Abstract
Purpose
We previously reported an IGF gene expression signature, based upon genes induced or repressed by IGF-I, which correlated with poor prognosis in breast cancer. We tested if the IGF signature was affected by anti-IGF-IR inhibitors, and if the IGF signature correlated with response to a dual anti-IGF-IR/InsR inhibitor BMS-754807.
Experimental Design
An IGF gene expression signature was examined in human breast tumors and cell lines, and changes noted following treatment of cell lines or xenografts with anti-IGF-IR antibodies or tyrosine kinase inhibitors. Sensitivity of cells to BMS-754807 was correlated with levels of the IGF signature. Human primary tumorgrafts were analyzed for the IGF signature and IGF-IR levels and activity, and MC1 tumorgrafts treated with BMS-754807 and chemotherapy.
Results
The IGF gene expression signature was reversed in three different models (cancer cell lines or xenografts) treated with three different anti-IGF-IR therapies. The IGF signature was present in triple-negative breast cancers (TNBC) and TNBC cell lines. TNBC cell lines were especially sensitive to BMS-754807, and sensitivity was significantly correlated to expression of the IGF gene signature. The TNBC primary human tumorgraft MC1 showed high levels of both IGF-IR expression and activity, and IGF gene signature score. Treatment of MC1 with BMS-754807 showed growth inhibition and in combination with docetaxel tumor regression occurred until no tumor was palpable. Regression was associated with reduced proliferation, increased apoptosis, and mitotic catastrophe.
Conclusion
These studies provide a clear biological rationale to test anti-IGF-IR/InsR therapy in combination with chemotherapy in patients with TNBC.
Keywords: IGF-IR, triple-negative breast cancer, BMS-754807, IGF, small molecule inhibitor
Introduction
Despite many advances in the prevention, detection, and targeted therapy of breast cancer, which has resulted in a 30% decline in annual breast cancer death rates since the mid 1990s (1), it is clear that better and more effective breast cancer therapies need to be developed.
Recent molecular classification of breast cancer identified breast cancer subtypes with divergent histopathological features, clinical outcomes, and therapeutic implications (2). Breast cancer is classified into two main subgroups: estrogen-receptor alpha (ER)-positive and ER-negative (3, 4). Triple-negative breast cancers (TNBC) are characterized by low to absent expression of ER, progesterone receptor (PR) and HER-2 (5, 6) and account for up to 20-25% of all breast cancers. 60-90% of TNBC consist of basal-like breast cancers expressing genes such as cytokeratins CK5 and CK14 which are characteristic of basal epithelial cells (7).
TNBC currently has no targeted therapies, and often responds poorly to chemotherapy (8). TNBC preferentially affects younger women and African-American women, and is associated with high histological grade and aggressive clinical behavior (9). TNBC has high unmet clinical need, and novel targeted therapies need to be discovered.
Statement of Translational Relevance.
This study shows that triple negative breast cancer (TNBC) cell lines and primary tumors show high levels of an IGF gene expression signature. Supporting this, cells lines with a high IGF signature are especially sensitive to a dual IGF-IR/InsR tyrosine kinase inhibitor (TKI). Treatment of a TNBC tumorgraft with an anti-IGF-IR/InsR dual TKI and chemotherapy resulted in mitotic catastrophe and complete tumor regression. This study provides strong preclincial evidence supporting the investigation of anti-IGF-IR/InsR therapy in combination with chemotherapy in TNBC.
The insulin-like growth factor (IGF) signaling pathway is a major regulator of growth, survival, migration, and invasion (10, 11). Experimental studies in vitro and in vivo have provided substantial evidence of a role for IGF-IR in human breast cancer. Overexpression of a constitutively active IGF-IR or inducible overexpression of wild-type IGF-IR, in the mouse mammary gland results in rapid mammary tumorigenesis (12, 13). Consistent with this, overexpression of IGF-IR transforms immortalized mammary epithelial cells (MCF10A) (14-16). Clinical studies support the importance of IGFs in breast cancer. In breast cancer specimens, IGF-IR is detected at very high frequency and levels and activity are increased compared to normal breast (17). High levels of phosphorylated IGF-IR/InsR are associated with poor patient prognosis (18). Studies have also shown that elevated levels of serum IGF-I are correlated with increased breast cancer risk (19).
Many preclinical studies targeting the IGF-IR have shown promising anti-neoplastic activity (17) and early phase 1 (20) and phase 2 (21) reports have been encouraging. Two predominant targeted strategies to inhibit IGF-IR function are in development: monoclonal antibodies, which are highly specific for the IGF-IR and cause downregulation of the receptor, and tyrosine kinase inhibitors, which are ATP-competitive inhibitors of the IGF-IR and insulin receptor (InsR) tyrosine kinase domains (22).
An important outstanding question in the clinical development of anti-IGF-IR therapy is to identify appropriate patient populations, allowing specific treatment of patients whose tumors show addiction to this pathway for continued survival and proliferation. Many studies showed that both IGF-IR and its downstream adaptor IRS1 are estrogen-regulated genes (23). Furthermore, IGF-I can activate the ER (24). This bi-directional positive feedback supported the concept of targeting both ER and IGF-IR in breast cancer, and many clinical trials are currently testing this strategy (25-27). However, there is also evidence for a role for IGF-IR in TNBC. A number of tumor suppressor genes such as p53 and BRCA1 represses the IGF-IR promoter. Mutations in these tumor suppressor genes in TNBC are associated with elevated IGF-IR levels (28). Furthermore, IGF-IR is amplified in basal breast cancer (29), and high levels of IGF-IR protein are seen in basal breast cancers (30).
In this study we show that IGF activity, as measured by a gene expression signature, is activated in TNBC patient tumors and cell lines in culture. This high level of the IGF signature correlates with sensitivity to a new dual IGF-IR/InsR small molecule inhibitor BMS-754807. Consistent with this, BMS-754807 alone is effective at causing growth inhibition of a TNBC tumorgraft, and in combination with chemotherapy results in tumor eradication. This preclinical study provides substantial support and rationale for a clinical trial using inhibitors against IGF-IR in combination with chemotherapy in TNBC.
Results
An IGF gene expression signature is present in triple-negative breast cancer (TNBC)
We previously reported the development of an ‘IGF-I gene signature’ consisting of pattern of genes up- or down-regulated by IGF-I (31). We found that this IGF signature was present in human breast cancers, specifically the subtypes luminal B and TNBC (31). To independently validate these results we analyzed the IGF gene signature in a recently published data set of 198 node negative breast cancers (Desmedt (32)) and confirmed the presence of the IGF signature in the majority of ER-negative tumors (Supplementary Fig.1A). We went on to apply the IGF signature to a more recent profile dataset from Hoadley et al. (33). This independent cohort of 248 tumors was previously classified into breast cancer subtypes using an intrinsic gene expression signature. The IGF gene signature was present in most basal tumors, luminal B tumors and a subset of Her2-positive tumors (Supplementary Fig. 1B) compared to other subtypes. This data suggests that the IGF signature is present in TNBC, and that these tumors may be candidates for anti-IGF-IR therapy.
The IGF gene expression signature is reversed by anti-IGF-IR inhibitors
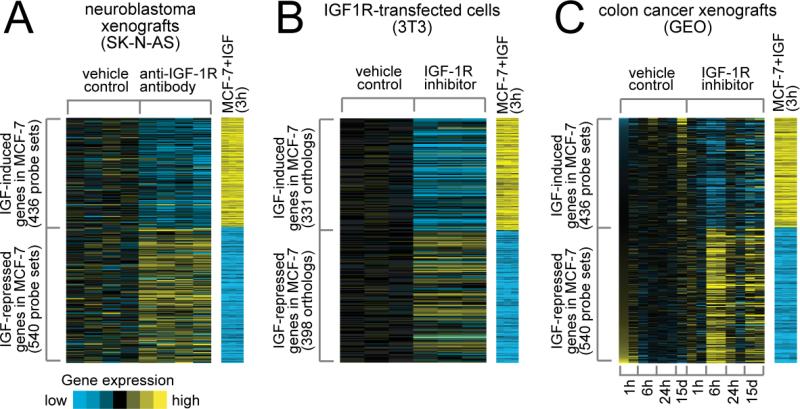
We next set out to examine the effect of IGF-IR inhibitors on the IGF signature. We reasoned that regulation of genes by IGF-I (which made up the IGF gene signature) should be reversed by anti-IGF-IR inhibitors. Consistent with this, when we examined the levels of genes we previously found to be induced or repressed by IGF-I in neuroblastoma xenografts treated with an anti-IGF-IR (h10H5, Genentech) antibody (34), we found that there was a striking reversion in their levels (Fig. 1A). Thus, genes induced by IGF-I in MCF-7 cells were repressed by h10H5 treatment of the neuroblastoma xenograft, and genes repressed by IGF-I were now induced. This result was highly significant (p~0). Highly similar results were obtained with an IGF-IR/InsR tyrosine kinase inhibitor (A-928605, Abbott) (35) that was administered in vitro to NIH3T3 fibroblasts transfected with the IGF-IR (Fig. 1B). Finally, we generated gene expression data from colon cancer xenografts (GEO) grown in vivo and then treated with an anti-IGF tyrosine kinase inhibitor BMS-754807 (12.5mg/kg/day) for various time-points of 1, 6 and 24 hours and 15 days. Treatment of these GEO xenografts resulted in gene expression values that were again reversed compared to the IGF-signature (Fig. 1C). Note that there was no change in the IGF signature after 1 hour, but after 6 hours of exposure to the drug a strong reversion of the IGF signature was seen, which is entirely consistent with the pharmacodynamics and pharmacokinetics of BMS-754807. This data gives us confidence that our signature can measure IGF activity, and furthermore, suggests that the IGF signature might indicate cells that have an active IGF pathway and thus may respond to an IGF-IR inhibitor.
Figure 1. An IGF gene signature is reversed by treatment of cancer xenografts and IGF-IR overexpressing NIH3T3 fibroblasts with anti-IGF-IR inhibitors.
