Abstract
Sequencing of bacterial and archaeal genomes has revolutionized our understanding of the many roles played by microorganisms1. There are now nearly 1,000 completed bacterial and archaeal genomes available2, most of which were chosen for sequencing on the basis of their physiology. As a result, the perspective provided by the currently available genomes is limited by a highly biased phylogenetic distribution3–5. To explore the value added by choosing microbial genomes for sequencing on the basis of their evolutionary relationships, we have sequenced and analysed the genomes of 56 culturable species of Bacteria and Archaea selected to maximize phylogenetic coverage. Analysis of these genomes demonstrated pronounced benefits (compared to an equivalent set of genomes randomly selected from the existing database) in diverse areas including the reconstruction of phylogenetic history, the discovery of new protein families and biological properties, and the prediction of functions for known genes from other organisms. Our results strongly support the need for systematic ‘phylogenomic’ efforts to compile a phylogeny-driven ‘Genomic Encyclopedia of Bacteria and Archaea’ in order to derive maximum knowledge from existing microbial genome data as well as from genome sequences to come.
Since the publication of the first complete bacterial genome, sequencing of the microbial world has accelerated beyond expectations. The inventory of bacterial and archaeal isolates with complete or draft sequences is approaching the two thousand mark2. Most of these genome sequences are the product of studies in which one or a few isolates were targeted because of an interest in a specific characteristic of the organism. Although large-scale multi-isolate genome sequencing studies have been performed, they have tended to be focused on particular habitats or on the relatives of specific organisms. This overall lack of broad phylogenetic considerations in the selection of microbial genomes for sequencing, combined with a cultivation bottleneck6, has led to a strongly biased representation of recognized microbial phylogenetic diversity3–5. Although some projects have attempted to correct this (for example, see ref. 5), they have all been small in scope. To evaluate the potential benefits of a more systematic effort, we embarked on a pilot project to sequence approximately 100 genomes selected solely for their phylogenetic novelty: the ‘Genomic Encyclopedia of Bacteria and Archaea’ (GEBA).
Organisms were selected on the basis of their position in a phylogenetic tree of small subunit (SSU) ribosomal RNA, the best sampled gene from across the tree of life7. Working from the root to the tips of the tree, we identified the most divergent lineages that lacked representatives with sequenced genomes (completed or in progress)8 and for which a species has been formally described9 and a type strain designated and deposited in a publicly accessible culture collection10. From hundreds of candidates, 200 type strains were selected both to obtain broad coverage across Bacteria and Archaea and to perform in-depth sampling of a single phylum. The Gram-positive bacterial phylum Actinobacteria was chosen for the latter purpose because of the availability of many phylogenetically and phenotypically diverse cultured strains, and because it had the lowest percentage of sequenced isolates of any phylum (1% versus an average of 2.3%)11. Of the 200 targeted isolates, 159 were designated as ‘high’ priority primarily on the basis of phylum-level novelty and the ability to obtain microgram quantities of high quality DNA. The genomes of these 159 are being sequenced, assembled, annotated (including recommended metadata12) and finished, and relevant data are being released through a dedicated Integrated Microbial Genomes database portal13 and deposited into GenBank. Currently, data from 106 genomes (62 of which are finished) are available.
