Abstract
Key challenges facing cancer therapy are the development of tumor-specific drugs and potent multimodal regimens. Oncolytic adenoviruses possess the potential to realize both aims by restricting virus replication to tumors and inserting therapeutic genes into the virus genome, respectively. A major effort in this regard is to express transgenes in a tumor-specific manner without affecting virus replication. Using both luciferase as a sensitive reporter and genetic prodrug activation, we show that promoter control of E1A facilitates highly selective expression of transgenes inserted into the late transcription unit. This, however, required multistep optimization of late transgene expression. Transgene insertion via internal ribosome entry site (IRES), splice acceptor (SA), or viral 2A sequences resulted in replication-dependent expression. Unexpectedly, analyses in appropriate substrates and with matching control viruses revealed that IRES and SA, but not 2A, facilitated indirect transgene targeting via tyrosinase promoter control of E1A. Transgene expression via SA was more selective (up to 1,500-fold) but less effective than via IRES. Notably, we also revealed transgene-dependent interference with splicing. Hence, the prodrug convertase FCU1 (a cytosine deaminase–uracil phosphoribosyltransferase fusion protein) was expressed only after optimizing the sequence surrounding the SA site and mutating a cryptic splice site within the transgene. The resulting tyrosinase promoter-regulated and FCU1-encoding adenovirus combined effective oncolysis with targeted prodrug activation therapy of melanoma. Thus, prodrug activation showed potent bystander killing and increased cytotoxicity of the virus up to 10-fold. We conclude that armed oncolytic viruses can be improved substantially by comparing and optimizing strategies for targeted transgene expression, thereby implementing selective and multimodal cancer therapies.
Quirin and colleagues analyze the mechanisms by which transcriptional targeting of adenovirus replication, via promoter control of E1A expression, mediates indirect targeting of transgenes expressed with late kinetics. Specifically, the authors sought to investigate how the method of late transgene expression affects such targeting. They compared late transgene expression mediated by IRES, S. sequence, or ribosomal skip with a viral 2A peptide.
Introduction
Oncolytic viruses facilitate targeted infection of tumor cells resulting in both tumor cell lysis and release of virus progeny for spreading tumor destruction. Success of viral oncolysis critically depends on replication selectivity and lytic potency of oncolytic viruses. However, physical barriers to viral spread, such as connective tissue or necrotic areas of the tumor, and antiviral immunity can prevent tumor eradication by oncolytic viruses in patients, even when lytic activity is high. Therefore, combination therapies of oncolytic viruses with conventional or other experimental anticancer regimens have come into focus for oncolytic virus research (Ottolino-Perry et al., 2010). One such strategy is viro-gene therapy by “arming” oncolytic viruses with therapeutic genes. This strategy facilitates the killing of noninfected tumor cells by therapeutic mechanisms determined by the inserted transgene, such as genetic prodrug activation, immunotherapy, or anti-angiogenesis.
Recombinant adenoviruses (Ads) are promising candidates for viro-gene therapy, as they enable both outstanding tumor selectivity of cell lysis and therapeutic gene insertion without loss of replication competency. For tumor-selective cell lysis, Ad gene functions dispensable for efficient replication in cancer cells, but required in healthy cells, have been mutated (Alemany et al., 2000; Fueyo et al., 2000; Heise et al., 2000). Alternatively, essential viral genes have been expressed from cell type-selective promoters (Rodriguez et al., 1997; Nettelbeck, 2008). For most reported viruses, such post-entry targeting is effective during early infection (i.e., by mutation of early genes or restricting their expression). In consequence, replication of the virus genome, expression of late genes, and infectious viral particle production are attenuated or ablated in healthy cells. An example for targeted Ad mutants is AdTyrΔ24, which has been exploited in the present study as a parental virus for generation of transgene-encoding derivatives. AdTyrΔ24 has been engineered by replacing the viral immediate early gene E1A promoter with an optimized pigment cell-selective human tyrosinase enhancer/promoter resulting in melanoma-selective E1A expression, virus replication, and cell lysis (Nettelbeck et al., 2002). Replication and cell lysis of AdTyrΔ24 are attenuated approximately two orders of magnitude in non-pigmented tumor cells or normal cells, but not in melanoma cells. Tumor selectivity of this and other transcriptionally targeted Ads was further increased by expressing additional viral genes from cell type-selective promoters (Johnson et al., 2002; Banerjee et al., 2004). Of note is that strategies that ensure tight tumor selectivity are of particular importance to allow the clinical investigation of future generations of oncolytic viruses with increased efficiency, as implemented, for example, by therapeutic gene insertion.
Different strategies have been pursued to achieve transgene expression by oncolytic Ads (Hermiston and Kuhn, 2002; Nettelbeck, 2008). Either transgene cassettes including promoter and polyadenylation signal were inserted into various positions of the Ad genome, or transgenes were inserted into viral transcription units. The latter strategy possesses two advantages: (1) It exploits viral mechanisms that ensure efficient gene expression, while nuclear export and translation of capped cellular mRNAs are suppressed during late Ad replication, and (2) it facilitates the timing of transgene expression within the viral replication cycle, for example, genes inserted into the late Ad transcription unit are expressed with late kinetics. Such late kinetics of therapeutic gene expression and activity are of interest to avoid interference with virus replication. Transgenes have been inserted into the late viral transcription unit either by replacing viral genes or as additional reading frames using internal ribosome entry sites (IRESs) or splice acceptor (SA) sites (Nettelbeck, 2008, and references therein). IRESs are highly structured viral sequences that facilitate bicistronic gene expression by internal translation initiation in addition to translation initiation at the 5' end of mRNAs. For example, the approximately 600-bp encephalomyocarditis virus (EMCV) IRES has been used for transgene insertion into both early and late Ad transcription units (Sauthoff et al., 2002; Rivera et al., 2004). Alternative splicing is widely utilized by Ads as a mechanism for expression of multiple proteins from one transcription unit. Indeed, splicing was first described for Ad late mRNAs (Berget et al., 1977; Chow et al., 1977). Artificial or Ad-derived SA sites have been used for splicing of reporter or therapeutic genes into the late Ad mRNA (thus constituting an L6 unit [Carette et al., 2005; Jin et al., 2005; Cascante et al., 2007]). The short sequence (<40 bp) of SA sites is of advantage, as larger insertions into the Ad genome necessitate deletions of viral genes in order to retain a packageable genome size. Viral 2A sequences are further short sequences that facilitate multiprotein expression from one open reading frame by skipping the formation of a peptide bond during translation, a process termed ribosomal skip (Donnelly et al., 2001; Szymczak et al., 2004). In the present study we explored a 2A sequence as a third tool for transgene insertion into the late transcription unit, in addition to IRES and SA sequence.
Genetic prodrug activation is a gene therapy approach of high interest for arming of oncolytic viruses (Portsmouth et al., 2007). In this approach, expression of a prodrug convertase in infected tumor cells and injection of the innocuous prodrug result in production of the chemotherapeutic drug specifically in the tumor. Cytosine deaminase (CD), a bacterial or fungal enzyme not present in mammalian cells, deaminates the prodrug 5-fluorocytosine (5-FC) to 5-fluorouracil (5-FU) (Portsmouth et al., 2007). 5-FU is an established chemotherapeutic drug that is converted by cellular enzymes into the active metabolites 5-fluoro-UTP and 5-fluoro-deoxyUMP, inhibitors of RNA processing and DNA synthesis. Of note is that a potent bystander effect has been reported for the CD prodrug convertase/5-FC system, which is due to the permeability of cell membranes for 5-FU (Portsmouth et al., 2007). The CD prodrug convertase has been previously improved by generating the FCU1 gene encoding an optimized yeast-derived CD fused to yeast uracil phosphoribosyltransferase (UPRT) (Erbs et al., 2000). UPRT converts 5-FU to its active metabolites and thereby sensitizes tumor cells that are otherwise resistant to 5-FU because they lack endogenous 5-FU metabolizing activity. For viro-gene therapy, the potent bystander effect of CD/5-FC is a key advantage because the killing of uninfected bystander cells is of critical importance to complement viral oncolysis. However, activity of the active 5-FU metabolites interferes also with Ad replication. Late expression of the prodrug convertase during the viral replication cycle is a strategy to reduce such effects. Moreover, viro-gene therapy can benefit from a unique feature of genetic prodrug activation, namely, that the time of therapeutic activity is not directly linked to therapeutic gene expression. Thus, by choosing the time of 5-FC injection, prodrug activation and 5-FU activity can be synchronized with maximal virus spread and expression of the prodrug convertase.
For many gene therapy approaches, such as genetic prodrug activation or apoptosis induction by death ligands, specific expression of the corresponding therapeutic genes in tumors is critical to avoid adverse side effects. Selectivity of therapeutic gene expression by oncolytic Ads is determined at three levels. First, conditional amplification of the virus genome increases the number of templates for transgene transcription selectively in targeted cells, in which the virus can replicate. Second, spread of progeny viruses in the tumor results in an increased number of transduced cells. For these two levels, selectivity of virus DNA replication and infectious viral particle production determine specificity of transgene expression. Third, for viruses that are targeted post-entry, as most oncolytic Ads are, expression of toxic genes in healthy cells should be prevented even though virus replication cannot take place in these cells. Note that this setting corresponds to standard gene therapy of replication-deficient viruses. Even more, in the context of viral oncolysis “spillover” infection by viruses released from infected tumor cells will increase side effects of gene expression on healthy tissues. Tumor selectivity of transgene expression from the unreplicated virus genome can be ensured by direct transcriptional control of the transgene from a tumor-selective promoter (Davis et al., 2006; Nettelbeck, 2008). Alternatively, indirect transcriptional targeting of transgene expression is possible by combining late, thus replication-dependent, expression of the transgene with promoter regulation of virus replication. This strategy seems of advantage as efficient viral gene expression mechanisms are exploited and therapeutic gene expression occurs with late kinetics (see above).
The late nature of transgene expression by oncolytic Ads has been frequently confirmed using inhibitors of virus genome replication. However, this is highly artificial. Whether the biological mechanism used for targeting viral replication (i.e., mutation or targeted expression of early genes) indeed prevents late expression and activity of therapeutic genes in infected healthy cells has been rarely shown. Therefore, the prime objective of the present study was to analyze more systematically how transcriptional targeting of Ad replication via promoter control of E1A expression, mediates indirect targeting of transgenes expressed with late kinetics. Specifically, we sought to investigate how the method of late transgene expression affects such targeting. For this purpose we exploited the optimized human tyrosinase enhancer/promoter because it mediates exceptionally “tight” control of transgene or E1A expression/virus replication (Siders et al., 1996; Nettelbeck et al., 1999, 2002). We compared late transgene expression mediated by IRES, SA sequence, or ribosomal skip with a viral 2A peptide. Finally, we verified the best strategy for indirect transcriptional targeting of transgene expression in a therapeutic setting using the FCU1/5-FC genetic prodrug activation system.