Genes in the IGF-I gene signature derived from MCF-7 cells stimulated with IGF-I (31) were examined in A. a gene expression profile of neuroblastoma (SK-N-AS) xenografts treated with vehicle or an anti-IGF-R antibody and in B. IGF-IR transfected NIH3T3 fibroblasts treated with an IGF-IR small molecule inhibitor and in C. colon cancer xenografts treated with the small molecule inhibitor BMS-754807. For A-C, relative gene expression is represented using a yellow–blue color scale; patterns for genes that are upregulated (yellow) in the IGF gene signature are separate from the patterns for genes that are downregulated (blue)..
When we originally developed the IGF signature, we attempted to reduce the importance of proliferative genes by 1) including only genes modulated by IGF-I at both 3 and 24hrs, 2) removing genes annotated as being associated with proliferation in Gene Ontology (GO), and 3) removing genes that were found to be induced in fibroblasts stimulated to proliferate by serum (36). In our previous report we confirmed that the IGF signature doesn't correlate with the proliferative rate of cell lines (36). Thus, this reversal of the signature by IGF-IR inhibitors (Figure 1) presumably doesn't simply reflect changes in cell growth, but rather highlights changes in metabolism and DNA repair (the major GO term-defined classes of genes represented in the IGF gene signature). To further examine the specificity of the IGF signature, and its reversion, we examined the levels of genes in the IGF signature on a data set of ovarian cancer cells (36M2) treated with chemotherapy (carboplatin). Importantly, chemotherapy had no discernable effect upon the IGF gene signature; indeed, it weakly induced the IGF signature perhaps due to a DNA damage response (Supplementary Fig 1D).
A recent study comparing genes regulated by growth factors (IGF-I, insulin, EGF and heregulin) in MCF-7 cells showed that there was a highly significant overlap (37), consistent with highly redundant nature of growth factor downstream signaling (e.g.. PI3K, ERK1/2 etc). We therefore examined whether the IGF-I signature would be modulated by inhibition of other growth factor receptors. We developed gene expression profiles from colon cancer (GEO) xenografts treated for various lengths of time with the EGFR inhibitor Erbitux (1mg/kg every three days). Interestingly, the IGF signature was reversed by Erbitux, indicating the IGF signature is likely a marker of active growth factor signaling and highlights pathways downstream of receptor tyrosine kinase inhibitors (Supplementary Fig. 1C).
Triple-negative/basal-like breast cancer cell lines show activation of the IGF gene signature
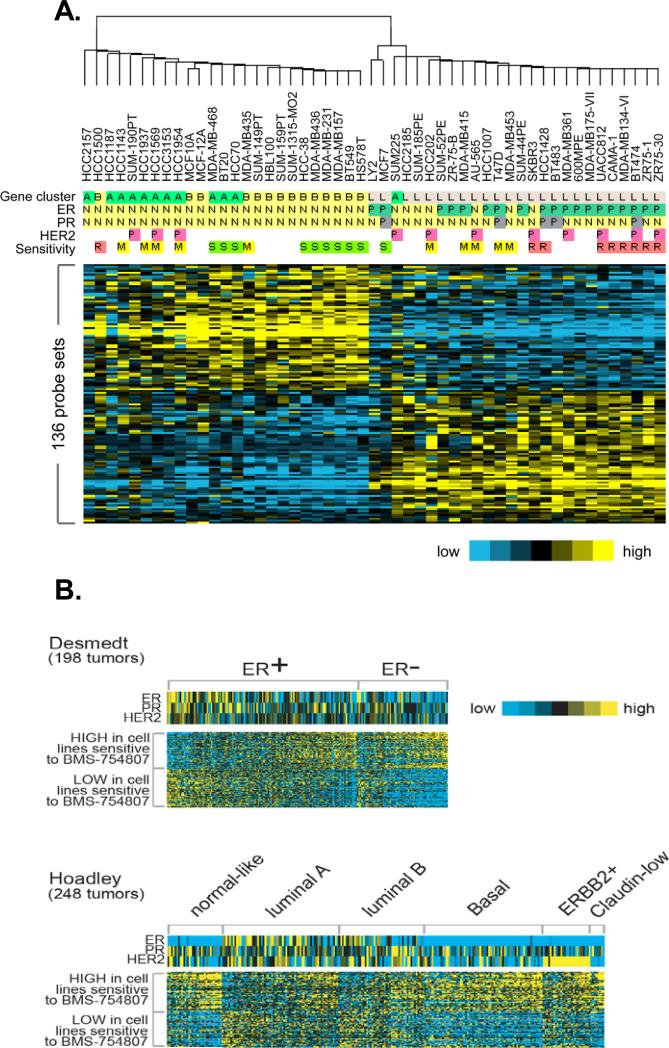
To examine whether the IGF gene signature is associated with response to inhibition of IGF-IR/InsR, we studied cell lines grown in culture, where one can rapidly assess response. Therefore, we examined the IGF-I gene signature in a publicly available dataset of gene expression profiles from a large panel of breast cancer cell lines (38). Figure 2A shows a panel of breast cancer cell lines arranged according to their intrinsic subtype (as defined using the Hoadley dataset) (38). The IGF signature is present in the majority of basal-like (and TNBC) breast cancer cell lines. We assigned each cell line a t-score based upon the similarity of its gene expression profile to the IGF signature and the positive t-score in the majority of TNBC cell lines highlights the presence of the signature in this subtype (Fig. 2B). This data is entirely consistent with Figure S1 and our previous report of the IGF signature in TNBC (31) suggesting that the IGF-IR pathway is highly active in the triple-negative/basal-like subtype of breast cancer.
Figure 2. IGF gene signature is present in triple-negative/basal-like breast cancer cell lines.
Breast cancer cell line profiles were classified using the Hoadley dataset, where intrinsic subtypes were previously defined. The correlation was computed between a given cell line gene expression profile and the mean centroid of each subtype; cell lines were then assigned to a subtype with the highest correlation. A. The heatmap represents the IGF gene signature in the cell lines according to subtype. ER, PR, and HER-2 mRNA levels are shown. In addition, the breast cancer subtype is indicated as defined in the original gene expression study by Neve et al. B. The graph represents the t-score for each cell line based on the similarity to the IGF signature.
An IGF-IR tyrosine kinase inhibitor BMS-754807 shows selective activity in TNBC cell lines
To test whether the presence of the IGF signature in basal-like/TNBC cell lines was associated with response to an IGF-IR/InsR inhibitor, the sensitivity of a new dual IGF-IR/InsR tyrosine kinase inhibitor (BMS-754807) currently in Phase 1/2 clinical trials was determined by MTS assay in a panel of 30 breast cancer cell lines. Among the different tumor cell lines, sensitivity presented as IC50 to BMS-754807 varied widely from 0.1μM to 25μM (Fig. 3A). When defining cell lines as sensitive or resistant based on the median IC50 (6.4μM), we found a clear correlation between the sensitive/resistant classification to BMS-754807 and specific breast cancer subtypes. The greatest response to BMS-754807 was in basal-like/TNBC cell lines (10/15), while luminal breast cancer cell lines and Her2 overexpressing cell lines were relatively resistant (13/15) (Fig 3A). Compared to all cell lines, the TNBC group was enriched for cell lines with low IC50. Importantly, when the IC50 is plotted against the t-score for the IGF gene signature for each cell line there is a significant inverse correlation (r=-0.41, p=0.014), with a higher t-score (indicating an active IGF pathway) being associated with a greater response (lower IC50) to BMS-754807 (Fig. 3B). These data strongly suggest that BMS-754807 is active in TNBC.
Figure 3. BMS-754807 is active in basal-like/triple-negative breast cancer cell lines.
A. The concentration of BMS-754807 required to reduce growth by 50% (IC50) was calculated for each cell line using monolayer proliferation and MTS assay. Breast cancer cells were seeded at 1,000 to 12,000 cells per well depending on the cell line in 96 well microtiter plates and incubated overnight. BMS-754807 was serially diluted and added. After 72 hr exposure, MTS assay was performed. Bars represent the average IC50 (μM) of each breast cancer cell line. For seven cells lines the IC50 was not reached (16.49μM and 25μM, respectively). Sensitive cell lines have an IC50 below the mean of the group of cells (6.4μM); resistant cell lines are above the mean. The graph shows cell lines ranked according to its IC50. Black bars represent cell lines that have a basal-like gene expression signature (TNBC) based upon the studies of Neve et al (38) and grey bars represent luminal or HER2 positive cell lines. B. IC50 is plotted alongside the t-score for the IGF signature for each cell line. Pearson correlation shows there is a significant correlation (r=-0.41, p=0.014), with a higher t-score (indicating an active IGF-IR pathway) being associated with a greater response (lower IC50) to BMS-754807.
114 differentially expressed genes identify TNBC cell lines as most responsive to BMS-754807
To examine genes and pathways associated with response to BMS-754807, we performed comparative gene expression analysis between the ten most sensitive cell lines with an IC50 below 4μM and the 9 most resistant cell lines with an IC50 above 14μM BMS-754807 using gene expression data published by Neve et al (38). As predicted from Figure 3B, these two panels of cell lines showed a statistically different IGF t-score (sensitive 3.59 +/- 1.64; resistant -3.91 +/- 2.57; p<0.05). Comparative gene expression analysis between these two sets of cell lines identified 136 probe sets corresponding to 114 genes (p < 0.001 and FDR < 5 %) that were differentially expressed between sensitive and resistant cell lines. The top 10 differentially expressed genes were validated in a panel of seven sensitive and six resistant breast cancer cell lines by qRT-PCR (Supplementary Fig 2). We analyzed the 114 genes in a panel of 51 breast cancer cell line profiles with known and unknown IC50 to BMS-754807 (38). Hierarchical clustering separated the cells lines into two major bins: ER-negative cell lines that were sensitive and ER-positive cell lines that were mostly resistant (Fig. 4A).
Figure 4. 114 differentially expressed genes identified triple negative breast cancer cell lines as most responsive to BMS-754807.