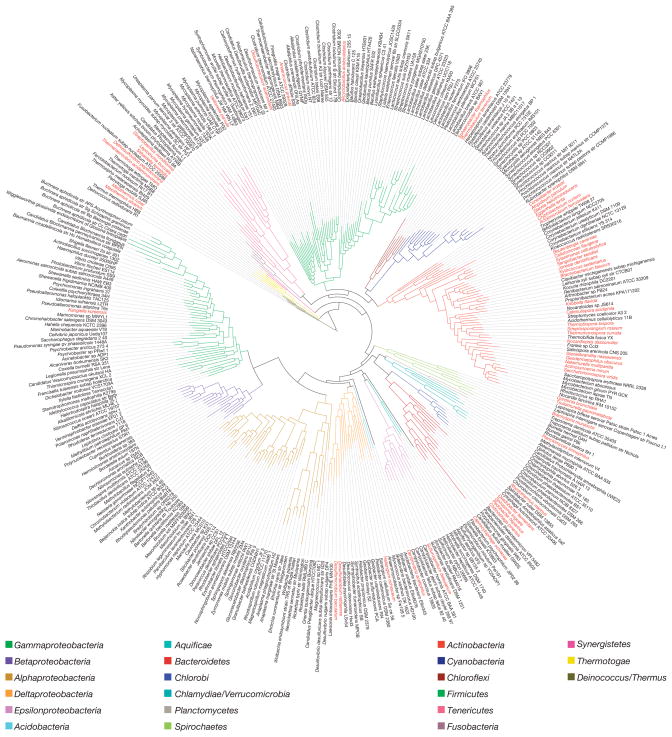
To assess the ramifications of this tree-based selection of organisms, we focused our analyses on the first 56 genomes for which the shotgun phase of sequencing was completed. The 53 bacteria and 3 archaea (Supplementary Table 1) represent both a broad sampling of bacterial diversity and a deeper sampling of the phylum Actinobacteria (26 GEBA genomes). An initial question we addressed was whether selection on the basis of phylogenetic novelty of SSU rRNA genes reliably identifies genomes that are phylogenetically novel on the basis of other criteria. This question arises because it is known that single genes, even SSU rRNA genes, do not perfectly predict genome-wide phylogenetic patterns14,15. To investigate this, we created a ‘genome tree’ (ref. 16) of completed bacterial genomes (Fig. 1) and then measured the relative contribution of the GEBA project using the phylogenetic diversity metric17. We found that the 53 GEBA bacteria accounted for 2.8–4.4 times more phylogenetic diversity than randomly sampled subsets of 53 non-GEBA bacterial genomes. A similar degree of improvement in phylogenetic diversity was seen for the more intensively sampled actinobacteria (Table 1). These analyses indicate that although SSU rRNA genes are not a perfect indicator of organismal evolution, their phylogenetic relationships are a sound predictor of phylogenetic novelty within the universal gene core present in bacterial genomes.
Figure 1.
Maximum-likelihood phylogenetic tree of the bacterial domain based on a concatenated alignment of 31 broadly conserved protein-coding genes16. Phyla are distinguished by colour of the branch and GEBA genomes are indicated in red in the outer circle of species names.
Table 1.
Effect of SSU rRNA tree-based selection of organisms on comparative genomic metrics
| Comparative genomic metric | GEBA set | Random sets (number of resamplings) | Fold improvement |
|---|---|---|---|
| Genome tree phylogenetic diversity17 | |||
| Bacteria (domain) | 11.0 | 3.2 ± 0.7 (100) | 2.8–4.4 |
| Actinobacteria (phylum) | 4.3 | 1.4 ± 0.3 (100) | 2.5–3.9 |
| New protein family links | 46 | 3 ± 4 (5) | 6.6 to >15.3 |
| Genes in new chromosomal cassettes | 71,579 | 16,579 ± 5,523 (20) | 3.2–6.5 |
| New gene fusions | 433 | 65 ± 31 (20) | 4.5–12.7 |
GEBA genomes were compared to equivalently sized random sets of reference genomes to quantify the effect of phylogenetic selection.
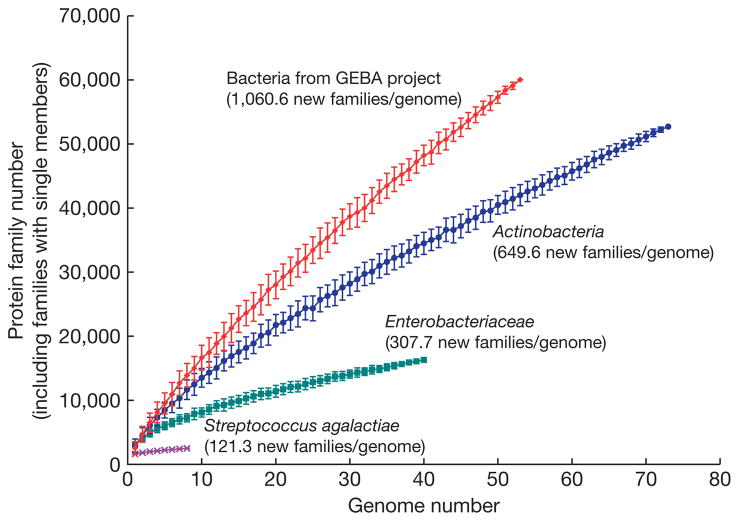
The discovery and characterization of new gene families and their associated novel functions provide one incentive for sequencing additional genomes, analysis of which has helped to redefine the protein family universe18. We explored the quantitative effect of tree-based genome selection on the pace of discovery of novel proteins and functions. Specifically, we compared the rate of discovery of novel protein families when progressively adding more closely related genomes versus when adding more distantly related ones (Fig. 2). Granted, many factors contribute to protein family diversity, such as ecological niche; nevertheless, higher rates of novel protein family discovery were found in the more phylogenetically diverse taxa (Fig. 2). In addition, of the 16,797 families identified in the 56 GEBA genomes, 1,768 showed no significant sequence similarity to any proteins, indicating the presence of novel functional diversity. These results highlight the utility of tree-based genome selection as a means to maximize the identification of novel protein families and argues against lateral gene transfer significantly redistributing genetic novelty between distantly related lineages.
