Abstract
In the mouse, each class of olfactory receptor neurons expressing a given odorant receptor converges their axons onto two specific glomeruli in the olfactory bulb (OB), thereby creating an odor map. How is this map represented in the olfactory cortex? Here we combine rabies virus-dependent retrograde mono-transsynaptic labeling with genetics to control the location, number and type of ‘starter’ cortical neurons, from which we trace their presynaptic neurons. We find that individual cortical neurons receive input from multiple mitral cells representing broadly distributed glomeruli. Different cortical areas represent the OB input differently. For example, the cortical amygdala preferentially receives dorsal OB input, whereas the piriform cortex samples the whole OB without obvious bias. These differences likely reflect different functions of these cortical areas in mediating innate odor preference or associative memory. The transsynaptic labeling method described here should be widely applicable to mapping connections throughout the mouse nervous system.
The functions of mammalian brains are based on activity patterns of large numbers of inter-connected neurons that form information processing circuits. Neural circuits consist of local connections, where pre- and post-synaptic partners reside within the same brain area, and long-distance connections, which link different areas. Local connections can be predicted by axon and dendrite reconstructions1, and confirmed by physiological recording and stimulation methods2. Long-distance connections are more difficult to map, as commonly used methods can only trace bulk projections with a coarse resolution. Most methods cannot distinguish axons in passing from those that form synapses, or pinpoint neuronal types to which connections are made1,2. Transsynaptic tracers can potentially overcome these limitations2. Here we combine a retrograde rabies virus (RV)-dependent mono-transsynaptic labeling technique3 with genetic control of the location, number and cell type of ‘starter’ neurons to trace their presynaptic partners. We systematically mapped long-distance connections between the first olfactory processing center, the olfactory bulb (OB), and its postsynaptic targets in the olfactory cortex (Supplementary Fig. 1).
Genetic Control of Transsynaptic Tracing
RV can cross synapses from postsynaptic to presynaptic neurons with high specificity4, without notable defects in the morphology or physiology of infected neurons for extended periods of time3,5. Recent genetic modifications of RV have permitted mono-transsynaptic labeling3. Specifically, the rabies envelope glycoprotein (G) required for viral spread was replaced with a fluorescent marker6. Additionally, the virus was pseudotyped with EnvA, an avian virus envelope protein that lacks an endogenous receptor in mammals, and thus cannot infect wild-type