Summary
Progression through the Caulobacter cell cycle is driven by the master regulator CtrA, an essential two-component signaling protein that regulates the expression of nearly 100 genes. CtrA is abundant throughout the cell cycle except immediately prior to DNA replication. However, the expression of CtrA-activated genes is generally restricted to S phase. We identify the conserved protein SciP (small CtrA inhibitory protein) and show that it accumulates during G1, where it inhibits CtrA from activating target genes. The depletion of SciP from G1 cells leads to the inappropriate induction of CtrA-activated genes and, consequently, a disruption of the cell cycle. Conversely, the ectopic synthesis of SciP is sufficient to inhibit CtrA-dependent transcription, also disrupting the cell cycle. SciP binds directly to CtrA without affecting stability or phosphorylation; instead, SciP likely prevents CtrA from recruiting RNA polymerase. CtrA is thus tightly regulated by a protein-protein interaction which is critical to cell-cycle progression.
Introduction
All dividing cells must ensure the correct timing and order of DNA replication, chromosome segregation, and cell division. These cell-cycle events are carefully orchestrated to ensure genome stability and cellular survival. Their execution and, hence, proper cell-cycle progression requires complex transcriptional programs, with batteries of genes turned on and off according to their times of action during the cell cycle (Breeden, 2003). These gene expression programs are controlled by master regulators whose activities are tightly regulated. For example, in metazoans the transcription factor E2F regulates the expression of hundreds of genes during late G1, many of which are critical to the ensuing S phase (Dyson, 1998). To prevent the premature, inappropriate expression of E2F targets in early G1, the retinoblastoma protein Rb binds directly to E2F and prevents it from stimulating gene expression, but without disrupting DNA binding (Ross et al., 1999). The regulated inactivation of Rb by phosphorylation in late G1 thus unleashes E2F activity and propels cells into S phase. In the yeast S. cerevisiae, the transcription factors SBF and MBF drive the expression of nearly 200 genes in late G1 (Iyer et al., 2001). Analogous to Rb, the yeast protein Whi5 binds directly to SBF and MBF to block transcription during early G1; the subsequent hyperphosphorylation of Whi5 relieves this inhibition and permits SBF and MBF to drive gene expression crucial to cell-cycle progression (Costanzo et al., 2004).
The regulation of gene expression during the bacterial cell cycle remains poorly understood. The Gram-negative bacterium Caulobacter crescentus is a tractable model for elucidating the mechanisms underlying the prokaryotic cell cycle (Laub et al., 2007; Ryan and Shapiro, 2003). The Caulobacter cell cycle begins with a G1-phased swarmer cell that is motile and cannot initiate DNA replication until it differentiates into a stalked cell. Concomitant with the swarmer-to-stalked cell transition, the cell enters S phase, during which it replicates its chromosome and partitions the new chromosomes to opposite poles of the predivisional cell. Each cell division is asymmetric, producing two different daughter cells—a G1 swarmer cell and a stalked cell that can immediately enter S phase. DNA replication in Caulobacter occurs in a once-and-only-once manner, resulting in distinct G1, S, and G2 phases. Additionally, Caulobacter cells can be easily synchronized, facilitating temporal analysis of the cell cycle.
In Caulobacter, cell-cycle progression requires the periodic activation and inactivation of the master regulator CtrA, which functions both as a transcription factor for nearly 100 genes and as a direct repressor of DNA replication initiation (Domian et al., 1997; Quon et al., 1996, 1998). CtrA is a response regulator and hence part of a two-component signaling pathway, the dominant form of signal transduction in bacteria (Laub and Goulian, 2007; Stock et al., 2000). These pathways typically initiate with a sensor histidine kinase that autophosphorylates and then transfers the phosphoryl group to a cognate response regulator. Phosphorylation of the response regulator activates an output domain that can drive changes in cellular behavior. Phosphorylation of CtrA enhances its ability to bind DNA (Reisenauer et al., 1999; Siam and Marczynski, 2000) and, consequently, to modulate transcription and silence the origin of replication. CtrA may also bind some atypical sites in a phosphorylation-independent manner (Spencer et al., 2009).
CtrA activity is controlled by a combination of regulated phosphorylation, proteolysis, and transcription (Biondi et al., 2006b; Domian et al., 1997, 1999; Hung and Shapiro, 2002; Jacobs et al., 1999; Wu et al., 1998). Phosphorylated CtrA is abundant in swarmer/G1 cells where it silences the origin of replication (Quon et al., 1998), likely by occluding the replication initation factor DnaA. At the G1-S transition, CtrA is dephosphorylated and degraded, freeing the origin and enabling DNA replication to commence. As cells proceed through S phase, ctrA is transcribed, and the newly synthesized CtrA is stabilized against proteolysis and activated by phosphorylation. CtrA then activates the expression of more than 60 target genes, many of which are important for late stages of the cell cycle and cell division (Laub et al., 2002). Following septation, CtrA is maintained in the phosphorylated state in daughter swarmer cells but proteolytically cleared from daughter stalked cells to permit another round of DNA replication.
These mechanisms ensure that CtrA is abundant and phosphorylated in both swarmer and predivisional cells. Consistent with this pattern of activity, CtrA-repressed genes are expressed only in stalked cells after CtrA has been degraded. However, most of the genes that are directly activated by CtrA are transcribed only in predivisional cells (Laub et al., 2002). A handful of CtrA-activated genes have mRNAs that are also abundant in swarmer cells, but this pattern may result from transcription in predivisional cells followed by stabilization of the mRNA in swarmer cells (Llewellyn et al., 2005; Milhausen and Agabian, 1983). These previous findings thus imply the existence of either a predivisional-specific transcriptional cofactor for CtrA or a swarmer-cell-specific inhibitor of CtrA-dependent gene expression. Neither such factor has been identified to date.
Here, we identify SciP and demonstrate that it binds directly to CtrA and inhibits its ability to activate transcription without disrupting DNA binding or the repression of certain target genes. SciP is restricted to G1 swarmer cells and thus plays a role in establishing the differential fates of daughter cells. SciP is critical for proper cell-cycle progression, and we show that SciP levels must be tightly regulated, as the ectopic production of SciP in predivisional cells is sufficient to block CtrA-dependent gene expression and, consequently, proper cell cycling. SciP is highly conserved and present in nearly every organism containing a CtrA ortholog, emphasizing the key role this molecule plays in cell-cycle progression in a wide range of bacteria. SciP represents a class of molecules that modulate the output of two-component signaling pathways without affecting phosphorylation, indicating that these critical signaling pathways are subject to even more complex regulation than previously appreciated.
Results
Identification of sciP as a Candidate Regulator of CtrA and the Cell Cycle
To identify putative regulators of CtrA transcriptional activity, we used a bioinformatic screen for genes that are coexpressed, coinherited, or colocated in bacterial genomes with ctrA (Srinivasan et al., 2006). We previously used a similar approach to identify the direct CtrA phosphodonor, ChpT (Biondi et al., 2006b). This screen identified CC0903, which we named sciP for small CtrA inhibitory protein. The sciP gene is annotated to encode a 93 amino acid protein of no known function. Orthologs of sciP are often found immediately adjacent to or within a few genes of ctrA in α-proteobacteria such as Rhodobacter sphaeroides, Sinorhizobium meliloti, Bartonella bacilliformis, Brucella melitensis, and Mesorhizobium loti (Figure 1A). Orthologs of sciP were found in virtually all α-proteobacteria, indicating strong coconservation with ctrA, with most orthologs nearly 70% identical and 85% similar to the C. crescentus SciP (see Figure S1 available online). Searching the PFAM database, we found that sciP also has some similarity to several large classes of helix-turn-helix proteins.
Figure 1. Identification of sciP as a Key Regulator of Cell-Cycle Progression.