A. 136 differentially expressed probesets, representing 114 genes, were analyzed in 51 breast cancer cell line profiles published by Neve et. al (38) with known and unknown IC50. S=sensitive (IC50<4μM), M=Medium (IC50 = 4μM<14μM), R=Resistant (IC50>14μM). In addition, the ER, PR and Her2 status is indicated (P = positive, N = negative). Gene-cluster indicates the breast cancer subtype as defined in the original gene expression study by Neve et al. (A = basal A, B = basal B, L = Luminal). B. 136 differentially expressed probesets, representing 114 genes, were analyzed in two published data sets of clinical breast tumors from Desmedt and Hoadley. Within the ER-positive and ER-negative as well as the distinct breast cancer subtype tumors are ordered from those with the least similarity to the 114 gene signature pattern to those with the highest similarity to the 114 gene signature.
We found that there was significant overlap between the 114 genes (136 probesets) and the IGF signature developed from MCF7 cells. Thus, of 66 probesets increase in sensitive cell lines, 13 were increased in the IGF signature (from a total of 436 (one-sided Fisher's exact test p=4.E-10). Of the 70 probesets that were decreased in sensitive cell lines, 7 were also decreased in the IGF signature from a total of 540 probeset (one-sided Fisher's exact test p=0.002).
Consistent with the IGF signature being present in TNBC and correlating with sensitivity to an IGF-IR inhibitor, when we examined the levels of the 136 probsets in human breast cancers, we found enrichment in TNBC (Figure 3B). When examining tumors subtyped according to Hoadley et al, we found the 114 genes enriched in basal, claudin-low, normal, and a subset of HER2 positive tumors. The patterns of enrichment of the 136 probesets (114 genes) strongly resembled the patterns seen with the IGF signature (Supplementary Fig. 1). We identified that many of these 114 genes were actually markers of the basal or luminal subtype with sensitive cell lines expressing basal markers such as CAV1 and CAV2 whereas resistant cell lines expressed luminal markers such as ErbB3 and SPDEF (Supplementary Fig. 2).
To examine the clustering data further, we determined the sensitivity to BMS-754807 of breast cancer cell lines with an unknown IC50 by MTS-assay. Among the cell lines tested the TNBC cell lines SUM149PT and MCF10A showed the greatest response to BMS-754807 whereas cell lines that are ER negative but overexpress Her2 (SUM225 and SUM190PT) are less sensitive to BMS-754807 (Fig. 4B). Luminal breast cancer cell lines such as ZR75B, MDA-MB-175VII and MDA-MB-316 showed the least response to BMS-754807. It is interesting to note that the sensitive cell lines required insulin for regular growth, whereas the resistant cells are routinely grown in the absence of insulin. This may highlight the growing evidence for a role for insulin in breast cancer which is being studied by many groups.
Novel tumorgraft models of human TNBC shows strong activation of IGF-IR/InsR
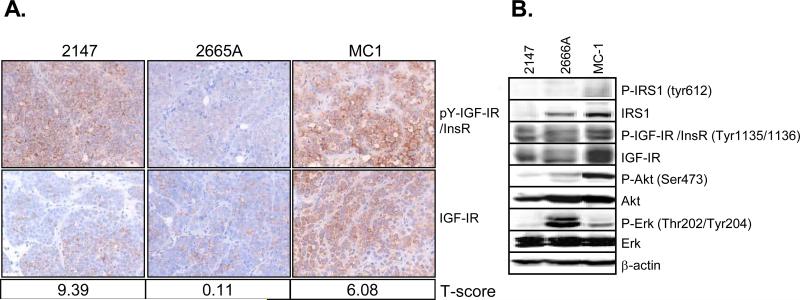
Preclinical cancer research has relied heavily upon cell lines grown in culture and then xenografted for growth in mice. However, recent work has shown that human cancers placed directly into the mouse (tumorgrafts) maybe a more appropriate model that is better at predicting response to drugs in humans (39). We have recently developed several new tumorgraft models of human TNBC. We screened seven TNBC tumorgrafts for activity of the IGF-IR and InsR by both immunohistochemistry (IHC) and immunoblotting (IB). Figure 5A shows three representative tumorgrafts expressed various levels of IGF-IR protein and had divergent levels of active phosphorylated pY-IGF-IR/InsR (antibodies are only available which detect both pY-IGF-IR and pY-InsR together). We generated gene expression data from the tumorgrafts and calculated an IGF signature t-score for each tumorgraft. Analysis of these profiles showed that tumorgraft MC1 (40) had the highest level of IGF-IR and pY-IGF-IR/InsR, and in addition had a high IGF signature t-score. Protein lysates from the same tumorgrafts as in Fig 5A confirmed that MC1 had the highest activation of IGF-IR/InsR (Fig. 5B). Activation of downstream signaling molecules varied among the tumorgraft models. MC1 had high levels of IRS1 and activated AKT whereas the tumorgraft 2665A showed activation of MAPK.
Figure 5. TNBC tumorgrafts express high levels of active IGF-IR.
A. Three different TNBC tumorgraft lines (2147, 2665A, MC1) were harvested and processed in paraffin. Immunohistochemistry was performed on five-micrometer sections for anti-pY-IGF-IR (Phospho-IGF-IR) and total IGF-IR. Microarray analysis was performed on these tumorgrafts. Based on the gene expression data an IGF signature t-score was calculated for each tumorgraft as indicated underneath the representative picture. B. The same three tumorgraft lines were lysed and analyzed by immunoblot and probed using IGF-IR specific antibodies as well as total/phospho-specific antibodies for IRS1, AKT, ERK1/2. β-actin was used as a loading control. Tumorgraft lines that were not relevant to this study were cropped out between 2665A and MC1.
BMS-754807 inhibits growth of MC1 TNBC tumorgrafts, and when combined with chemotherapy causes complete regression
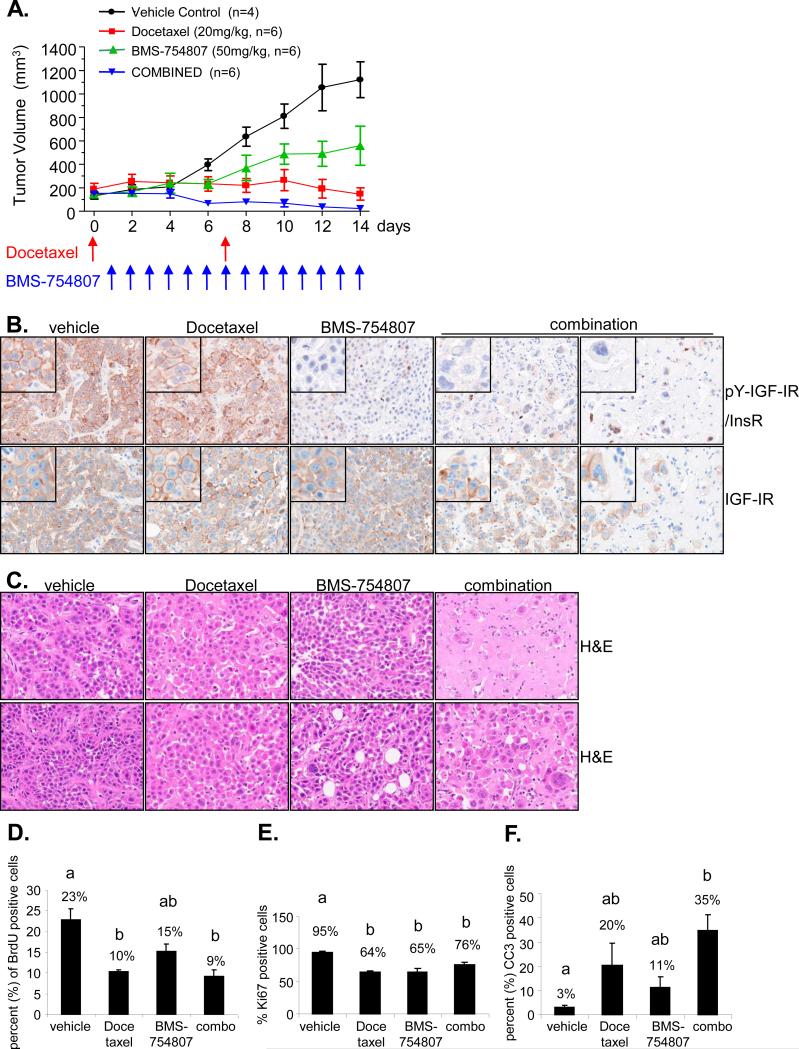
Our studies identified TNBC and TNBC cell lines as having an active IGF pathway. As we previously showed that BMS-754807 is most active in TNBC cell lines in vitro, we directly examined the effectiveness of the dual IGF-IR/InsR inhibitor BMS-754807 in the MC1 tumorgraft model of TNBC breast cancer alone as a single agent or in the presence of chemotherapy (docetaxel). We chose the tumorgraft MC1 for this preclinical study as it showed the highest levels of active and total IGF-IR and also a high IGF t-score (Figure 5). A recent study examining timing of anti-IGF-IR therapy and chemotherapy in cells in culture showed that most efficacious combination was chemotherapy followed by anti-IGF-IR therapy (41). We thus administered docetaxel followed by BMS-754807 the next day. Single agent BMS-754807 achieved a statistically significant (p<0.001) reduction in tumor growth when compared with the control group (Fig. 6A). Docetaxel stabilized MC1 tumor growth. Strikingly, combined treatment with BMS-754807 and docetaxel showed superior tumor growth inhibition to either single agent alone (p<0.001), and four out of six mice receiving the combined agents had tumors regress until no tumor was palpable.
Figure 6. BMS-754807 inhibits growth of TNBC tumorgrafts and causes regression in combination with chemotherapy.
A. 4 to 6-week-old female NOD/scid mice were transplanted with a 1mm3 piece of tumorgraft into a cleared number four mammary fat pad. When tumors reached a volume between 100-200mm3 they were randomized to receive the following treatments: vehicle, 50mg/kg BMS-754807 daily by oral gavage, 20mg/kg docetaxel weekly by intraperitoneal injection or the combination of 50mg/kg BMS-754807 daily and 20mg/kg docetaxel weekly by intraperitoneal injection. Tumor volume and body weight was measured daily. B. Untreated and treated tumors were processed in a tissue microarray (TMA) for immunohistochemical analysis. Immunohistochemistry was performed for phospho and total IGF-IR. Representative IHC staining of the treatment groups is taken at 40x magnification. C. Representative tumor sections of the treatment groups stained with hematoxylin and eosin are taken at 40x magnification. D. Quantification of BrdU incorporation per cell was done by image analysis. Data represents means ± SE of 12 representative pictures per treatment group. E and F. The percentage of Ki67/CC3-positive cells within the tumor was scored. Values represent the means ± SE of 12 representative pictures per treatment group. D-F Bars with different letters are significantly different (p<0.05) by ANOVA analyses and pairwise comparisons with Holm adjustment for multiple comparisons.