Figure 2. Rate of discovery of protein families as a function of phylogenetic breadth of genomes.
For each of four groupings (species, different strains of Streptococcus agalactiae; family, Enterobacteriaceae; phylum, Actinobacteria; domain, GEBA bacteria), all proteins from that group were compared to each other to identify protein families. Then the total number of protein families was calculated as genomes were progressively sampled from the group (starting with one genome until all were sampled). This was done multiple times for each of the four groups using random starting seeds; the average and standard deviation were then plotted.
Novel proteins also can serve to link distantly related homologues whose relatedness would otherwise go undetected. Forty-six such links were identified in the 56 GEBA genomes compared to an average of only three new links in equivalent sets of randomly sampled non-GEBA genomes (Table 1). A useful complement to homology-based predictions of gene function are ‘non-homology methods’ (ref. 19) such as gene context-based inference that relies on the conserved clustering of functionally related genes across multiple genomes, often in operons or as gene fusions20. We identified over 70,000 genes in new chromosomal cassettes of two or more genes in the GEBA genomes. This represents a three- to sixfold increase over equivalent sets of non-GEBA genomes (Table 1). Similarly, the number of new gene fusions identified in the GEBA genomes is 4 to ~13 times greater than in randomly selected genome sets (Table 1). Because the GEBA data set produced a several-fold improvement over random sets for all metrics examined (Table 1), we predict that other aspects of sequence-based biological discovery will similarly benefit from tree-based genome sequencing.
The GEBA genomes also show significant phylogenetic expansions within known protein families. For example, although only two of the 56 GEBA organisms are known cellulose degraders, we identified in the set of genomes a variety of glycoside hydrolase (GH) genes that may participate in the breakdown of cellulose and hemicelluloses. Among these are 28 and 7 phylogenetically divergent members of the endoglucanase- and processive exoglucanase-containing GH6 and GH48 families, respectively. Halorhabdus utahensis, a halophilic archaeon known to have β-xylanase and β-xylosidase activities21, has a chromosomal cluster including two GH10 family β-xylanases and six novel GH5 family proteins of unknown specificity.
The enrichment of genetic diversity is also seen within families of non-coding RNAs, transposable elements, and other cellular components. For example, the genome of the marine myxobacterium Haliangium ochraceum contains 807 CRISPR (clustered regularly interspaced short palindromic repeats) units including the largest single CRISPR array known, comprising 382 spacer/repeat units. CRISPR is a newly recognized, but ancient and widespread, system in bacteria and archaea that confers resistance to viruses and other invading foreign DNAs22.