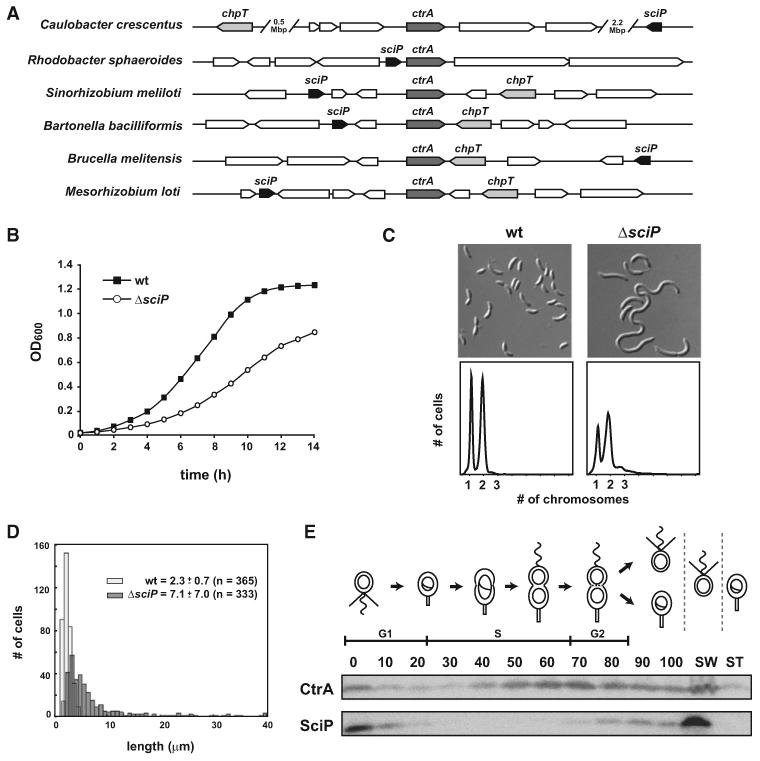
(A) Genomic organization of ctrA, chpT, and sciP orthologs in C. crescentus and other α-proteobacteria.
(B) Growth curve of ΔsciP and wild-type grown in PYE.
(C) Cellular morphology (top) and flow cytometry analysis (bottom) of ΔsciP compared to wild-type. Samples were treated with rifampin, and 50,000 cells from each population were analyzed.
(D) Quantification of cell lengths for wild-type and ΔsciP.
(E) Cell-cycle abundance of SciP. Synchronized wild-type swarmer cells were released into PYE and allowed to proceed through one cell cycle with samples collected every 10 min for western blot analysis with CtrA or SciP antisera. After 100 min, the culture was again synchronized to separate swarmer and stalked daughter cells (lanes labeled SW and ST). The cell-cycle diagram shown on top indicates relative stage of the cell cycle for each time point.
To test whether sciP is important for cell-cycle progression, we deleted the chromosomal copy of the gene from the wild-type (CB15N) and replaced it with a tetracycline-resistance cassette to produce the strain ΔsciP. In the C. crescentus genome, sciP is located between two operons that encode flagellar proteins, but is transcribed independently (Mullin et al., 2001), so the deletion of sciP is unlikely to affect nearby genes. Although the ΔsciP strain is viable, it exhibited significant growth and cell-cycle defects with a doubling time of ∼120 min in rich PYE media compared to ∼80 min for the wild-type (Figure 1B). Cells from a mid-log phase culture of ΔsciP were filamentous with lengths ranging from slightly elongated to extremely filamentous (Figures 1C and 1D). The filamentous cells were either smooth and unpinched or had a single constriction, which is likely the site of active cell division. Flow cytometry analysis of the ΔsciP strain revealed a modest but reproducible accumulation of cells with 3N chromosomal content compared to the wild-type (Figure 1C). These growth, morphological, and chromosomal content defects were rescued by providing a wild-type copy of sciP in trans at the chromosomal xylX promoter (see below).
SciP Is Present Only in G1 Swarmer
Cells To determine when SciP functions, we measured its abundance during the cell cycle using a polyclonal antibody specific to SciP. Wild-type swarmer cells were then harvested and allowed to proceed synchronously through the cell cycle with samples isolated every 10 min for immunoblot analysis (Figure 1E). We found that SciP was abundant in swarmer cells but rapidly disappeared at the G1-S transition and did not accumulate again until very late in the cell cycle, coincident with the first appearance of new swarmer cells (Figure S2). This temporal pattern suggests that SciP may be restricted primarily to the G1/swarmer stage of the cell cycle. To test this conclusion, we used density centrifugation to separate new swarmer cells from new stalked cells immediately following cell division. Immunoblots of samples from each population revealed that SciP was present only in the swarmer fraction (Figure 1E). Together, these data indicate that SciP is present primarily, and perhaps exclusively, in G1-phased swarmer cells. We note, however, that sciP mRNA was previously found to be cell-cycle regulated, reaching peak levels in predivisional cells (Chen et al., 1986). The delay in SciP accumulation following its transcription suggests sciP expression may be posttranscriptionally regulated.
sciP Is Required for Proper Swarmer Cell Development and Cell-Cycle Progression
To better characterize the cell-cycle function of SciP, we created a depletion strain in which the chromosomal copy of sciP was deleted and a wild-type copy of sciP was placed at the chromosomal xylX locus under the control of the xylose-inducible, glucose-repressible promoter Pxyl. When grown in PYE supplemented with xylose, the depletion strain exhibited morphology, chromosome content, and SciP protein levels similar to wild-type (Figures 2A–2C). Upon shifting to media supplemented with glucose, SciP was rapidly depleted, with no protein detected after 1 hr (Figure 2B). We then examined the morphology and chromosomal content of cells following a shift from xylose to glucose. After 2 hr, the culture showed a significant increase in cells with one chromosome, suggesting that the loss of SciP may cause a transient arrest in G1 (Figure 2C). After 12 hr, the depletion strain had overcome this delay and reached a steady state in which the chromosomal content profile was nearly identical to the sciP deletion strain. By 12 hr, cells also exhibited an unpinched, filamentous morphology similar to the ΔsciP strain (Figure 2A).
Figure 2. SciP Depletion Phenotype.
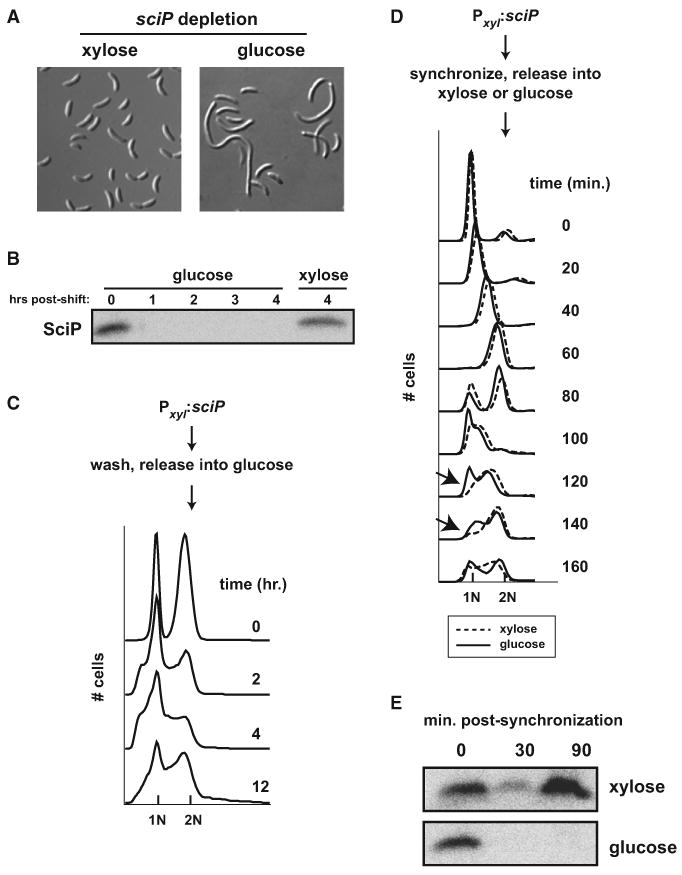
(A) Cellular morphology of the sciP depletion strain grown in PYE supplemented with xylose or shifted to PYE with glucose for 12 hr to deplete SciP.
(B) Western blot analysis showing SciP levels post-shift to glucose or maintained in xylose at the times indicated.
(C) Flow cytometry analysis of chromosomal content in the sciP depletion strain grown in PYE supplemented with xylose and then shifted to PYE with glucose.
(D) Flow cytometry analysis of synchronized swarmer cells from the depletion strain released into PYE supplemented with glucose or xylose. Arrows indicate the delay in DNA replication initiation seen after the first cell division for the strain depleted of sciP.
(E) Western blot analysis of SciP levels in cultures synchronized as in (D), at the time points indicated.
To further examine whether sciP affects the G1-S transition, we synchronized the depletion strain grown in xylose and then released swarmer cells into media containing either xylose or glucose to maintain or inhibit SciP production, respectively. Cell-cycle progression for each population of cells was monitored by flow cytometry (Figure 2D) and SciP levels examined by western blotting (Figure 2E). As expected, for cells released into xylose, SciP was initially abundant but eliminated at the G1-to-S transition (30 min time point), as seen with wild-type cells. SciP accumulated again in these cells during the latest stages of the cell cycle, at a time coincident with cell division and the generation of new swarmer cells. Although sciP is constitutively expressed, SciP levels still drop after G1 (Figure 2E), further demonstrating that sciP expression is posttranscriptionally regulated. Following cell division, daughter cells from the culture grown in xylose initiated a new round of DNA replication, manifest in the flow cytometry profiles by a shift of the entire 1N chromosomal peak toward 2N by the 100 min time point.