To confirm the ability of BMS-754807 to inhibit IGF-IR activity in triple negative breast cancer, tyrosine phosphorylation and total levels of IGF-IR/InsR were examined in the various treatment groups. Both pY-IGF-IR/InsR and IGF-IR showed membrane staining with a small amount of cytoplasmic staining (Fig. 6B). There was no change in the levels of pY-IGF-IR/InsR between tumors treated with docetaxel or vehicle (Fig. 6B). In contrast, BMS-754807 completely blocked IGF-IR/InsR phosphorylation. There was no change in levels of total IGF-IR between the different treatment groups.
Toxicity associated with BMS-754807
Despite the potent inhibitory activity of BMS-754807 on MC1 TNBC tumorgrafts, minimal toxicity was observed in animals at doses that show significant antitumor activity. Mice treated with docetaxel or BMS-754807 as a single agent lost a maximum of 5% body weight (Supplementary Fig. 4B). Combination therapy with BMS-754807 plus docetaxel resulted in a body weight loss of 15%.
BMS-754807 was developed as a dual IGF-IR/InsR small molecule inhibitor that inhibits IGF-IR and insulin receptor (IR) with a similar affinity. Because glucose homeostasis in vivo is maintained through insulin-mediated uptake of glucose in skeletal muscle and suppression of glucose production in the liver (42), the effect of BMS-754807 on InsR inhibition was determined. As expected, blood glucose levels remained unaltered in mice treated with chemotherapy. However, mice treated once daily with BMS-754807 alone showed a significant two-fold increase in glucose levels when compared with vehicle-treated mice (adjusted p-value =0.035, Supplementary Fig. 4C). Interestingly, mice treated with the combination of BMS-754807 and docetaxel showed a lower elevation of glucose levels, but this was not significantly different compared to vehicle or docetaxel treated mice. Blood insulin levels were also monitored in mice after 14 days of treatment. No changes in insulin were observed in chemotherapy treated mice compared to vehicle treated mice. However, insulin levels increased from 0.5 to 110ng/mL after 14 days of treatment with BMS-754807 alone (adjusted p-value <0.0001, Supplementary Fig. 4D). Consistent with the reduction of elevated glucose in the combination treatment group, BMS-754807 and chemotherapy induced insulin levels (57ng/mL) to a level lower than BMS-754807 alone (114ng/ml) which was not significant, however the hyperinsulinemia produced by the combination treatment was still significantly elevated compared to vehicle or chemotherapy alone (adjusted p-value < 0.0001).
BMS-754807 blocks growth and induces apoptosis, and sensitizes MC1 tumorgrafts to docetaxel-induced mitotic catastrophe
Several reports have shown that chemotherapy agents such as docetaxel affect the stability of the microtubules and in doing so induce mitotic catastrophe (43, 44). Mitotic catastrophe results from aberrant mitosis, or missegregation of chromosomes followed by cell division which results in the formation of multinucleated giant cells leading to cell death. Cell death through mitotic catastrophe may occur through apoptosis as well as necrosis (44). MC1 tumorgrafts treated with Docetaxel showed relatively few multinucleated giant cells (Fig. 6C). However, the addition of BMS-754807 dramatically increased docetaxel-induced mitotic catastrophe (Fig. 6C). Analysis of H&E staining showed that normal breast cancer cells were still present in Docetaxel treated tumors whereas only multinucleated cells were present in the combination treated tumors.
Since multinucleated cells may be temporarily viable and mitotic catastrophe may be a process leading to death (44), we analyzed treated tumors for replication, proliferation, and apoptosis. All treatment groups showed a reduction in replication as indicated by less BrdU incorporation into DNA compared to untreated tumor cells (Fig. 6D). BMS-754807 alone reduced replication by 36% (22% BrdU positive cells in vehicle tumors versus 16% in treated tumors) but this was not significant (adjusted p-value=0.13). Chemotherapy significantly reduced replication by 55% (adjusted p-value=0.015) and the combination by 59% compared to vehicle (adjusted p-value=0.0018). We next analyzed proliferation as assessed by Ki67 positive cells (Fig. 6E, Supplementary Fig. 4D). Whereas both single agents alone were able to significantly reduce proliferation by 30-32% (adjusted p-values < 0.0001), combination treatment resulted in only a 17% reduction in proliferation (adjusted p-value < 0.005, compared to vehicle). We then investigated whether single agents alone or the combination of BMS-754807 and Docetaxel induced cell death. BMS-754807 caused a 4-fold elevation in apoptosis as measured by cleaved caspase 3 (CC3) from 3% apoptotic cells in vehicle treated tumors to 11% in BMS-754807 treated tumors (Fig. 6F, Supplementary Fig 4D), however this elevation was not significant (adjusted p-value=0.36). Chemotherapy resulted in a 6.7-fold induction of apoptosis compared to vehicle (adjusted p-value=0.1) which again wasn't significant, whereas the combination of BMS-754807 and chemotherapy caused a striking 12-fold induction of apoptosis compared to vehicle treated tumors (Fig. 6F, Supplementary Fig 4D) which was highly significant (adjusted p-value=0.005). In addition, combination therapy resulted in massive cell destruction through necrosis as shown in Figure 6C.
Discussion
An important component in the clinical development of targeted therapies against cancer is to identify appropriate patient populations in which tumors show addiction to a particular pathway for continued survival and proliferation, and in which these tumors are susceptible to the drug. In this study, we confirmed that an IGF-gene signature can measure IGF activity, as based on reversion of the signature in three different cancer models treated with different anti-IGF-IR therapies. We found that the majority of human TNBC and TNBC cell lines have an active IGF signature. Consistent with this, TNBC cells lines are most sensitive to the dual anti-IGF-IR/InsR tyrosine kinase inhibitor BMS-754807, and sensitivity correlates with expression of the IGF signature. Finally, new tumorgraft models of TNBC show activation of IGF-IR and treatment with BMS-754807 in combination with chemotherapy results in complete tumor regression. This preclinical study provides a strong clear biological rationale to test anti-IGF-IR therapy in combination with chemotherapy in patients with TNBC.
Although many studies identified ER-positive breast cancer as an important target for anti-IGF-IR therapy, the significance of IGF-IR as a target in triple-negative breast cancer has been poorly addressed. However, there is a growing literature indicating a role for IGF-IR in the aggressive subtype of breast cancer. IGF-IR is amplified (albeit in a small number of tumors) in TNBC (29) and IGF-IR protein is detected in triple negative breast cancer (30). Moreover, activated (phosphorylated) IGF-IR and IR are also found in TNBC (18). Growth inhibition of the TNBC cell line SUM149 in vitro with BMS-536924 suggests that this subtype of breast cancer may be inhibited by targeting IGF-IR/InsR (18).
There has been recent considerable interest in the identification of biomarkers for anti-IGF-IR therapies. Several in vitro cell culture studies have recently been reported. Studies of an IGF-IR tyrosine kinase inhibitor, NVP-AEW541, reported high levels of IGF-IR in all breast cancer cell lines (45). IGF-IR expression alone provided insufficient information to select cell lines sensitive to NVP-AEW541 but sensitivity was limited to those that express both IGF-IR and IRS1 (45). Huang et al examined response to a panel of neuroblastoma and sarcoma cells lines to BMS-536924 and identified genes differentially expressed between sensitive and resistant cell lines. Interestingly, IGF-IR mRNA was a weak predictive marker of response to BMS-536924, but prediction improved when the levels of IGF-IR, IGF-I, and IGF-II were also considered (46). We found IGF-IR mRNA levels didn't predict response to BMS-536924 in breast cancer cell lines, but in contrast that IGF-IR protein was weakly associated with response (47). Similarly, a study in lung cancer cells also showed that IGF-IR protein predicts response to a monoclonal antibody R1507 (48). A study of a large panel of breast cancer cell lines in response to the monoclonal antibody h10H5 showed that IGF-IR mRNA levels correlate with response, with IGF-IR mRNA being an excellent negative predictive factor (low levels being associated with resistance) but mRNA levels being a relatively poor positive predictive factor. Importantly, inclusion of IGF-II, IRS1 and IRS2 mRNA levels increase the potential to predict which cells respond to h10H5 (49). We recently performed the first pilot analysis in a small number of human NSCLC tumors treated in a Phase 2 clinical trial with the anti-IGF-IR antibody Figitumumab (Pfizer) and found that IGF-IR was a weak predictor of response (50) . To date, the majority of data suggest that IGF-IR may be a relatively strong negative predictive factor, similar to the estrogen receptor for hormone therapy and HER-2 for anti-HER-2 therapy, but that IGF-IR alone is a weak positive predictive factor. Interestingly, ER seems to be a strong negative predictor of response to BMS-754807, and correlates more with response than the actual IGF signature. This is likely due to the fact that ER is a major driver of proliferation and survival in many ER+ cell lines, and inhibition of ER would likely sensitize some of these cell lines to an IGF-IR inhibitor. Supporting this, co-treatment with anti-estrogen and anti-IGF-IR inhibitors shows additive and synergistic effects both in vitro and in vivo (51, 52).
We developed the IGF signature to learn more about transcriptional events downstream of IGF-IR, and to examine the role of IGF-IR regulated genes in breast cancer. However, the obvious question as to the potential role of the IGF signature in predicting response to anti-IGF-IR therapy arose. We show here that the IGF signature can measure IGF activity in tumors, however it also measures other related growth factor pathways. Not surprisingly, the IGF signature is only weakly correlated (r=0.41) with response of cells to an IGF-IR inhibitor. However, the strength of the signature may come when used in combination with IGF-IR protein levels and activity. Thus IGF-IR alone, or the IGF signature alone, maybe insufficient to indicate an active IGF pathway, but the combination may better indicate an active IGF pathway. To this end, we used this strategy to select a TNBC tumorgraft for study and found dramatic effects of an IGF-IR inhibitor. Further comprehensive studies are required to definitively prove whether the combination of IGF-IR levels and downstream gene transcripts is a useful method to identify patients who may respond to anti-IGF-IR therapy.