Materials and Methods
Cell culture
Human cell lines SK-MEL-28 (melanoma) (American Type Culture Collection, Manassas, VA), C8161 (melanoma) (kindly provided by D. Welch, Birmingham, AL), HaCat (immortalized keratinocyte cell line) (Boukamp et al., 1988), and 293 were cultivated in Dulbecco's Modified Eagle's Medium (Invitrogen, Karlsruhe, Germany). The human melanoma cell lines Mel888, Mel624 (kindly provided by J. Schlom, Bethesda, MD), Colo829 (kindly provided by J. Banchereau, Baylor, TX), and A375M (kindly provided by I.J. Fidler, Houston, TX) were cultivated in RPMI 1640 medium (Invitrogen), and HFF cells (primary normal foreskin fibroblast) (kindly provided by M. Marschall, Erlangen, Germany) were cultivated in Minimal Essential Medium (Invitrogen). Media were supplemented with 10% heat-inactivated fetal bovine serum (FBS) (PAA, Cölbe, Germany), 100 IU/ml penicillin, and 100 μg/ml streptomycin (both from Invitrogen). Cells were grown at 37°C in a humidified atmosphere of 5% CO2.
Recombinant Ads
For a schematic outline of the Ad genomes generated and used in this study, see Figs. 1 and 5. Ad5Δ24E3-, Ad5IL, Ad5Luc1, and Ad5CMVFCU were described previously (Kransnykh et al., 1996; Erbs et al., 2000; Suzuki et al., 2002; Rivera et al., 2004). Ad5CMVLuc is a replication-deficient Ad with a pGL3-derived luciferase gene under control of the cytomegalovirus (CMV) promoter. Ad5TL was generated by fusion of the luciferase gene to the Ad5 fiber gene via a GSG linker and a codon-optimized self-cleaving 2A motif of Thosea asigna virus (EGRGSLLTCGDVEENPGP [Szymczak et al., 2004]). To introduce an SA sequence upstream of the luciferase gene, pGL3BPSA was generated by inserting the BPS sequence containing a branchpoint, polypyrimidine tract, and SA element (Jin et al., 2005) into the BglII/NcoI sites of pGL3basic (Promega, Madison, WI). Next, pSΔ24BPSA was generated by inserting the BPS–luciferase fragment of pGL3BPSA as the MluI/SalI fragment into a derivative of pShuttleΔ24 (Suzuki et al., 2002) into which the corresponding restriction sites had been inserted between the E4 genes and the right inverted terminal repeat (ITR) before (between nucleotides 35,774 and 35,775 of the Ad5 genome). pSΔ24BPSA was recombined with pAdEasy-1 to generate the virus genome of Ad5SL. The corresponding targeted viruses Ad5TyrIL, Ad5TyrSL, and Ad5TyrTL were generated by replacing the Ad E1A promoter with an artificial human tyrosinase enhancer/promoter (hTyr2E/P) and upstream polyA as described by Nettelbeck et al. (2002). Plasmid pGL3S_FCU1 was cloned by replacing the luciferase cDNA of pGL3BPSA with the FCU1 cDNA (Genbank accesssion number AAG33626) of plasmid pCIneoFCU1 (Erbs et al., 2000). The MluI/SalI fragment of pGL3SA-FCU1 was inserted into pSΔ24BPSA and pSTyrΔ24BSA, generating pSS_FCU and pSTyrS_FCU, respectively. For pGL3Ssp_FCU, a spacer consisting of nucleotides 4–54 of the luciferase gene was inserted into the Nhe/NcoI sites of pGL3S_FCU. The shuttle plasmids pSSsp_FCU and pSTyrSsp_FCU were constructed by inserting the MluI/SalI fragment of pGL3Ssp_FCU into the corresponding restriction sites of pSΔ24BPSA or pSTyrΔ24BPSA, respectively. pSSsp_mFCU and pSTyrSsp_mFCU were generated by replacing the cryptic SA sequence TAC TCC GAT AGA ATC ATC AGA at positions 588–608 of the FCU gene of pSSsp_FCU and pSTyrSsp_FCU with the sequence TAT AGC GAC CGG ATT ATT CGG by polymerase chain reaction (PCR) cloning. pSS_FCU, pSTyrS_FCU, pSSsp_FCU, pSTyrSsp_FCU, pSSsp_mFCU, and pSTyrSsp_mFCU were recombined with pAdEasy-1 to generate virus genomes. Plasmids containing the genomes of the recombinant Ads were generated by homologous recombination in BJ5183 bacteria as described (He et al., 1998). Virus particles were produced by transfection of A549 cells or of Mel888 cells for viruses containing the tyrosinase promoter with PacI-digested genome plasmids using Lipofectamine (Invitrogen). The recombinant viruses were amplified in A549 or Colo829 cells. Viruses were purified by two rounds of CsCl equilibrium density gradient ultracentrifugation. Verification of viral genomes and exclusion of wild-type contamination were performed by PCR. Physical particle concentration (viral particles/ml) was determined by reading the optical density at 260 nm; infectious viral particle titers were determined by 50% tissue culture infective dose (TCID50) assay on 293 cells. The ratios of virus particles to infectious virus particles for virus preparations were as follows (two ratios are given in the case of independent virus preparations): Ad5IL, 124 and 135; Ad5TyrIL, 300 and 378; Ad5TL, 26 and 75; Ad5TyrTL, 62 and 229; Ad5SL, 20 and 29; Ad5TyrSL, 25 and 32; Ad5CMVLuc, 13; Ad5Δ24E3-, 5; Ad5Ssp_mFCU, 16; Ad5TyrSsp_mFCU, 32; Ad5TyrSsp_FCU, 2; Ad5TyrSFCU, 15; and Ad5CMVFCU, 24.
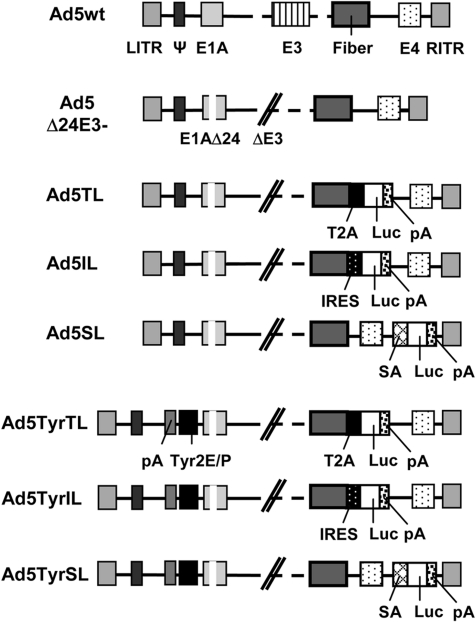
FIG. 1.
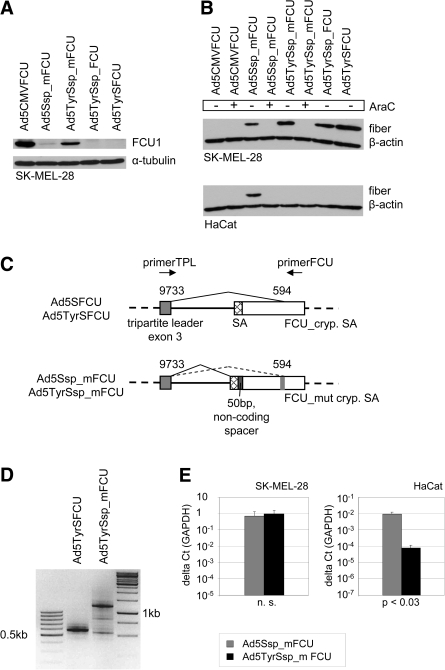
Ad constructs with luciferase reporter gene inserted into the late transcription unit via IRES, SA, or T2A: Schematic outline of the genomes of the six oncolytic luciferase reporter viruses investigated in this study; of the parental E3-deleted recombinant Ad without transgene insertion, Ad5Δ24E3-; and of wild-type Ad serotype 5 (Ad5wt). ΔE3, E3 region deleted; E1A, E3, and E4, early Ad genes; LITR/RITR, left/right ITR; Luc, firefly luciferase gene; pA, polyadenylation signal; Tyr2E/P, artificial tyrosinase promoter consisting of two enhancers and the core promoter of the human tyrosinase gene; Ψ, packaging signal.
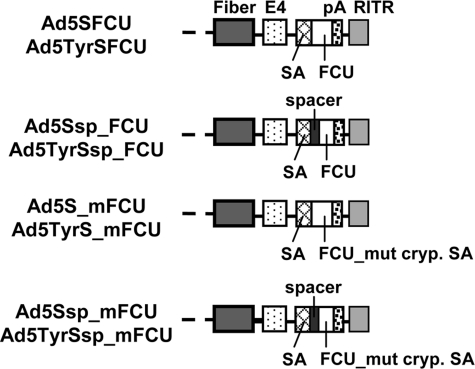
FIG. 5.
Ad constructs with CD-UPRT fusion gene FCU1 inserted into the late transcription unit via SA: Schematic outline of the genomes of oncolytic viruses with insertion of the FCU1 gene encoding an optimized yeast CD-UPRT fusion protein. Shown are the right termini of the genomes (see Fig. 1 for left termini of Ad5 and Ad5Tyr genomes). E4, early Ad gene; FCU_mut cryp. SA, FCU1 gene with mutation of the cryptic SA site; pA, polyadenylation signal; RITR, right IRT.
Virus-mediated spread and cytotoxicity
Cells (3 × 104) were seeded in 48-well plates and were infected the next day in 200 μl of growth medium containing 2% FBS. Four hours post-infection, growth medium containing 10% FBS was added. When cell lysis was observed at the lowest virus titers, cells were fixed and stained with 1% crystal violet in 70% ethanol for 10 min, followed by washing with tap water to remove excess color. Plates were dried, and images were captured with an Epson (Long Beach, CA) Perfection V500 Photo scanner.
Luciferase assay
Cells (5 × 104) were seeded in 24-well plates. The next day, cells were infected in 250 μl of growth medium containing 2% FBS. One hour post-infection, growth medium containing 10% FBS was added with or without the viral replication inhibitor cytosine β-d-arabinofuranoside (AraC) (Sigma, Deisenhofen, Germany) at a concentration of 2 μM. AraC was replenished every 12 hr. Luciferase activity of cell lysates was determined at indicated time points using a luciferase assay system (Promega, Mannheim, Germany).