When the swarmer cells of the depletion strain were released into glucose, SciP was also initially abundant and then rapidly eliminated at the G1-S transition (Figure 2E). However, in these cells SciP remained undetectable throughout the remainder of the first cell cycle and after cell division, indicating a complete repression of sciP expression. These cells divided at approximately the same time as the control cells grown in xylose, demon-strating that SciP is not required for progression through the late stages of the cell cycle or for cell division (Figure 2D). However, following the first cell division (80 min time point), approximately half of the daughter cells were delayed in initiating DNA replication for up to 60 min, relative to the control cells, (Figure 2D). This result supports our conclusion that sciP is important for the G1-S transition and proper cell-cycle progression.
We also resynchronized the culture of cells released into glucose after the first cell division. However, this double synchrony produced a low yield of swarmer cells in contrast to cells released into xylose. Based on flow cytometry (Figure 2D) and time-lapse microscopy (data not shown), we concluded that the lack of swarmer cells was not due to a defect in cell division. Instead, cells lacking sciP likely do not undergo the change in buoyant density that facilitates synchronization by centrifugation. The swarmer cells that were harvested, though, showed a significant delay in initiating DNA replication.
Depletion of sciP Affects CtrA-Dependent Gene Expression
Next, we tested whether the lack of sciP in G1 swarmer cells affected gene expression, particularly the CtrA regulon. Again, we synchronized the depletion strain and released swarmer cells into media supplemented with either glucose or xylose to repress or maintain SciP levels, respectively. After 100 min, once all cells in each culture had divided, we isolated RNA from each culture and compared the samples by competitive hybridization on DNA microarrays. These postdivision samples include both swarmer and stalked cells because, as noted, the yield of swarmer cells following a second synchronization of the depletion strain is low. However, because SciP is only abundant in swarmer cells (see above), differences in gene expression will only reflect differences in swarmer cell gene expression.
Nearly all of the genes showing significant changes in expression level were CtrA-dependent genes (for complete data, see the Supplemental Information). Figure 3 shows the changes in expression for genes that are directly regulated by CtrA. Strikingly, nearly every CtrA-activated gene was expressed at significantly higher levels in the cells depleted of sciP. In contrast, the expression of CtrA-repressed genes was unchanged. These data indicate that SciP is required to silence the expression of CtrA-activated genes in swarmer cells, thereby restricting their expression to predivisional cells.
Figure 3. Effects of sciP Overexpression and Depletion on CtrA-Dependent Gene Expression.
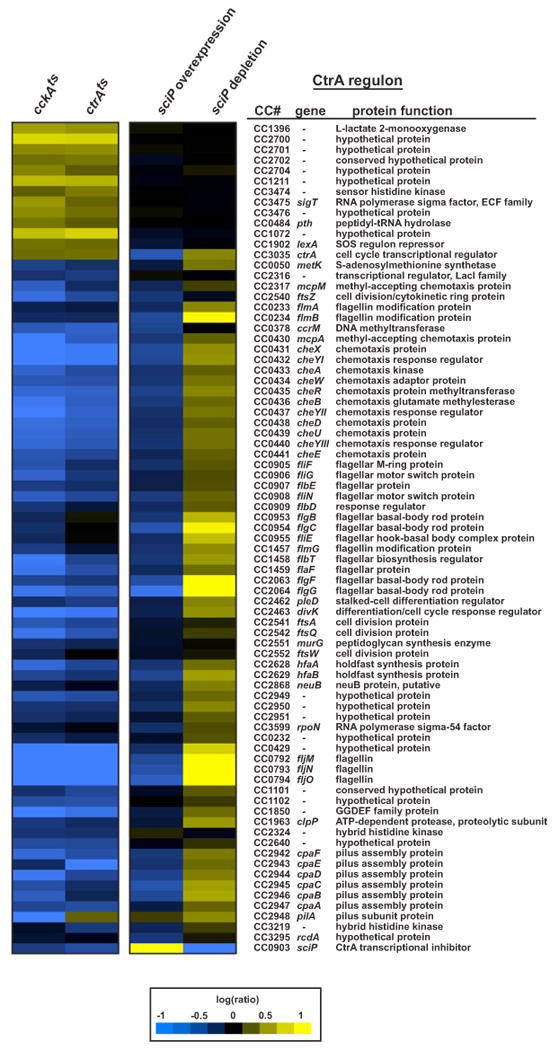
Whole-genome DNA microarrays were used to examine gene expression changes in the strains indicated. The cckAts and ctrAts mutants were grown in PYE at 28° C, shifted to 37° C for 2 hr, and compared to wild-type grown under the same conditions (data are from Jacobs et al., 2003; Laub et al., 2002). For sciP overexpression, strain ML1749, which harbors pHXM-sciP, was grown in PYE supplemented with xylose for 2 hr and compared to wild-type grown under identical conditions. For sciP depletion, strain ML1750 was grown in PYE supplemented with xylose, synchronized, and released into PYE supplemented with glucose or xylose. Each population of cells was grown for 100 min and then compared to each other. Expression changes are color coded according to the legend at the bottom. The genes shown include those previously reported to be direct CtrA targets based on ChIP-chip analysis and whose expression changed significantly in both cckAts and ctrAts strains after a shift to the restrictive temperature (Jacobs et al., 2003; Laub et al., 2002). Note, the divK/pleD operon was not identified by ChIP-chip analysis but was subsequently confirmed as a direct CtrA target (Hung and Shapiro, 2002). sciP is included at the bottom of the list.
Overexpressing sciP Disrupts the Cell Cycle and Drives the Downregulation of CtrA-Dependent Genes
The data thus far are consistent with SciP being a G1-specific inhibitor of CtrA. To test whether SciP is sufficient to inhibit CtrA transcriptional activity, we examined the consequence of forcing the synthesis of SciP in predivisional cells when it is normally absent. For this experiment we used pHXM-sciP, a high-copy vector for expressing sciP with an N-terminal M2-epitope tag. Although strains overexpressing either sciP or M2-sciP gave similar phenotypes, inclusion of the tag enabled us to distinguish plasmid and chromosomally encoded SciP, as needed below.
The strain harboring pHXM-sciP exhibited normal cellular morphology and chromosome content when grown in glucose to repress expression (Figure 4A). To overproduce SciP, cells were shifted to media containing xylose. Immunoblots showed that SciP levels increased approximately 7-fold after 4 hr of growth in xylose (data not shown). After 4 hr postinduction, cells had become filamentous and accumulated up to four chromosomes per cell, phenotypes consistent with a disruption of cell division (Figures 4A and 4B). Immunoblot analysis of synchronized cells demonstrated that SciP was now abundant throughout the cell cycle, overcoming the regulatory mechanisms that usually limit it to swarmer cells (Figure 4C).
Figure 4. Phenotypic Consequences of sciP Overexpression.
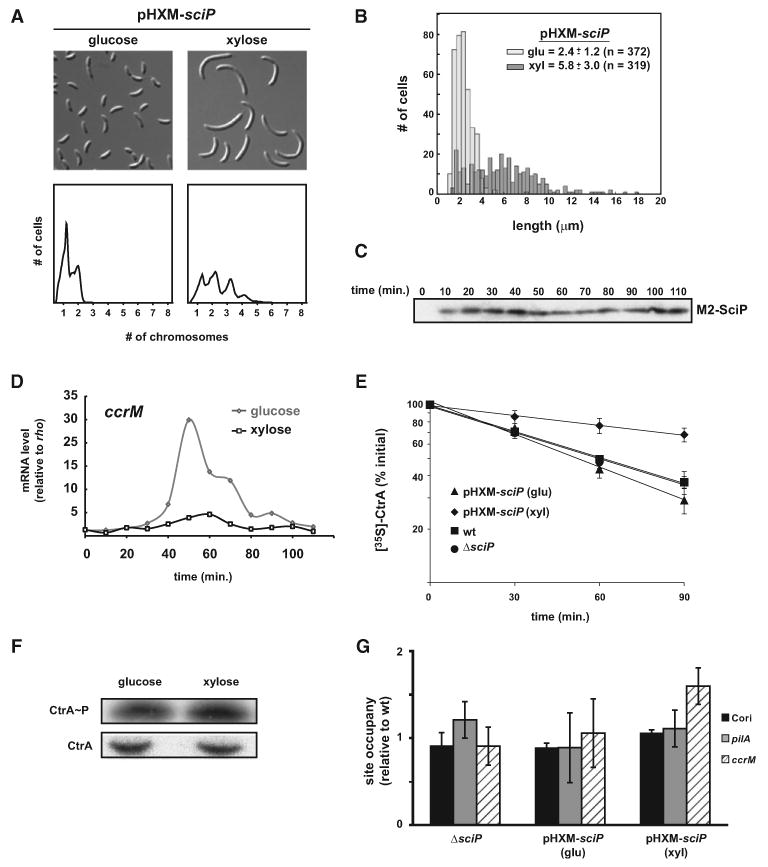
(A) Cellular morphology (top) and flow cytometry analysis (bottom) of cells overexpressing sciP from a high-copy plasmid (strain ML1749). A culture was grown in PYE glucose and sciP expression induced by the addition of xylose for 4 hr.