Standard xenografts using permanent cell lines poorly predict how a drug works in patients with the same type of tumor (39). A study showed that tumorgrafts correctly predict response in 90% of patients (19 of 21 tumors) and resistance in 97% (57 of 59) tumors (53). A clinical trial in pediatric neuroblastoma testing the drug topotecan was consistent with that reported in preclinical tumorgraft models of neuroblastoma (54). Therefore, we chose to study recently developed tumorgrafts, in which tumors taken from patients and small pieces directly implanted into immunodeficient mice. We found a dramatic effect of BMS-754807 against MC1 tumorgrafts, with complete regression when the drug was combined with chemotherapy. Complete regression of standard cell line xenografts has been rarely reported, and we found that BMS-754807 showed only minimal activity against a range of breast cancer cell lines grown as xenografts (data not shown), again indicating that the tumorgraft model maybe more appropriate for determining response.
IGF-IR has been linked to resistance to hormone therapy, anti-HER-2 therapy, chemotherapy, and radiation therapy (25). As such, there is great interest in combing anti-IGF-IR therapies with currently anti-cancer agents. IGF-IR signals to potent survival pathways, and has been shown to confer resistance to chemotherapy-induced death (55, 56). Consistent with this, inhibition of the IGF-IR sensitizes breast cancer cells to chemotherapy (57, 58). In this study we combined BMS-754807 with docetaxel as it has previously been shown that IGF-IR is a mediator of survival to taxanes, and the combination of an IGF-IR antibody (A12) and docetaxel showed excellent anti-tumor activity in prostate cancer (59). We found a similar effect with the combination therapy. Why anti-IGF-IR therapy is so active in combination with an anti-microtubule therapy is intriguing. As anti-IGF therapy is thought to mainly acts as a G1 block, and docetaxel causes a G2/M block, it is possible that the double blockade is especially effective. However, a study showed that anti-IGF-IR blockade causes a G2/M block (59). Indeed, in our analysis of the IGF signature, ingenuity pathway analysis identified G2M checkpoint as the major IGF regulated process after 24 hours, with IGF upregulating numerous genes involved in G2/M transition including AURKA, AURKB, BUB1, CCNB, CENPE, CENPA and CDCA8. Thus, is it possible that IGF-IR normally confers G2/M progression and that in combination with docetaxel this leads to mitotic catastrophe. However, further intrigue is provided by the finding that BMS-754807 actually inhibits AURKA (60), albeit with lower affinity than IGF-IR. It is thus possible that BMS-754807 and docetaxel synergize due to a concomitant action on the G2M checkpoint.
In summary, this preclinical study shows that an IGF-IR/InsR inhibitor is active in TNBC and provides substantial support and rationale for a clinical trial using BMS-754807 in ER-negative and HER2-negative breast cancer.
Methods
BMS-754807
The chemical structure of BMS-754807 and its activity against IGF-IR has been recently described (60). For in vitro studies, BMS-754807 was dissolved in DMSO to 10mM. For in vivo studies, BMS-754807 was formulated in PEG400:H2O (80:20).
Cell culture
Breast cancer cell lines were obtained from American Type Culture Collection (Manassas, VA), except H3396, which was obtained from the Pacific Northwest Institute (Seattle, WA). The MCF7/Her2 cell line was made by stable transfection of the HER2 gene into MCF7 cells. Cells were grown using the recommended culture conditions according to Neve et al. (33). Of note, the cell lines used to test the IC50 of BMS-754807 didn't require insulin to be added to the medium. MCF10A cells were cultured in DMEM/F12 medium supplemented with 5% horse serum, 20ng/ml epidermal growth factor, 10μg/ml insulin, 0.5μg/ml hydrocortisone, 100ng/ml cholera toxin, and 100 units/ml penicillin-streptomycin. SUM breast cancer cell lines also required the addition of insulin.
Monolayer growth assay
For breast cancer cell line studies, cells were plated at 1,000 to 12,000 cells per well depending on the growth properties of each cell line in 96 well microtiter plates and incubated overnight. The next day, the start absorbance (cell number at the beginning of the experiment, i.e. at time of drug addition), was measured in which no compound was added and the plate immediately developed by the method below. BMS-754807 was serially diluted and added. After 72 hr exposure, cell viability was determined by MTS assay (Promega). Percentage of cell growth inhibition was calculated as % of control = end Absorbance - start Absorbance × 100%. The growth curves (expressed as a percentage of growth observed in untreated controls) were used to determine the IC50. The IC50 value is the drug concentration at which 50% of maximal growth inhibition was observed.
Comparative gene expression analysis
Comparative gene expression analysis was performed by determining genes with significantly different expression between sensitive cell lines with an IC50<4μM (n=10) and resistant cell lines with an IC50>14μM (n=9) using dChip software (61, 62). We utilized publicly available gene expression data published by Neve et al (38). Differentially expressed genes were found using t-test. False discovery rate (FDR) was estimated by permutation method. We used p-value = 0.001, which resulted in 136 differentially expressed probesets representing 114 genes, with FDR < 5%. Hierarchical clustering and expression values were visualized as heat-maps.
To assign cell lines to breast cancer subtypes, we scored the cell lines for subtype, essentially as previously described by using the dataset from Hoadley et al (33). In brief, for each gene common to the Hoadley platform and the other breast-array dataset platform, we computed the mean centroid of the subtypes in the Hoadley dataset and centered each group average on the centroid. We then took the Pearson correlation (using all genes common to both array datasets) between the Hoadley centered averages and the expression values of each profile in the independent dataset.
RNA isolation and quantitative real-time PCR
To validate the comparative gene expression analysis, seven sensitive and six resistant cell lines were cultured in triplicates. The sensitive cell lines MCF7, BT20, MDA-MB-468, HS758T, MDA-MB-231, HCC38, MDA-MB-436 and the resistant cell lines ZR75-1, SKBR3, BT474, CAMA-1, MDA-MB-134, UACC812 were used. RNA was isolated using RNeasy mini kit (QIAGEN). mRNA was converted to cDNA in a final volume of 100μl using 0.5μg RNA, 0.5μl random primers, and 25mM desoxynucleotide triphosphate. After the samples were heated for 5 min at 65°C, 20μl of 5x first strand buffer (Invitrogen), 5μl 0.1M dithiothreitol and 0.5μl of Superscript II RNase H reverse transcriptase (Invitrogen) were added. Reverse transcription was performed at 25°C for 5min, 48°C for 30min and 70°C for 10min. Quantitative reverse transcription-PCR analysis was performed with the following primers: Cav2-fwd: 5’ ATCCCCACCGGCTCAACT, Cav2-rev: 5 ’CCGGCTCTGCGATCACAT, Cav1-fwd: 5’GGTCAACCGCGACCCTAAA, Cav1-rev: 5’ CCTTCCAAATGCCGTCAAA, Spdef-fwd: 5’ TGGATGAAAGAGCGGACTTCA, Spdef-rev: 5’ TCGGTCCAGCTCTCCTCACT, ErbB3-fwd: 5’ CGGTTATGTCATGCCAGATAC, ErbB3-rev: 5’GAACTGAGACCCACTGAAG AAAGG. qRT-PCR was performed in a ABI Prism 7900 sequence detector using the power SYBR green PCR master mix (Applied Biosystems). Each reaction contained 5μl of template cDNA, a final concentration of 0.15μM forward and reverse primer, 12.50μl of 2X SYBR Green Buffer and RNAse free H2O to make a final volume of 25μl. The PCR was performed at 50°C for 2 minutes, denaturing at 95°C for 10 minutes, 40 cycles of 95°C for 15 seconds and 60°C for 30 seconds. Analysis was done using the ΔΔCT method, normalizing first to the average of the housekeeping β-actin. The expression values were log10 transformed and graphs were represented as the mean and error bars represent the SEM. Data points were compared using a two-tailed t-test.
Immunohistochemistry
A tissue microarray (TMA) was generated for immunohistochemical analysis. To account for cancer tissue heterogeneity, 0.6mm inch diameter cores were punched from regions selected on the original tumor slides by H&E to include all patterns of differentiation. The TMA was sectioned at 5μm onto Superfost Plus slides (Fisher Scientific, Fair Lawn, NJ), deparaffinized and rehydrated using a Shandon-Lipshaw Varistain (program 2). Subsequently, slides were stained with hematoxylin and eosin (H&E) and then examined microscopically by a pathologist Immunohistochemistry was performed after heat-induced citrate based antigen retrieval for p-IGFIR/Y1161 (Abcam, ab39398-100), BrdU (5-bromo-2-deoxyuridine), cleaved caspase 3 (CC3, Covance, Berkeley, CA), Ki67 (Dako) using a Vectastain ABC peroxidase immunodetection kit. Slides were counterstained with hematoxylin, dehydrated, and mounted using cytoseal. Immunohistochemistry for total IGF-IR (Ventana) was performed on the Ventana Benchmark XT according to the manufacturer's protocol. Images were taken with a Nuance camera at 20x and 40x magnification.
