SUMMARY
The majority of trypanosomal mitochondrial pre-mRNAs undergo massive uridine insertion/deletion editing which creates open reading frames. Although the pre-editing addition of short 3′ A-tails is known to stabilize transcripts during and after the editing, the processing event committing the fully-edited mRNAs to translation remained unknown. Here we show that a heterodimer of pentatricopeptide repeat-containing (PPR) proteins, termed kinetoplast polyadenylation/ uridylation factors (KPAFs) 1 and 2, induces the post-editing addition of A/U-heteropolymers by KPAP1 poly(A) polymerase and RET1 terminal uridyltransferase. Edited transcripts bearing 200–300 nucleotide-long A/U-tails, but not short A-tails, were enriched in translating ribosomal complexes and affinity-purified ribosomal particles. KPAF1 repression led to a selective loss of A/U-tailed mRNAs and concomitant inhibition of protein synthesis. These results establish A/U extensions as the defining cis-elements of translation-competent mRNAs. Furthermore, we demonstrate that A/U-tailed mRNA preferentially interacts with the small ribosomal subunit, whereas edited substrates and complexes bind to the large subunit.
Keywords: Trypanosoma, mitochondria, mitochondrial ribosome, RNA editing, polyadenylation, translation, TUTase, PPR proteins
INTRODUCTION
Trypanosomes are characterized by the presence of a “kinetoplast,” a disc-shaped DNA structure located in the mitochondrion adjacent to the flagellar basal body. The mitochondrial DNA (kDNA) is composed of catenated maxicircles and minicircles; the former contain genes for ribosomal RNAs and subunits of respiratory complexes while the latter encode guide RNAs (gRNAs). Polycistronic precursors are transcribed from the maxicircles and then processed into rRNA and pre-mRNAs through an unknown nucleolytic pathway. In Trypanosoma brucei, six of the 18 annotated mRNAs encode predicted polypeptides and are referred to as “never-edited.” The remaining 12 pre-mRNAs are termed “pre-edited,” as they must undergo RNA editing to acquire open reading frames. The mRNA cleavage, U-insertion/deletion and ligation editing reactions are catalyzed by the RNA editing core complex (RECC, the 20S editosome) (Aphasizhev et al., 2003a; Panigrahi et al., 2003). Each step of the editing cascade is directed by gRNAs (Blum et al., 1990; Seiwert et al., 1996). These 50–60 nucleotide (nt) molecules are uridylated by RNA editing TUTase 1 (RET1) (Aphasizhev et al., 2002; Aphasizhev et al., 2003b) and stabilized via association with the gRNA binding complex (GRBC) (Weng et al., 2008).
Mitochondrial mRNAs possess short 5′ untranslated regions (UTRs) lacking apparent ribosome binding sites and are 3′ polyadenylated (Bhat et al., 1990). In the procyclic (insect) (PF) actively-respiring developmental form, the length of the 3′ extension correlates with the mRNA’s editing status: pre-edited and partially-edited mRNAs possess short (20–30 nt) A-tails, while in fully-edited mRNAs the short tail is often extended into 200–300 nt A/U- heteropolymer (Etheridge et al., 2008). Similar to fully-edited mRNAs, the major fraction of never-edited mRNAs bears long 3′ extensions while the minor portion terminates with short A-tail. Interestingly, RNA editing continues in the oxidative phosphorylation-deficient mitochondrion of bloodstream form (BF) parasites and is essential for their viability (Schnaufer et al., 2001). In the bloodstream form, ATP is produced via substrate level phosphorylation and is hydrolyzed by the ATP synthase to generate mitochondrial transembrane potential, thus maintaining the organelle (Schnaufer et al., 2005; Brown et al., 2006). Therefore, editing and translation of at least one mitochondrial mRNA encoding subunit 6 of the F1F0-ATPase complex is expected to occur.
Adenylated partially- or fully-edited mRNAs are relatively stable in mitochondrial extracts, while non-adenylated molecules are rapidly degraded (Ryan et al., 2003; Kao and Read, 2005). Along with knockdown studies of the mitochondrial poly(A) polymerase KPAP1, available evidence suggests that the short A-tail is required for maintenance of partially- and fully-edited mRNAs. Formation of the long A/U-tail is temporally separated from initial adenylation event and occurs after completion of editing (Etheridge et al., 2008). In addition to KPAP1, this process may involve RET1 TUTase, which also adds continuous 15–20 nt 3′ U-tails to gRNAs and ribosomal RNAs (Aphasizheva and Aphasizhev, 2010). The mechanism of postediting addition of a long A/U-heteropolymer to a pre-existing short A-tail and the function of A/U-structure remained unclear.
We hypothesized that the 3′ A/U-tail constitutes a key cis-element which enables translation of mitochondrial mRNAs and set out to explore the mechanism of A/U-addition, interactions of the A/U-tailed mRNA with ribosome and the effects of the A/U-tail removal on protein synthesis. Here we demonstrate that a heterodimer of pentatricopeptide repeat-containing (PPR) proteins, termed kinetoplast polyadenylation/uridylation factors (KPAFs) 1 and 2, is required for mRNA adenylation/uridylation in vivo. Furthermore, reconstituted KPAF1–2 heterodimer is sufficient to induce the A/U-heteropolymer synthesis by recombinant KPAP1 and RET1 in vitro. The fully-edited RPS12 mRNA with long A/U-extension, but not with a short A-tail, was enriched in translating mitochondrial ribosomes. KPAF1 RNAi knockdown led to selective loss of long A/U-tailed mRNAs and accumulation of short-tailed transcripts. Finally, concomitant with long tail ablation, we observed inhibition of mitochondrial protein synthesis which was followed by severe growth phenotype.
Hundreds of PPR proteins control virtually every aspect of organellar RNA processing in terrestrial plants (Schmitz-Linneweber and Small, 2008), while trypanosomes possess possibly the largest set of PPRs among the non-plant organisms. Surprisingly, we found that the majority of nearly 40 mitochondrial PPRs and related tetratricopeptide repeat-containing (TPR) proteins associate with ribosomes. Preferential binding of editing substrates (pre-edited mRNAs and gRNAs) and editing complexes (RECC and GRBC) to the large ribosomal subunit and fully-edited A/U-tailed mRNA to the small subunit suggests a functional coupling between mRNA editing, polyadenylation and translation machineries. Collectively, our findings unravel a distinct post-editing 3′ mRNA processing event which generates translation-competent mRNAs in mitochondria of trypanosomes.
RESULTS
Heterodimer of Kinetoplast Polyadenylation Factors 1 and 2 Interacts with KPAP1, Editing and Ribosomal Complexes
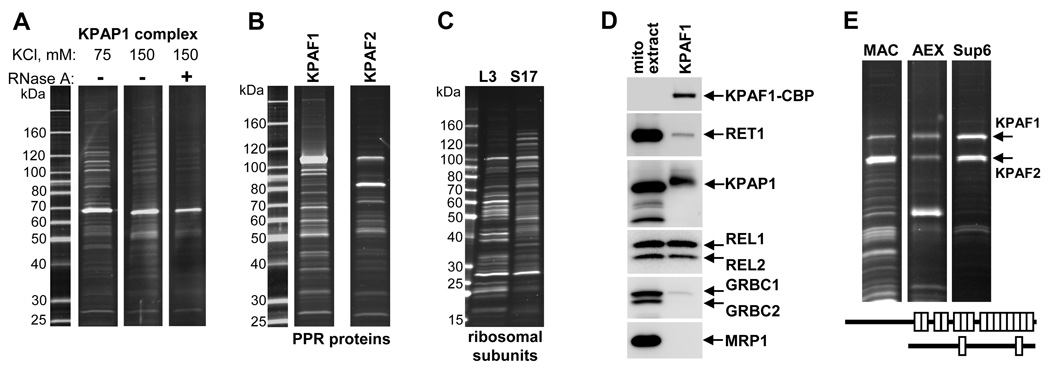
Previous mass spectrometric analyses of KPAP1 complexes from T. brucei (Etheridge et al., 2008) and a related kinetoplastid protozoan L. tarentolae (Weng et al., 2008) revealed partially overlapping sets of proteins including PPR-containing polypeptides, GRBC1 and 2, RGG-box RNA binding proteins, and proteins without discernable functional domains. To obtain a more comprehensive view of the polyadenylation complex and to identify factors required for the mRNA polyadenylation/uridylation in T. brucei, we have purified KPAP1 by tandem affinity chromatography under different conditions (Fig. 1A). LC MS/MS analysis of tryptic digests identified more than 150 proteins in each preparation (Table S1). Three broadly defined groups were apparent among the candidates with possible functions in RNA processing: ∼25 PPR proteins, the components of the mitochondrial RNA binding complex 1 (MRB1) (Panigrahi et al., 2007), which is similar in composition to GRBC (Weng et al., 2008), and ribosomal proteins. To further dissect these datasets, we expressed and affinity-purified two PPR proteins, Tb927.2.3180 and Tb11.01.5980, whose association with KPAP1 was reduced by RNase A treatment (Fig. 1B). To reflect the transient association of Tb927.2.3180 and Tb11.01.5980 PPR proteins with polyadenylation complex, these were renamed kinetoplast polyadenylation/uridylation factors (KPAFs) 1 and 2, respectively. To distinguish polyadenylation complex-associated proteins from ribosomal polypeptides, small subunit (SSU) protein 17 (S17, Tb09.211.2580) and the large subunit (LSU) protein 3 (L3, Tb927.3.5610) have been also purified (Fig. 1C).