DNA/RNA quantification by real-time PCR
For quantification of viral genome copy numbers or RNA expression, 5 × 104 cells were seeded in 24-well plates. The next day, cells were infected in 250 μl of growth medium containing 2% FBS. One hour post-infection, growth medium containing 10% FBS was added with or without 2 μM AraC. AraC was replenished every 12 hr. Samples were harvested at indicated time points, and DNA was purified from cell lysates with the QIAamp Blood Mini Kit (Qiagen, Chatsworth, CA); RNA was purified with the RNeasy kit including DNase digest (Qiagen) following the manufacturer's instructions. Oligonucleotides used for quantification of viral genomes, viral E1A, E4, or fiber mRNA, cellular DNA, and cellular RNA were as in Rivera et al. (2004). Oligonucleotides for quantification of FCU1 mRNA were FCU forward (5'-GAA ACT GAC ACC AAC GAA AAC) and FCU reverse (5'-TTT TAC CGA TAC GCA CAG AC). Real-time PCR was performed with the 7300 Real Time PCR system (Applied Biosystems, Darmstadt, Germany) using MicroAmp 96-well reaction plates (Applied Biosystems) in a total volume of 25 μl for each PCR assay. Each probe contained 23 μl of Power SYBR Green Master Mix (Applied Biosystems), 2 μl of template mRNA or DNA, and 10 pmol of each oligonucleotide. For mRNA quantification, reverse transcriptase and RNase inhibitor (both from Applied Biosystems) were added. Quantitative PCR (qPCR) was performed with an initial denaturation step of 10 min at 95°C, followed by 50 cycles of 15 sec of denaturation at 95°C, 10 sec of annealing at 60°C, and 15 sec of elongation at 72°C. At the end of each cycle, the fluorescence emitted by the SYBR Green was measured. After completion of the cycling process, samples were subjected to melting curve analysis. pTG3602 (108, 106, 104, and 102 copies/μl) was amplified for each reaction series to generate a standard curve for quantification of the copy numbers of viral genomes or viral mRNA. Data were normalized with cellular genomic DNA or cellular RNA for each sample individually. Cellular RNA was quantified using glyceraldehyde 3-phosphate dehydrogenase oligonucleotides and 200, 20, 2, and 0.2 ng/μl human RNA isolated from A549 cells as the standard. Cellular DNA was quantified using β-actin oligonucleotides and 200, 20, 2, and 0.2 ng/μl human DNA isolated from A549 cells as the standard. Data were analyzed with 7300 System SDS software (Applied Biosystems). Negative controls with no template were carried out for each reaction series.
Western blot analysis
Cells (5 × 105) were plated in six-well plates and infected with indicated viruses in 1 ml of growth medium containing 2% FBS. One hour post-infection 2 ml of growth medium containing 10% FBS was added with or without 2 μM AraC. AraC was replenished every 12 hr. At indicated time points cells were lysed in 10 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1% Nonidet P-40 (Igepal), 1% sodium desoxylcholate, 0.1% dodium dodecyl sulfate, 1 mM phenylmethylsulfonyl fluoride, 20 mM sodium fluoride, and 2 mM sodium orthovanadate. The membrane was probed with 4D2 monoclonal antibody specific for Ad serotype 5 fiber (Abcam, Cambridge, UK) or with a polyclonal yeast CD antibody (AbD Serotec, Oxford, UK). Antibodies to α-tubulin and β-actin (both from Sigma) were used as the loading control for CD-UPRT and fiber, respectively. Different loading controls were necessary to avoid overlapping protein sizes. Antibody binding was visualized using chemiluminescence (Pierce ECL, Thermo Fisher Scientific, Bonn, Germany).
Reverse transcription for FCU1
Total cellular RNA was isolated from 5 × 105 cells at 24 hr post-infection with Ad at 100 TCID50 per cell using the Qiagen RNeasy Mini RNA Extraction Kit according to the manufacturer's instructions. To remove cell debris and shear genomic DNA from lysate, QIAshredder (Qiagen) columns were used. Three micrograms of total RNA was reverse-transcribed into single-strand cDNA using the SuperScript II Reverse Transcriptase Kit (Invitrogen) and oligo(dT) primers (Invitrogen) as described by the manufacturer. PCR assays were performed using 2 μl of cDNA in a final volume of 25 μl, containing 1.5 mM MgCl2, 0.2 mM deoxynucleotide triphosphate mixture each, 1.25 U of Taq polymerase (Invitrogen), and 10 pmol of forward (5'-CTG AGC GAG TCC GCA TCG) and reverse (5'-TAT CTG TCA CCA AAG TCA) primers. PCR products were analyzed by agarose gel electrophoresis.
Determination of 50% inhibitory concentration values for 5-FC and 5-FU
Cells were plated in 96-well plates at 5,000 cells per well (Colo829, 10,000 cell per well; C8161, 25,000 cells per well) in 100 μl of growth medium. The next day, SK-MEL-28 and Colo829 cells were transduced with Ad5CMVFCU or Ad5Luc1 at 2 TCID50 per cell or were mock-transduced. After 24 hr, cells were treated with 5-FC (catalog number F7129-IG, Sigma) (100 mM–0.01 μM in 10-fold dilutions) or 5-FU (catalog number A13456, Alfa Aesar, Karlsruhe, Germany) (10 mM–0.01 μM in 10-fold dilutions) in 200 μl of growth medium or were incubated with growth medium alone. Five days later, cells were fixed and stained with 1% crystal violet in 70% ethanol for 10 min, followed by washing with tap water to remove excess color. The plates were dried, and the optical density at 595 nm was measured using a Lab Systems Multiscan MS (Thermo Fisher Scientific). The absorbance of wells with mock-treated cells defined 100% viability. The 5% inhibitory concentration (IC50) values were determined using Origin8 software (OriginLab, Northampton, MA).
Burst assay
Cells (5 × 104) were plated in 24-well plates. The next day, cells were infected at 1 TCID50 per cell in a volume of 250 μl of growth medium containing 2% FBS. Two hours post-infection the medium was removed, and cells were washed twice with phosphate-buffered saline to remove unbound viruses. Then 1 ml of growth medium was added. Where indicated, 10 mM 5-FC was added 1 day post-infection. Three days post-infection, supernatants and cells were harvested, and viruses were released from cell pellets by three cycles of freezing and thawing. Infectious virus particles were determined by TCID50 assay on 293 cells.
Combination therapy of oncolysis and prodrug activation
Cells were plated in 96-well plates at a concentration of 10,000 cells per well in 100 μl of growth medium containing 2% FBS. After 24 hr, cells were infected with serial dilutions of viruses added in 50 μl of medium containing 2% FBS or were mock-infected. Two days post-infection either 5-FC or medium was added in a volume of 50 μl to the cells. Six days post-infection, cells were stained with crystal violet, and viability was calculated as described above.
Bystander effect
For the determination of the bystander effect, 5 × 104 SK-MEL-28 cells were seeded in 24-well plates. The next day, cells were infected at 10 TCID50 per cell in 500 μl of growth medium containing 2% FBS. Two days post-infection, infection medium was removed, and cells were incubated either with 5-FC at a concentration of 10 mM or with medium alone. The next day, supernatants were collected and heated for 10 min at 50°C. Serial dilutions of the supernatants were added to SK-MEL-28 cells, which were plated in 48-well plates at 30,000 cells per well the day before. As controls, serial dilutions of 10 mM 5-FU in medium, heated for 10 min at 50°C or unheated, were incubated with the cells. Cytotoxicity was determined by crystal violet staining 4 days later.
Statistical analysis
Differences between indicated groups were analyzed using Student's t test. Values of p < 0.05 were considered statistically significant.
Results
Transcriptionally targeted oncolytic Ads with transgenes inserted via IRES, SA, or T2A into the late transcription unit
To investigate indirect promoter control of late transgene expression by oncolytic Ads, we generated a set of six oncolytic luciferase reporter viruses (Fig. 1). The luciferase gene was inserted into the late Ad transcription unit by three different strategies: For IL viruses, the 588-bp EMCV IRES sequence was used for bicistronic expression of the viral fiber and luciferase genes as we described before (Rivera et al., 2004). The SL viruses were generated by inserting the luciferase gene between the E4 genes and the right ITR using a 26-bp artificial SA site consisting of a branchpoint, polypyrimidine tract, and SA element following the strategy reported by Hermiston and colleagues (Jin et al., 2005). TL viruses were generated by fusion of the fiber and luciferase genes via the 54-bp 2A sequence of the insect virus T. asigna (T2A) with upstream GSG-linker for expression of both proteins from a single open reading frame by ribosomal skip (Szymczak et al., 2004 with addendum). The recombinant viruses Ad5IL, Ad5SL, and Ad5TL contained the endogenous E1A promoter. For Ad5TyrIL, Ad5TyrSL, and Ad5TyrTL, this promoter was replaced by the optimized human tyrosinase enhancer/promoter, for which we previously reported tight control of E1A expression and virus replication (Nettelbeck et al., 2002). All six viruses contained the E1AΔ24 mutant gene for attenuation of virus replication in quiescent normal cells (Fueyo et al., 2000; Heise et al., 2000).
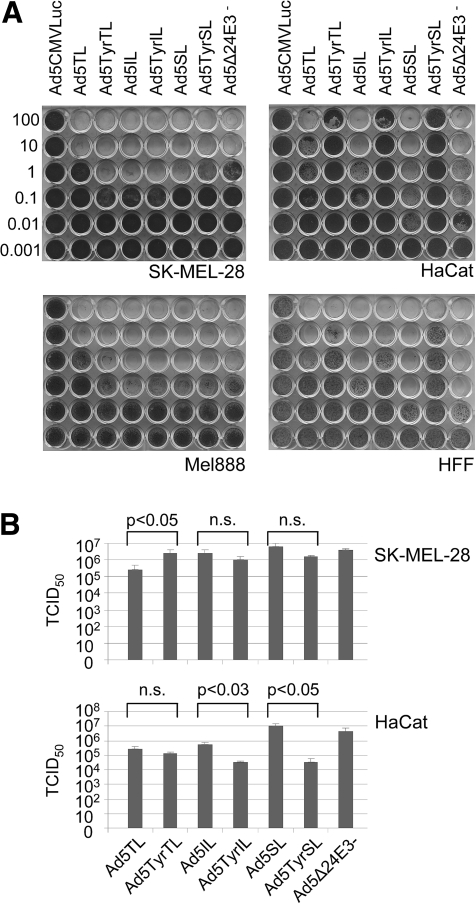
Selectivity of cell lysis and spread by Ad5TyrIL/SL/TL
In melanoma cells, the IL, SL, and TL viruses, with the exception of Ad5TL, showed oncolytic activity and replication efficiency similar to the parental virus Ad5Δ24E3− (Fig. 2A and B). Ad5TL was attenuated approximately 10-fold. In accord with these results, E1A expression was only marginally affected by tyrosinase promoter control for the IL and SL viruses, whereas Ad5TL, but not Ad5TyrTL, showed a substantial reduction of E1A expression in melanoma cells (Supplementary Fig. S1A; Supplementary Data are available at www.liebertonline.com/hum). In consequence, fiber and hexon expressions were also delayed for Ad5TL (Supplementary Fig. S1B). In normal fibroblasts (HFF) and keratinocytes (HaCat), spread-dependent lytic activity of the tyrosinase promoter–regulated viruses was attenuated relative to the matching viruses with endogenous E1A promoter (Fig. 2A), demonstrating transcriptional targeting of Ad replication. However, the degree of tyrosinase promoter–dependent attenuation of oncolysis in normal cells was dependent on the strategy of transgene insertion. For Ad5TyrSL, lytic activity was 3 and >4 orders of magnitude attenuated relative to Ad5SL in HFF and HaCat cells, respectively, demonstrating a remarkable selectivity. The IL viruses showed intermediate and the TL viruses showed the lowest promoter-dependent selectivity of viral lysis in HFF and HaCat cells (1–2 orders of magnitude). Likewise, we found that the tyrosinase promoter–mediated attenuation of virus burst size in HaCat cells was highest for the SL viruses (291-fold), followed by the IL viruses (17-fold), and was only marginal for TL viruses (Fig. 2B). The analysis of viral gene expression and genome replication kinetics corroborated these results: Tyrosinase promoter control resulted in strongly reduced expression of E1A and fiber genes and reduced viral genome copy numbers in HaCat cells for IL and SL viruses (several orders of magnitude at 1 day post-infection), but not for TL viruses (Supplementary Fig. S1C and D). It is notable that E1A and fiber mRNA copy numbers and genome copy numbers were considerably higher for Ad5TyrTL than for Ad5TyrIL and Ad5TyrSL in HaCat cells. In conclusion, melanoma selectivity of cell lysis/spread and replication of tyrosinase promoter–regulated oncolytic Ads were retained when the transgene was inserted into the late transcription unit via the IRES and SA sequence but was reduced when the T2A sequence was used.