Immunoblot analysis
Human tumorgraft tissues were lysed by electric homogenizer for 8 seconds in RPPA lysis buffer (1% Triton X-100, 50mM HEPES (pH 7.4), 150mM NaCl, 1.5mM MgCl2, 1mM EGTA, 100mM NaF, 10mM NaPPi, 10% glycerol, 1mM Na3VO4, and protease inhibitor (Roche Applied Science, Indianapolis, IN, USA) and incubated on ice with occasional shaking for 20 minutes. The supernatant was collected by centrifugation at 14,000rpm, 4°C for 10 minutes and quantified by BCA protein assay (Pierce, Rockford, IL, USA). Total protein extract (50-75μg) was resuspended in denaturing sample loading buffer, separated by 8% SDS-PAGE, and transferred to a nitrocellulose membrane overnight at 4°C. The membrane was blocked with phosphate buffered saline plus 0.05% Tween-20 (PBST) containing 5% bovine serum albumin for 1 hour. The following antibodies were utilized: anti-phospho-IRS1 (tyr612,44-816G, Biosource, 1:500), anti-IRS1 (#sc-7200, Santa Cruz Biotechnology, 1:500) anti-phospho-IGF-IR/InsR (Tyr1135/1136, #3024, Cell Signaling, 1:500), anti-IGF-IRß (#sc-713, Santa Cruz Biotechnology, 1:500), anti-phospho-Erk1/2 (#9101, Cell Signaling, 1:1000), anti-Erk1/2 (#06-182, Upstate/Millipore 1:1000), anti-phospho-AKT (Ser473, #9271, Cell Signaling, 1:1000), anti-AKT (#9272, Cell Signaling, 1:1000), and anti-β-actin (#A1978, Sigma, 1:4000). Antibodies were incubated in blocking solution for 4 hours. Subsequently, the membrane was washed three times with PBST for 5min and then incubated with a horseradish peroxidase-linked secondary antibody (Amersham Pharmacia Biotech, Piscataway, NJ) at a dilution of 1:4000 in blocking solution. Bands were visualized by enhanced chemiluminescence (Pierce Biotechnology, Rockford, IL) and captured using an Alpha Innotech 7000 Imager (Alpha Innotech, San Leandro, CA).
Tumorgraft study
All procedures were conducted in accordance with the NIH Guide for the Care and Use of Laboratory Animals and were approved by the IACUC of Baylor College of Medicine. Human breast tumors were obtained as core biopsies or pieces of tumors after surgery and implanted in humanized cleared fat pads of NOD/scid mice for establishing human tumorgrafts. Tumorgraft lines are maintained by transplantation of small pieces (1mm3) of tumorgraft into 4 to 6-week-old female NOD/scid mice (Harlan Sprague-Dawley Inc., Madison, WI). When tumorgrafts reached 1cm3 they were then re-transplanted. For the testing the efficacy of BMS-754807 with and without chemotherapy 4 to 6-week-old female NOD/scid mice were transplanted with a 1mm3 piece of tumorgraft into a cleared number four mammary fat pad. Tumor volume was measured weekly with a digital caliper according to the formula for an ellipsoid sphere: (long dimension) × (short dimension)2/2 = mm3. When tumors reached a volume between 100-200mm3, they were randomized to receive the following treatments: vehicle, 50mg/kg BMS-754807 daily by oral gavauge, 20mg/kg docetaxel weekly by intraperitoneal injection or the combination of 50mg/kg BMS-754807 daily and 20mg/kg docetaxel weekly by intraperitoneal injection. Tumor volume and body weight was measured daily. All mice we sacrificed when control tumors reached 1000mm3. Plots of tumor growth curves and body weight measurements were generated using Graphpad (Prism).
Measurement of serum glucose
Whole blood from mice was collected three hours after the final treatment with single agents or combination of BMS-754807 and docetaxel, inserted into BD microtainer serum separator tubes (VWR, #VT365956), and allowed to clot at room temperature for 1 hour. Samples were then centrifuged for 7min at 12000rpm. The top layer containing the serum was collected from each tube and stored at -80°C. Serum glucose was measured using a Glucose Assay Kit (BioVision - Mountain View, CA). Samples (1μl) were diluted in supplied assay buffer and tested in duplicate using the protocol provided by the supplier. Serum glucose was measured from the standard curve of the supplied glucose positive control. Standard deviation was calculated using the duplicate values in the assay.
Measurement of serum insulin
Serum samples were stored at -80°C and thawed on ice prior to being tested. Serum insulin was measured using the Ultra Sensitive Mouse Insulin ELISA kit (Crystal Chem, Inc – Downers Grove, IL). Samples (0.5 and 1μl) were diluted in kit supplied assay diluent and tested in duplicate. Vendor supplied assay protocol was followed and insulin concentration was measured from the standard curve of the supplied insulin standard. Standard deviation was derived from the duplicate samples.
IGF signature t-score
In order to score each breast cancer cell line within a set for similarity to the IGF-gene signature, a “t-score” for each breast cancer cell line in relation to the IGF-gene signature was derived, similar to previously published analyses (31). The t-score was defined as the two-sided t-statistic comparing the average of the IGF-induced genes with that of the IGF-repressed genes within each breast cancer cell line. The gene expression values in the breast cancer cell line dataset (n=51 breast cancer cell lines, Neve et al.) were first normalized to the median before computing the t-score. For each gene transcription profile dataset, we assigned intrinsic molecular subtypes to the cell lines, essentially as previously described (63), using the human tumor dataset from Hoadley et al. (33) to define the subtype-specific expression patterns.
Statistical methods
To compare the tumor growth pattern among the treatment groups in the tumorgraft study, data were analyzed by a mixed model with treatment group, polynomial terms of day up to the third order, and interactions as fixed-effects; and subject as a random-effect factor. Tumor volumes at day 14 were then compared pairwise among the groups using model-based contrasts. The overall growth curves were also compared by examining the interaction terms in the model. ANOVA analyses and pairwise comparisons with Holm adjustment for multiple comparisons were performed for comparisons of glucose and insulin levels, BrdU, Ki67 and CC3 between the 4 groups: vehicle, BMS-754807, Docetaxel, and combination. To achieve normality, log-transformation was used for glucose and insulin data; Arcsine square-root transformation was used for BrdU, Ki67, and CC3 data because data were expressed as proportion from 0 to 1.
Supplementary Material
Supplementary Figure 1: IGF-induced gene expression signature is present in triple-negative breast cancer A. and B. Genes in the IGF gene signature were examined in two published data sets of clinical breast tumors from Desmedt and Hoadley. Within the ER-positive and ER-negative as well as the distinct breast cancer subtype tumors are ordered from those with the least similarity to the IGF signature pattern to those with the highest similarity to the IGF signature. C. Genes in the IGF-I gene signature were examined in a gene expression profile of colon cancer xenografts treated with the IGF-IR small molecule inhibitor BMS-754807 and the EGFR small molecule inhibitor Erbitux for 1h, 6h, 24h and 15 days. Patterns for genes that are upregulated (yellow) in the IGF gene signature are separate from the patterns for genes that are downregulated (blue). In both data sets, treatment with IGF-IR and EGFR inhibitors reversed expression of the IGF regulated genes. D. Gene expression analysis of ovarian cancer cells 36M2 treated with the chemotherapy agent carboplatin does not result in reversion of the IGF gene signature.
Supplementary Figure 2: Sensitive cell lines express basal markers whereas resistant cell lines express luminal markers. A. The expression of differentially expressed genes were validated by qRT-PCR in a panel of seven sensitive cell lines (MCF7, BT20, MDA-MB-468, HS758T, MDA-MB-231, HCC38, MDA-MB-436) and six resistant cell lines (ZR75-1, SKBR3, BT474, CAMA-1, MDA-MB-134, UACC812). Analysis was done using the ΔΔCT method, normalizing first to the average of the housekeeping β-actin. The results are presented as log10 transformed transcript levels. Graphs represent the mean and error bars represent the SEM. Data points were compared by the two-tailed t-test. B. Protein quantification values for each cell line in A were taken from Neve et al. (38). Bars represent the mean and error bars represent the SEM. Data values were compared by the two-tailed t-test.
Supplementary Figure 3: Triple negative breast cancer cell lines are most responsive to BMS-754807. Breast cancer cells representing each subtype of breast cancer with unknown sensitivity to BMS-754807 were plated in 96 well microtiter plates and incubated overnight. The next day, the start absorbance (cell number at the beginning of the experiment, i.e. at time of drug addition), was measured by MTS assay in which no compound was added. BMS-754807 was serially diluted and added. After 72 hr exposure, MTS assay was performed. Percentage of cell growth inhibition was calculated as % of control = end Absorbance - start Absorbance × 100%. The growth curves are expressed as the percentage of growth observed in untreated controls. Sensitive cell lines: IC50<4μM, moderate: IC50 between 4μM and 14μM, resistant: IC50>14μM.
Supplementary Figure 4: Minimal toxicity was observed in animals at doses that show significant antitumor activity. A. Statistical analysis of tumor volumes at day 14 that were compared pairwise. B. Body weight was measured daily. Graph represents the percentage of body weight compared to treatment initiation. Control mice weigh 104% compared to treatment initiation. Docetaxel and BMS-754807 treated mice weigh 95% and 94%, respectively. Combination treated mice weigh 85% compared to treatment initiation C. After 14 days of treatment serum glucose was measured using a Glucose Assay Kit from BioVision. D. Serum insulin was measured using the Ultra Sensitive Mouse Insulin ELISA kit. E. Representative tumor sections of the treatment groups stained with Ki67 and CC3 (cleaved caspase 3) are taken at 40x magnification.
Acknowledgements
We thank Dr. Max Wicha (University of Michigan Comprehensive Cancer Center, Ann Arbor, MI) for providing the MC1 tumor model and Dr. Melissa Landis, Ricardo Moraes, and Lacey Dobrolecki, and others in the Breast Center at Baylor College of Medicine for technical assistance and helpful suggestions. Thanks to the pathology core at the Lester and Sue Smith Breast Center for immunohistochemistry staining and Stephen Hillerman at Bristol-Myers Squibb for measuring serum glucose and insulin levels. The study was supported in part by a research grants from the National Institute of Health/National Cancer Institute NIH/NCI R01CA94118 (AVL), P50CA58183 (AVL/JC/MTL) and R01-CA138197 (JC/MTL). Beate Litzenburger is a recipient of the Department of Defense Predoctoral Traineeship Award (DAMD-W81XWH-08-1-0220).
Footnotes
Conflict of Interest:
BCL, CJC, AT, BC, SG, TW, JCC, MTL, AVL have no conflict of interest.
JMC, MMG, and FH are employees of Bristol Myers Squibb (BMS).