Figure 1. Identification of Kinetoplast Polyadenylation Factors 1 and 2.
(A) Tandem affinity purification of KPAP1 complex in the presence of 75 or 150 mM of KCl, and 150 mM KCl plus 0.1 mg/ml of RNase A. Final fractions were separated on 8%–16% SDS gel and stained with Sypro Ruby.
(B) Purification of PPR proteins identified in the polyadenylation complex. Gene identification numbers for KPAF1 and KPAF2 in Ttitryp Database (www.tritrypdb.org) were Tb927.2.3180 and Tb11.01.5980, respectively. Re-sequenced coding regions contained several silent mutations and were deposited to GenBank under accession codes HQ191435 and HQ191436, respectively.
(C) Purification of ribosomal particles via TAP-tagging of large subunit L3 (Tb927.3.5610) and small subunit S17 (Tb09.211.2580) proteins.
(D) Immunoblotting of the affinity-purified KPAF1 fraction with anti-calmodulin binding peptide antibody to detect a tagged protein, KPAF1-CBP. Replica membranes were probed with polyclonal antibodies against RET1, KPAP1, GRBC1–2 and MRP1. KPAP1 undergoes rapid proteolysis in mitochondrial extract (Etheridge et al., 2008) which likely caused its faster gel migration. RNA editing core complex was visualized by self-adenylation of RNA editing ligases REL1 and REL2.
(E) Reconstitution and purification of the KPAF1–2 heterodimer. The N-terminally 6-His-tagged KPAF2 was co-expressed with KPAF1 in bacteria and purified via metal affinity (MAC), anion exchange (AEX) and size exclusion (Sup6) chromatographic steps. Identities of protein bands were confirmed by in-gel digestion and LC-MS/MS analysis. Repeat structures of KPAF1 and 2 are diagrammed under gel panels.
The mass spectrometry data were used to construct a visual representation of the co-complex protein interactions network among KPAP1 and four other sources (Fig. S1). Translation factors were distributed among ribosomal subunits as expected: initiation factor 2 (IF2) (Charriere et al., 2005) was detected exclusively in the small subunit, while elongation factor Tu (EF-Tu) was found in both small and large subunits (Table S1). The discrete sets of PPRs, including two repeat-containing proteins originally identified in the KPAP1 complex (Tb11.02.3180 and Tb927.8.3170, (Etheridge et al., 2008)), were enriched in either small or large ribosomal subunits (Tables S1 and S3). To typify a remarkably high amount of PPR proteins in mitochondrial ribosomes, Tb11.02.3180 was termed KRIPP1 (Kinetoplast Ribosomal PPR-repeat containing protein 1) with other ribosomal PPRs numbered sequentially (Table S2).
Although co-complex interactions indicated their association with ribosomes, KPAF1 and KPAF2 were undetectable by mass spectrometry in affinity-purified ribosomal subunits. Likewise, while both proteins appeared as prominent polyadenylation complex components (Table S1), KPAP1 was not found by MS analysis of either KPAF1 or KPAF2 fractions (Fig. 1B, Table S1). However, immunochemical screening of TAP-fractions shown in Fig. 1B and C revealed KPAP1 co-purification only with KPAF1 (Fig. 1D and data not shown). The detection in the same fraction of trace amounts of RET1 TUTase indicated its weak association with KPAF1. Similarly, the subunits of the gRNA binding complex which was shown to interact with the polyadenylation complex (Etheridge et al., 2008; Weng et al., 2008), were barely detectable in the KPAF1 fraction. Finally, the presence of self- adenylating RNA editing ligases REL1 and REL2 indicated transient interaction of KPAF1 with RECC. The specificity of these contacts was confirmed by the lack of immunoblotting signal for the abundant mitochondrial RNA binding protein MRP1 (Fig. 1D).
Remarkably, both KPAF1 and KPAF2 were highly represented in reciprocal cross-tagging experiments (48 unique KPAF1-derived peptides in KPAF2 fraction and 34 peptides vice versa,Table S1). This observation led us to inquire if these polypeptides interact directly. Indeed, co-expression in bacteria and purification by three chromatographic steps yielded a particle with an apparent molecular mass of ∼200 kDa (determined by gel filtration, not shown) (Fig. 1E). The stoichiometric presence of both proteins suggests a heterodimer as the most probable quaternary structure of the KPAF1–2 unit. Thus, our experiments demonstrate that KPAF1–2 heterodimer is engaged in co-complex interactions with mRNA editing and polyadenylation complexes, and, to a lesser extent, with gRNA processing and translation components.
KPAF1 is Essential for Parasite Viability and Formation of Long Tails
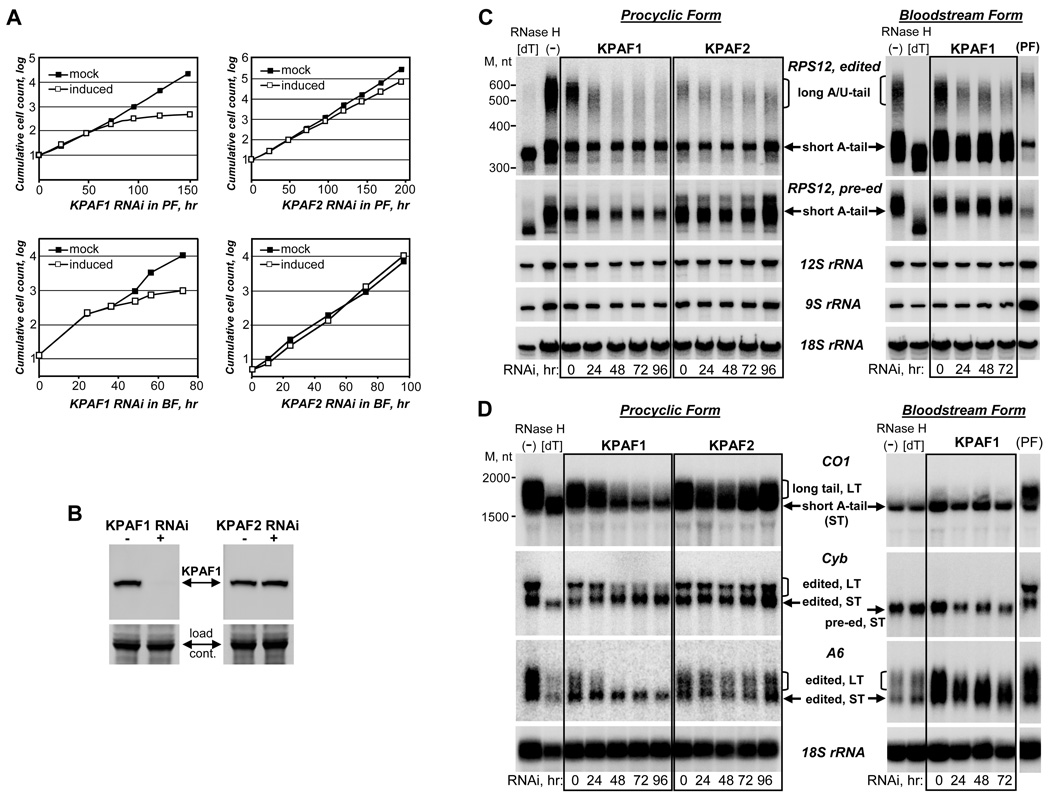
The roles of KPAF1 and KPAF2 in mitochondrial mRNA 3′ processing were examined by RNAi knockdowns in procyclic and bloodstream developmental forms of T. brucei. KPAF1 RNAi induced a severe growth inhibition phenotype indicating an essential function of this protein in both actively-respiring procyclic and oxidative-phosphorylation-deficient bloodstream parasites (Fig. 2A). KPAF2 knockdown in PF caused a lesser growth inhibition while BF remained unaffected. KPAF1 protein depletion and specificity of RNAi were confirmed by immunoblotting in procyclic cells (Fig. 2B). Quantitative RT-PCR was used to verify knockdowns of nuclear-encoded mRNAs and to evaluate changes in relative abundance of mitochondrial transcripts (Fig. S2A). Although RNAi efficiently abrogated KPAF1 and KPAF2 mRNAs, the relative abundance of kDNA-encoded never-edited, pre-edited and edited mRNAs and rRNAs (Fig. S2A) and gRNAs (not shown) remained mostly unaltered.
Figure 2. Effects of KPAF1 and KPAF2 Repression on Cell Viability and 3′ Modification Status of Mitochondrial mRNAs.
(A) Growth kinetics of procyclic (PF) and bloodstream (BF) parasite cultures after mock (closed squares) or RNAi induction (open squares) of KPAF1 and KPAF2.
(B) Immunoblotting of mock (−) or RNAi-induced for 48 hours (+) PF cell lysates with polyclonal antibody against KPAF1. Loading control: Coomassie-stained gel.