FIG. 2.
Cytotoxicity and replication of transcriptionally targeted oncolytic Ads with luciferase reporter gene inserted into the late transcription unit. (A) Melanoma cells (SK-MEL-28, Mel888), keratinocytes (HaCat), or primary fibroblasts (HFF) were infected with Ads, and cytotoxicity was determined by crystal violet staining of surviving cells. Numbers are viral titers in TCID50 per cell used for infection. Ad5CMVLuc is an E1/E3-deleted replication-deficient Ad. (B) Infectious virus particle production in SK-MEL-28 or HaCat cells at 3 days after infection of 5 × 104 cells. n.s., not significant.
Late transgene expression by Ad5TyrIL/SL/TL
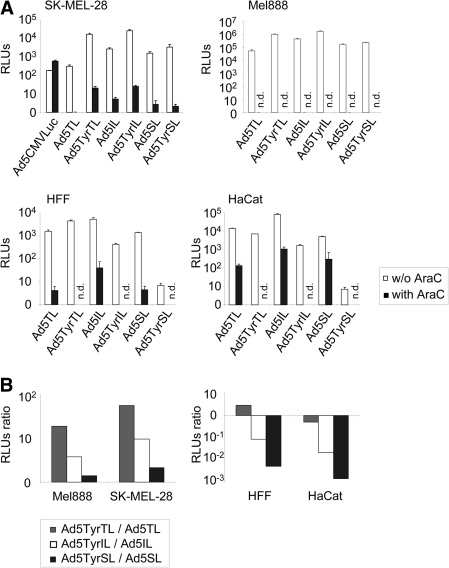
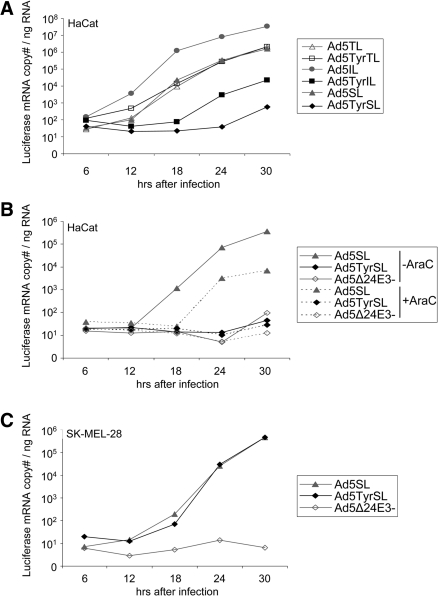
We next investigated transgene expression by the IL, SL, and TL viruses and dependence of transgene expression on virus replication by infection of the melanoma cell line SK-MEL-28 in the absence or presence of the viral replication inhibitor AraC (Fig. 3A). At 2 days post-infection, these viruses resulted in luciferase activity that was superior to that of replication-deficient Ad5CMVLuc in the absence of AraC. Luciferase activity for Ad5TyrTL was nearly as high as for Ad5TyrIL and was higher than for Ad5TyrSL, revealing that the T2A element is functional and efficient. Indeed, no fiber–luciferase fusion protein was detected (Supplementary Fig. S1B). However, Ad5TL showed the weakest luciferase activity of the six replication-competent viruses, which is consistent with its attenuated viral gene expression, replication, and cytotoxicity. It is interesting that, also for the IRES and SA strategy of transgene insertion, luciferase activity was superior for the tyrosinase viruses in SK-MEL-28 (Fig. 3). These results were confirmed in a second melanoma cell line Mel888 (Fig. 3). All IL, SL, and TL viruses showed similar replication dependence of transgene expression in SK-MEL-28 cells, as AraC treatment caused a reduction in luciferase activity of approximately 3 orders of magnitude for these viruses, but not for Ad5CMVLuc. Ad5TyrSL showed the highest block of transgene expression by AraC of approximately 1,600-fold. These results prove that all three strategies of transgene insertion into the late transcription unit—IRES, SA, and T2A—facilitate late transgene expression kinetics. Transgene expression in the absence of AraC was superior for the tyrosinase promoter–regulated viruses.
FIG. 3.
Transgene expression by transcriptionally targeted oncolytic Ads with luciferase reporter gene inserted into the late transcription unit. Melanoma cells (SK-MEL-28, Mel888), keratinocytes (HaCat), or primary fibroblasts (HFF) were infected with Ads at 1 TCID50 per cell in the presence or absence (w/o) of AraC, and luciferase activity was determined 2 days post-infection. Ad5CMVLuc is an E1/E3-deleted replication-deficient Ad with luciferase expressed from the CMV enhancer/promoter. (A) Columns show mean relative luminescence units (RLU) values; error bars show SDs of triplicate experiments. (B) Columns show the ratio of RLU values for Ad5Tyr viruses to RLUs for Ad5 viruses. Note the difference in scale between the left and right panels. n.d., not determined.
Indirect transcriptional targeting of late transgene expression by the tyrosinase promoter is dependent on the transgene insertion strategy
We next explored whether E1A expression from the tyrosinase promoter causes a loss of transgene expression for IL, SL, and TL viruses in normal cells. For IL and SL viruses, tyrosinase promoter control of the E1A gene mediated a reduction of luciferase expression of >10-fold and >100-fold, respectively, in both HaCat and HFF cells (Fig. 3). Ad5TyrSL-mediated transgene expression in the absence of AraC was similar (HFF) or even lower (HaCat) than for Ad5SL in the presence of AraC. However, it should be noted that we did not observe a complete block of Ad replication by AraC treatment in HaCat cells (Supplementary Fig. S1E). In contrast, luciferase activity after infection with Ad5TyrTL was higher in HFF cells and only slightly reduced in HaCat cells compared with Ad5TL and also substantially higher than for Ad5TyrIL and Ad5TyrSL. Quantification of luciferase mRNA expression kinetics corroborated these results (Fig. 4A): Regulation of E1A expression from the tyrosinase promoter caused a substantial reduction in luciferase mRNA copy numbers in HaCat cells for IL and SL viruses, but not for TL viruses. Specifically, luciferase mRNA expression by Ad5TyrTL was similar to that by Ad5TL and substantially higher than those for Ad5TyrIL and Ad5TyrSL. For Ad5TyrSL, luciferase mRNA copy numbers were less than for Ad5SL after AraC treatment (Fig. 4B). The kinetics of luciferase mRNA expression nicely correlated with fiber mRNA expression (Supplementary Fig. S1C), which was expected as expression of these genes was linked for all three transgene insertion strategies. Ad5SL showed lower luciferase activity than Ad5IL in HFF and HaCat cells (Fig. 3A). Correspondingly, luciferase mRNA copy numbers were lower for the SL viruses than for the matching IL viruses (Fig. 4A). However, this was not the case for fiber mRNA copy numbers (Supplementary Fig. S1C), indicating that the SA strategy does not harm viral replication and gene expression but mediates less efficient transgene expression than the IRES strategy. The latter was also observed in melanoma cells, however, to a lower extent (Fig. 3A). When comparing melanoma and normal cells, the selectivity of transgene expression was between 280-fold and 1,530-fold for Ad5TyrSL and between 45-fold and 450-fold for Ad5TyrIL (Fig. 3B, compare left panel with right panel). Note that the melanoma selectivity of transgene expression by the TL viruses reflects attenuated replication and transgene expression of Ad5TL in melanoma cells and not indirect transcriptional targeting, as Ad5TyrTL causes strong transgene expression in HaCat and HFF cells.
FIG. 4.
Luciferase mRNA expression kinetics by transcriptionally targeted oncolytic Ads with luciferase reporter gene inserted into the late transcription unit. Keratinocytes ([A and B] HaCat) or melanoma cells ([C] SK-MEL-28) were infected with Ads at 10 TCID50 per cell in the presence or absence of AraC. Cells were harvested at indicated time points post-infection, and luciferase mRNA was quantified by qPCR. The results were reproduced in an independent experiment.
These results show that indirect transcriptional targeting of transgene expression by the tyrosinase promoter is lost when the transgene is fused via the T2A sequence to the fiber gene, even though late expression is observed when virus replication is blocked artificially. Indirect transcriptional targeting was effective for the IRES and SA strategies for transgene insertion. The IRES strategy mediated the strongest transgene expression, whereas the SA strategy implemented the highest degree of specificity for indirect promoter regulation. As the Ad5TyrSL virus also showed the highest selectivity of oncolysis (Fig. 2), we decided to further investigate indirect transcriptional targeting of transgene expression with the SL viruses. We showed by qPCR and immunoblot analyses that E1A, fiber, and E4 expression (note that the E4 locus is located next to the transgene insertion site) and viral genome replication are very similar for Ad5SL and parental Ad5Δ34E3− (Supplementary Fig. S1D–F). Moreover, expressions of luciferase, E1A, and fiber genes as well as viral DNA replication were similar for Ad5TyrSL and Ad5SL in melanoma cells (Fig. 4C and Supplementary Fig. S1D and E). These results are in agreement with the results of the oncolysis and burst assays (Fig. 2) demonstrating that virus replication in melanoma cells is not affected by transgene insertion via the SA site into the late transcription unit and also not or only minimally by replacement of the E1A promoter with the tyrosinase promoter. In consideration of the aforementioned results, we propose the insertion of therapeutic genes via SA into oncolytic Ads as an effective strategy for indirect transcriptional targeting of therapeutic gene activity. To evaluate this approach in a therapeutic setting, we next analyzed transcriptional targeting of genetic prodrug activation to melanoma cells using a tyrosinase promoter–driven oncolytic Ad with the FCU1 suicide gene inserted via the SA site.