(B) Quantification of cell lengths for populations in (A).
(C) Western blot for SciP in samples taken from a synchronous population of cells overexpressing sciP.
(D) The overexpression strain was synchronized and released into glucose or xylose to repress or overexpress, respectively, sciP. ccrM mRNA levels as a function of cell-cycle time were measured using qRT-PCR and expressed relative to a control, rho.
(E) Pulse-chase analysis of CtrA stability in the strains indicated. For the overexpression strain harboring pHXM-sciP, cells were grown in M2G or shifted to M2G supplemented with xylose for 1 hr to induce sciP expression. Each experiment was performed in triplicate except for wild-type, which was done in duplicate; error bars represent standard error of the mean.
(F) Phosphorylation levels of CtrA in vivo. Cells harboring pHXM-sciP were grown in media supplemented with glucose and then shifted to media with xylose for 2 hr. In each case, CtrA was immunoprecipitated from cells after labeling with [γ32P]-ATP and then separated by SDS-PAGE followed by exposure to a phosphor-image screen (top panel), with CtrA levels measured by western blotting (bottom panel).
(G) CtrA occupancy of the chromosomal origin, Cori, and pilA and ccrM promoters was measured by ChIP of CtrA from ΔsciP and cells overexpressing sciP. Each measurement was done in triplicate and divided by the average triplicate percent input for wild-type; error bars represent the standard error of the mean.
To test whether the overproduction of SciP was sufficient to downregulate CtrA-activated genes, we again used whole-genome DNA microarrays. Cells harboring pHXM-sciP were shifted to xylose for 2 hr, and mRNA from these cells was compared to mRNA from wild-type cells grown in identical conditions (for complete data, see the Supplemental Information). Nearly every CtrA-activated gene showed significant downregulation in the cells overproducing SciP (Figure 3). The magnitudes of these drops in expression were comparable to, although slightly less than, those seen in ctrAts and cckAts strains (Jacobs et al., 2003; Laub et al., 2002) grown at the restrictive temperature relative to wild-type (Figure 3). As with the sciP depletion strain, genes repressed by CtrA were not significantly affected by the overproduction of SciP.
Notably, the expression of sciP itself was strongly decreased in ctrAts and cckAts (Figure 3), and overproducing M2-SciP from a high-copy plasmid led to a significant drop in the levels of native, chromosomally encoded SciP (see Figure 5B). Previous ChIP-chip studies did not include the sciP promoter on the microarrays used, but the predicted regulatory region has a near-consensus CtrA-binding site, suggesting sciP is a direct CtrA target.
Figure 5. SciP and CtrA Interact Directly in a Yeast Two-Hybrid Assay.
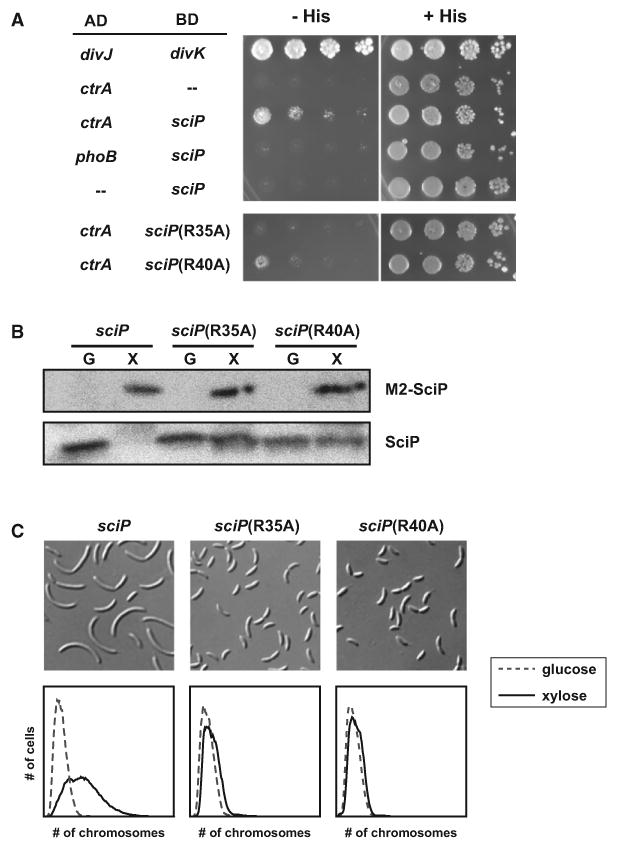
(A) Yeast strain PJ69-4A was cotransformed with plasmids harboring fusions of the indicated genes to either the activation or DNA-binding domain of Gal4, as indicated, and spotted onto histidine deplete or replete solid media. Each row contains serial 10-fold dilutions of overnight cultures normalized to OD 0.5. Growth after 72 hr is shown. Growth on media lacking histidine indicates reconstitution of Gal4 activity by protein-protein interaction.
(B) Western blot analysis of plasmid-encoded M2-SciP and chromosomally encoded SciP for strains grown in glucose or xylose to repress or overexpress, respectively, the plasmid-borne M2-sciP.
(C) Cellular morphology (top) and flow cytometry analysis (bottom) of Caulobacter strains overexpressing sciP or the indicated alanine mutants of sciP. Cultures were induced with xylose for 4 hr.
To confirm that the downregulation of CtrA-activated genes was due to the ectopic production of SciP in predivisional cells, we synchronized the overexpression strain, released cells into media with either glucose or xylose, and monitored cell-cycle-dependent expression of ccrM, a CtrA-activated gene, using quantitative PCR. Cells grown in glucose showed the expected peak in ccrM expression in predivisional cells, while cells grown in xylose never induced ccrM above background levels (Figure 4D). In sum, our gene expression studies demonstrate that SciP is sufficient to inhibit CtrA from activating its target genes.
SciP Does Not Affect CtrA Degradation or Phosphorylation
How does SciP inhibit CtrA-dependent transcription? To begin addressing this question, we first examined the steady-state levels of CtrA following both sciP depletion and overexpression and found that they did not change significantly in either case (data not shown). We also measured the stability of CtrA in the sciP deletion strain and in a strain overproducing SciP. In ΔsciP, the half-life of CtrA was 62 min, similar to the value measured in wild-type cells (Figure 4E). With the overexpression strain, CtrA was more stable in cells grown in xylose to induce SciP than in cells grown in glucose (50 versus 173 min) (Figure 4E). These data indicate that SciP does not downregulate CtrA activity by stimulating degradation but may stabilize it. To test the effect of SciP on the phosphorylation of CtrA, we first reconstituted the CckA-ChpT-CtrA phosphorelay in vitro. The addition of SciP had no significant effect on the rate or steady-state levels of CtrA phosphorylation (data not shown). SciP also had no substantial effect on the dephosphorylation of CtrA∼P by phosphorelay reversal. We also measured the phosphorylation level of CtrA in vivo in cells over-expressing sciP by immunoprecipating CtrA after growth in the presence of [γ32P]-ATP. CtrA∼P levels did not change significantly after overexpressing sciP for 2 hr (Figure 4F).
Finally, we tested whether sciP affects DNA binding by CtrA using chromatin immunoprecipitation (ChIP) to examine the occupancy of CtrA at two target gene promoters and at the origin of replication in vivo. Neither the overexpression nor the depletion of sciP had a significant effect on CtrA occupancy of the origin of replication, or the pilA and ccrM promoters (Figure 4G). These findings also corroborate our global gene expression data showing that overexpressing and depleting sciP affects CtrA-activated, but not CtrA-repressed, genes. Additionally, these data are consistent with the observation that cells overproducing SciP exhibited relatively moderate chromosomal accumulation. If SciP disrupted CtrA phosphorylation or DNA binding, we would have expected more significant chromosomal accumulation, as seen with ctrAts and cckAts mutants (Jacobs et al., 1999; Quon et al., 1998). The mild accumulation of chromosomes following sciP overexpression is likely due to the disruption of cell division that results from SciP's inhibition of CtrA target genes, many of which are important for cell division. Consistent with this interpretation, we found that cells depleted of the essential cell division gene ftsZ showed a similar mild chromosomal accumulation phenotype (Figure S3).