mammalian cells. However, it can infect cells expressing the EnvA receptor TVA, and can subsequently produce infectious particles if TVA-expressing cells also express G to complement the ΔG RV (Fig. 1a, bottom). The new viral particles can cross synapses to label presynaptic partners of ‘starter’ neurons. As transsynaptically infected neurons do not express G, the modified virus cannot spread from them to other neurons. Paired recordings in cultured brain slices support the efficacy and specificity of this strategy3.
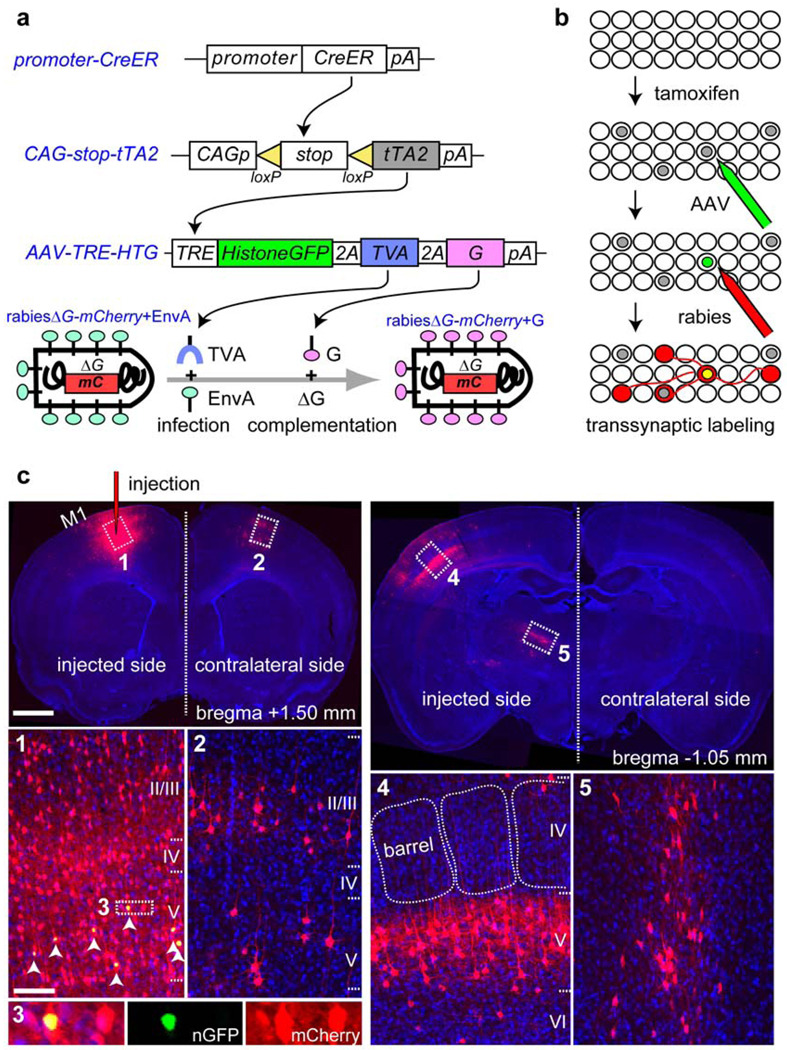
Figure 1. Genetic control of rabies-mediated neural circuit tracing.
a-b, Schematic representation of the methodology used to control the location, number and type of starter cells for RV-mediated transsynaptic labeling. tTA2 is expressed in a small subset of CreER(+) cells (grey nuclei in b). tTA2 activates an AAV-delivered transgene to express: 1) a histone-GFP marker to label the nuclei of starter cells in green, 2) EnvA receptor (TVA) to enable subsequent infection by EnvA-pseudotyped RV (rabies ΔG-mCherry+EnvA), and 3) rabies glycoprotein (G) to initiate transsynaptic labeling. c, Top left, a 60-µm coronal section that includes the injection site in the motor cortex (M1). Cells labeled with both histone-GFP (n-GFP) and mCherry (arrowheads in c1, magnified in c3) can be distinguished from cells labeled with mCherry alone, which are found near the injection site (c1), in the contralateral motor cortex (c2), in the somatosensory barrel cortex (top right; magnified in c4), and in the motor-specific ventrolateral nucleus of the thalamus (c5). Scale bars, 1 mm for low-magnification images on top, 100 µm for high-magnification images at the bottom.
To extend this method to a limited number of ‘starter’ cells of a defined type and at a precise location in vivo, we combined mouse genetics and viral infections (Fig. 1a-b). We created a transgenic mouse (CAG-stop-tTA2) that conditionally expresses the tetracycline transactivator tTA2 under the control of a ubiquitous CAG promoter only upon Cre-mediated excision of a transcriptional stop cassette. After crossing these mice with transgenic mice expressing the tamoxifen-inducible Cre (CreER), a small fraction of CreER(+) cells also express tTA2 following tamoxifen induction. We then used stereotactic injections to deliver into specific regions of the brain an adeno-associated virus (AAV) serotype 2 expressing three proteins: histone-GFP, TVA and G, under the control of a tetracycline-response element (TRE). Expression of TVA and G allows infected, tTA2(+) cells to be receptive to infection by the modified RV, which we injected into the same location two weeks later. We define starter cells as those infected by both AAV and RV, and therefore labeled by both histone-GFP and mCherry; their presynaptic partners are infected only by RV and therefore express only mCherry.