Expression of therapeutic genes by oncolytic Ads via alternative splicing depends on the transgene and might require sequence optimization
We first investigated whether the FCU1/5-FC prodrug activation system is effective in melanoma cells. Six tested melanoma cell lines showed sensitivity to 5-FU with IC50 values between 3 μM and 10 μM (Table 1). Melanoma cells were strongly sensitized to 5-FC by transduction of a fraction of cells with replication-deficient Ad5CMVFCU, a replication-deficient FCU1-encoding Ad (Erbs et al., 2000). IC50 values were similar to direct treatment with 5-FU. IC50 values for Ad5CMVFCU + 5-FU were lower than for 5-FU or Ad5Luc1 + 5-FU, confirming UPRT activity of FCU1. Ad5CMVFCU + 5-FC showed higher cytotoxicity than 5-FU or Ad5Luc1 + 5-FU in one of two tested cell lines. Overall, these results confirm the efficiency of prodrug activation by FCU1 in melanoma cells. Next, we generated replication-competent viruses Ad5SFCU and Ad5TyrSFCU by inserting FCU1 via SA into the late Ad transcription unit (replacing the luciferase gene of Ad5SL and Ad5TyrSL, respectively, Fig. 5). To our surprise, neither Ad5TyrSFCU (Fig. 6A) nor Ad5SFCU (data not shown) showed any detectable expression of the FCU1 fusion protein after infection of melanoma cells, even though the fiber gene was expressed (Fig. 6B for Ad5TyrSFCU) and lytic activity was not affected (see Fig. 7 for Ad5TyrSFCU). As their genomes outside the FCU1 gene matched those of Ad5SL and Ad5TyrSL, we speculated that the different nucleotide sequence downstream of the SA in Ad5SFCU and Ad5TyrSFCU interfered with proper splicing. Therefore, we generated Ad5Ssp_FCU and Ad5TyrSsp_FCU, which contained an SA nucleotide environment more similar to the Ad5SL and Ad5TyrSL viruses, by inserting a corresponding 51-bp spacer between the SA and the FCU1 start codon (Figs. 5 and 6C). However, these viruses also showed no or marginal FCU1 protein expression (Fig. 6A and data not shown). We then analyzed FCU1 mRNA expression and splicing in Ad5TyrSFCU-infected SK-MEL-28 cells. By RT-PCR using a forward primer binding in the tripartite leader and a reverse primer binding in the 3' end of the FCU1 gene, a 1,185-bp product was expected if splicing was correct; however, we obtained a band of approximately 550 bp (Fig. 6C and D). Sequencing of the PCR products revealed a cryptic SA site at position 594 of the FCU1 gene that prevented synthesis of the proper fusion protein-encoding mRNA. This position was also found in a search for splice sites using EMBnet/Scientific Computing Service software. In light of these results we generated Ad5Ssp_mFCU and Ad5TyrSsp_mFCU by changing the sequence surrounding the cryptic SA site within the FCU1 gene with silent mutations (Figs. 5 and 6C). After infection of SK-MEL-28 cells, these viruses expressed the FCU1 fusion protein (Fig. 6A). FCU1 expression was stronger for Ad5TyrSsp_mFCU than for Ad5Ssp_mFCU, which corresponded to the strength of luciferase expression observed for Ad5TyrSL and Ad5SL (Fig. 3). Viruses with mutated FCU1 gene but without spacer (Ad5TyrS_mFCU and Ad5S_mFCU, Fig. 5) showed lower FCU1 expression than Ad5TyrSsp_mFCU and Ad5Ssp_mFCU (data not shown), indicating that the combination of cryptic splice site mutation and spacer ensured the strongest transgene expression. As expected, expression of fiber protein by Ad5TyrSsp_mFCU and Ad5Ssp_mFCU was dependent on virus replication and was specific for melanoma cells (Fig. 6A and B). Immunoblot analysis was not sensitive enough to detect FCU1 protein in HaCat cells; however, we could demonstrate by quantitative RT-PCR that FCU1 mRNA expression by Ad5TyrSsp_mFCU was highly melanoma-specific (Fig. 6E). We conclude that insertion of the prodrug convertase fusion gene FCU1 via SA into the late transcription unit of the oncolytic Ad Ad5Tyr requires sequence optimization but ultimately results in transcriptionally targeted FCU1 expression.
Table 1.
IC50 Values of 5-FU and 5-FC for Melanoma Cells Transduced with Ad5Luc1 or Ad5CMVFCU-1 or Mock-Infected
| |
IC50 (μM) |
|||||
|---|---|---|---|---|---|---|
| SK-MEL-28 | Colo829 | Mel624 | Mel888 | A375M | C8161 | |
| 5-FU | 10.1 | 10 | 3.3 | 7.8 | 5.7 | 8.6 |
| 5-FC | >10,000 | >10,000 | >10,000 | >10,000 | >10,000 | >10,000 |
| Ad5Luc1/5-FU | 9.7 | 14.5 | ||||
| Ad5Luc1/5-FC | >10,000 | >10,000 | ||||
| Ad5CMVFCU/5-FU | 3.9 | 1.6 | ||||
| Ad5CMVFCU/5-FC | 45 | 2.5 | ||||
Titers of Ad vectors were 2 TCID50 per cell, resulting in transduction of <18% or <6% of cells for SK-MEL-28 or Colo829, respectively.
FIG. 6.
Expression of FCU1 inserted into the late transcription unit via SA requires sequence optimization. (A) SK-MEL-28 cells were infected at 100 TCID50 per cell, and expression of FCU1 was detected by immunoblot 2 days post-infection. As a loading control α-tubulin was detected. Ad5CMVFCU is an E1/E3-deleted replication-deficient Ad with a CMV-FCU1 expression cassette. (B) SK-MEL-28 cells were infected at 10 TCID50 per cell in the absence or presence of AraC, and fiber expression was detected by immunoblot 1 day post-infection. As a loading control β-actin was detected. (C) Schematic outline of the FCU1 gene insertion via SA into the late virus transcription unit and sequence optimization in Ad5Ssp_mFCU/Ad5TyrSsp_mFCU. Numbers indicate nucleotide positions in the Ad5 genome or FCU1 gene. The non-coding spacer corresponds to nucleotides 4–54 of the luciferase gene. The original FCU1 gene contains the nucleotide sequence of the cryptic SA site TACTCCGATAGAATCATCAGA (FCU_cryp. SA) that was mutated to TATAGCGACCGGATTATTCGG (FCU_ mut cryp. SA). (D) RT-PCR analysis of FCU1 mRNA expression by SK-MEL-28 harvested 1 day post-infection with Ads at 100 TCID50 per cell and using the primer pair indicated in (C). The correct FCU1-encoding splice product is approximately 1,200 bp. (E) qPCR analysis of FCU1 mRNA expression in melanoma cells and keratinocytes harvested 1 day post-infection with Ads at 10 TCID50 per cell. GAPDH, glyceraldehyde 3-phosphate dehydrogenase; n.s., not significant.
FIG. 7.
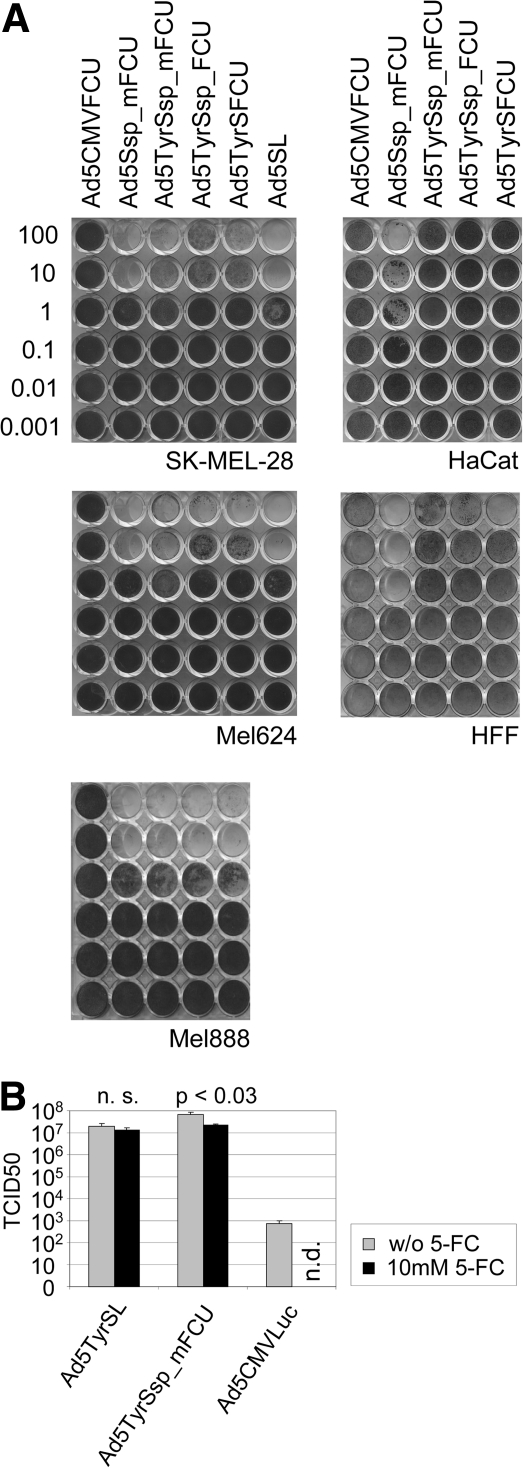
Oncolytic activity and prodrug effect on virus burst of Ad constructs with the FCU1 gene inserted via SA into the late transcription unit. (A) Melanoma cells (SK-MEL-28, Mel624, Mel888), keratinocytes (HaCat), or primary fibroblasts (HFF) were infected with Ads, and cytotoxicity was determined by crystal violet staining of surviving cells. Numbers are viral titers in TCID50 per cell used for infection. Ad5CMVFCU is an E1/E3-deleted replication-deficient Ad with a CMV-FCU1 expression cassette. (B) Infectious virus particle production by 5 × 104 SK-MEL-28 cells at 3 days post-infection with Ad5TyrSL or Ad5TyrSsp_FCU in the presence or absence (w/o) of the prodrug 5-FC. Ad5CMVLuc is an E1/E3-deleted replication-deficient Ad used as the control. n.d., not determined; n.s., not significant.