SciP Binds Directly to CtrA
Our data suggest that SciP could bind to CtrA to prevent it from interacting productively with or recruiting RNA polymerase (RNAP) to drive transcription of target genes. To test whether SciP regulates CtrA via a direct protein-protein interaction, we first tested whether CtrA and SciP interact in a yeast two-hybrid system. CtrA was fused to the activating domain of Gal4 and SciP to the DNA-binding domain of Gal4. Together, these constructs reconstituted Gal4 activity and drove expression of GAL1-HIS3 at a level sufficient to enable growth on minimal medium lacking histidine (Figure 5A). Neither CtrA nor SciP alone was sufficient to activate HIS3 expression. Also, SciP did not interact with the response regulator PhoB, supporting the notion that SciP binds specifically to CtrA.
To identify amino acids important for the CtrA-SciP interaction, we constructed a series of alanine substitutions in SciP at sites that are (1) perfectly conserved in sciP orthologs and (2) solvent exposed in an NMR structure of an ortholog from R. sphaeroides solved by a structural genomics project (PDB, 2JRT) (Figure S4). In total we made five mutants, R35A, R40A, E57A, Y62A, and E68A. We tested the activity of these alanine mutants in vivo by overexpressing them in Caulobacter cells using the high-copy vector pHXM. Immunoblots of the overexpression strains indicated that only the R35A and R40A mutants accumulated to levels equivalent to overexpressed wild-type sciP, suggesting that the other mutations may affect SciP stability (Figure 5B). Both R35A and R40A completely eliminated the sciP overexpression phenotype (Figure 5C). In addition, both mutations completely disrupted the CtrA-SciP interaction in the yeast two-hyrbid assay (Figure 5A). Finally, we found that each allele of sciP failed to fully rescue the morphological and growth phenotypes of a sciP deletion (Figure S5). These results suggest that the severe phenotype of sciP overexpression requires a direct interaction between SciP and CtrA, and that residues R35 and R40 are critical for this interaction.
SciP Binds CtrA without Disrupting DNA Binding
Next, we used electrophoretic mobility shift assays (EMSAs) to test whether SciP could interact with CtrA when CtrA is bound to target promoters. We added purified CtrA∼P to radiolabeled, 50 base pair fragments of the ccrM, pilA, and fliF promoters, which contain CtrA-binding sites (Quon et al., 1996; Skerker and Shapiro, 2000; Wu et al., 1998). As expected, CtrA bound these DNA fragments, leading to a decrease in unbound DNA and the appearance of lower-mobility bands (Figures 6A–6C). The intensity of these latter bands increased with increasing CtrA concentration and were correlated with decreases in unbound DNA. CtrA did not bind to a DNA fragment from the vanA promoter, which lacks a CtrA-binding site (data not shown). We then added SciP to each reaction and found that it led to a dramatic enhancement of DNA binding as evidenced by the nearly complete disappearance of free, unbound DNA at the lowest concentration of CtrA and the appearance of low-mobility bands at lower concentrations of CtrA than seen without SciP (Figures 6A–6C). The shift observed with the pilA fragment was larger than with ccrM and fliF, likely due to the presence of multiple CtrA half-sites in the pilA fragment compared to the two half-sites in ccrM and fliF. We found that SciP alone did not bind any of the probes tested, even at concentrations up to 10 μM, suggesting it does not bind DNA alone but may do so in the presence of CtrA. To test if SciP requires contact with DNA to interact with CtrA, we also examined whether SciP caused a supershift when CtrA was added to radiolabeled DNA fragments of only 25 base pairs. As CtrA protects ∼25 base pairs of DNA in footprinting assays (Skerker and Shapiro, 2000), these smaller probes should have little or no free DNA exposed. With a 25 base pair fliF probe, SciP did not cause a significant supershift (Figure 6D). With a 25 base pair probe for pilA, SciP caused a slight shift, but much less than seen with a larger probe (Figure 6E). These data suggest that the binding of SciP to DNA normally stabilizes its interaction with CtrA. DNA-binding by SciP is, however, likely nonspecific, as it interacts strongly with CtrA bound to three DNA fragments that share no similarity outside the CtrA binding sites (Figures 6A–6C). Finally, we also found that SciP(R35A) and SciP(R40A) did not supershift the 50 base pair fliF probe, as observed with wild-type SciP (Figure 6F). In sum, these data support the conclusion that SciP interacts directly with CtrA, but without disrupting CtrA's ability to bind DNA.
Figure 6. Electrophoretic Mobility Shift Assays with CtrA and SciP.
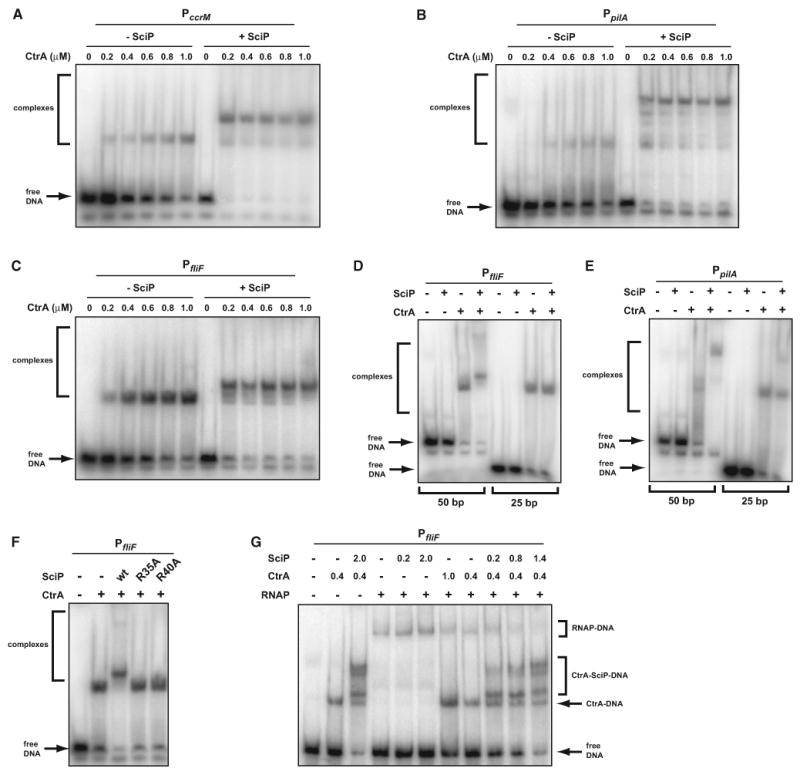
(A–C) CtrAP∼ binding to a 50 bp fragment of the (A) ccrM, (B) pilA, or (C) fliF promoters, each containing a CtrA-binding site. SciP, if present, was added at a final concentration of 1 μM. The concentration of CtrA in each lane is indicated.
(D and E) CtrA∼P binding to 25 bp fragments of the fliF and pilA promoters compared to the 50 bp probes also used in (B) and (C). CtrA was present at 1 μM and SciP at 1 μM.
(F) Binding of CtrA∼P to the fliF promoter in the presence of wild-type SciP or the mutant indicated. When included, CtrA was present at 1 μM and SciP at 1 μM.
(G) CtrA∼P and SciP prevent RNAP from binding the fliF promoter. CtrA∼P and SciP were added at the concentrations indicated. TAP-tagged RNAP was included, where indicated, at a final concentration of 50 nM (see the Experimental Procedures for details on RNAP preparation). (A)–(F) and (G) use 6% or 5%, respectively, polyacrylamide gels for separation.