(C) High-resolution Northern blot of mitochondrial RNAs. RNAi was induced for indicated periods of time in procyclic (KPAF1 and KPAF2, left panels) and bloodstream (KPAF1, right panels) cell lines. Total RNA was separated on a 5% polyacrylamide/ 8M urea gel, transferred onto membrane and probed for fully-edited and pre-edited RPS12 mRNAs, 9S and 12S mitochondrial rRNAs, and cytosolic 18S rRNA as loading control. [dT], RNA was treated with RNase H in the presence of 18-mer [dT] to remove poly(A) tails; (−), RNase H digestion without DNA oligo. M, RNA length marker in nucleotides.
(D) Northern blot of never-edited cytochrome c oxidase subunit 1 (CO1), moderately-edited cytochrome b (Cyb) and pan-edited ATPase subunit 6 (A6) mRNAs. The same RNA samples as in (C) were resolved on 1.8% agarose/formaldehyde gels. Hybridization probe for Cyb mRNA, which undergoes insertion of 34 Us at the 5′ region, was designed to recognize both pre-edited and edited isoforms. RNA from PF was separated alongside BF samples (right panel).
Because the KPAP1-catalyzed adenylation is required for maintenance of never-edited and edited mRNAs (Etheridge et al., 2008), we hypothesized that the short A-tail represents the necessary and sufficient cis-stability element, and its synthesis does not involve KPAF1 or KPAF2. To test this hypothesis and to investigate whether PPR factors may be instead required for a long A/U-tail formation, we analyzed the 3′-modification status of select mitochondrial mRNAs. A high-resolution Northern blot of pan-edited small ribosomal subunit protein 12 (RPS12) mRNA showed a rapid loss of the long A/U-tail with progression of KPAF1 RNAi in procyclic parasites; the effect of KPAF2 knockdown was less pronounced (Fig, 2C, left panels). Neither individual nor simultaneous KPAF1 and KPAF2 knockdowns affected pre-edited or fully-edited short-A-tailed mRNAs, which confirms a specific role of these factors in the A/U-tail formation (Fig. 2C and S2B). This finding also establishes the KPAP1-catalyzed addition of the short A-tail as necessary and sufficient for stabilizing mitochondrial mRNAs.
The kDNA-encoded ribosomal protein RPS12 is likely incorporated into mitochondrial ribosomes in both developmental stages of T. brucei. The translation product of ATPase subunit 6 mRNA is also expected to be required in procyclic and bloodstream forms, notably in the latter as a component of the ATPase complex essential for maintaining mitochondrial membrane potential (Schnaufer et al., 2005). Conversely, the mitochondrion of the bloodstream form is devoid of cytochrome-containing complexes (Hannaert et al., 2003). Therefore, translation of apocytochrome b (Cyb) and cytochrome c oxidase subunit 1 (CO1) mRNAs should not be critical for bloodstream parasites. We next investigated the KPAF1–2-dependent presence of long A/U-tails in transcripts encoding proteins predicted to be essential in both developmental forms (edited RPS12 and A6), and those possibly dispensable for viability of the bloodstream form (never-edited CO1 and edited Cyb). Edited RPS12 and A6 mRNAs were indeed present in both PF and BF as short- and long-tailed molecules, and the KPAF1 repression led to specific elimination of the latter mRNA population (Fig. 2C and D, right panels). In agreement with the hypothesis that the long tail constitutes a hallmark of the translation-competent mRNA, this structure was not detected in either CO1 or Cyb mRNAs in the bloodstream form.
KPAF1-KPAF2 Complex Induces A/U-tail Synthesis by KPAP1 and RET1 in vitro
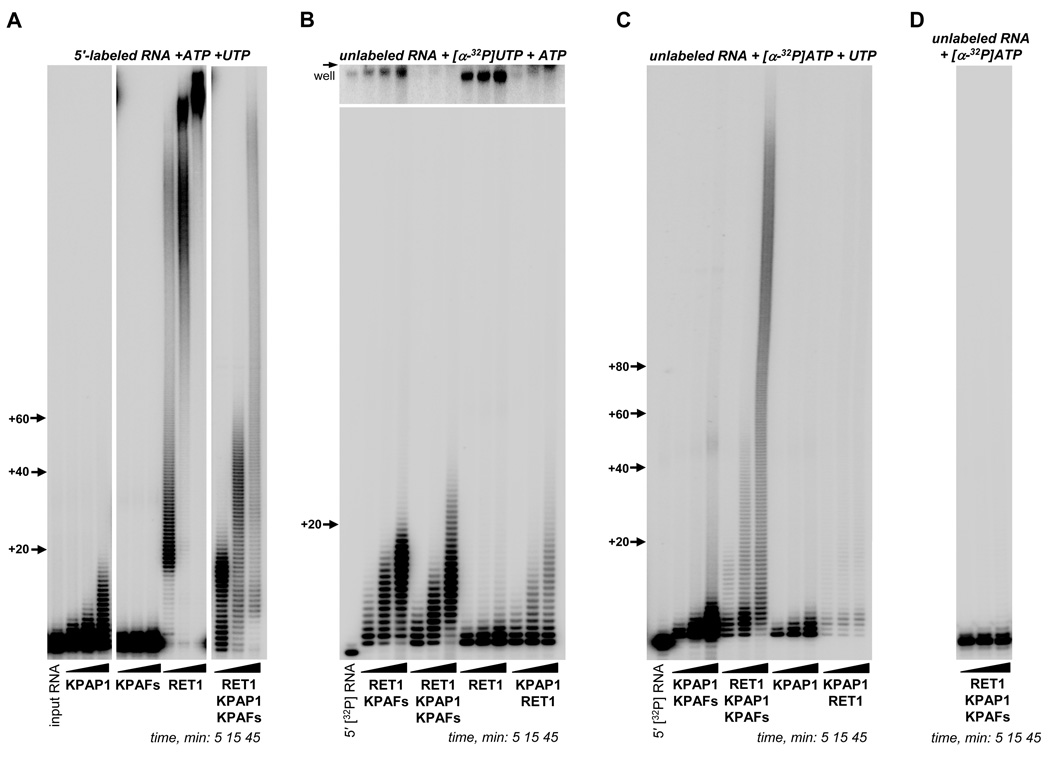
Our previous experiments suggested that KPAP1 poly(A) polymerase and RET1 TUTase are the most probable catalysts of the A/U-tail formation, but this reaction could not be reproduced with the purified KPAP1 complex, or recombinant enzymes. The short A-tail length is consistent with intrinsically limited processivity of the recombinant KPAP1 (Etheridge et al., 2008). In contrast, RET1 adds continuous 15–25 nt U-tails to ribosomal RNAs, guide RNAs, and a few unstable mRNAs (Aphasizheva and Aphasizhev, 2010) while the recombinant enzyme processively polymerizes hundreds of Us (Aphasizhev et al., 2002; Aphasizheva et al., 2004). Sequencing of cloned A/U-tails from PRS12 mRNA demonstrated that stretches of 5–10 As are typically interspersed by 2–3 Us, a pattern which is not characteristic for either nucleotidyl transferase. To investigate whether KPAF1–2 heterodimer can induce A/U-synthesis by the poly(A) polymerase and TUTase, we have established an in vitro system composed of purified recombinant proteins (Figs. 1E and S3A) and synthetic RNA resembling a 3′ fragment of the edited RPS12 mRNA.
Elongation patterns produced by KPAP1 and RET1 in the presence of 5′ radiolabeled mRNA, UTP and ATP (Fig. 3A) were consistent with those reported for generic RNA substrates (Aphasizhev et al., 2002; Etheridge et al., 2008). Omission of UTP from KPAP1- or ATP from RET1-catalyzed reactions did not significantly affect extension patterns (Fig. S3B). In the presence of KPAFs, however, reaction products differed from those produced by either individual enzyme (Fig. 3A). To distinguish the predominantly AMP- from UMP-containing extensions, reconstitution reactions were set up with unlabeled RNA and equal concentrations of ATP and UTP laced with [α32-P]UTP (Fig. 3B). Purified RET1 displayed a distributive pattern of adding 2–3 Us and processive polymerization with products compressed at the top of the gel. In the presence of KPAFs, the UMP incorporation by RET1 was limited to addition of ∼18 nts. Addition of KPAP1 also inhibited processive UTP polymerization by RET1 likely because of competition between the two enzymes for the RNA substrate.
Figure 3. Reconstitution of Adenylation-Uridylation Reaction in vitro.
(A) Purified KPAP1, RET1 or KPAFs were incubated with 5′ labeled RNA 80-mer representing the 3′ region of edited RPS12 mRNA in the presence of ATP and UTP for indicated periods of time. Products were resolved on 8% polyacrylamide/8M urea gel.
(B) Proteins were pre-incubated for 15 min and added to the reaction mixture containing unlabeled 80-mer RNA, [α-32P]UTP and ATP. Reactions and product analysis were carried out as in (A). Upper inset shows portion of the gel adjacent to the loading well. 5′[32P]RNA, the 5′-labeled 80-mer was separated alongside to visualize input RNA without 3′ additions.
(C) Indicated protein combinations were added to the reaction mixture containing unlabeled 80-mer RNA, [α-32P]ATP and UTP.
(D) RET1, KPAP1 and KPAFs were pre-incubated for 15 min and added to the reaction mixture containing unlabeled 80-mer RNA and [α-32P]ATP but lacking UTP.