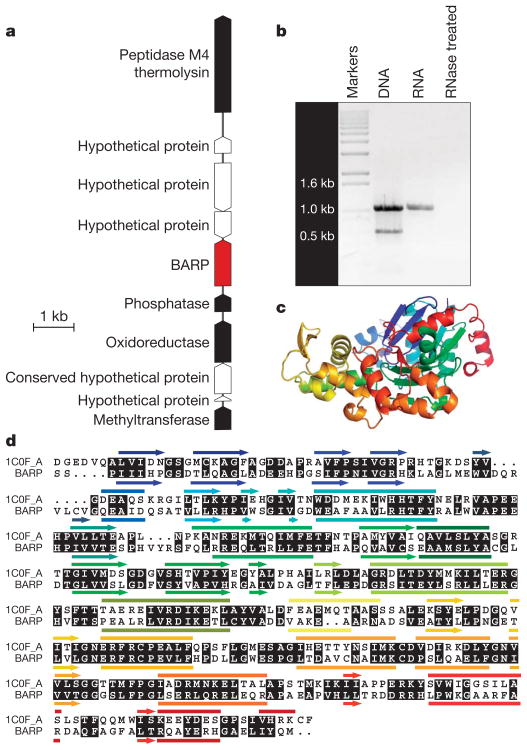
Results from the GEBA pilot project challenge our current understanding for the taxonomic distribution of known gene families. The most striking example of which is the discovery of an actin homologue in H. ochraceum. Actin and its close relatives are structural components of the eukaryotic cytoskeleton that are found in every eukaryote and only in eukaryotes. Bacteria and archaea encode instead the shape-determining protein MreB. Although MreBs have some functional and structural similarities to eukaryotic actins, they are regarded, at best, distantly related homologues23 and possibly not even homologous. Like other bacteria, H. ochraceum encodes a bona fide MreB protein, but in addition, it encodes a protein that is clearly a member of the actin family, which we have named BARP (bacterial actin-related protein; Fig. 3). Although we do not yet have evidence for its precise function, BARP is expressed in H. ochraceum (Fig. 3b). Assuming that the H. ochraceum mreB orthologue performs the same function as in other bacteria, and given that the myxobacteria, to which this species belongs, are known to synthesize actin-targeting toxins24, we propose that this BARP may be a dominant-negative inhibitor of eukaryotic actin polymerization. Regardless of its precise function, this first—and so far only—discovery of an expressed homologue of eukaryotic actin in a member of the Bacteria highlights the potential for novel and surprising biological discoveries given a wider genomic sampling of the tree of life.
Figure 3. A bacterial homologue of actin.
a, Genomic context of the bacterial actin-related protein (BARP) gene within the genome of the marine Delta proteobacterium H. ochraceum. Red, gene encoding BARP; white, genes encoding hypothetical proteins; black, genes with functional annotations. b, RT–PCR demonstration of expression of the gene encoding BARP in H. ochraceum. c, Ribbon plot of the putative structure of BARP. d, Alignment of BARP with actin from Dictyostelium discoideum29 with similarities in black shaded text. Secondary structure elements (arrows, beta-strands; bars, alpha-helices) are colour-coded as in c. A phylogenetic tree including this protein is in Supplementary Figure 1.
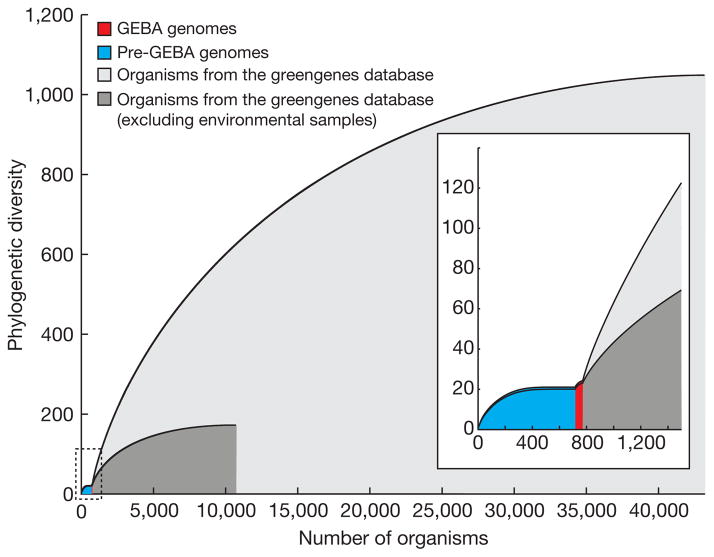
We conclude that targeting microorganisms for genome sequencing solely on the basis of phylogenetic considerations offers significant far-reaching benefits in diverse areas. Furthermore, the benefits of phylogenetically driven genome sequencing show no sign of saturating with these first 56 genomes. A key question then lies in determining how much bacterial and archaeal diversity remains to be sampled. Using SSU rRNA gene sequences as a proxy for organismal diversity (Fig. 4), we estimate that sequencing the genomes of only 1,520 phylogenetically selected isolates could encompass half of the phylogenetic diversity represented by known cultured bacteria and archaea. Given the continuing reductions in both the cost and difficulty in sequencing genomes25, this is certainly a tractable target in the next few years.
Figure 4. Phylogenetic diversity of bacteria and archaea on the basis of SSU rRNA genes.