References
- 1.Early Breast Cancer Trialists’ Collaborative Group Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet. 2005;365:1687–717. doi: 10.1016/S0140-6736(05)66544-0. [DOI] [PubMed] [Google Scholar]
- 2.Pusztai L. Current Status of Prognostic Profiling in Breast Cancer. Oncologist. 2008;13:350–60. doi: 10.1634/theoncologist.2007-0216. [DOI] [PubMed] [Google Scholar]
- 3.Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–74. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sorlie T, Tibshirani R, Parker J, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A. 2003;100:8418–23. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kreike B, van Kouwenhove M, Horlings H, et al. Gene expression profiling and histopathological characterization of triple-negative/basal-like breast carcinomas. Breast Cancer Research. 2007;9:R65. doi: 10.1186/bcr1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rakha E, Tan D, Foulkes W, et al. Are triple-negative tumours and basal-like breast cancer synonymous? Breast Cancer Research. 2007;9:404. doi: 10.1186/bcr1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rakha E, Reis-Filho J, Ellis I. Basal-like breast cancer: a critical review. J Clin Oncol. doi: 10.1200/JCO.2007.13.1748. [DOI] [PubMed] [Google Scholar]
- 8.Cleator S, Heller W, Coombes RC. Triple-negative breast cancer: therapeutic options. Lancet Oncol. 2007;8:235–44. doi: 10.1016/S1470-2045(07)70074-8. [DOI] [PubMed] [Google Scholar]
- 9.Carey LA, Perou CM, Livasy CA, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA. 2006;295:2492–502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 10.Pollak MN, Schernhammer ES, Hankinson SE. Insulin-like growth factors and neoplasia. Nat Rev Cancer. 2004;4:505–18. doi: 10.1038/nrc1387. [DOI] [PubMed] [Google Scholar]
- 11.Kurmasheva RT, Houghton PJ. IGF-I mediated survival pathways in normal and malignant cells. Biochim Biophys Acta. 2006;1766:1–22. doi: 10.1016/j.bbcan.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 12.Carboni JM, Lee AV, Hadsell DL, et al. Tumor development by transgenic expression of a constitutively active insulin-like growth factor I receptor. Cancer Res. 2005;65:3781–7. doi: 10.1158/0008-5472.CAN-04-4602. [DOI] [PubMed] [Google Scholar]
- 13.Jones RA, Campbell CI, Gunther EJ, et al. Transgenic overexpression of IGF-IR disrupts mammary ductal morphogenesis and induces tumor formation. Oncogene. 2006 doi: 10.1038/sj.onc.1209955. [DOI] [PubMed] [Google Scholar]
- 14.Irie HY, Pearline RV, Grueneberg D, et al. Distinct roles of Akt1 and Akt2 in regulating cell migration and epithelial-mesenchymal transition. J Cell Biol. 2005;171:1023–34. doi: 10.1083/jcb.200505087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim HJ, Litzenburger BC, Cui X, et al. Constitutively Active IGF-IR Causes Transformation and Xenograft Growth of Immortalized Mammary Epithelial Cells, and is Accompanied by an Epithelial to Mesenchymal Transition Mediated by NF-kappaB and Snail. Molecular and Cellular Biology. 2007 doi: 10.1128/MCB.01315-06. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yanochko GM, Eckhart W. Type I insulin-like growth factor receptor over-expression induces proliferation and anti-apoptotic signaling in a three-dimensional culture model of breast epithelial cells. Breast Cancer Res. 2006;8:R18. doi: 10.1186/bcr1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Resnik JL, Reichart DB, Huey K, Webster NJ, Seely BL. Elevated insulin-like growth factor I receptor autophosphorylation and kinase activity in human breast cancer. Cancer Research. 1998;58:1159–64. [PubMed] [Google Scholar]
- 18.Law JH, Habibi G, Hu K, et al. Phosphorylated insulin-like growth factor-i/insulin receptor is present in all breast cancer subtypes and is related to poor survival. Cancer Res. 2008;68:10238–46. doi: 10.1158/0008-5472.CAN-08-2755. [DOI] [PubMed] [Google Scholar]
- 19.Key TJ, Appleby PN, Reeves GK, Roddam AW. Insulin-like growth factor 1 (IGF1), IGF binding protein 3 (IGFBP3), and breast cancer risk: pooled individual data analysis of 17 prospective studies. Lancet Oncol. 11:530–42. doi: 10.1016/S1470-2045(10)70095-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haluska P, Shaw HM, Batzel GN, et al. Phase I dose escalation study of the anti insulin-like growth factor-I receptor monoclonal antibody CP-751,871 in patients with refractory solid tumors. Clin Cancer Res. 2007;13:5834–40. doi: 10.1158/1078-0432.CCR-07-1118. [DOI] [PubMed] [Google Scholar]
- 21.Karp DD, Paz-Ares LG, Novello S, et al. Phase II study of the anti-insulin-like growth factor type 1 receptor antibody CP-751,871 in combination with paclitaxel and carboplatin in previously untreated, locally advanced, or metastatic non-small-cell lung cancer. J Clin Oncol. 2009;27:2516–22. doi: 10.1200/JCO.2008.19.9331. [DOI] [PubMed] [Google Scholar]
- 22.Sachdev D, Yee D. Disrupting insulin-like growth factor signaling as a potential cancer therapy. Molecular Cancer Therapeutics. 2007;6:1–12. doi: 10.1158/1535-7163.MCT-06-0080. [DOI] [PubMed] [Google Scholar]
- 23.Lee AV, Jackson JG, Gooch JL, et al. Enhancement of insulin-like growth factor signaling in human breast cancer: estrogen regulation of insulin receptor substrate-1 expression in vitro and in vivo. Molecular Endocrinology. 1999;13:787–96. doi: 10.1210/mend.13.5.0274. [DOI] [PubMed] [Google Scholar]
- 24.Lee AV, Weng CN, Jackson JG, Yee D. Activation of estrogen receptor-mediated gene transcription by IGF-I in human breast cancer cells. J Endocrinol. 1997;152:39–47. doi: 10.1677/joe.0.1520039. [DOI] [PubMed] [Google Scholar]
- 25.Casa AJ, Dearth RK, Litzenburger BC, Lee AV, Cui X. The type I insulin-like growth factor receptor pathway: a key player in cancer therapeutic resistance. Front Biosci. 2008;13:3273–87. doi: 10.2741/2925. [DOI] [PubMed] [Google Scholar]
- 26.Sachdev D, Yee D. Inhibitors of insulin-like growth factor signaling: a therapeutic approach for breast cancer. J Mammary Gland Biol Neoplasia. 2006;11:27–39. doi: 10.1007/s10911-006-9010-8. [DOI] [PubMed] [Google Scholar]
- 27.Weroha S, Haluska P. IGF-1 Receptor Inhibitors in Clinical Trials—Early Lessons. Journal of Mammary Gland Biology and Neoplasia. 2008;13:471–83. doi: 10.1007/s10911-008-9104-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sarfstein R, Maor S, Reizner N, Abramovitch S, Werner H. Transcriptional regulation of the insulin-like growth factor-I receptor gene in breast cancer. Molecular and Cellular Endocrinology. 2006;252:241–6. doi: 10.1016/j.mce.2006.03.018. [DOI] [PubMed] [Google Scholar]
- 29.Adelaide J, Finetti P, Bekhouche I, et al. Integrated Profiling of Basal and Luminal Breast Cancers. Cancer Res. 2007;67:11565–75. doi: 10.1158/0008-5472.CAN-07-2536. [DOI] [PubMed] [Google Scholar]
- 30.Lerma E, Peiro G, Ramon T, et al. Immunohistochemical heterogeneity of breast carcinomas negative for estrogen receptors, progesterone receptors and Her2//neu (basal-like breast carcinomas). Mod Pathol. 2007;20:1200–7. doi: 10.1038/modpathol.3800961. [DOI] [PubMed] [Google Scholar]
- 31.Creighton CJ, Casa A, Lazard Z, et al. Insulin-like growth factor-I activates gene transcription programs strongly associated with poor breast cancer prognosis. J Clin Oncol. 2008;26:4078–85. doi: 10.1200/JCO.2007.13.4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Desmedt C, Piette F, Loi S, et al. Strong Time Dependence of the 76-Gene Prognostic Signature for Node-Negative Breast Cancer Patients in the TRANSBIG Multicenter Independent Validation Series. Clinical Cancer Research. 2007;13:3207–14. doi: 10.1158/1078-0432.CCR-06-2765. [DOI] [PubMed] [Google Scholar]
- 33.Hoadley K, Weigman V, Fan C, et al. EGFR associated expression profiles vary with breast tumor subtype. BMC Genomics. 2007;8:258. doi: 10.1186/1471-2164-8-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shang Y, Mao Y, Batson J, et al. Antixenograft tumor activity of a humanized anti-insulin-like growth factor-I receptor monoclonal antibody is associated with decreased AKT activation and glucose uptake. Molecular Cancer Therapeutics. 2008;7:2599–608. doi: 10.1158/1535-7163.MCT-07-2401. [DOI] [PubMed] [Google Scholar]
- 35.Pappano W, Jung P, Meulbroek J, et al. Reversal of oncogene transformation and suppression of tumor growth by the novel IGF1R kinase inhibitor A-928605. BMC Cancer. 2009;9:314. doi: 10.1186/1471-2407-9-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Whitfield ML, Sherlock G, Saldanha AJ, et al. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol Biol Cell. 2002;13:1977–2000. doi: 10.1091/mbc.02-02-0030.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loboda A, Nebozhyn M, Cheng C, et al. Biomarker Discovery: Identification of a Growth Factor Gene Signature. Clin Pharmacol Ther. 2009;86:92–6. doi: 10.1038/clpt.2009.48. [DOI] [PubMed] [Google Scholar]
- 38.Neve RM, Chin K, Fridlyand J, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–27. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garber K. From human to mouse and back: ‘tumorgraft’ models surge in popularity. J Natl Cancer Inst. 2009;101:6–8. doi: 10.1093/jnci/djn481. [DOI] [PubMed] [Google Scholar]