We tested our strategy by using a ubiquitously expressing actin-CreER7 in combination with CAG-stop-tTA2 in the neocortex. Starter cells could be unambiguously identified by histone-GFP expression (Supplementary Fig. 2). In all but one case, we observed more than one starter cell (Supplementary Fig. 3 shows the example of a single starter cell). In a typical example, 35 starter cells in the motor cortex expressed histone-GFP and mCherry (Fig. 1c3), demonstrating that AAV and RV can infect the same cells in vivo. In addition to many locally labeled cells (Fig. 1c1), mCherry(+) cells were enriched in layer II, III and V in the contralateral motor cortex (Fig. 1c2), consistent with layer specificity of callosal projections8. mCherry(+) cells were also found in layer III and V of the ipsilateral somatosensory cortex (Fig. 1c4) and motor-specific thalamic nuclei (Fig. 1c5), which are known sources of monosynaptic inputs to the motor cortex9.
In all experiments, histone-GFP(+) cells were found within 450 µm from injection sites, consistent with a previous report that AAV serotype 2 predominantly infects neurons locally10. Omitting AAV or tamoxifen yielded no transsynaptically labeled neurons (Supplementary Fig. 4). Moreover, our strategy labeled neurons only through synaptic connections but not through axons in passage (Supplementary Fig. 5). Finally, RV spread was restricted to neurons directly connected to starter cells, and only in the retrograde direction (Supplementary Fig. 6). Together, these experiments validated our genetic strategy for retrograde mono-transsynaptic labeling in vivo.
AON Maintains Dorsal-Ventral Topography
In the mouse, olfactory receptor neurons (ORNs) that express a single type of odorant receptor send convergent axonal projections to a specific pair of glomeruli in the lateral and medial OB11–13. Odorants are detected by combinations of ORN classes14, and are represented as spatiotemporal activity patterns of glomeruli15. Each mitral cell sends its apical dendrite to a single glomerulus and thus receives direct input from a single ORN class. Mitral cell axons relay information to the olfactory cortex (Supplementary Fig. 1a). Previous axon tracing studies showed that individual mitral cells send axons to distinct cortical areas, and that small cortical regions receive broad input from the OB16–19. However, understanding the principles underlying odor perception and odor-mediated behaviors requires systematic and quantitative analysis of connection patterns of mitral cells with their cortical target neurons.
We first established that mitral cells throughout the OB can be infected by RV via their axons (Supplementary Fig. 7). We then applied our strategy (Fig. 1a-b) to specific areas of the anterior olfactory nucleus (AON), piriform cortex, and cortical amygdala (Supplementary Fig. 1b), and examined the distribution of transsynaptically labeled mitral cells. In a typical example, 11 clustered starter cells in the AON (Fig. 2a) resulted in 69 labeled mitral cells distributed widely across the OB (Supplementary Fig. 8; Supplementary Movie 1). Bright mCherry fluorescence from RV allowed us to unequivocally follow the primary dendrites of the labeled mitral cells to single target glomeruli (Fig. 2b). Each mitral cell sent its apical dendrite into a single glomerulus. Four glomeruli were each innervated by two labeled mitral cells (Fig. 2b, right; Supplementary Table 1).