Using a phylogenetic tree of unique SSU rRNA gene sequences7, phylogenetic diversity was measured for four subsets of this tree: organisms with sequenced genomes pre-GEBA (blue), the GEBA organisms (red), all cultured organisms (dark grey), and all available SSU rRNA genes (light grey). For each subtree, taxa were sorted by their contribution to the subtree phylogenetic diversity30 and the cumulative phylogenetic diversity was plotted from maximal (left) to the least (right). The inset magnifies the first 1,500 organisms. Comparison of the plots shows the phylogenetic ‘dark matter’ left to be sampled.
However, the great majority of recognized bacterial and archaeal diversity is not represented by pure cultures and an additional 9,218 genome sequences from currently uncultured species would be required to capture 50% of this recognized diversity (Fig. 4). Such an undertaking will require new approaches to culturing or processing of multi-species samples using methods such as metagenomics26 or physical isolation of cells from mixed populations followed by whole genome amplification methods27. Obtaining reference genomes for the uncultured microbial majority will be a natural extension of the GEBA project, the ultimate goal of which is to provide a phylogenetically balanced genomic representation of the microbial tree of life. The pilot study presented here is a dedicated first step in this direction.
METHODS SUMMARY
Starting with a phylogenetic tree of SSU rRNA genes7, we identified major branches that had no available genome sequences but for which cultured isolates were available in the DSMZ or ATCC culture collections. Selected isolates (Supplementary Table 1a, b) from these branches were grown and DNA isolated (Supplementary Table 1c) and quality checked. DNA was then used for shotgun genome sequencing by Sanger/ABI, Roche/454 and/or Illumina/Solexa technologies (Supplementary Table 2). Sequence reads were assembled separately with different assembly methods and the best draft assembly was used for annotation and as a starting point for genome completion (current genome status is in Supplementary Table 2). Annotation (gene identification, functional prediction, etc.) was performed using the IMG system (http://img.jgi.doe.gov/geba); this was done both after shotgun sequencing and again after genome completion. For ‘whole genome tree’ analysis, a PHYML maximum likelihood phylogenetic tree of a concatenated alignment of 31 marker genes was built using AMPHORA16. Phylogenetic diversity was calculated as the sum of branch lengths in this and other trees. Protein families were built for various genome sets by using the Markov clustering algorithm (MCL)28 to group proteins on the basis of ‘all versus all’ blastp searches. For analysis of phylogenetic diversity of organisms, a phylogenetic tree was built for a combined alignment of SSU rRNA sequences from published genomes and a non-redundant subset of greengenes SSU rRNA7. Further analysis of the genomes was done using IMG database queries and new computational analyses as described in the main text, legends and Supplementary Methods.
Supplementary Material
Acknowledgments
We thank the following people for assistance in aspects of the project including planning and discussions (R. Stevens, G. Olsen, R. Edwards, J. Bristow, N. Ward, S. Baker, T. Lowe, J. Tiedje, G. Garrity, A. Darling, S. Giovannoni), analysis of genomes whose work could not be included in this report (B. Henrissat, G. Xie, J. Kinney, I. Paulsen, N. Rawlings, M. Huntemann), project management (M. Miller, M. Fenner, M. McGowen, A. Greiner), sequencing and finishing (K. Ikeda, M. Chovatia, P. Richardson, T. Glavinadelrio, C. Detter), culture growth, DNA extraction, and metadata (D. Gleim, E. Brambilla, S. Schneider, M. Schröder, M. Jando, G. Gehrich-Schröter, C. Wahrenburg, K. Steenblock, S. Welnitz, M. Kopitz, R. Fähnrich, H. Pomrenke, A. Schütze, M. Rohde, M. Göker), and manuscript editing (M. Youle). This work was performed under the auspices of the US Department of Energy’s Office of Science, Biological and Environmental Research Program, and by the University of California, Lawrence Berkeley National Laboratory under contract no. DE-AC02-05CH11231, Lawrence Livermore National Laboratory under contract no. DE-AC52-07NA27344, and Los Alamos National Laboratory under contract no. DE-AC02-06NA25396. Support for J.A.E., D.W. and M.W. was provided by the Gordon and Betty Moore Foundation Grant no. 1660 to J.A.E. Support for work at DSMZ was provided under DFG INST 599/1-1.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions D.W. (rRNA analysis, gene families, actin tree, manuscript preparation), P.H. (selection of strains, analysis, manuscript preparation, project coordination), L.G. and D.B. (project management), R.P., B.J.T., E.L., S.G., S.S. (strain curation and growth), K.M., N.N.I., I.J.A., S.D.H., A.P., A.Ly. (annotation, genome analysis), V.K. (CRISPRs, actin), M.W. (whole genome tree), P.D., C.K., A.Z. and M.S. (actin studies), M.N., S.L., J.-F.C., F.C. and E.D. (sequencing), C.H., A.La., M.N. and A.C. (finishing), P.C. (analysis), E.M.R. (manuscript preparation), N.C.K. (selection of strains, annotation, analysis), H.-P.K. (strain selection and growth, DNA preparation, manuscript preparation), J.A.E. (project lead and coordination, analysis, manuscript preparation).