To test whether synthesis of long adenosine-rich 3′ extensions (in vivo A:U ratio for RPS12 mRNA is ∼7:3 (Etheridge et al., 2008)) can be induced by KPAFs, we next carried out reactions with radioactive ATP and non-labeled UTP (Fig. 3C). Under these conditions, KPAP1 incorporated only a few adenosines (Fig. 3C), but in contrast to RET1, its activity was stimulated ∼5-fold by addition of KPAFs. In agreement with results obtained in the presence of [α32-P]UTP (Fig. 3B), the mixture of two enzymes was the least active combination in polymerizing ATP (Fig. 3C). Remarkably, including KPAFs into the KPAP1-RET1 mixture induced A-rich extensions exceeding 200 nts and similar to those observed with 5′ labeled RNA (Fig. 3A). Such products were no longer generated in the same tripartite (KPAP1, RET1 and KPAFs) reaction without UTP (Fig. 3D). Therefore, the limited UMP incorporation by RET1 is required for the synthesis of A-rich long A/U-tails. To confirm that the A/U-tails indeed contain UMP residues, we have digested reconstruction reaction products with RNase A (Fig. S3C). Collectively, these data define KPAP1 poly(A) polymerase, RET1 TUTase and KPAF1–2 heterodimer as factors necessary and sufficient for the A/U-tail addition in vitro.
A/U-tailed RPS12 mRNA is Enriched in Large Ribosomal Complexes
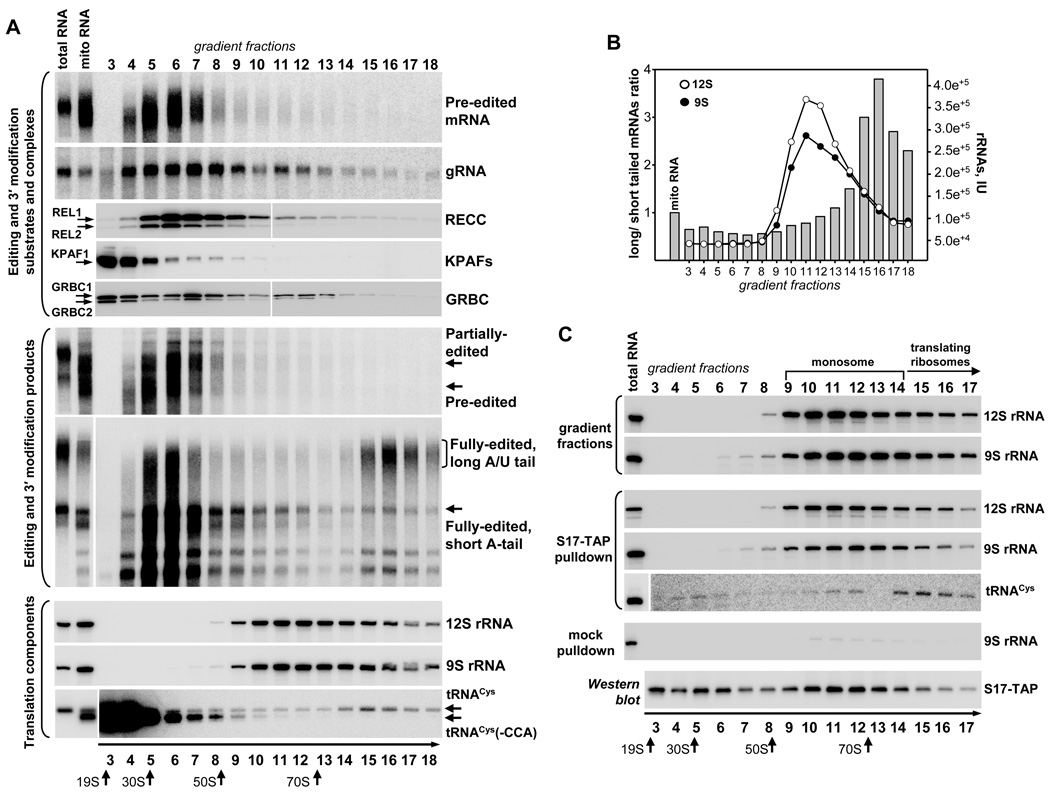
A/U-tails have been detected in fully-edited mRNAs while partially-edited transcripts, even those with just few unedited sites at the 5′ end, possessed short A-tails (Etheridge et al., 2008). This observation suggested a functional coupling between completion of mRNA editing at the 5′ end and KPAF1–2-induced A/U-addition at the 3′ end. Although expendable in terms of mRNA stability, the A/U-tailing is developmentally regulated to match the requirements for respiratory complex components in procyclic and bloodstream parasites (Fig. 2C, D). Furthermore, mass spectrometric and immunochemical analyses demonstrated that KPAP1 and KPAF1–2 are engaged in transient interactions with ribosomal subunits and RNA editing complexes (Figs.1 and S1). These findings position the A/U-tail formation at the interface of RNA editing and translation and indicate a critical role of this structure in mitochondrial protein synthesis. To better understand functional coupling between mRNA editing, 3′ modification and translation, we analyzed the distribution of respective complexes and their substrates and products during gradient fractionation of the mitochondrial extract (Fig. 4A). The pre-edited RPS12 mRNA co-sedimented with major gRNA, RECC and GRBC peaks in the 25S-50S region while minor fractions of editing components were detected in the >60S sector. Partially-edited mRNAs were confined to 25–50S fractions. In contrast, fully-edited mRNAs were clearly segregated: a size-heterogeneous fraction sedimented in 25S–50S region, while a predominantly long-tailed form was enriched in the >70S region. The ratio between long- and short-tailed mRNAs in gradient fraction 16 was approximately 4-fold higher as compared to input mitochondrial RNA (Fig. 4B).
Figure 4. Sedimentation Analysis of mRNA Editing, 3′ modification and Translation Substrates and Complexes.
(A) Glycerol gradient fractionation of the mitochondrial lysate from procyclic parasites. Detergent-extracted mitochondrial fraction was separated on 10%–30% glycerol gradient. RNA was extracted from each fraction and separated on 5% polyacrylamide/8M urea gels for Northern blotting of mRNAs and rRNAs, or 10% gels for hybridization with gRNA RPS12[64] and tRNACys-specific probes. RECC was detected by self-adenylation of RNA ligases. KPAFs and GRBC were visualized with antibodies against KPAF1 and GRBC1–2, respectively. Total RNA, RNA isolated from freshly-collected cells; mito RNA, RNA isolated from mitochondrial extract. Thyroglobulin (19S), bacterial ribosomal subunits and ribosomes were separated in parallel experiments. Arrows indicate peaks of optical density for S value markers.
(B) The signal intensity for long-tailed mRNA band was divided by value corresponding to the short tailed mRNA migration region in each fraction and plotted as histogram. Sedimentation of ribosomal RNAs is represented by overlaying the histogram with arbitrary intensity units (IU) graphs for 9S and 12S rRNAs.
(C) Mitochondrial extract was purified from a cell line overexpressing TAP-tagged S17 protein and fractioned as in (A). Affinity pulldowns were carried out with ¾ of each fraction with IgG-Sepharose (rRNA detection) or IgG-coated magnetic beads (tRNA detection). RNA was extracted from beads and remaining parts of gradient fractions, and separated alongside total cell RNA and probed for indicated RNA species. A background co-precipitation of rRNA constituted less than 5%, as determined by pulldowns in gradient fractions from the parental cell line.
The peak of long tailed mRNA co-sedimented with a heavier fraction of ribosomal RNA-containing particles while the balk of rRNA was detected in the 60S-70S region. To distinguish translating ribosomes among various particles present within the rRNA sedimentation spectrum, we next analyzed the distribution of tRNAs. In trypanosomes, the entire tRNA set is imported from the cytoplasm, often followed by additional modifications and CCA-end repair (Alfonzo and Soll, 2009). Northern blotting showed that approximately 80% of tRNACys molecules lost their CCA-ends during mitochondrial lysate preparation, as determined by comparison of total RNA isolated from freshly-collected cells and RNA isolated from gradient input material (Fig. 4A, bottom panel). Fractionation of the mitochondrial extract, however, led to a specific enrichment of the full-length tRNACys, together with long-tailed mRNAs, in fractions 15–18.
The co-sedimentation of A/U-tailed mRNA, full-length tRNA and minor fraction of rRNAs in the >70S region led us to investigate whether in mitochondrial extract small and large subunits exist predominantly as assembled monosomes or as dissociated particles. Mitochondrial extract obtained from cells expressing TAP-tagged small subunit protein S17 was fractionated in a glycerol gradient followed by affinity pull-down with IgG-coated beads in each fraction. RNA was eluted from washed beads and also directly extracted from gradient fractions, and analyzed by Northern blotting. A mock pull-down was carried out with gradient fractions obtained from the parental 29–13 cell line (Fig. 4C). The virtually identical distribution of 9S and 12S rRNAs in gradient fractions and S17-TAP pull-downs demonstrates that in the mitochondrial extract, the greater part of small and large ribosomal subunits associate into a 60S–70S complex. The rRNA content in gradient fractions also correlated well with proteomic overlap between TAP-purified large and small subunits (Fig. S1,Table S1). The tRNACys, however, co-precipitated with S17-TAP predominantly in fractions 15–17 (>70S complex) closely resembling sedimentation patterns of long-tailed mRNA (Fig. 4A). In conclusion, our data demonstrate that ribosomal particles sedimenting at > 70S may be associated with fully-edited long-tailed mRNA and tRNAs, thus representing translating ribosomes.