Given that CtrA binds near the − 35 region of target promoters, we hypothesized that the binding of SciP to CtrA may prevent CtrA from recruiting RNAP to activate gene expression. To test this hypothesis, we purified RNAP from C. crescentus and examined binding to an 80 base pair probe encompassing the +60 to − 20 region of fliF. As expected, 50 nM RNAP bound this fliF probe and produced a band that ran more slowly than CtrA-DNA or CtrA-SciP-DNA complexes (Figure 6G). Adding CtrA caused a smearing of the RNAP-DNA complex but did not diminish the integrated intensity of the low-mobility species, indicating that CtrA and RNAP can bind the same probe, consistent with previous studies (Wu et al., 1998). Adding SciP alone did not significantly alter the RNAP-DNA band. In contrast, adding both SciP and CtrA disrupted the binding of RNAP to the fliF promoter fragment. We titrated SciP against fixed CtrA and RNAP and observed a concentration-dependent loss of the low-mobility, RNAP-containing bands with a corresponding increase in higher-mobility CtrA-SciP-DNA complexes (Figure 6G). All together, our data indicate that the binding of SciP to CtrA blocks the binding of RNAP, which in turn prevents the activation of CtrA-dependent target genes.
Discussion
As with all organisms, cell-cycle progression in Caulobacter requires the just-in-time transcription of large batteries of genes (Laub et al., 2000). At the heart of this transcriptional program is CtrA, which directly controls the expression of nearly 100 genes (Laub et al., 2002) and indirectly many more. CtrA both activates and represses genes; for activated genes, CtrA binds near the − 35 site and probably recruits or binds RNAP, while for repressed genes, CtrA binds near the +1 site and presumably occludes RNAP (Domian et al., 1999; Spencer et al., 2009). A long-standing conundrum has been why CtrA represses genes and the origin of replication in both swarmer and predivisional cells but activates most of its target genes only in predivisional cells. Here, we identified SciP and showed that it temporally restricts the transcriptional activity of CtrA by inhibiting CtrA-activated transcription in G1 swarmer cells.
SciP plays a crucial role in regulating the activity of CtrA and, consequently, in regulating the cell cycle and the establishment of asymmetric daughter cells in Caulobacter. CtrA and SciP collaborate to create three distinct states corresponding to the three major Caulobacter cell types (Figure 7). In G1 swarmer cells, CtrA∼P and SciP levels are both high such that CtrA can repress genes and silence the origin of replication but cannot activate gene expression. At the G1-S transition, stalked cells are cleared of CtrA and SciP, thereby permitting DNA replication and the expression of CtrA-repressed genes. As cells progress through S phase, CtrA∼P again accumulates to high levels but SciP levels remain low, enabling CtrA to activate the expression of target genes and propel cells through the ensuing cell division.
Figure 7. Regulation of CtrA-Dependent Gene Expression during Cell-Cycle Progression.
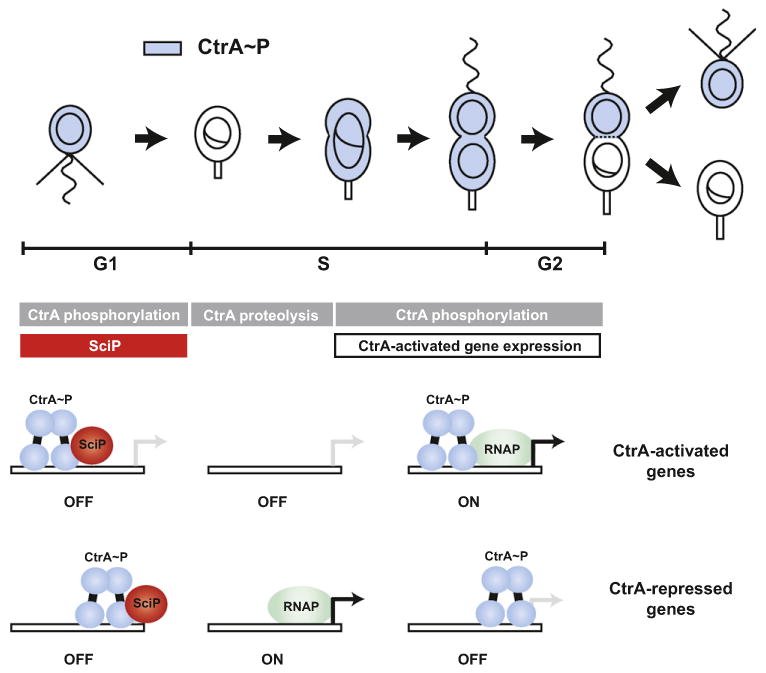
CtrA transcriptional activity is controlled by temporally regulated proteolysis, phosphorylation, and protein-protein interaction with SciP. The timing of each regulatory event is shown beneath a schematic of the Caulobacter cell cycle. These layers of regulation restrict CtrA-activated gene expression to the predivisional stage. A model for how SciP and CtrA combine to regulate gene expression during cell-cycle progression is shown at the bottom.
Why block transcriptional activition by CtrA in G1 cells? The majority of CtrA target genes are involved in polar morphogenesis, including flagellar biogenesis, chemotaxis, and pili biogenesis. CtrA also activates the expression of key cell division genes and ccrM, an essential DNA methyltransferase. These processes—polar morphogenesis, cell division, and DNA methylation—occur exclusively, or at least primarily, in predivisional cells. Constitutive expression of many of these genes, such as ftsA and ccrM, has deleterious consequences to cell-cycle progression (Martin et al., 2004; Zweiger et al., 1994). SciP thus helps fulfill the important task of preventing the execution of these cell-cycle processes at the wrong time.
Some CtrA-activated genes produce transcripts that are abundant in swarmer cells (Laub et al., 2002) and seemingly exceptions to our model for SciP. However, DNA microarrays measure steady-state mRNA levels and reflect both transcription and mRNA decay. Thus, some genes may be transcribed in predivisional cells but yield mRNAs that are unstable until the swarmer phase, as is the case for some flagellin filament genes (Llewellyn et al., 2005). For instance, the CtrA-activated gene rcdA is transcribed predominantly in predivisional cells, but RcdA protein does not accumulate until the swarmer phase, suggesting it may be posttranscriptionally regulated. Alternatively, some genes could have promoter architectures that prevent SciP from inhibiting CtrA-dependent transcription.
Regulation of SciP
Given the importance of SciP to cell-cycle progression and the consequences of misexpression, it comes as no surprise that SciP itself is tightly regulated and restricted to G1. sciP mRNA is detectable in predivisional and swarmer cells. However, SciP protein only accumulates in swarmer cells, suggesting either SciP is rapidly degraded in predivisional cells or the translation of sciP is blocked in predivisional cells and/or activated in swarmer cells. The rapid disappearance of SciP at the G1-S transition suggests it could be subject to temporally regulated degradation. Notably, CtrA is rapidly degraded by the ClpXP protease at precisely this stage of the cell cycle (Jenal and Fuchs, 1998). Consistent with posttranscriptional regulation of sciP, cells constitutively expressing sciP from a xylose-inducible promoter on the chromosome still only accumulated protein during G1 (Figure 2E). However, the regulation could be overcome by constitutively expressing sciP from a high-copy number plasmid that forced expression of sciP throughout S phase (Figure 4C). Importantly, the overexpression of sciP was sufficient to disrupt CtrA-dependent gene activation and, consequently, the cell cycle.
Our results outline a feedback loop important for cell-cycle progression in Caulobacter. sciP appears to be a direct transcriptional target of CtrA as sciP expression drops significantly in ctrAts strains and the regulatory region upstream of sciP has a near-consensus CtrA binding site. In predivisional cells, active CtrA likely drives the expression of sciP. Following cell division, SciP accumulates in swarmer cells, where it feeds back to inhibit CtrA as a transcriptional activator. This feedback is reminiscent of two other feedback loops, involving the cell-cycle regulators RcdA and DivK. Both rcdA and divK are direct transcriptional targets of CtrA (Laub et al., 2002), and each feeds back to inhibit CtrA. RcdA somehow contributes to the regulated degradation of CtrA at the G1-S transition (McGrath et al., 2006), although its function is unknown, while DivK inhibits the phosphorelays that drive the phosphorylation and proteolytic stabilization of CtrA (Biondi et al., 2006a; Chen et al., 2009). The combination of multiple feedback loops is likely critical to maintaining robust, sustained cell cycling in Caulobacter.
Mechanism of Inhibiting CtrA
We found no evidence that SciP decreased the steady-state levels, stability, or phosphorylation of CtrA. Overexpressing SciP did lead to a modest stabilization of CtrA, which could result from the decreased expression of divK and rcdA, which both help trigger CtrA proteolysis, or an increase in DNA-bound CtrA in the presence of SciP could physically protect CtrA from degradation. However, with regard to SciP's inhibition of CtrA, our data suggest that SciP does so by directly binding to CtrA. SciP does not, however, prevent CtrA from binding to DNA, consistent with our finding that altering sciP expression does not affect CtrA-repressed genes. Instead, the simplest model is that SciP blocks CtrA from recruiting RNAP and consequently activating transcription of target genes. The interaction of SciP and CtrA in the presence of DNA was particularly striking and suggestive of a highly cooperative interaction. At concentrations of CtrA that yield only minimal DNA binding, the addition of SciP stimulated very strong binding. SciP did not bind DNA on its own, although it appears to bind DNA nonspecifically in the presence of CtrA (Figures 6D and 6E). Together, CtrA and SciP prevented the binding of RNAP to target promoters (Figure 6G).