Combining Ad oncolysis with tumor-directed chemotherapy by indirect transcriptional targeting of genetic prodrug activation
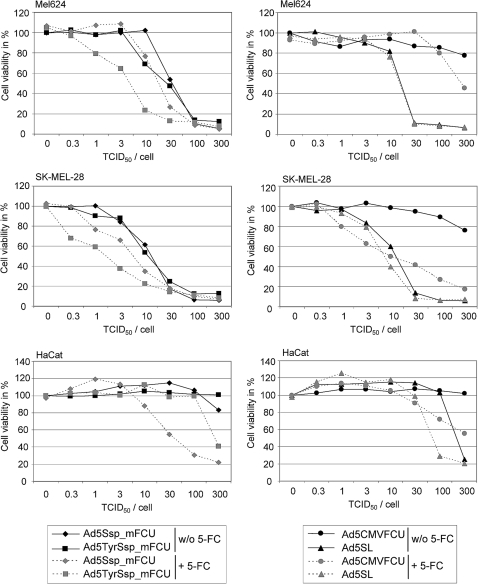
In a cytotoxicity assay we first assessed melanoma selectivity of cell lysis and spread by Ad5TyrSsp_mFCU (Fig. 7). Lysis and spread in melanoma cells were similar for Ad5TyrSsp_mFCU, Ad5Ssp_mFCU, and Ad5SL. In HFF and HaCat cells, lysis and spread by Ad5TyrSsp_mFCU were attenuated >2 orders of magnitude compared with Ad5Ssp_mFCU. These results show that insertion of the mutated FCU1 gene, SA, and spacer did not interfere with selective replication and cell lysis of the virus. Furthermore, results of a virus burst assay (Fig. 7B) show that application of 5-FC was compatible with the production of infectious particles of Ad5TyrSsp_mFCU at high titers in melanoma cells, although virus yields were reduced approximately threefold (1.5-fold for control virus Ad5TyrSL not encoding FCU1). We next investigated a combination therapy of viral oncolysis and tumor-targeted prodrug activation mediated by Ad5TyrSsp_mFCU/5-FC. Therefore, we infected melanoma cells SK-MEL-28 and Mel624 and HaCat cells with dilutions of Ad5TyrSsp_mFCU, Ad5Ssp_mFCU, Ad5SL, or replication-deficient Ad5CMVFCU, with the latter expressing FCU1 from the strong CMV enhancer/promoter. The prodrug 5-FC or buffer was added 2 days post-infection, thus before virus spread, and cytotoxicity was determined 4 days later (Fig. 8). As expected, the replication-competent Ads Ad5Ssp_mFCU, Ad5TyrSsp_mFCU, and Ad5SL showed similar dose-dependent cytotoxicity in melanoma cells, which was not observed for replication-deficient AdCMVFCU. Addition of 5-FC resulted in increased cytotoxicity for all FCU1-encoding Ads, but not for Ad5SL. In both melanoma cells, the strongest cytotoxicity was observed for Ad5TyrSsp_mFCU in the presence of 5-FC, which reduced the viral IC50 dose approximately 10-fold. The stronger prodrug effect in comparison with Ad5Ssp_mFCU corresponds with the higher expression of FCU1 by Ad5TyrSsp_mFCU (Fig. 6A). In contrast to melanoma cells, the untargeted Ad5Ssp_mFCU + 5-FC showed the strongest cytotoxicity in HaCat cells, which was in large part prodrug-dependent. For Ad5TyrSsp_mFCU no cytotoxicity was observed in the absence of 5-FC, and only at the highest virus titer did we observe a prodrug effect, which was much reduced compared with Ad5Ssp_mFCU + 5-FC. Note that in HaCat cells 5-FC mediated also an increase in cytotoxicity of Ad5SL, which does not encode the prodrug convertase. We conclude that Ad5TyrSsp_mFCU induces melanoma-selective oncolysis and selectively enhanced cytotoxicity in combination with 5-FC.
FIG. 8.
Combined toxicity of oncolysis and prodrug activation by Ad constructs with the FCU1 gene inserted via SA into the late transcription unit. Melanoma cells (SK-MEL-28, Mel624) or keratinocytes (HaCat) were infected with Ads at indicated titers or were mock-infected in triplicates. 5-FC or medium was added 2 days post-infection at 10 mM (SK-MEL-28, HaCat) or at 5 mM (Mel624). Cytotoxicity was determined 6 days post-infection by staining of surviving cells with crystal violet. Cell content was determined by measuring optical density at 595 nm, and cell viability was plotted in percentage of mock-infected cells that did not obtain 5-FC. Mean values of triplicates are shown. For clarity of presentation, SDs are not shown and were below 16.6%. In HaCat cells, cytotoxicity of Ad5TyrSsp_mFCU was significantly lower (p < 0.05) than for Ad5Ssp_mFCU at titers higher than 3 TCID50 and in the presence of 5-FC. In contrast, cytotoxicity of Ad5TyrSsp_mFCU was significantly higher (p < 0.05) than for Ad5Ssp_mFCU in SK-MEL-28 and Mel624 at titers between 0.3 and 30 TCID50 and 1 and 30 TCID50, respectively, in the presence of 5-FC. Cell viability was significantly reduced by 5-FC (p < 0.05) for Ad5CMVFCU at 100 and 300 TCID50 (HaCat), at titers higher than 1 TCID50 (SK-MEL-28), or at 300 TCID50 (Mel624); for Ad5SL at 30 and 100 TCID50 (HaCat) or at 30 TCID50 (SK-MEL-28); for Ad5Ssp_mFCU at titers >3 TCID50 (HaCat), at 1–10 and 100 TCID50 (SK-MEL-28), or at 10 and 30 TCID50 (Mel624); and for Ad5TyrSsp_mFCU at 300 TCID50 (HaCat), 0.3– 30 TCID50 (SK-MEL-28), or 1, 3, 30, and 300 TCID50 (Mel624).
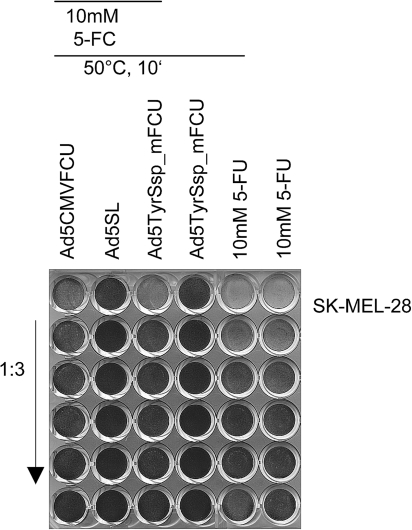
When combined with viral oncolysis, a bystander effect of genetic prodrug activation is essential in order to kill those tumor cells that the lytic virus cannot reach, for example, because of connective tissue barriers in the tumor. We therefore investigated whether Ad5TyrSsp_mFCU + 5-FC has the potential to induce bystander killing in melanoma cells (Fig. 9). Supernatants of Ad5TyrSsp_mFCU-infected and 5-FC-treated SK-MEL-28 cells, after virus inactivation by heat treatment, showed a clear cytotoxicity for uninfected SK-MEL-28 cells. Cytotoxicity of supernatants was nearly as efficient as the drug 5-FU at 10 mM. As the prodrug 5-FC was added at 10 mM to the Ad5TyrSsp_mFCU-infected cells, these results demonstrate efficient activation of the prodrug and release of the drug into the media by Ad5TyrSsp_mFCU-infected SK-MEL-28 cells. No bystander killing was observed for Ad5SL/5-FC or for Ad5TyrSsp_mFCU without 5-FC, demonstrating that virus particles in the supernatants were effectively inactivated by heat treatment and that the detected cytotoxicity was dependent on prodrug activation.
FIG. 9.
Bystander effect of prodrug activation by Ad constructs with the FCU1 gene inserted via SA into the late transcription unit. SK-MEL-28 cells were infected with Ad5TyrSsp_mFCU, Ad5CMVFCU, or Ad5SL at 10 TCID50 per cell. 5-FC (10 mM) was added 2 days post-infection. As a control, further Ad5TyrSsp_mFCU-infected cells were treated with medium alone. Supernatants were harvested 3 days post-infection and were incubated for 10 min at 50°C for virus inactivation. Then, each heat-treated supernatant was added in a threefold dilution series to fresh SK-MEL-28 cell monolayers. In parallel, SK-MEL-28 cells were treated with a threefold dilution series of 10 mM 5-FU (with or without heat pretreatment). The arrow indicates increasing dilutions of supernatants and 5-FU. Cytotoxicity was determined 4 days after adding the supernatant by staining of surviving cells with crystal violet.
Discussion
The development of tumor-specific drugs and the implementation of potent multimodal treatment regimens are of critical importance for cancer therapy. Oncolytic Ads have the potential to realize both tumor specificity and multimodality by restricting virus replication to tumor cells and by insertion of therapeutic genes into the virus genome, respectively. Using the luciferase gene as a sensitive reporter for monitoring transgene expression, we show in this study that expression of E1A from a cell type-specific promoter not only mediates targeted virus replication, but also facilitates the expression of transgenes inserted in the late transcription unit with remarkable selectivity (up to 1,500-fold). Notably, we reveal that selectivity and efficiency of transgene expression depend on the strategy of transgene insertion into the late transcription unit and might require optimization for individual transgenes. Specifically, our results demonstrate that both the IRES and SA strategies of transgene insertion facilitate indirect transcriptional targeting of transgene expression by tyrosinase promoter–dependent expression of E1A without interfering with virus replication. When comparing melanoma cells with fibroblasts and keratinocytes, the SA strategy allowed for the highest selectivity of up to 1,500-fold; for the IRES strategy, selectivity was up to 450-fold. However, the IRES strategy caused approximately 10-fold stronger transgene expression. In consequence, we suggest the IRES strategy for expression of therapeutic genes for which high expression levels are necessary in order to reach therapeutic efficacy, whereas the SA strategy is especially advantageous whenever side effects of therapeutic genes are limiting. Moreover, these “add-in” strategies for transgene insertion into the Ad genome, in contrast to replacing late viral genes with transgenes (Nettelbeck, 2008), in principle allow for retaining all viral genes—at least if transgene sequences are short (the viruses used in this study were E3-deleted because of the large size of the luciferase gene). This is especially true for the SA strategy, because of the short SA sequences. Both the EMCV IRES and different SA sequences have been previously used for late therapeutic gene expression (Sauthoff et al., 2002, 2006; Fuerer and Iggo, 2004; Carette et al., 2005; Lukashev et al., 2005; Cascante et al., 2007; Robinson et al., 2007). Our results suggest that comparing both strategies in therapeutic studies might indeed be of interest to improve a given therapeutic approach.
Lower transgene expression by the SA strategy observed in our study might reflect the locale of transgene insertion between the E4 genes and right ITR of the Ad genome. Other groups have inserted transgenes with different SA sites directly downstream of the fiber gene (Fuerer and Iggo, 2004; Cascante et al., 2007). Fuerer and Iggo (2004) reported that therapeutic gene expression by the EMCV IRES resulted in stronger gene expression than the SA sequence of the Ad41 long fiber, when both were inserted at the end of the L5 transcript. It remains to be investigated in direct comparisons whether this fiber locale results in more efficient transgene expression than the E4 site and whether the selectivity we report in the present study is dependent on the specific SA sequence used. However, in a transposon-based scan for insertion sites for SA–transgene cassettes in the Ad genome, Hermiston and colleagues reported that an E4 insertion site, similar to the one we used, afforded strongest transgene expression and implemented late transgene expression kinetics best (Jin et al., 2005).
An additional important finding of our study with respect to the necessity to optimize transgene insertion into oncolytic Ad genomes is that transgenes can interfere in a sequence-specific manner with splicing. Thus, efficient transgene expression was lost by replacing the luciferase cDNA of SL viruses with the FCU1 cDNA. Interestingly, this was not only due to insufficient activity of the inserted SA sequence (caused by a different sequence downstream of the SA site), but was also resulting from usage of a cryptic SA site within the transgene. In this regard, it is important to note that the SA sequence used in our study, containing branchpoint and polypyrimidine tract elements, was already optimized for more “tight” splicing than shorter sequences (Jin et al., 2005). Having revealed the reasons for ineffective splicing, we could restore FCU1 expression by insertion of a luciferase gene-derived spacer downstream of the SA sequence in combination with switching codons in and around the identified cryptic splice site. The former might restore a favorable sequence environment around the SA site. One could envision that cryptic splicing of transgenes could also interfere with viral gene expression and thus with virus replication, although this was not the case for the viruses generated in this study. These results suggest that, when “arming” viruses with therapeutic genes, it is advisable to perform splicing analyses.