Mitochondrial Ribosome Selectively Binds the A/U-tailed Edited mRNA
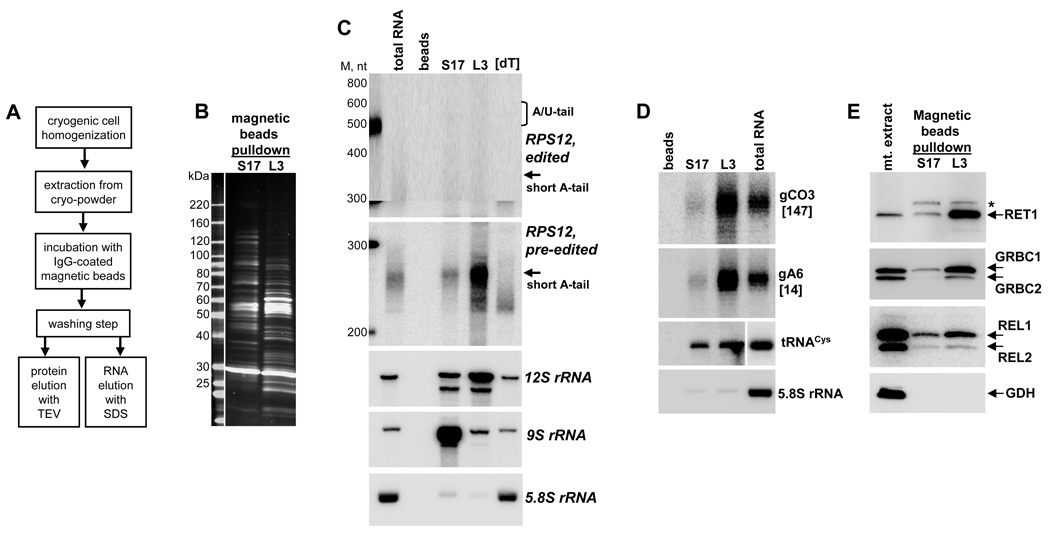
The sedimentation analysis showed that mRNA is not required for ribosomal subunits association into the 60–70S complex. Conversely, co-sedimentation of the A/U-tailed fully-edited RPS12 mRNA with tRNA and ribosomal particles as high-molecular weight complexes exceeding 70S provided a compelling argument that the long A/U-tail enhances mRNA affinity for the ribosome. However, our repeated attempts to detect mRNAs in tandem affinity purified S17 and L3 fractions (Fig. 1C), or pulldown material obtained from gradient fractions (Fig. 4C), were unsuccessful. It seems likely that the mRNA-ribosome interaction does not withstand mitochondrial isolation and TAP procedures. To directly test whether the ribosome preferentially binds A/U-tailed mRNA versus short-tailed mRNA, we developed a rapid purification method (Fig. 5A). This technique takes advantage of protein A binding to non-porous IgG-coated magnetic beads (Oeffinger et al., 2007), and does not require mitochondrial isolation. Mass spectrometric analysis of the rapidly-purified SSU and LSU fractions (Fig. 5B) showed a substantial proteomic overlap between the subunits, essentially mirroring the results obtained by a conventional TAP procedure (Table S3). Lower relative amounts of the 12S RNA in the S17 fraction and the 9S RNA in the L3 fraction indicate that in addition to ribosomes these fractions also contain individual subunits. Accordingly, purification in presence of puromycin resulted in a noticeable reduction of ribosomes present in each preparation (Fig. S4).
Figure 5. Differential Binding of Editing Substrates and Products to Ribosomal Subunits.
(A) A schematic diagram of the rapid affinity isolation strategy. TEV, Tobacco Etch Virus protease.
(B) Purified ribosomes were separated on 8%–16% SDS PAGE and stained with Sypro Ruby.
(C) Northern blot of rRNAs and ribosome-bound mRNAs. RNA was extracted from magnetic beads and separated on a 5% polyacrylamide/8M urea gel. The membrane was probed for fully-edited and pre-edited RPS12 mRNAs, 12S and 9S mitochondrial rRNAs, and cytosolic 5.8S rRNA as contamination control. [dT], total RNA was treated with RNase H in the presence of 18-mer [dT] to remove poly(A) tails. Beads, IgG-coated magnetic beads were incubated with extract from the parental cell line. M, RNA length marker in nucleotides.
(D) Guide RNAs required for editing of cytochrome c oxidase subunit 3 and ATPase subunit 6 mRNAs and tRNACys were detected by Northern blotting. The same RNA samples as in (C) were separated on 10% polyacrylamide/8M urea gel.
(E) Immunoblotting of affinity-purified ribosomal fractions with polyclonal antibodies against RET1 and GRBC1–2. RNA editing core complex was visualized by self-adenylation of RNA editing ligases REL1 and REL2. GDH, mitochondrial glutamate dehydrogenase. Asterisk indicates a cross-reacting band.
Although the ribosomal protein and ribosomal RNA contents were similar between fractions obtained by conventional two-step and rapid affinity procedures, mRNA was detectable only in the latter (Fig. 5C). In comparison to steady-state cellular levels, the long-tailed edited RPS12 mRNA was enriched by ∼5-fold in the S17 fraction, which is similar to enrichment observed in >70S complexes (Fig. 4A,B). Lesser quantities of edited RPS12 mRNA in the L3 fraction suggests a higher A/U-tailed mRNA affinity for the small subunit. However, the large subunit may contribute to A/U-tailed mRNA selection or stabilization of the bound mRNA: partial dissociation of the ribosomes during purification in the presence of puromycin resulted in only 2-fold enrichment of long-tailed mRNA (Fig. S4). In striking contrast with the edited mRNA, RNA substrates of editing reactions (pre-edited mRNA and gRNA), gRNA processing (RET1) and RNA editing (RECC and GRBC) complexes preferentially associate with the large ribosomal subunit (Fig. 5 C–E). The second peak of gRNA and GRBC sedimenting at ∼60–70S (fractions 11–12, Fig. 4A) provides additional evidence for the higher-order association of mRNA editing substrates and complexes with ribosomes. Finally, tRNACys was detected in both S17 and L3 fractions. Collectively, these results provide evidence for selective binding of the A/U-tailed edited mRNA to mitochondrial ribosome via contact with small subunit and demonstrate physical interactions of mRNA editing and translation machineries.
Loss of A/U-tails Inhibits Mitochondrial Translation
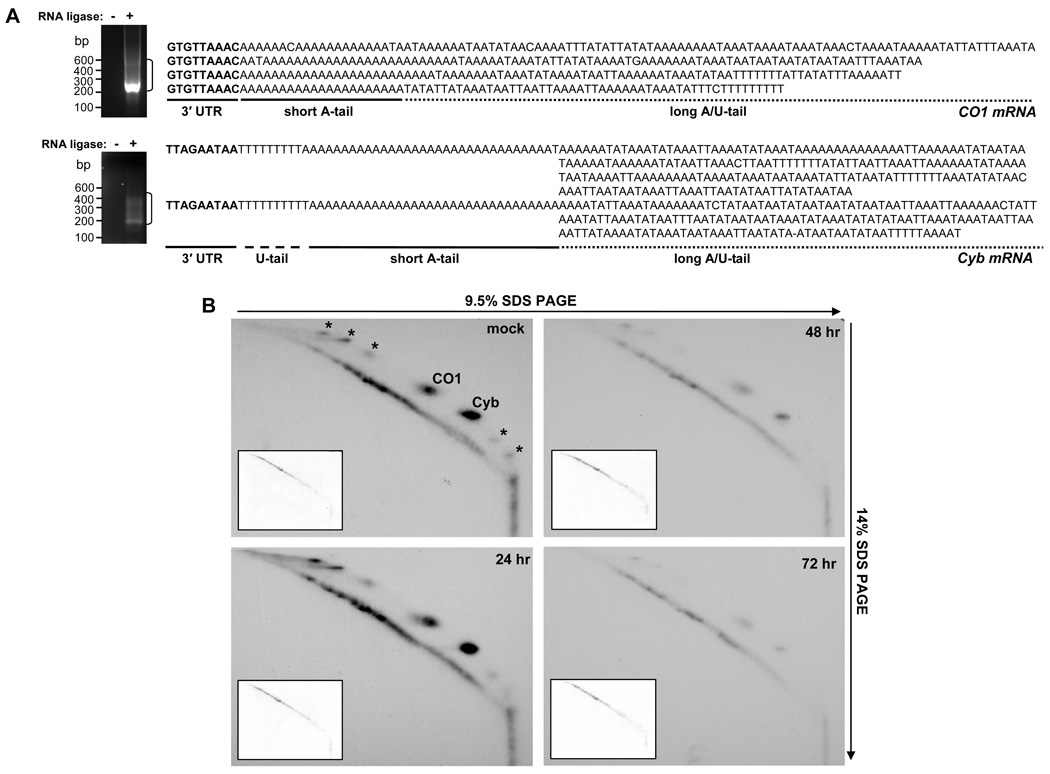
Most predicted products of mitochondrial translation are likely to be extremely hydrophobic, and therefore prone to aggregation and abnormal electrophoretic mobility. Hence, the existing techniques (Horváth et al., 2000; Nebohacova et al., 2004) are limited to detecting apocytochrome b and cytochrome c oxidase subunit I proteins, which are encoded by edited and never-edited mRNAs, respectively. Although in the procyclic form both mRNAs possess long tails, the primary structure of these extensions remained unknown. To determine whether bimodal structures (short A-tail plus A/U-extension) exist in Cyb and CO1 mRNA, we cloned and sequenced 3′ extensions from these transcripts. In CO1 mRNA, the majority of clones contained only short A-tails while in approximately 15% of cloned mRNAs the short tails were extended into A/U-heteropolymers (Fig. 6A). In fully-edited Cyb mRNA, more that 50% of clones contained A/U-tails with some exceeding 300 nt. Surprisingly, a non-encoded stretch of Us separating the 3′ UTR from the short A-tail was found in some clones, which resembles RET1-catalyzed 3′ uridylation of never-edited ND1 mRNA (Aphasizheva and Aphasizhev, 2010).