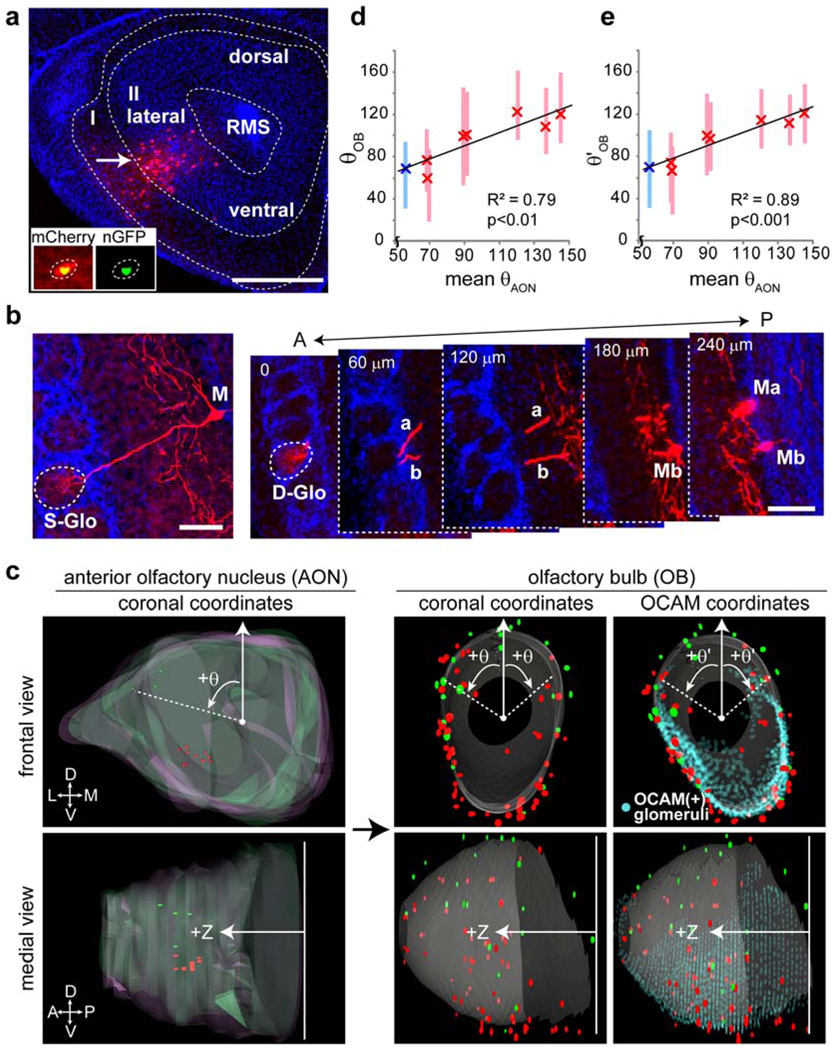
Figure 2. The OB to AON connections display a dorsal-ventral topography.
a, A 60 µm coronal section with two starter cells located in layer II of the ventrolateral AON, one of which (arrow) is magnified in the inset. RMS, rostral migratory stream. Scale, 500 µm. b, Typical examples of transsynaptically labeled mitral cells from cortical starter cells. Left, a 60 µm coronal section that captures both the cell body and the apical dendrite of a mitral cell. Right, more frequently, a mitral cell apical dendrite spans several consecutive 60 µm coronal sections. S-Glo/D-Glo, glomerulus innervated by a single or two (Ma, Mb) labeled mitral cells. A, anterior; P, posterior. Scale bar, 100 µm. c, Superimposed 3D-reconstructions of the AONs and the OBs from two injected brains. 11 red and 4 green starter cells from two AONs labeled red and green glomeruli, respectively. Light red and green, contours of two superimposed AONs. d-e, Correlations between θAON and θOB (d) and θAON and θ’OB (e). Crosses represent mean θAON (x-axis) and mean θOB or θ’OB (y-axis). Bars represent 50% of the distribution surrounding the mean θOB or θ’OB. R, Pearson’s correlation coefficient; p, statistical significance tested against the null hypothesis assuming no correlation between θAON and θOB or θ’OB. Red and blue, experiments using actin-CreER and CaMKII-CreER48, respectively.
To quantitatively compare the patterns of labeled glomeruli from different animals, we established a 3D-reconstruction protocol for the OB, and aligned each OB to a standard OB model (Fig. 2c). To test the accuracy of this procedure, we reconstructed and aligned OBs from three P2-IRES-tauGFP transgenic mice20. These GFP-labeled glomeruli were located within a distance of a few glomeruli from each other (Supplementary Fig. 9), consistent with the natural variability of ORN axon targeting21. This precision of our 3D reconstruction enables the comparison of OBs from different animals.
The AON has been proposed to provide feedforward modification of information from the OB to the piriform cortex22. Little is known about its organization except for a small and distinct AON pars externa, which maintains dorsal-ventral (DV) OB topography23–25. We injected AAV and RV to different areas of the AON (Supplemental Table 1), and established an AON 3D-reconstruction protocol analogous to that for the OB (Fig. 2c, left). Labeled glomeruli from AON injections were distributed widely in the OB (Fig. 2c, middle). However, starter cells from the ventral and dorsal AON preferentially labeled ventral and dorsal glomeruli, respectively (Fig 2c). To quantify the spatial distributions of starter cells in the AON and transsynaptically labeled glomeruli in the OB, we introduced a cylindrical coordinate system into the OB and AON models, where Z represents the position along the anterior-posterior axis and θ represents the angle from the polar axis (Fig. 2c). No correlations were found between ZAON and θOB, ZAON and ZOB, or θAON and ZOB (Supplementary Fig. 10a). However, we found a strong positive correlation (R2=0.79) between θAON and θOB (Fig. 2d), which correspond to the DV axes of AON and OB, respectively. Thus, the AON maintains the DV topography of the OB.