Regulating Response Regulators
SciP represents a previously undescribed mechanism for modulating the output of a two-component signaling pathway. Response regulators are usually regulated at the level of phosphorylation by cognate kinases and phosphatases. There are also regulatory proteins that can bind and stabilize the aspartyl-phosphate linkage. For example, in Salmonella enterica the protein PmrD binds the response regulator PmrA to block dephosphorylation (Kato and Groisman, 2004). There are only a few examples of proteins that regulate response regulators without affecting phosphorylation, such as the B. subtilis proteins RapC and RapG, which prevent ComA and DegU, respectively, from binding DNA (Ogura et al., 2003; Solomon et al., 1996), and the C. crescentus protein FliX, which inhibits FlbD from binding DNA (Dutton et al., 2005). The E. coli protein TorI was reported to inhibit transcriptional activation by the regulator TorR in vitro, but torI null mutants were not shown to affect TorR-dependent gene expression (Ansaldi et al., 2004), TorI is not expressed during exponential growth phase (M. Ansaldi and V. Mejean, personal communication), and recent data indicate it functions primarily as a prophage excisionase (Panis et al., 2007).
Perhaps the protein most analogous to SciP is Spx in B. subtilis. Spx is a small protein that binds to the α subunit of RNAP at the same site contacted by the response regulators ComA and ResD (Nakano et al., 2003). Spx thereby prevents these regulators from binding RNAP to stimulate the transcription of target genes. This may not be a common regulatory strategy, though, as binding to RNAP polymerase runs an inherent risk of having pleiotropic effects on transcription.
SciP's inhibition of CtrA-dependent expression thus appears to represent a new mechanism for regulating gene expression that may be used throughout the bacterial kingdom. SciP exhibits strict coconservation with CtrA and has an ortholog in nearly every α-proteobacterial genome. More generally, SciP shows moderate homology to several other large families of proteins of unknown function. We speculate that some of these proteins could inhibit other response regulators by a similar mechanism to SciP.
Concluding Remarks
SciP's function underscores the notion that bacterial gene expression is often not dictated simply by the activation of a single transcription factor, but instead involves the combinatorial action of multiple regulators. The identification of SciP also highlights the importance of regulated protein-protein interactions to cell-cycle control in organisms from bacteria to metazoans. As noted earlier, the master transcription factor in metazoans, E2F, is regulated by the binding of Rb, while in budding yeast the cell-cycle transcription factors SBF and MBF are regulated through the binding of Whi5. Other key cell-cycle regulators, such as cyclin-dependent kinases, are also regulated by protein-protein interactions (Morgan, 2007; Sherr and Roberts, 1999). For instance, SIC1 in S. cerevisiae accumulates to maximal levels in G1 cells, where it inhibits S phase-specific CDKs, while Rum1p and Rux play similar roles in S. pombe and D. melanogaster, respectively. In mammalian cells, two major classes of inhibitors, exemplified by p27 and p16INK4, directly inhibit different classes of CDKs. Regulated protein-protein interactions involving master regulators are thus critical to cell-cycle progression at many levels and in all organisms.
Experimental Procedures
A detailed description of bacterial strain construction and growth can be found in the Supplemental Information.
Immunoblots
Cell pellets were resuspended in 1× SDS sample buffer, resolved on 15% Tris-HCl gels (Bio-Rad), run at 150 V for 1 hr at room temperature, transferred to PVDF membrane, and probed with a 1:2000 dilution of SciP antisera, 1:1000 anti-M2 affinity-purified antibody (Sigma), or 1:10000 of CtrA antisera in 1× TBS plus 0.05% Tween-20.
Microscopy
Cells were fixed with 0.5% paraformaldehyde, centrifuged at 6800 g, washed with PBS, and resuspended in a small volume of PBS. Differential interference contrast images were taken with a Zeiss Axiovert 200 microscope with a Zeiss αPlan-Fluar 100×/1.45 oil objective and an Orca II camera (Hamatsu) controlled by MetaMorph 7.1.3.0 software.
Flow Cytometry
Samples for flow cytometry were prepared and analyzed as described previously (Chen et al., 2009).
DNA Microarray Analysis
Expression analyses were conducted as described previously (Laub et al., 2000), but using custom Agilent arrays. Analysis of the depletion and overex-pression strains was done in duplicate and triplicate, respectively, and results averaged in each case. Complete data are deposited in GEO GSE22062.
Yeast Two-Hybrid Assays
Protein-protein interactions were assayed in the yeast two-hybrid system described previously (James et al., 1996), except using the Gateway modified pAD-DEST and pBD-DEST vectors.
Electrophoretic Mobility Shift Assays
EMSAs were performed using His6-SciP, His6-CtrA, His6-RpoD, and TAP-RNA polymerase. For details, including information on protein purification, see the Supplemental Information.
Quantitative Real-Time PCR
Quantitative real-time PCR for ChIP and expression analyses was performed using the DNA Engine Opticon 2 system (MJ Research) and the QuantiFast SYBR Green PCR Kit (QIAGEN), with full details provided in the Supplemental Information.
Pulse-Chase and In Vivo Phosphorylation Analyses
CtrA stability and phosphorylation were assayed as described previously (Domian et al., 1997), with details provided in the Supplemental Information.
Supplementary Material
Acknowledgments
We thank A. Yuan, J. Modell, K. Milaninia, B. Pando, and A. Reinke for help with experimental techniques. We thank F. Solomon, M. Neiditch, A. Fan, A. Grossman, and members of the Laub lab for comments on the manuscript. M.T.L. is an Early Career Scientist at the Howard Hughes Medical Institute. This work was supported by a National Institutes of Health (NIH) grant (5R01GM082899) to M.T.L.
Footnotes
Supplemental Information: Supplemental Information includes five figures, three tables, Supplemental Experimental Procedures, and Supplemental References and can be found with this article online at doi:10.1016/j.molcel.2010.06.024.