Author Information Genome sequence and annotation data is available at the JGI IMG-GEBA page http://img.jgi.doe.gov/geba and has been submitted to GenBank with accessions ABSZ00000000, ABTA00000000, ABTB00000000, ABTC00000000, ABTD00000000, CP001618, ABTF00000000, ABTG00000000, ABTH00000000, ABTI00000000, ABTJ00000000, ABTK00000000, ABTM00000000, NZ_ABTN00000000, NZ_ABTO00000000, ABTP00000000, ABTQ00000000, NZ_ABTR00000000, NZ_ABTS00000000, NZ_ABTT00000000, NZ_ABTU00000000, NZ_ABTV00000000, NZ_ABTW00000000, NZ_ABTX00000000, NZ_ABTY00000000, NZ_ABTZ00000000, NZ_ABUA00000000, NZ_ABUB00000000, NZ_ABUC00000000, NZ_ABUD00000000., NZ_ABUE00000000, NZ_ABUF00000000, NZ_ABUG00000000, NZ_ABUH00000000, NZ_ABUI00000000, NZ_ABUJ00000000, NZ_ABUK00000000, ABUL00000000, NZ_ABUM00000000, NZ_ABUO00000000, NZ_ABUP00000000, ABUQ00000000, NZ_ABUR00000000, ABUS00000000, NZ_ABUT00000000, NZ_ABUU00000000, NZ_ABUV00000000, NZ_ABUW00000000, NZ_ABUX00000000, NZ_ABUZ00000000, NZ_ABVA00000000, NZ_ABVB00000000 and NZ_ABVC00000000. All strains that have been sequenced are available from the DSMZ culture collection and culture accessions are available in Supplementary Information. This paper is distributed under the terms of the Creative Commons Attribution-Non-Commercial-Share Alike licence, and is freely available to all readers at www.nature.com/nature. Reprints and permissions information is available at www.nature.com/reprints. Further details on sequencing and genome properties of each organism are being published in the journal Standards in Genomic Sciences (SIGS) (http://standardsingenomics.org/).
References
- 1.Fraser CM, Eisen JA, Salzberg SL. Microbial genome sequencing. Nature. 2000;406:799–803. doi: 10.1038/35021244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liolios K, Mavromatis K, Tavernarakis N, Kyrpides NC. The Genomes On Line Database (GOLD) in 2007: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res. 2008;36 (database issue):D475–D479. doi: 10.1093/nar/gkm884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hugenholtz P. Exploring prokaryotic diversity in the genomic era. Genome Biol. 2002;3:REVIEWS0003.1–REVIEWS0003.8. doi: 10.1186/gb-2002-3-2-reviews0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eisen JA. Assessing evolutionary relationships among microbes from whole-genome analysis. Curr Opin Microbiol. 2000;3:475–480. doi: 10.1016/s1369-5274(00)00125-9. [DOI] [PubMed] [Google Scholar]
- 5.Wu D, et al. Complete genome sequence of the aerobic CO-oxidizing thermophile Thermomicrobium roseum. PLoS One. 2009;4:e4207. doi: 10.1371/journal.pone.0004207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pace NR. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 7.DeSantis TZ, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bernal A, Ear U, Kyrpides N. Genomes OnLine Database (GOLD): a monitor of genome projects world-wide. Nucleic Acids Res. 2001;29:126–127. doi: 10.1093/nar/29.1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lapage SP, et al. International Code of Nomenclature of Bacteria, 1990 Revision. American Society for Microbiology; 1992. [PubMed] [Google Scholar]
- 10.Ward N, Eisen J, Fraser C, Stackebrandt E. Sequenced strains must be saved from extinction. Nature. 2001;414:148. doi: 10.1038/35102737. [DOI] [PubMed] [Google Scholar]
- 11.Hugenholtz P, Kyrpides NC. A changing of the guard. Environ Microbiol. 2009;11:551–553. doi: 10.1111/j.1462-2920.2009.01888.x. [DOI] [PubMed] [Google Scholar]
