wells R. D. Unusual DNA structures. J Biol Chem. 1988 Jan 25;263(3):1095–1098. [PubMed] [Google Scholar]
- Williamson J. R., Boxer S. G. Multinuclear NMR studies of DNA hairpins. 1. Structure and dynamics of d(CGCGTTGTTCGCG). Biochemistry. 1989 Apr 4;28(7):2819–2831. doi: 10.1021/bi00433a012. [DOI] [PubMed] [Google Scholar]
- Wu M., McDowell J. A., Turner D. H. A periodic table of symmetric tandem mismatches in RNA. Biochemistry. 1995 Mar 14;34(10):3204–3211. doi: 10.1021/bi00010a009. [DOI] [PubMed] [Google Scholar]
- Xodo L. E., Manzini G., Quadrifoglio F., van der Marel G. A., van Boom J. H. The B-Z conformational transition in folded oligodeoxynucleotides: loop size and stability of Z-hairpins. Biochemistry. 1988 Aug 23;27(17):6327–6331. doi: 10.1021/bi00417a019. [DOI] [PubMed] [Google Scholar]
- Xodo L. E., Manzini G., Quadrifoglio F., van der Marel G. A., van Boom J. H. The duplex-hairpin conformational transition of d(CGCGCGATCGCGCG) and d(CGCGCGTACGCGCG): a thermodynamic and kinetic study. J Biomol Struct Dyn. 1988 Aug;6(1):139–152. doi: 10.1080/07391102.1988.10506487. [DOI] [PubMed] [Google Scholar]
- Xodo L. E., Manzini G., Quadrifoglio F., van der Marel G., van Boom J. H. Hairpin structures in synthetic oligodeoxynucleotides: sequence effects on the duplex-to-hairpin transition. Biochimie. 1989 Jul;71(7):793–803. doi: 10.1016/0300-9084(89)90042-4. [DOI] [PubMed] [Google Scholar]
- el antri S., Bittoun P., Mauffret O., Monnot M., Convert O., Lescot E., Fermandjian S. Effect of distortions in the phosphate backbone conformation of six related octanucleotide duplexes on CD and 31P NMR spectra. Biochemistry. 1993 Jul 20;32(28):7079–7088. doi: 10.1021/bi00079a003. [DOI] [PubMed] [Google Scholar]