Figure 6. KPAF1 Repression Inhibits Mitochondrial Translation.
(A) Cloning and sequencing of A/U-tails in CO1 and Cyb mRNAs. Total RNA was circularized with T4 RNA ligase (+), or incubated without ligase (−), and subjected to a single-tube RT-PCR amplification with outward-directed primers specific for 5′ and 3′ mRNA termini. PCR products were separated on 1.4% agarose gel and DNA was extracted from regions indicated by brackets, cloned and sequenced. Representative A/U-tails are shown in 5′-3′ polarity.
(B) Analysis of translation products in KPAF1 RNAi-induced parasites. RNAi was induced for indicated periods of time and cells were labeled with EasyTag labeling mix (PerkinElmer Life Sciences) in isotonic buffer supplemented with 0.1 mg/ml of cycloheximide. Cells were collected by centrifugation, dissolved in SDS gel loading buffer and fractionated by two-dimensional electrophoresis. Gels were stained with Coomassie Brilliant Blue R250 (inset panels) and exposed to X-ray film (large panels). Based on previous protein identifications (Nebohacova et al., 2004), major spots indicated by arrows represent CO1 and Cyb subunits. Cycloheximide-resistant translation produced several additional spots of unknown identity (marked by asterisks) which were also abolished by KPAF1 RNAi.
We next tested a temporal correlation between the decline of A/U-tails in CO1 and Cyb mRNAs (Fig. 2D) and inhibition of de novo protein synthesis. KPAF1 RNA interference was induced in procyclic parasites, cells were collected at 24 h intervals and cytosolic translation was blocked with cycloheximide. Mitochondrial proteins were labeled by incubating cells with [35S]-amino acids, extracted and separated by two-dimensional SDS PAGE. In this system, some hydrophobic inner membrane proteins migrate off the diagonal in the second-dimension (Fig. 6B). In agreement with steady levels of A/U-tailed mRNAs, the intensity of radioactively-labeled spots normalized to the total protein loading remained virtually unchanged for the first 24 h of RNAi induction. Continued RNAi expression led to a concomitant loss of approximately 80% of long-tailed mRNAs and mitochondrial translation products by 48 h, and continued to decline up to 72 h (Fig. 6B). The inhibition of mitochondrial translation in KPAF1 RNAi preceded the appearance of a cell grown inhibition phenotype (Fig. 2A) by at lest 24–36 h. It is noted that we have not been able to detect CO1 and Cyb proteins in bloodstream parasites (Fig. S5). This experiment indicates an inability of CO1 mRNA, which is prominently-present but lacks long tails in bloodstream parasites (Fig. 2D), to support the synthesis of the encoded polypeptide. To conclude, KPAF1 repression inhibits A/U-tail synthesis by KPAP1 and RET1 thereby impeding mitochondrial translation. These findings establish mRNA 3′ adenylation/ uridylation as a final step in biogenesis of translation-competent mRNAs in trypanosomal mitochondria.
DISCUSSION
Our findings reveal the existence of a discrete post-editing processing event that enables translation of mitochondrial mRNAs in Trypanosoma brucei. We report that 200–300 nt long 3′ extensions are characteristic of mRNAs expected to be translated in bloodstream and procyclic developmental stages. In both edited and never-edited mRNAs, these 3′ extensions consist of a short A-tail continued into a long A/U-heteropolymer. The bimodal organization reflects distinct mechanisms of synthesis and functions for these two constituents. The short A-tail is added by KPAP1 prior to extensive editing; dispensable for the pre-edited mRNA maintenance, this stretch of 20–40 A’s becomes a required and sufficient cis-stability element once editing is initiated. In edited mRNAs, additions of short A-tails and A/U-heteropolymers are temporally separated: the latter event is triggered only after completion of the editing process. In never-edited mRNAs, these reactions are likely to occur sequentially. The mitochondrial poly(A) polymerase KPAP1 and RET1 TUTase have been previously implicated in A/U-tail addition (Etheridge et al., 2008; Aphasizheva and Aphasizhev, 2010). However, a clear disengagement of the A/U-tail synthesis from other 3′ modifications carried out by KPAP1 and RET1 was unattainable, thus impeding functional and mechanistic understanding of the A/U-structure.
We have identified a heterodimer of two pentatricopeptide-repeat proteins, KPAF1 and 2, as essential for mRNA 3′ adenylation/uridylation in vivo. Furthermore, the A/U-synthesis was reconstituted in vitro with KPAF1–2, KPAP1 and RET1. Identification of PPR factors involved specifically in the A/U-tail formation distinguished KPAP1-catalyzed addition of short A-tail from contributing adenosines into A/U-tails. After extensive investigation, we concluded that RNA classes targeted by RET1 (Aphasizheva and Aphasizhev, 2010), other then A/U-tailed mRNAs, are not affected by KPAF1–2 knockdowns. Therefore, KPAF1 and KPAF2 represent mRNA-specific 3′ adenylation/uridylation factors. In the steady-state only about 50% of fully-edited mRNAs and never-edited mRNAs are maintained as long-tailed forms; this observation implicates the 3′ A/U-addition as the rate limiting step of mRNA processing. Indeed, the PPR-induced reaction regulating availability of translation-competent mRNAs may be the key regulatory step of mitochondrial gene expression.
Pentatricopeptide-repeat containing RNA binding proteins are defined by the presence of 35-amino acid tandem repeats (Small and Peeters, 2000). In terrestrial plants, hundreds of non-redundant PPR factors control various aspects of organellar RNA processing including editing, splicing, processing and stability (Schmitz-Linneweber and Small, 2008). In T. brucei, 28 PPR proteins, including KPAF1 and KPAF2, were predicted by data mining (Pusnik et al., 2007) and 16 were detected experimentally in purified mitochondrial ribosomes (Zikova et al., 2008). Our proteomic and motif analyses of mitochondrial ribosomal and polyadenylation complexes expanded this family to 36 members, including 21 ribosomal PPRs and three proteins with related tetratricopeptide repeat (TPR) motifs (Table S2). At least some ribosome-associated PPRs (Tables S2 and S3) are essential for rRNA biogenesis or stability, as evidenced by the loss of either 9S or 12S rRNAs in respective RNAi knockdowns (Pusnik et al., 2007).
The mechanism by which mitochondrial ribosomes select fully-edited mRNAs while pre-edited mRNAs are prevented from translation remained puzzling since the discovery of RNA editing in CO2 mRNA (Benne et al., 1986). In this particular instance, the pre-edited and edited mRNAs differ by only four uridines which are inserted approximately 500 nt from the 5′ end to correct the frame shift. Therefore, the unbiased recognition of the 5′ UTR and/or initiation codon would inevitably produce truncated protein potentially acting as dominant negative mutant. Our findings indicate that the 3′ A/U-tail represents a key feature enabling translation via increasing affinity of edited mRNA for the small ribosomal subunit. As shown by cryo-electron microscopy, the overall architecture of the mitochondrial ribosome from L. tarentolae (Lmr) is similar to eubacterial ribosome. Significant portions of Lmr’s highly-porous structure are composed of Lmr-specific (i.e., not present in bacterial ribosomes) proteins to compensate for much smaller rRNAs (Sharma et al., 2009). Remarkably, the surfaces of the mRNA binding channel around the small subunit’s head, as well as the entrance and the exit, are lined up with Lmr-specific proteins. To this end, the co-complex interactions point to KRIPP1, 2, 5 and 8 (Table S2, File S1) as probable mediators between the polyadenylation complex and SSU. Conversely, KPAF1–2 interactions with the polyadenylation complex and RET1 TUTase appear to be transient and likely RNA-mediated (Tables S1–S3).
Ribosomal RNAs, pre-edited and edited mRNAs have been detected by end-point RT-PCR in the purified L. tarentolae RNA editing core complex (Osato et al., 2009). A combination of rapid purification and quantitative techniques described here extended these results revealing the association of RNA editing substrates (pre-edited mRNA and gRNA) and editing core and gRNA binding complexes with the large ribosomal subunit. These results suggest that editing and translation machineries are physically associated, but translation initiation requires an interface event: the mRNA 3′ A/U-tailing reaction. Overall, our findings bridged the gap between the mRNA editing and translation leading to a revised model of mitochondrial gene expression (Fig. 7). A hypothesis that some ribosome-associated PPR proteins function as transcript- or sequence-specific factors enabling adenylation/uridylation of certain mRNAs is among possible outcomes of this model awaiting experimental verification.