References
- Ansaldi M, Théraulaz L, Méjean V. TorI, a response regulator inhibitor of phage origin in Escherichia coli. Proc Natl Acad Sci USA. 2004;101:9423–9428. doi: 10.1073/pnas.0401927101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biondi EG, Reisinger SJ, Skerker JM, Arif M, Perchuk BS, Ryan KR, Laub MT. Regulation of the bacterial cell cycle by an integrated genetic circuit. Nature. 2006a;444:899–904. doi: 10.1038/nature05321. [DOI] [PubMed] [Google Scholar]
- Biondi EG, Skerker JM, Arif M, Prasol MS, Perchuk BS, Laub MT. A phosphorelay system controls stalk biogenesis during cell cycle progression in Caulobacter crescentus. Mol Microbiol. 2006b;59:386–401. doi: 10.1111/j.1365-2958.2005.04970.x. [DOI] [PubMed] [Google Scholar]
- Breeden LL. Periodic transcription: a cycle within a cycle. Curr Biol. 2003;13:R31–R38. doi: 10.1016/s0960-9822(02)01386-6. [DOI] [PubMed] [Google Scholar]
- Chen LS, Mullin D, Newton A. Identification, nucleotide sequence, and control of developmentally regulated promoters in the hook operon region of Caulobacter crescentus. Proc Natl Acad Sci USA. 1986;83:2860–2864. doi: 10.1073/pnas.83.9.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YE, Tsokos CG, Biondi EG, Perchuk BS, Laub MT. Dynamics of two Phosphorelays controlling cell cycle progression in Caulobacter crescentus. J Bacteriol. 2009;191:7417–7429. doi: 10.1128/JB.00992-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M, Nishikawa JL, Tang X, Millman JS, Schub O, Breitkreuz K, Dewar D, Rupes I, Andrews B, Tyers M. CDK activity antagonizes Whi5, an inhibitor of G1/S transcription in yeast. Cell. 2004;117:899–913. doi: 10.1016/j.cell.2004.05.024. [DOI] [PubMed] [Google Scholar]
- Domian IJ, Quon KC, Shapiro L. Cell type-specific phosphorylation and proteolysis of a transcriptional regulator controls the G1-to-S transition in a bacterial cell cycle. Cell. 1997;90:415–424. doi: 10.1016/s0092-8674(00)80502-4. [DOI] [PubMed] [Google Scholar]
- Domian IJ, Reisenauer A, Shapiro L. Feedback control of a master bacterial cell-cycle regulator. Proc Natl Acad Sci USA. 1999;96:6648–6653. doi: 10.1073/pnas.96.12.6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutton RJ, Xu Z, Gober JW. Linking structural assembly to gene expression: a novel mechanism for regulating the activity of a sigma54 transcription factor. Mol Microbiol. 2005;58:743–757. doi: 10.1111/j.1365-2958.2005.04857.x. [DOI] [PubMed] [Google Scholar]
- Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- Hung DY, Shapiro L. A signal transduction protein cues proteolytic events critical to Caulobacter cell cycle progression. Proc Natl Acad Sci USA. 2002;99:13160–13165. doi: 10.1073/pnas.202495099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer VR, Horak CE, Scafe CS, Botstein D, Snyder M, Brown PO. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature. 2001;409:533–538. doi: 10.1038/35054095. [DOI] [PubMed] [Google Scholar]
- Jacobs C, Domian IJ, Maddock JR, Shapiro L. Cell cycle-dependent polar localization of an essential bacterial histidine kinase that controls DNA replication and cell division. Cell. 1999;97:111–120. doi: 10.1016/s0092-8674(00)80719-9. [DOI] [PubMed] [Google Scholar]
- Jacobs C, Ausmees N, Cordwell SJ, Shapiro L, Laub MT. Functions of the CckA histidine kinase in Caulobacter cell cycle control. Mol Microbiol. 2003;47:1279–1290. doi: 10.1046/j.1365-2958.2003.03379.x. [DOI] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenal U, Fuchs T. An essential protease involved in bacterial cell-cycle control. EMBO J. 1998;17:5658–5669. doi: 10.1093/emboj/17.19.5658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato A, Groisman EA. Connecting two-component regulatory systems by a protein that protects a response regulator from dephosphorylation by its cognate sensor. Genes Dev. 2004;18:2302–2313. doi: 10.1101/gad.1230804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laub MT, Goulian M. Specificity in two-component signal transduction pathways. Annu Rev Genet. 2007;41:121–145. doi: 10.1146/annurev.genet.41.042007.170548. [DOI] [PubMed] [Google Scholar]
- Laub MT, McAdams HH, Feldblyum T, Fraser CM, Shapiro L. Global analysis of the genetic network controlling a bacterial cell cycle. Science. 2000;290:2144–2148. doi: 10.1126/science.290.5499.2144. [DOI] [PubMed] [Google Scholar]
- Laub MT, Chen SL, Shapiro L, McAdams HH. Genes directly controlled by CtrA, a master regulator of the Caulobacter cell cycle. Proc Natl Acad Sci USA. 2002;99:4632–4637. doi: 10.1073/pnas.062065699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laub MT, Shapiro L, McAdams HH. Systems biology of Caulobacter. Annu Rev Genet. 2007;41:429–441. doi: 10.1146/annurev.genet.41.110306.130346. [DOI] [PubMed] [Google Scholar]
- Llewellyn M, Dutton RJ, Easter J, O'Donnol D, Gober JW. The conserved flaF gene has a critical role in coupling flagellin translation and assembly in Caulobacter crescentus. Mol Microbiol. 2005;57:1127–1142. doi: 10.1111/j.1365-2958.2005.04745.x. [DOI] [PubMed] [Google Scholar]
- Martin ME, Trimble MJ, Brun YV. Cell cycle-dependent abundance, stability and localization of FtsA and FtsQ in Caulobacter crescentus. Mol Microbiol. 2004;54:60–74. doi: 10.1111/j.1365-2958.2004.04251.x. [DOI] [PubMed] [Google Scholar]
- McGrath PT, Iniesta AA, Ryan KR, Shapiro L, McAdams HH. A dynamically localized protease complex and a polar specificity factor control a cell cycle master regulator. Cell. 2006;124:535–547. doi: 10.1016/j.cell.2005.12.033. [DOI] [PubMed] [Google Scholar]
- Milhausen M, Agabian N. Caulobacter flagellin mRNA segregates asymmetrically at cell division. Nature. 1983;302:630–632. doi: 10.1038/302630a0. [DOI] [PubMed] [Google Scholar]
- Morgan DO. The Cell Cycle: Principles of Control. London: New Science Press Ltd; 2007. [Google Scholar]
- Mullin DA, Ohta N, Mullin AH, Newton A. Organization, expression, and function of Caulobacter crescentus genes needed for assembly and function of the flagellar hook. Mol Genet Genomics. 2001;265:445–454. doi: 10.1007/s004380000432. [DOI] [PubMed] [Google Scholar]
- Nakano S, Nakano MM, Zhang Y, Leelakriangsak M, Zuber P. A regulatory protein that interferes with activator-stimulated transcription in bacteria. Proc Natl Acad Sci USA. 2003;100:4233–4238. doi: 10.1073/pnas.0637648100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura M, Shimane K, Asai K, Ogasawara N, Tanaka T. Binding of response regulator DegU to the aprE promoter is inhibited by RapG, which is counteracted by extracellular PhrG in Bacillus subtilis. Mol Microbiol. 2003;49:1685–1697. doi: 10.1046/j.1365-2958.2003.03665.x. [DOI] [PubMed] [Google Scholar]
- Panis G, Mejean V, Ansaldi M. Control and regulation of KplE1 prophage site-specific recombination: a new recombination module analyzed. J Biol Chem. 2007;282:21798–21809. doi: 10.1074/jbc.M701827200. [DOI] [PubMed] [Google Scholar]
- Quon KC, Marczynski GT, Shapiro L. Cell cycle control by an essential bacterial two-component signal transduction protein. Cell. 1996;84:83–93. doi: 10.1016/s0092-8674(00)80995-2. [DOI] [PubMed] [Google Scholar]
- Quon KC, Yang B, Domian IJ, Shapiro L, Marczynski GT. Negative control of bacterial DNA replication by a cell cycle regulatory protein that binds at the chromosome origin. Proc Natl Acad Sci USA. 1998;95:120–125. doi: 10.1073/pnas.95.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisenauer A, Quon K, Shapiro L. The CtrA response regulator mediates temporal control of gene expression during the Caulobacter cell cycle. J Bacteriol. 1999;181:2430–2439. doi: 10.1128/jb.181.8.2430-2439.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross JF, Liu X, Dynlacht BD. Mechanism of transcriptional repression of E2F by the retinoblastoma tumor suppressor protein. Mol Cell. 1999;3:195–205. doi: 10.1016/s1097-2765(00)80310-x. [DOI] [PubMed] [Google Scholar]
- Ryan KR, Shapiro L. Temporal and spatial regulation in prokaryotic cell cycle progression and development. Annu Rev Biochem. 2003;72:367–394. doi: 10.1146/annurev.biochem.72.121801.161824. [DOI] [PubMed] [Google Scholar]
- Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- Siam R, Marczynski GT. Cell cycle regulator phosphorylation stimulates two distinct modes of binding at a chromosome replication origin. EMBO J. 2000;19:1138–1147. doi: 10.1093/emboj/19.5.1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skerker JM, Shapiro L. Identification and cell cycle control of a novel pilus system in Caulobacter crescentus. EMBO J. 2000;19:3223–3234. doi: 10.1093/emboj/19.13.3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon JM, Lazazzera BA, Grossman AD. Purification and characterization of an extracellular peptide factor that affects two different developmental pathways in Bacillus subtilis. Genes Dev. 1996;10:2014–2024. doi: 10.1101/gad.10.16.2014. [DOI] [PubMed] [Google Scholar]
- Spencer W, Siam R, Ouimet MC, Bastedo DP, Marczynski GT. CtrA, a global response regulator, uses a distinct second category of weak DNA binding sites for cell cycle transcription control in Caulobacter crescentus. J Bacteriol. 2009;191:5458–5470. doi: 10.1128/JB.00355-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan BS, Novak A, Flannick J, Batzoglou S, McAdams HH, editors. Integrated Protein Interaction Networks for 11 Microbes. New York: Springer; 2006. [Google Scholar]
- Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- Wu J, Ohta N, Newton A. An essential, multicomponent signal transduction pathway required for cell cycle regulation in Caulobacter. Proc Natl Acad Sci USA. 1998;95:1443–1448. doi: 10.1073/pnas.95.4.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweiger G, Marczynski G, Shapiro L. A Caulobacter DNA methyltransferase that functions only in the predivisional cell. J Mol Biol. 1994;235:472–485. doi: 10.1006/jmbi.1994.1007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