To optimize transgene insertion into oncolytic Ad genomes, we also investigated a viral 2A sequence as a tool for co-expression of two or more proteins from one transcription unit (Szymczak et al., 2004). We show that the short 2A sequence of the T. asigna virus facilitates replication-dependent co-expression of Ad fiber and luciferase proteins as observed via inhibition of virus replication with AraC, with no fusion protein detectable. However, our data also reveal that indirect promoter control was not effective using the 2A strategy, as Ad5TyrTL, in contrast to Ad5TyrIL and Ad5TyrSL, showed strong luciferase expression in fibroblasts and keratinocytes. Thus, an important result of our study is that replication dependence of transgene expression, as demonstrated, for example, by AraC block, does not always correctly predict selectivity of transgene expression by targeted oncolytic Ads. In consequence, it is necessary to investigate selectivity of armed oncolytic Ads in a biologically relevant system (e.g., in both target and non-target cells) with an appropriate control of a matching but non-targeted, transgene-encoding virus. This was frequently not done in previous studies. For reasons that remain to be determined, the 2A strategy also resulted in reduced E1A expression, replication, and transgene expression for the Ad5TL virus, but not the Ad5TyrTL virus. Iggo and co-workers recently reported feasibility of 2A sequences derived from the foot-and-mouth disease virus (F2A) and from the porcine teschovirus-1 (P2A) for co-expression of green fluorescent protein with Ad pIX (Funston et al., 2008). They also observed reduced replication and spread for the 2A viruses, which they attributed to interference with pIX activity of the 2A-derived amino acids that remain in the pIX protein. In our study both Ad5TL and Ad5TyrTL feature the same fiber modification but a different replication phenotype. Thus, interference of the 2A-derived amino acids with capsid stability or virus entry into cells is unlikely. We conclude that the 2A strategy, facilitating efficient co-expression in other systems (Szymczak et al., 2004; Fang et al., 2005), can affect both replication and targeted transgene expression in armed oncolytic Ads.
Iggo and co-workers previously investigated selective expression of therapeutic genes inserted via IRES or splice site into the viral late transcription unit of an Ad transcriptionally targeted to colon cancer via TCF4 transcription factor binding sites in E1 and E4 promoters (Fuerer and Iggo, 2004; Lukashev et al., 2005). Comparing tumor and normal cells, they could also demonstrate tumor-selective expression of the therapeutic protein by immunoblot; however, they did not quantify the level of specificity. Other studies reporting late therapeutic gene insertion into transcriptionally targeted oncolytic Ads frequently lacked a proof of indirect promoter regulation of transgene expression.
We report that the SA strategy, after optimizing splice-dependent FCU1 expression, facilitates the transcriptionally targeted combination therapy of efficient oncolysis and molecular chemotherapy. For Ad5TyrSsp_mFCU, production of infectious virus particles was only modestly reduced (less than fourfold) by 5-FC, showing that late expression of FCU1 in the presence of 5-FC is compatible with virus replication, which is in agreement with previous reports in other tumor cells (Fuerer and Iggo, 2004). It is important that cytotoxicity of the oncolytic virus was strongly increased by 5-FC in a melanoma-specific manner and was superior to Ad5CMVFCU treatment in melanoma cells; for Mel624 cells this difference was quite remarkable, more than 30-fold. It is interesting that we observed that Ad5TyrSsp_mFCU showed higher FCU1 fusion protein expression in melanoma cells than Ad5Ssp_mFCU, which translated in stronger sensitization to the prodrug 5-FC. This was in agreement with the results of luciferase reporter assays for IL and SL viruses, which also showed stronger luciferase expression when E1A was expressed from the tyrosinase promoter. As E1A expression kinetics of Ad5SL and Ad5TyrSL were similar during the first 30 hr after infection of melanoma cells, the reason for this difference is not clear and might be due to different expression kinetics at later times. Our results clearly establish the efficiency and specificity of the Ad5TyrSsp_mFCU/5-FC therapy for melanoma, but there is further opportunity for improvement. We always added the prodrug during the first virus replication cycle after infection in order to show the selectivity of the system. However, an important advantage of the genetic prodrug activation therapy is that the timing of gene transfer and prodrug application can be optimized. This is especially of interest for armed oncolytic viruses: Both efficacy and selectivity of oncolytic virus-delivered genetic prodrug therapy can be improved by adding the prodrug later after infection, allowing for (tumor-specific) virus spread and thus more widespread expression of the prodrug convertase. Our observation that supernatants of Ad5TyrSsp_mFCU/5-FC-treated melanoma cells, after virus inactivation, can efficiently kill uninfected melanoma cells demonstrates the potential of the system for effective prodrug conversion and for killing of uninfected “bystander” tumor cells. This bystander effect might be brought about by diffusion of membrane-permeable 5-FU, produced by CD activity of FCU1, and/or by viral cell lysis-dependent release of 5-FU and 5-fluoro-deoxyUMP, the membrane-impermeable product of FCU1 UPRT activity. It is notable that a potent bystander effect is of critical importance for combination therapies with “armed” oncolytic Ad, as the killing of uninfected cells is the goal of the arming strategy. Together with a previous report on a vaccinia virus encoding FCU1 (Foloppe et al., 2008) and a recent article on treatment of head and neck cancer with a FCU1-encoding oncolytic Ad (Dias et al., 2010), our study demonstrates the potency of FCU1-mediated genetic prodrug activation for “arming” of oncolytic viruses. It is the first study to show tumor cell selectivity of FCU1 expression by oncolytic Ads, of prodrug activation, and of combination therapy.
Previous studies investigated oncolytic Ads expressing Escherichia coli CD directly from tyrosinase promoters, either by expressing E1A and CD from two identical copies of the promoter (Liu and Deisseroth, 2006) or by expressing a CD-IRES-E1A fusion gene (Liu et al., 2006). In agreement with our results, these studies also reported synergy between Ad oncolysis and CD/5-FC therapy. However, the virus described in the present study possesses qualities that differ from these viruses in several aspects. It encodes the optimized yeast CD-UPRT fusion protein FCU1, which has been reported to be a more effective prodrug convertase (Erbs et al., 2000), which might be the basis of the efficient bystander effect we observed. This protein is expressed with late kinetics, which is advantageous to prevent interference with viral replication. A single copy of the tyrosinase promoter ensures stability of the virus genome by avoiding homologous recombination. Direct control and thus strong expression of E1A ensure efficient virus replication and oncolysis, whereas indirect control of late gene expression still facilitates melanoma-specific expression of the CD-UPRT fusion protein FCU1.
5-FU is not used in clinical routine for chemotherapy of malignant melanoma. However, we believe that local application of the drug at high doses, as we propose by targeted prodrug activation after infection with “armed” oncolytic Ads and further conversion to the active form 5-fluoro-deoxyUMP by FCU1, holds promise as a novel regimen for targeted treatment of malignant melanoma. This is in accord with previous reports on CD/5-FC gene therapy by nonviral, viral, bacterial, or cellular vectors (Szala et al., 1996; Cao et al., 1998; Aboody et al., 2006; Kucerova et al., 2008; Stritzker et al., 2008). Also, several studies indicate that treatment of melanoma with 5-FU improves cellular natural and therapeutic immune responses to the tumor (Ryan et al., 1988; Neefe and Glass, 1991; Gazit et al., 1992; Cao et al., 1998; Yang and Haluska, 2004). In this regard, activation of antitumor immunity by oncolytic viruses is considered a promising strategy to complement viral tumor cell lysis for increased and prolonged therapeutic activity. Finally, the Ad5TyrSA–transgene format developed in this study represents a platform for efficient and targeted delivery also of other genetic prodrug activation systems or different therapeutic genes to malignant melanoma.
In summary, our study establishes Ad5TyrSsp_mFCU/5-FC as an effective strategy for targeted killing of melanoma cells by combined viral oncolysis and molecular chemotherapy. We report several steps for genetic engineering of the therapeutic virus that were required to optimize late therapeutic gene expression. This, in turn, facilitated targeting of both viral replication and therapeutic gene expression to melanoma cells from the cellular tyrosinase promoter.
Supplementary Material
Acknowledgments
We thank Jacques Banchereau (Baylor University, Dallas, TX), David T. Curiel (University of Alabama at Birmingham, Birmingham, AL), Isaiah J. Fidler (Houston, TX), Jeffrey Schlom (National Institutes of Health, Bethesda, MD), M. Marschall (University of Erlangen, Erlangen, Germany), Bert Vogelstein (Johns Hopkins University, Baltimore, MD), and Danny Welch (University of Alabama at Birmingham) for research materials and Stefanie Sandmann for technical assistance. This work was supported by the Helmholtz Association of National Research Centers (Helmholtz-University Group grant), the Deutsche Krebshilfe (German Cancer Aid, grant 10-7946), the Wilhelm Sander-Stiftung (grant 2003.118.1), the German-Israeli Foundation (grant I-817-38.11/2004), and the Monika Kutzner-Stiftung.
Author Disclosure Statement
For C.Q., S.R., I.F-U., A.H., S.E., M.B., A.H.E., and D.M.N. no competing financial interests exist. P.E. is an employee of Transgene, S.A., owns stock options in this company, and is an inventor on patents and patent applications assigned to Transgene, S.A. Transgene, S.A. holds a patent on FCU1 with P.E. as an inventor.