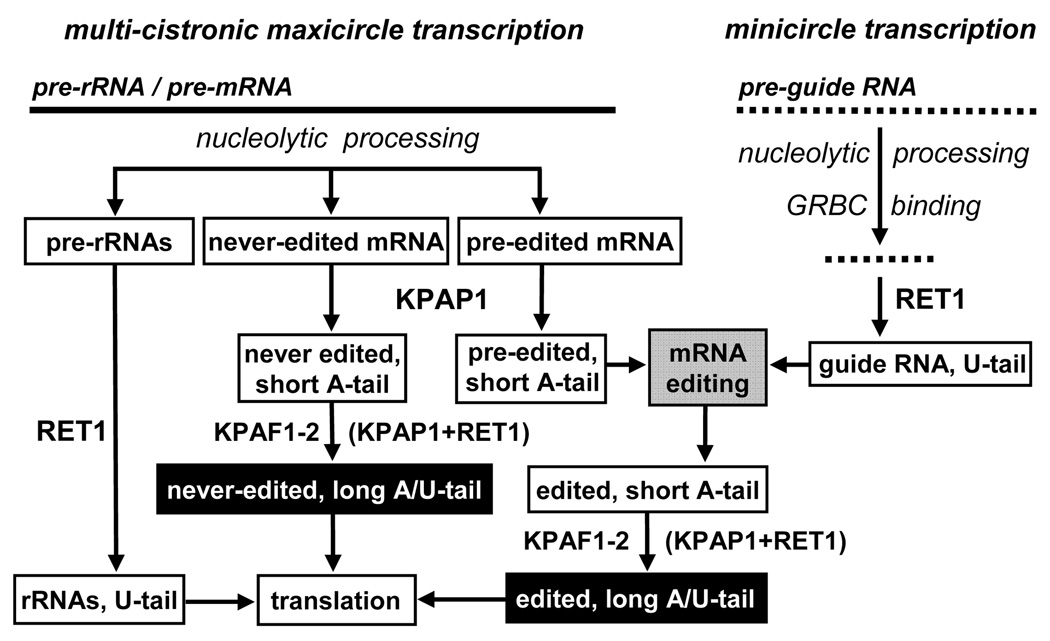
Figure 7. A Revised Model for mRNA Processing and Translation in Trypanosome Mitochondria.
Polycistronic transcripts are produced by the mitochondrial RNA polymerase from maxicircles and minicircles. Pre-gRNAs undergo 3′ nucleolytic processing by a cryptic nuclease via a pathway requiring RET1 activity (Aphasizheva and Aphasizhev, 2010). Processed gRNAs are stabilized by binding to the gRNA binding complex (GRBC, (Weng et al., 2008; Aphasizheva and Aphasizhev, 2010)) and 3′ uridylated by RET1 TUTase (Aphasizhev et al., 2002; Aphasizhev et al., 2003b). Pre-rRNAs are also trimmed at the 3′ end and uridylated by RET1 (Aphasizheva and Aphasizhev, 2010). Guide RNA hybridization with pre-edited mRNA activates the editing process. Editing events in the 3′ region confer a requirement for the short A-tail as cis-element necessary (Etheridge et al., 2008) and sufficient (this study) for mRNA stability. The completion of editing, typically at the 5′ region, triggers extension of the preexisting A-tail into A/U-heteropolymer by the KPAF1–2-coordinated, KPAP1/ RET1-catalyzed reaction. This final processing event facilitates mRNA binding to the small ribosomal subunit leading to translation initiation.
EXPERIMENTAL PROCEDURES
Trypanosome Culture, RNAi and Protein Expression
The RNAi plasmids were generated by cloning ∼500-bp gene fragments into p2T7–177 vector for tetracycline-inducible expression (Wickstead et al., 2002). DNA manipulations are described in Supplemental Information. The constructs were transfected into procyclic 29–13 or bloodstream “single marker” T. brucei strains (Wirtz et al., 1999). RNAi was performed as described (Weng et al., 2008). For inducible protein expression in PF T. brucei, full-length genes were cloned into pLEW-MHTAP vector (Jensen et al., 2007).
Mitochondrial Isolation, Glycerol Gradients, Affinity Purification and Western Blotting
Mitochondrial fraction was isolated as described except omitting the Percoll density gradient (Pelletier et al., 2007). Fresh mitochondrial pellets were mixed with three parts (wet weight) of buffer containing 25 mM HEPES (pH 7.6), 125 mM KCl, 12 mM MgCl2 and 1.2 % Nonidet P-40 (NP-40). After 20 minutes incubation on ice, the extract was clarified at 18,000 g for 10 min and fractionated on 10%–30% glycerol gradient in SW41 rotor for 4 hours at 178,000 g and 580 µl fractions were collected from the top. Self-adenylation of RNA ligases was carried out with 1 µCi of [α-32P]ATP per 10 µl of gradient fraction. Western blotting was performed with rabbit antigen-purified polyclonal antibodies or with anti-CBP antibodies (GenScript) to detect tagged proteins. Affinity pulldowns were performed with 10 µl of beads (GE Healthcare) or 2 mg of IgG-coated magnetic beads in the presence of 1 mg/ml of albumin and 0.2% of NP-40. Beads were washed three times for 10 min in the extraction buffer with 0.2 % NP-40. RNA was extracted from beads with phenol/chloroform and precipitated. Quantitative chemiluminescent images were acquired with LAS-4000 digital analyzer (Fuji). The conventional TAP procedure was performed as described (Aphasizhev and Aphasizheva, 2007). For the rapid pulldown, rabbit IgG was coupled to Dynabeads M-270 Epoxy (Invitrogen) according to (Oeffinger et al., 2007) and used in total cell extract. See Supplemental Information for detailed protocols.
RNA Analysis and circular RT-PCR
Total RNA was isolated using modified procedure (Chomczynski and Sacchi, 1987). Methods for qRT-PCR and Northern blotting are provided in the Supplemental Information. The change in relative abundance was calculated assuming the ratio between analyzed transcripts and loading control in mock-induced cells as 100%. Membranes and gels were exposed to phosphor storage screens and volume quantitation was performed with Quantity One software (BioRad). For cRT-PCR, 5 µg of total RNA (qRT-PCR-grade) was circularized with 25U of T4 RNA ligase in 25 µl at 37°C for 45 min in manufacturer-supplied buffer (NEB). Approximately 2.5 µg of purified circularized RNA was used for cDNA synthesis and amplification with SuperScrip III One-Step RT-PCR system (Invitrogen). PCR products were gel-purified, cloned and at least 96 clones sequenced.
KPAF1–2 Complex Purification and the A/U-synthesis Reaction
KPAF1 and 2 genes were cloned into pDUET-1 vector in which KPAF2 was expressed as N-terminally 6His-tagged protein. Complex was purified via sequential Talon metal affinity (BD Biosciences), Sepharose HQ anion exchange and Superose 6 size exclusion (GE Healthcare) columns per manufactures’ instructions. For reconstitution reactions, the RPS12 mRNA fragment (5′- ggguggugguuuuguugauuuacccgguguaaaguauuauacacguauuguaaguuagauuuagauauaagauatguuuu-3′) was purified on 10% polyacrylamide/ 8M urea gel. For control reactions, the same RNA was labeled with T4 polynucleotide kinase in the presence of [γ-32P]ATP. Reconstitutions were set up with 10 nM of RET1, 100 nM of KPAP1, ∼100 nM of KPAFs, and 100 nM of RNA substrate in 20 µl reaction mixture containing 20 mM HEPES (pH 7.6), 25 mM KCl, 10 mM MgCl2, 1 mM DTT, 0.05% of NP-40 and 50 µM of each nucleotide triphosphate. Radioactive NTP mixtures were prepared by combining 50 µM of unlabelled and 0.1 µM of [α-32P]-NTP (6000Ci/mmol, PerkinElmer).
Mitochondrial Translation
Cell labeling and gel separations were performed as described (Horváth et al., 2000; Nebohacova et al., 2004) with modifications specified in the Supplemental Information.