A coarse topography exists between ORN cell body positions in the olfactory epithelium and target glomeruli in the OB along the DV axis26. Specifically, the olfactory cell adhesion molecule (OCAM) is expressed in a subset of ORNs27 that project to the ventral ~55% glomeruli in the OB. In the OB, ~25° clockwise rotation of the polar axis around the Z axis maximized the separation of OCAM(+) and OCAM(−) glomeruli (Supplementary Fig. 11). In this new OCAM coordinate system represented by θ'OB (Fig. 2c, right), the correlation coefficient between θ’OB and θAON increased to R2=0.89 (Fig. 2e), showing that adjusting the DV axis of the OB according to a biological marker improved the AON and OB topographic correspondence. Thus, the topography between the olfactory epithelium and the OB further extends to the AON.
Dorsally Biased OB Input to Amygdala
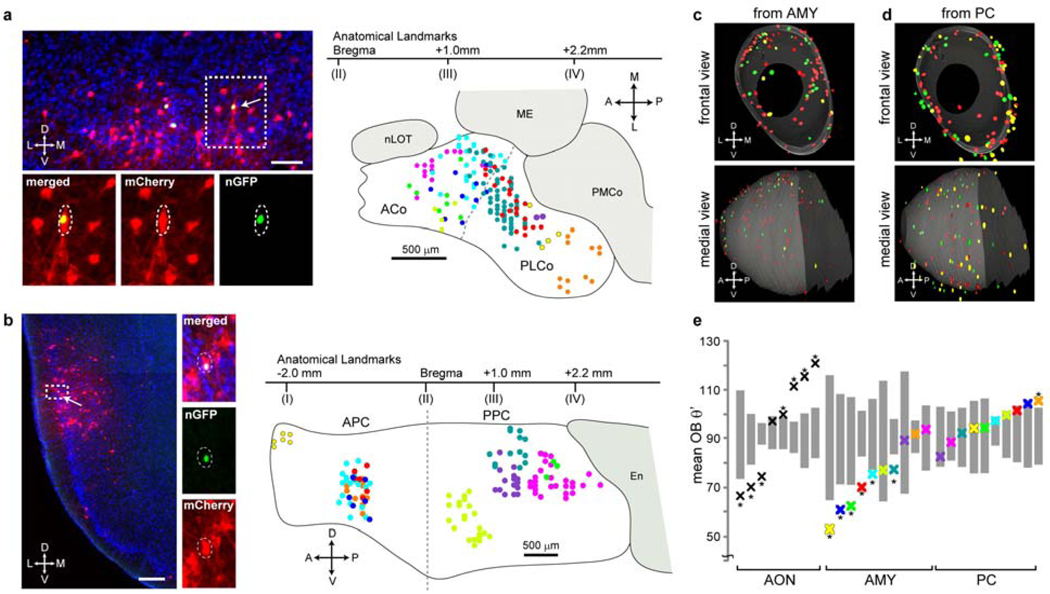
Mitral cell axons project to the anterior and posterolateral cortical amygdala (ACo and PLCo)28,29. The organization of this axonal input is unknown. We injected AAV and RV to small areas within these regions, and mapped starter cells onto a common schematic drawing based on anatomical landmarks (Fig. 3a). Transsynaptically labeled mitral cells and glomeruli from amygdala starter cells were broadly distributed in the OB. However, the labeled glomeruli were enriched in the dorsal OB (Fig. 3c). For quantification, we compared the mean experimental θ’OB for each injection with mean θ’OB values produced by computer simulation from the same number of glomeruli distributed randomly throughout the OB (simθ’OB). For the AON experiments, the mean experimental θ’OB values for the majority of the samples were significantly larger or smaller than the corresponding mean simθ’OB values (Fig. 3e, left), reflecting the DV topography between the OB and the AON. By contrast, none of the mean θ’OB from amygdala was significantly larger than the corresponding mean simθ’OB (Fig. 3e, middle). Six out of ten mean θ’OB values from cortical amygdala fell significantly below the corresponding mean simθ’OB values. For these dorsally biased samples, the density of labeled glomeruli gradually decreased along the DV axis without a sharp boundary (Supplementary Fig. 12). Simple spatial correspondence between starter cell locations and the degree of dorsal bias was not evident (Supplementary Fig. 10b). In summary, the cortical amygdala overall receives biased input from the dorsal OB.