References
- Aboody K.S. Najbauer J. Schmidt N.O., et al. Targeting of melanoma brain metastases using engineered neural stem/progenitor cells. Neuro-Oncol. 2006;8:119–126. doi: 10.1215/15228517-2005-012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alemany R. Balague C. Curiel D.T. Replicative adenoviruses for cancer therapy. Nat. Biotechnol. 2000;18:723–727. doi: 10.1038/77283. [DOI] [PubMed] [Google Scholar]
- Banerjee N.S. Rivera A.A. Wang M., et al. Analyses of melanoma-targeted oncolytic adenoviruses with tyrosinase enhancer/promoter-driven E1A, E4, or both in submerged cells and organotypic cultures. Mol. Cancer Ther. 2004;3:437–449. [PubMed] [Google Scholar]
- Berget S.M. Moore C. Sharp P.A. Spliced segments at the 5' terminus of adenovirus 2 late mRNA. Proc. Natl. Acad. Sci. U.S.A. 1977;74:3171–3175. doi: 10.1073/pnas.74.8.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boukamp P. Petrussevska R.T. Breitkreutz D., et al. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J. Cell Biol. 1988;106:761–771. doi: 10.1083/jcb.106.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X. Ju D.W. Tao Q., et al. Adenovirus-mediated GM-CSF gene and cytosine deaminase gene transfer followed by 5-fluorocytosine administration elicit more potent antitumor response in tumor-bearing mice. Gene Ther. 1998;5:1130–1136. doi: 10.1038/sj.gt.3300727. [DOI] [PubMed] [Google Scholar]
- Carette J.E. Graat H.C. Schagen F.H., et al. Replication-dependent transgene expression from a conditionally replicating adenovirus via alternative splicing to a heterologous splice-acceptor site. J. Gene Med. 2005;7:1053–1062. doi: 10.1002/jgm.754. [DOI] [PubMed] [Google Scholar]
- Cascante A. Abate-Dage D. Garcia-Rodriguez L., et al. GCV modulates the antitumoural efficacy of a replicative adenovirus expressing the Tat8-TK as a late gene in a pancreatic tumour model. Gene Ther. 2007;14:1471–1480. doi: 10.1038/sj.gt.3303008. [DOI] [PubMed] [Google Scholar]
- Chow L.T. Gelinas R.E. Broker T.R. Roberts R.J. An amazing sequence arrangement at the 5' ends of adenovirus 2 messenger RNA. Cell. 1977;12:1–8. doi: 10.1016/0092-8674(77)90180-5. [DOI] [PubMed] [Google Scholar]
- Davis J.J. Wang L. Dong F., et al. Oncolysis and suppression of tumor growth by a GFP-expressing oncolytic adenovirus controlled by an hTERT and CMV hybrid promoter. Cancer Gene Ther. 2006;13:720–723. doi: 10.1038/sj.cgt.7700944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias J.D. Liikanen I. Guse K., et al. Targeted chemotherapy for head and neck cancer with a chimeric oncolytic adenovirus coding for bifunctional suicide protein FCU1. Clin. Cancer Res. 2010;16:2540–2549. doi: 10.1158/1078-0432.CCR-09-2974. [DOI] [PubMed] [Google Scholar]
- Donnelly M.L. Luke G. Mehrotra A., et al. Analysis of the aphthovirus 2A/2B polyprotein 'cleavage' mechanism indicates not a proteolytic reaction, but a novel translational effect: A putative ribosomal 'skip.'. J. Gen. Virol. 2001;82:1013–1025. doi: 10.1099/0022-1317-82-5-1013. [DOI] [PubMed] [Google Scholar]
- Erbs P. Regulier E. Kintz J., et al. In vivo cancer gene therapy by adenovirus-mediated transfer of a bifunctional yeast cytosine deaminase/uracil phosphoribosyltransferase fusion gene. Cancer Res. 2000;60:3813–3822. [PubMed] [Google Scholar]
- Fang J. Qian J.J. Yi S., et al. Stable antibody expression at therapeutic levels using the 2A peptide. Nat. Biotechnol. 2005;23:584–590. doi: 10.1038/nbt1087. [DOI] [PubMed] [Google Scholar]
- Foloppe J. Kintz J. Futin N., et al. Targeted delivery of a suicide gene to human colorectal tumors by a conditionally replicating vaccinia virus. Gene Ther. 2008;15:1361–1371. doi: 10.1038/gt.2008.82. [DOI] [PubMed] [Google Scholar]
- Fuerer C. Iggo R. 5-Fluorocytosine increases the toxicity of Wnt-targeting replicating adenoviruses that express cytosine deaminase as a late gene. Gene Ther. 2004;11:142–151. doi: 10.1038/sj.gt.3302148. [DOI] [PubMed] [Google Scholar]
- Fueyo J. Gomez M.C. Alemany R., et al. A mutant oncolytic adenovirus targeting the Rb pathway produces anti-glioma effect in vivo. Oncogene. 2000;19:2–12. doi: 10.1038/sj.onc.1203251. [DOI] [PubMed] [Google Scholar]
- Funston G.M. Kallioinen S.E. de Felipe P., et al. Expression of heterologous genes in oncolytic adenoviruses using picornaviral 2A sequences that trigger ribosome skipping. J. Gen. Virol. 2008;89:389–396. doi: 10.1099/vir.0.83444-0. [DOI] [PubMed] [Google Scholar]
- Gazit Z. Weiss D.W. Shouval D., et al. Chemo-adoptive immunotherapy of nude mice implanted with human colorectal carcinoma and melanoma cell lines. Cancer Immunol. Immunother. 1992;35:135–144. doi: 10.1007/BF01741861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He T.C. Zhou S. da Costa L.T., et al. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. U.S.A. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heise C. Hermiston T. Johnson L., et al. An adenovirus E1A mutant that demonstrates potent and selective systemic anti-tumoral efficacy. Nat. Med. 2000;6:1134–1139. doi: 10.1038/80474. [DOI] [PubMed] [Google Scholar]
- Hermiston T.W. Kuhn I. Armed therapeutic viruses: Strategies and challenges to arming oncolytic viruses with therapeutic genes. Cancer Gene Ther. 2002;9:1022–1035. doi: 10.1038/sj.cgt.7700542. [DOI] [PubMed] [Google Scholar]
- Jin F. Kretschmer P.J. Hermiston T.W. Identification of novel insertion sites in the Ad5 genome that utilize the Ad splicing machinery for therapeutic gene expression. Mol. Ther. 2005;12:1052–1063. doi: 10.1016/j.ymthe.2005.07.696. [DOI] [PubMed] [Google Scholar]
- Johnson L. Shen A. Boyle L., et al. Selectively replicating adenoviruses targeting deregulated E2F activity are potent, systemic antitumor agents. Cancer Cell. 2002;1:325–337. doi: 10.1016/s1535-6108(02)00060-0. [DOI] [PubMed] [Google Scholar]
- Krasnykh V.N. Mikheeva G.V. Douglas J.T. Curiel D.T. Generation of recombinant adenovirus vectors with modified fibers for altering viral tropism. J. Virol. 1996;70:6839–6846. doi: 10.1128/jvi.70.10.6839-6846.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucerova L. Matuskova M. Pastorakova A., et al. Cytosine deaminase expressing human mesenchymal stem cells mediated tumour regression in melanoma bearing mice. J. Gene Med. 2008;10:1071–1082. doi: 10.1002/jgm.1239. [DOI] [PubMed] [Google Scholar]
- Liu Y. Deisseroth A. Oncolytic adenoviral vector carrying the cytosine deaminase gene for melanoma gene therapy. Cancer Gene Ther. 2006;13:845–855. doi: 10.1038/sj.cgt.7700962. [DOI] [PubMed] [Google Scholar]
- Liu Y. Ye T. Sun D., et al. Tumor-specific therapeutic effect induced by an oncolytic adenoviral vector containing heat shock protein 70 and prodrug activation genes. Gene Ther. 2006;13:1235–1243. doi: 10.1038/sj.gt.3302776. [DOI] [PubMed] [Google Scholar]
- Lukashev A.N. Fuerer C. Chen M.J., et al. Late expression of nitroreductase in an oncolytic adenovirus sensitizes colon cancer cells to the prodrug CB1954. Hum. Gene Ther. 2005;16:1473–1483. doi: 10.1089/hum.2005.16.1473. [DOI] [PubMed] [Google Scholar]
- Neefe J.R. Glass J. Abrogation of interferon-induced resistance to interferon-activated major histocompatibility complex-unrestricted killers by treatment of a melanoma cell line with 5-fluorouracil. Cancer Res. 1991;51:3159–3163. [PubMed] [Google Scholar]
- Nettelbeck D.M. Cellular genetic tools to control oncolytic adenoviruses for virotherapy of cancer. J. Mol. Med. 2008;86:363–377. doi: 10.1007/s00109-007-0291-1. [DOI] [PubMed] [Google Scholar]
- Nettelbeck D.M. Jerome V. Muller R. A dual specificity promoter system combining cell cycle-regulated and tissue-specific transcriptional control. Gene Ther. 1999;6:1276–1281. doi: 10.1038/sj.gt.3300943. [DOI] [PubMed] [Google Scholar]
- Nettelbeck D.M. Rivera A.A. Balague C., et al. Novel oncolytic adenoviruses targeted to melanoma: Specific viral replication and cytolysis by expression of E1A mutants from the tyrosinase enhancer/promoter. Cancer Res. 2002;62:4663–4670. [PubMed] [Google Scholar]
- Ottolino-Perry K. Diallo J.S. Lichty B.D., et al. Intelligent design: Combination therapy with oncolytic viruses. Mol. Ther. 2010;18:251–263. doi: 10.1038/mt.2009.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portsmouth D. Hlavaty J. Renner M. Suicide genes for cancer therapy. Mol. Aspects Med. 2007;28:4–41. doi: 10.1016/j.mam.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Rivera A.A. Wang M. Suzuki K., et al. Mode of transgene expression after fusion to early or late viral genes of a conditionally replicating adenovirus via an optimized internal ribosome entry site in vitro and in vivo. Virology. 2004;320:121–134. doi: 10.1016/j.virol.2003.11.028. [DOI] [PubMed] [Google Scholar]
- Robinson K.A. Ball L.E. Buse M.G. Reduction of O-GlcNAc protein modification does not prevent insulin resistance in 3T3-L1 adipocytes. Am. J. Physiol. Endocrinol. Metab. 2007;292:E884–E890. doi: 10.1152/ajpendo.00569.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez R. Schuur E.R. Lim H.Y., et al. Prostate attenuated replication competent adenovirus (ARCA) CN706: A selective cytotoxic for prostate-specific antigen-positive prostate cancer cells. Cancer Res. 1997;57:2559–2563. [PubMed] [Google Scholar]
- Ryan R.F. Krementz E.T. Litwin M.S. A role for topical 5-fluorouracil therapy in melanoma. J. Surg. Oncol. 1988;38:250–256. doi: 10.1002/jso.2930380409. [DOI] [PubMed] [Google Scholar]
- Sauthoff H. Pipiya T. Chen S., et al. Modification of the p53 transgene of a replication-competent adenovirus prevents mdm2- and E1b-55kD-mediated degradation of p53. Cancer Gene Ther. 2006;13:686–695. doi: 10.1038/sj.cgt.7700936. [DOI] [PubMed] [Google Scholar]
- Sauthoff H. Pipiya T. Heitner S., et al. Late expression of p53 from a replicating adenovirus improves tumor cell killing and is more tumor cell specific than expression of the adenoviral death protein. Hum. Gene Ther. 2002;13:1859–1871. doi: 10.1089/104303402760372954. [DOI] [PubMed] [Google Scholar]
- Siders W.M. Halloran P.J. Fenton R.G. Transcriptional targeting of recombinant adenoviruses to human and murine melanoma cells. Cancer Res. 1996;56:5638–5646. [PubMed] [Google Scholar]
- Stritzker J. Pilgrim S. Szalay A.A. Goebel W. Prodrug converting enzyme gene delivery by L. monocytogenes. BMC Cancer. 2008;8:94. doi: 10.1186/1471-2407-8-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki E. Nishimatsu H. Satonaka H., et al. Angiotensin II induces myocyte enhancer factor 2- and calcineurin/nuclear factor of activated T cell-dependent transcriptional activation in vascular myocytes. Circ. Res. 2002;90:1004–1011. doi: 10.1161/01.res.0000017629.70769.cc. [DOI] [PubMed] [Google Scholar]
- Szala S. Missol E. Sochanik A. Strozyk M. The use of cationic liposomes DC-CHOL/DOPE and DDAB/DOPE for direct transfer of Escherichia coli cytosine deaminase gene into growing melanoma tumors. Gene Ther. 1996;3:1026–1031. [PubMed] [Google Scholar]
- Szymczak A.L. Workman C.J. Wang Y., et al. Correction of multi-gene deficiency in vivo using a single 'self-cleaving' 2A peptide-based retroviral vector. Nat. Biotechnol. 2004;22:589–594. doi: 10.1038/nbt957. [DOI] [PubMed] [Google Scholar]
- Yang S. Haluska F.G. Treatment of melanoma with 5-fluorouracil or dacarbazine in vitro sensitizes cells to antigen-specific CTL lysis through perforin/granzyme- and Fas-mediated pathways. J. Immunol. 2004;172:4599–4608. doi: 10.4049/jimmunol.172.7.4599. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.