Supplementary Material
AKNOLEDGMENTS
We thank members of our laboratories and Juan Alfonzo for helpful discussions. We also thank Bert Semler and Yongsheng Shi for reading the manuscript. We appreciate the contributions of materials and data by Drew Etheridge and Greg Bonello. We thank George Cross for kind gifts of trypanosomal strains and plasmids. This work was supported by NIH grants AI091914 to RA, GM74830 to LH and AI088292 to DM.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Alfonzo JD, Soll D. Mitochondrial tRNA import--the challenge to understand has just begun. Biol. Chem. 2009;390:717–722. doi: 10.1515/BC.2009.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aphasizhev R, Aphasizheva I. RNA Editing Uridylyltransferases of Trypanosomatids. Methods Enzymol. 2007;424:51–67. doi: 10.1016/S0076-6879(07)24003-0. [DOI] [PubMed] [Google Scholar]
- Aphasizhev R, Aphasizheva I, Nelson RE, Gao G, Simpson AM, Kang X, Falick AM, Sbicego S, Simpson L. Isolation of a U-insertion/deletion editing complex from Leishmania tarentolae mitochondria. EMBO J. 2003a;22:913–924. doi: 10.1093/emboj/cdg083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aphasizhev R, Aphasizheva I, Simpson L. A tale of two TUTases. Proc. Natl. Acad. Sci. U. S. A. 2003b;100:10617–10622. doi: 10.1073/pnas.1833120100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aphasizhev R, Sbicego S, Peris M, Jang SH, Aphasizheva I, Simpson AM, Rivlin A, Simpson L. Trypanosome Mitochondrial 3' Terminal Uridylyl Transferase (TUTase): The Key Enzyme in U-insertion/deletion RNA Editing. Cell. 2002;108:637–648. doi: 10.1016/s0092-8674(02)00647-5. [DOI] [PubMed] [Google Scholar]
- Aphasizheva I, Aphasizhev R. RET1-catalyzed Uridylylation Shapes the Mitochondrial Transcriptome in Trypanosoma brucei. Mol. Cell. Biol. 2010;30:1555–1567. doi: 10.1128/MCB.01281-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aphasizheva I, Aphasizhev R, Simpson L. RNA-editing terminal uridylyl transferase 1: identification of functional domains by mutational analysis. J. Biol. Chem. 2004;279:24123–24130. doi: 10.1074/jbc.M401234200. [DOI] [PubMed] [Google Scholar]
- Benne R, Van den Burg J, Brakenhoff J, Sloof P, Van Boom J, Tromp M. Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell. 1986;46:819–826. doi: 10.1016/0092-8674(86)90063-2. [DOI] [PubMed] [Google Scholar]
- Bhat GJ, Koslowsky DJ, Feagin JE, Smiley BL, Stuart K. An extensively edited mitochondrial transcript in kinetoplastids encodes a protein homologous to ATPase subunit 6. Cell. 1990;61:885–894. doi: 10.1016/0092-8674(90)90199-o. [DOI] [PubMed] [Google Scholar]
- Blum B, Bakalara N, Simpson L. A model for RNA editing in kinetoplastid mitochondria: "Guide" RNA molecules transcribed from maxicircle DNA provide the edited information. Cell. 1990;60:189–198. doi: 10.1016/0092-8674(90)90735-w. [DOI] [PubMed] [Google Scholar]
- Brown SV, Hosking P, Li J, Williams N. ATP synthase is responsible for maintaining mitochondrial membrane potential in bloodstream form Trypanosoma brucei. Eukaryot. Cell. 2006;5:45–53. doi: 10.1128/EC.5.1.45-53.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charriere F, Tan TH, Schneider A. Mitochondrial initiation factor 2 of Trypanosoma brucei binds imported formylated elongator-type tRNA(Met) J. Biol. Chem. 2005;280:15659–15665. doi: 10.1074/jbc.M411581200. [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Etheridge RD, Aphasizheva I, Gershon PD, Aphasizhev R. 3' adenylation determines mRNA abundance and monitors completion of RNA editing in T. brucei mitochondria. EMBO J. 2008;27:1596–1608. doi: 10.1038/emboj.2008.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannaert V, Bringaud F, Opperdoes FR, Michels PA. Evolution of energy metabolism and its compartmentation in Kinetoplastida. Kinetoplastid. Biol. Dis. 2003;2:11. doi: 10.1186/1475-9292-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horváth A, Berry EA, Maslov DA. Translation of the edited mRNA for cytochrome b in trypanosome mitochondria. Science. 2000;287:1639–1640. doi: 10.1126/science.287.5458.1639. [DOI] [PubMed] [Google Scholar]
- Jensen BC, Kifer CT, Brekken DL, Randall AC, Wang Q, Drees BL, Parsons M. Characterization of protein kinase CK2 from Trypanosoma brucei. Mol Biochem Parasitol. 2007;151:28–40. doi: 10.1016/j.molbiopara.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao CY, Read LK. Opposing effects of polyadenylation on the stability of edited and unedited mitochondrial RNAs in Trypanosoma brucei. Mol. Cell Biol. 2005;25:1634–1644. doi: 10.1128/MCB.25.5.1634-1644.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebohacova M, Maslov DA, Falick AM, Simpson L. The effect of RNA interference Down-regulation of RNA editing 3'-terminal uridylyl transferase (TUTase) 1 on mitochondrial de novo protein synthesis and stability of respiratory complexes in Trypanosoma brucei. J. Biol. Chem. 2004;279:7819–7825. doi: 10.1074/jbc.M311360200. [DOI] [PubMed] [Google Scholar]
- Oeffinger M, Wei KE, Rogers R, Degrasse JA, Chait BT, Aitchison JD, Rout MP. Comprehensive analysis of diverse ribonucleoprotein complexes. Nat. Methods. 2007;4:951–956. doi: 10.1038/nmeth1101. [DOI] [PubMed] [Google Scholar]
- Osato D, Rogers K, Guo Q, Li F, Richmond G, Klug F, Simpson L. Uridine insertion/deletion RNA editing in trypanosomatid mitochondria: In search of the editosome. RNA. 2009;15:1338–1344. doi: 10.1261/rna.1642809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi AK, Schnaufer A, Ernst NL, Wang B, Carmean N, Salavati R, Stuart K. Identification of novel components of Trypanosoma brucei editosomes. RNA. 2003;9:484–492. doi: 10.1261/rna.2194603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi AK, Zikova A, Dalley RA, Acestor N, Ogata Y, Anupama A, Myler PJ, Stuart KD. Mitochondrial complexes in trypanosoma brucei: a novel complex and a unique oxidoreductase complex. Mol. Cell Proteomics. 2007;7:534–545. doi: 10.1074/mcp.M700430-MCP200. [DOI] [PubMed] [Google Scholar]
- Pelletier M, Read LK, Aphasizhev R. Isolation of RNA Binding Proteins Involved in Insertion/deletion Editing. Methods Enzymol. 2007;424:69–96. doi: 10.1016/S0076-6879(07)24004-2. [DOI] [PubMed] [Google Scholar]
- Pusnik M, Small I, Read LK, Fabbro T, Schneider A. Pentatricopeptide repeat proteins in Trypanosoma brucei function in mitochondrial ribosomes. Mol. Cell Biol. 2007;27:6876–6888. doi: 10.1128/MCB.00708-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan CM, Militello KT, Read LK. Polyadenylation regulates the stability of Trypanosoma brucei mitochondrial RNAs. J. Biol. Chem. 2003;278:32753–32762. doi: 10.1074/jbc.M303552200. [DOI] [PubMed] [Google Scholar]
- Schmitz-Linneweber C, Small I. Pentatricopeptide repeat proteins: a socket set for organelle gene expression. Trends Plant Sci. 2008;13:663–670. doi: 10.1016/j.tplants.2008.10.001. [DOI] [PubMed] [Google Scholar]
- Schnaufer A, Panigrahi AK, Panicucci B, Igo RP, Salavati R, Stuart K. An RNA ligase essential for RNA editing and survival of the bloodstream form of Trypanosoma brucei. Science. 2001;291:2159–2161. doi: 10.1126/science.1058955. [DOI] [PubMed] [Google Scholar]
- Schnaufer A, Clark-Walker GD, Steinberg AG, Stuart K. The F1-ATP synthase complex in bloodstream stage trypanosomes has an unusual and essential function. EMBO J. 2005;24:4029–4040. doi: 10.1038/sj.emboj.7600862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiwert SD, Heidmann S, Stuart K. Direct visualization of uridylate deletion in vitro suggests a mechanism for kinetoplastid RNA editing. Cell. 1996;84:831–841. doi: 10.1016/s0092-8674(00)81062-4. [DOI] [PubMed] [Google Scholar]
- Sharma MR, Booth TM, Simpson L, Maslov DA, Agrawal RK. Structure of a mitochondrial ribosome with minimal RNA. Proc. Natl. Acad. Sci. U. S. A. 2009;106:9637–9642. doi: 10.1073/pnas.0901631106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small ID, Peeters N. The PPR motif - a TPR-related motif prevalent in plant organellar proteins. Trends Biochem Sci. 2000;25:46–47. doi: 10.1016/s0968-0004(99)01520-0. [DOI] [PubMed] [Google Scholar]
- Weng J, Aphasizheva I, Etheridge RD, Huang L, Wang X, Falick AM, Aphasizhev R. Guide RNA-Binding Complex from Mitochondria of Trypanosomatids. Mol. Cell. 2008;32:198–209. doi: 10.1016/j.molcel.2008.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickstead B, Ersfeld K, Gull K. Targeting of a tetracycline-inducible expression system to the transcriptionally silent minichromosomes of Trypanosoma brucei. Mol. Biochem. Parasitol. 2002;125:211–216. doi: 10.1016/s0166-6851(02)00238-4. [DOI] [PubMed] [Google Scholar]
- Wirtz E, Leal S, Ochatt C, Cross GA. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 1999;99:89–101. doi: 10.1016/s0166-6851(99)00002-x. [DOI] [PubMed] [Google Scholar]
- Zikova A, Panigrahi AK, Dalley RA, Acestor N, Anupama A, Ogata Y, Myler PJ, Stuart K. Trypanosoma brucei mitochondrial ribosomes: affinity purification and component identification by mass spectrometry. Mol Cell Proteomics. 2008;7:1286–1296. doi: 10.1074/mcp.M700490-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.