- 12.Field D, et al. The minimum information about a genome sequence (MIGS) specification. Nature Biotechnol. 2008;26:541–547. doi: 10.1038/nbt1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Markowitz VM, et al. The integrated microbial genomes (IMG) system in 2007: data content and analysis tool extensions. Nucleic Acids Res. 2008;36 (database issue):D528–D533. doi: 10.1093/nar/gkm846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Achtman M, Wagner M. Microbial diversity and the genetic nature of microbial species. Nature Rev Microbiol. 2008;6:431–440. doi: 10.1038/nrmicro1872. [DOI] [PubMed] [Google Scholar]
- 15.Beiko RG, Doolittle WF, Charlebois RL. The impact of reticulate evolution on genome phylogeny. Syst Biol. 2008;57:844–856. doi: 10.1080/10635150802559265. [DOI] [PubMed] [Google Scholar]
- 16.Wu M, Eisen JA. A simple, fast, and accurate method of phylogenomic inference. Genome Biol. 2008;9:R151. doi: 10.1186/gb-2008-9-10-r151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pardi F, Goldman N. Resource-aware taxon selection for maximizing phylogenetic diversity. Syst Biol. 2007;56:431–444. doi: 10.1080/10635150701411279. [DOI] [PubMed] [Google Scholar]
- 18.Kunin V, Cases I, Enright AJ, de Lorenzo V, Ouzounis CA. Myriads of protein families, and still counting. Genome Biol. 2003;4:401. doi: 10.1186/gb-2003-4-2-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marcotte EM, et al. Detecting protein function and protein-protein interactions from genome sequences. Science. 1999;285:751–753. doi: 10.1126/science.285.5428.751. [DOI] [PubMed] [Google Scholar]
- 20.Enright AJ, Iliopoulos I, Kyrpides NC, Ouzounis CA. Protein interaction maps for complete genomes based on gene fusion events. Nature. 1999;402:86–90. doi: 10.1038/47056. [DOI] [PubMed] [Google Scholar]
- 21.Wainø M, Ingvorsen K. Production of β-xylanase and β-xylosidase by the extremely halophilic archaeon Halorhabdus utahensis. Extremophiles. 2003;7:87–93. doi: 10.1007/s00792-002-0299-y. [DOI] [PubMed] [Google Scholar]
- 22.Barrangou R, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 23.Doolittle RF, York AL. Bacterial actins? An evolutionary perspective. Bioessays. 2002;24:293–296. doi: 10.1002/bies.10079. [DOI] [PubMed] [Google Scholar]
- 24.Sasse F, Kunze B, Gronewold TM, Reichenbach H. The chondramides: cytostatic agents from myxobacteria acting on the actin cytoskeleton. J Natl Cancer Inst. 1998;90:1559–1563. doi: 10.1093/jnci/90.20.1559. [DOI] [PubMed] [Google Scholar]
- 25.Shendure J, Ji H. Next-generation DNA sequencing. Nature Biotechnol. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 26.Kunin V, Copeland A, Lapidus A, Mavromatis K, Hugenholtz P. A bioinformatician’s guide to metagenomics. Microbiol Mol Biol Rev. 2008;72:557–578. doi: 10.1128/MMBR.00009-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ishoey T, Woyke T, Stepanauskas R, Novotny M, Lasken RS. Genomic sequencing of single microbial cells from environmental samples. Curr Opin Microbiol. 2008;11:198–204. doi: 10.1016/j.mib.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsuura Y, et al. Structural basis for the higher Ca2+-activation of the regulated actin-activated myosin ATPase observed with Dictyostelium/Tetrahymena actin chimeras. J Mol Biol. 2000;296:579–595. doi: 10.1006/jmbi.1999.3467. [DOI] [PubMed] [Google Scholar]
- 30.Moulton V, Semple C, Steel M. Optimizing phylogenetic diversity under constraints. J Theor Biol. 2007;246:186–194. doi: 10.1016/j.jtbi.2006.12.021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.