- 40.Ginestier C, Hur MH, Charafe-Jauffret E, et al. ALDH1 Is a Marker of Normal and Malignant Human Mammary Stem Cells and a Predictor of Poor Clinical Outcome. Cell Stem Cell. 2007;1:555–67. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zeng X, Sachdev D, Zhang H, Gaillard-Kelly M, Yee D. Sequencing of type I insulin-like growth factor receptor inhibition affects chemotherapy response in vitro and in vivo. Clin Cancer Res. 2009;15:2840–9. doi: 10.1158/1078-0432.CCR-08-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leney SE, Tavare JM. The molecular basis of insulin-stimulated glucose uptake: signalling, trafficking and potential drug targets. J Endocrinol. 2009;203:1–18. doi: 10.1677/JOE-09-0037. [DOI] [PubMed] [Google Scholar]
- 43.Morse DL, Gray H, Payne CM, Gillies RJ. Docetaxel induces cell death through mitotic catastrophe in human breast cancer cells. Molecular Cancer Therapeutics. 2005;4:1495–504. doi: 10.1158/1535-7163.MCT-05-0130. [DOI] [PubMed] [Google Scholar]
- 44.Vakifahmetoglu H, Olsson M, Zhivotovsky B. Death through a tragedy: mitotic catastrophe. Cell Death Differ. 2008;15:1153–62. doi: 10.1038/cdd.2008.47. [DOI] [PubMed] [Google Scholar]
- 45.Mukohara T, Shimada H, Ogasawara N, et al. Sensitivity of breast cancer cell lines to the novel insulin-like growth factor-1 receptor (IGF-1R) inhibitor NVP-AEW541 is dependent on the level of IRS-1 expression. Cancer Letters. 2009;282:14–24. doi: 10.1016/j.canlet.2009.02.056. [DOI] [PubMed] [Google Scholar]
- 46.Huang F, Greer A, Hurlburt W, et al. The mechanisms of differential sensitivity to an insulin-like growth factor-1 receptor inhibitor (BMS-536924) and rationale for combining with EGFR/HER2 inhibitors. Cancer Res. 2009;69:161–70. doi: 10.1158/0008-5472.CAN-08-0835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Litzenburger BC, Kim H-J, Kuiatse I, et al. BMS-536924 Reverses IGF-IR-Induced Transformation of Mammary Epithelial Cells and Causes Growth Inhibition and Polarization of MCF7 Cells. Clinical Cancer Research. 2009;15:226–37. doi: 10.1158/1078-0432.CCR-08-0801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gong Y, Yao E, Shen R, et al. High expression levels of total IGF-1R and sensitivity of NSCLC cells in vitro to an anti-IGF-1R antibody (R1507). PLoS One. 2009;4:e7273. doi: 10.1371/journal.pone.0007273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zha J, O'Brien C, Savage H, et al. Molecular predictors of response to a humanized anti–insulin-like growth factor-I receptor monoclonal antibody in breast and colorectal cancer. Molecular Cancer Therapeutics. 2009;8:2110–21. doi: 10.1158/1535-7163.MCT-09-0381. [DOI] [PubMed] [Google Scholar]
- 50.Gualberto A, Dolled-Filhart M, Gustavson M, et al. Molecular analysis of non-small cell lung cancer identifies subsets with different sensitivity to insulin-like growth factor I receptor inhibition. Clin Cancer Res. 16:4654–65. doi: 10.1158/1078-0432.CCR-10-0089. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 51.Lisztwan J, Pornon A, Chen B, Chen S, Evans DB. The aromatase inhibitor letrozole and inhibitors of insulin-like growth factor I receptor synergistically induce apoptosis in in vitro models of estrogen-dependent breast cancer. Breast Cancer Res. 2008;10:R56. doi: 10.1186/bcr2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cohen BD, Baker DA, Soderstrom C, et al. Combination therapy enhances the inhibition of tumor growth with the fully human anti-type 1 insulin-like growth factor receptor monoclonal antibody CP-751,871. Clin Cancer Res. 2005;11:2063–73. doi: 10.1158/1078-0432.CCR-04-1070. [DOI] [PubMed] [Google Scholar]
- 53.Fiebig HH. 13 Patient and cell line derived human tumor xenograft models - preclinical/clinical correlations. European Journal of Cancer. 2004;2:8. [Google Scholar]
- 54.Santana VM, Furman WL, Billups CA, et al. Improved response in high-risk neuroblastoma with protracted topotecan administration using a pharmacokinetically guided dosing approach. J Clin Oncol. 2005;23:4039–47. doi: 10.1200/JCO.2005.02.097. [DOI] [PubMed] [Google Scholar]
- 55.Gooch JL, Van Den Berg CL, Yee D. Insulin-like growth factor (IGF)-I rescues breast cancer cells from chemotherapy-induced cell death--proliferative and anti-apoptotic effects. Breast Cancer Res Treat. 1999;56:1–10. doi: 10.1023/a:1006208721167. [DOI] [PubMed] [Google Scholar]
- 56.Miglietta A, Panno ML, Bozzo F, Gabriel L, Bocca C. Insulin can modulate MCF-7 cell response to paclitaxel. Cancer Lett. 2004;209:139–45. doi: 10.1016/j.canlet.2003.12.023. [DOI] [PubMed] [Google Scholar]
- 57.Fowler CA, Perks CM, Newcomb PV, Savage PB, Farndon JR, Holly JM. Insulin-like growth factor binding protein-3 (IGFBP-3) potentiates paclitaxel-induced apoptosis in human breast cancer cells. Int J Cancer. 2000;88:448–53. [PubMed] [Google Scholar]
- 58.Beech DJ, Parekh N, Pang Y. Insulin-like growth factor-I receptor antagonism results in increased cytotoxicity of breast cancer cells to doxorubicin and taxol. Oncol Rep. 2001;8:325–9. doi: 10.3892/or.8.2.325. [DOI] [PubMed] [Google Scholar]
- 59.Wu JD, Haugk K, Coleman I, et al. Combined in vivo effect of A12, a type 1 insulin-like growth factor receptor antibody, and docetaxel against prostate cancer tumors. Clin Cancer Res. 2006;12:6153–60. doi: 10.1158/1078-0432.CCR-06-0443. [DOI] [PubMed] [Google Scholar]
- 60.Carboni JM, Wittman M, Yang Z, et al. BMS-754807, a small molecule inhibitor of insulin-like growth factor-1R/IR. Molecular Cancer Therapeutics. 2009;8:3341–9. doi: 10.1158/1535-7163.MCT-09-0499. [DOI] [PubMed] [Google Scholar]
- 61.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: Expression index computation and outlier detection.. Proceedings of the National Academy of Sciences of the United States of America; 2001. pp. 31–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li C, Hung Wong W. Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biology. 2001;2:research0032.1–research.11. doi: 10.1186/gb-2001-2-8-research0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Creighton CJ, Li X, Landis M, et al. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc Natl Acad Sci U S A. 2009;106:13820–5. doi: 10.1073/pnas.0905718106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: IGF-induced gene expression signature is present in triple-negative breast cancer A. and B. Genes in the IGF gene signature were examined in two published data sets of clinical breast tumors from Desmedt and Hoadley. Within the ER-positive and ER-negative as well as the distinct breast cancer subtype tumors are ordered from those with the least similarity to the IGF signature pattern to those with the highest similarity to the IGF signature. C. Genes in the IGF-I gene signature were examined in a gene expression profile of colon cancer xenografts treated with the IGF-IR small molecule inhibitor BMS-754807 and the EGFR small molecule inhibitor Erbitux for 1h, 6h, 24h and 15 days. Patterns for genes that are upregulated (yellow) in the IGF gene signature are separate from the patterns for genes that are downregulated (blue). In both data sets, treatment with IGF-IR and EGFR inhibitors reversed expression of the IGF regulated genes. D. Gene expression analysis of ovarian cancer cells 36M2 treated with the chemotherapy agent carboplatin does not result in reversion of the IGF gene signature.
Supplementary Figure 2: Sensitive cell lines express basal markers whereas resistant cell lines express luminal markers. A. The expression of differentially expressed genes were validated by qRT-PCR in a panel of seven sensitive cell lines (MCF7, BT20, MDA-MB-468, HS758T, MDA-MB-231, HCC38, MDA-MB-436) and six resistant cell lines (ZR75-1, SKBR3, BT474, CAMA-1, MDA-MB-134, UACC812). Analysis was done using the ΔΔCT method, normalizing first to the average of the housekeeping β-actin. The results are presented as log10 transformed transcript levels. Graphs represent the mean and error bars represent the SEM. Data points were compared by the two-tailed t-test. B. Protein quantification values for each cell line in A were taken from Neve et al. (38). Bars represent the mean and error bars represent the SEM. Data values were compared by the two-tailed t-test.
Supplementary Figure 3: Triple negative breast cancer cell lines are most responsive to BMS-754807. Breast cancer cells representing each subtype of breast cancer with unknown sensitivity to BMS-754807 were plated in 96 well microtiter plates and incubated overnight. The next day, the start absorbance (cell number at the beginning of the experiment, i.e. at time of drug addition), was measured by MTS assay in which no compound was added. BMS-754807 was serially diluted and added. After 72 hr exposure, MTS assay was performed. Percentage of cell growth inhibition was calculated as % of control = end Absorbance - start Absorbance × 100%. The growth curves are expressed as the percentage of growth observed in untreated controls. Sensitive cell lines: IC50<4μM, moderate: IC50 between 4μM and 14μM, resistant: IC50>14μM.
Supplementary Figure 4: Minimal toxicity was observed in animals at doses that show significant antitumor activity. A. Statistical analysis of tumor volumes at day 14 that were compared pairwise. B. Body weight was measured daily. Graph represents the percentage of body weight compared to treatment initiation. Control mice weigh 104% compared to treatment initiation. Docetaxel and BMS-754807 treated mice weigh 95% and 94%, respectively. Combination treated mice weigh 85% compared to treatment initiation C. After 14 days of treatment serum glucose was measured using a Glucose Assay Kit from BioVision. D. Serum insulin was measured using the Ultra Sensitive Mouse Insulin ELISA kit. E. Representative tumor sections of the treatment groups stained with Ki67 and CC3 (cleaved caspase 3) are taken at 40x magnification.