Figure 3. Representations of OB input in the amygdala and piriform cortex.
a-b, Starter cells from the cortical amygdala and piriform cortex. Left, single coronal sections at the injection sites in the posterolateral cortical amygdala (a) and the posterior piriform cortex (b). Arrows point to starter cells magnified in insets. Scale bars, 100 µm in a, 200 µm in b. Right, schematic representations of 10 independent injections each into amygdala (a) or piriform cortex (b). Starter cells from each injection are labeled with a specific color. The dotted line denotes the rough border between ACo (anterior cortical amygdala) and PLCo (posterolateral cortical amygdala) based on anatomical landmarks according to a mouse brain atlas49. nLOT, nucleus of lateral olfactory tract; ME, medial amygdala; PMCo, posteromedial cortical amygdala; APC, anterior piriform cortex; PPC, posterior piriform cortex; En, lateral entorhinal cortex. c-d, Superposition of three independent 3D-reconstructions of glomerular maps with starter cells from the cortical amygdala (c) or the piriform cortex (d). e, Mean θ’OB values (crosses) from each experiment are plotted in the same column with the 95% confidence intervals for corresponding simθ’OB values (gray bars). Samples with experimental mean θ’OB outside the 95% confidence intervals are labeled with asterisks (*, p<0.05). Colors in a (scheme), c and e (AMY) are matched to represent the same samples, and so are the colors in b (scheme), d and e (PC).
Less Organized OB Input to Piriform Cortex
The piriform cortex is the largest cortical area in the olfactory cortex. Recent physiological analysis30,31 found that neurons activated by specific odors are apparently not spatially organized; the underlying anatomical basis is unclear. We injected AAV and RV into several areas in the anterior and posterior piriform cortex, and mapped starter cells from different brains onto a common schematic drawing of the entire piriform cortex based on anatomical landmarks (Fig. 3b). Labeled glomeruli were broadly distributed throughout the OB, regardless of starter cell locations in the piriform cortex (Fig. 3d). In sharp contrast to transsynaptic labeling from AON or amygdala, where different samples showed highly variable mean θ’OB, mean θ’OB values from the piriform cortex tracings were much less variable and closely resembled a random distribution (Fig. 3e). Only one out of ten samples had a mean θ’OB slightly above the 95th percentile of the mean simθ’OB. Furthermore, no strong spatial correspondence was evident in correlation analyses of θ’OB, ZOB and the location of starter cells in the piriform cortex (Supplementary Fig. 10c). These data indicate that highly restricted areas of the piriform cortex receive direct mitral cell input representing glomeruli that are distributed throughout the OB with no apparent spatial organization.
Convergence of Mitral Cell Input
Convergent inputs from different glomeruli to individual cortical neurons could allow the olfactory cortex to integrate combinatorial odor representations in the OB. In support of this, previous studies have shown that odor receptive ranges of AON cells are broader than those of mitral cells32, and that some piriform cortex neurons are activated by a binary odor mix but not individual components31. However, a large fraction of inputs in these studies could come from other cortical neurons through extensive recurrent connections (Fig. 2a, 3a-b). Direct convergence of mitral cell axons onto individual cortical neurons is implied in physiological studies of piriform cortical neurons in slices33–35. Our transsynaptic labeling enabled a direct examination of mitral cell convergence to individual cortical neurons in vivo.
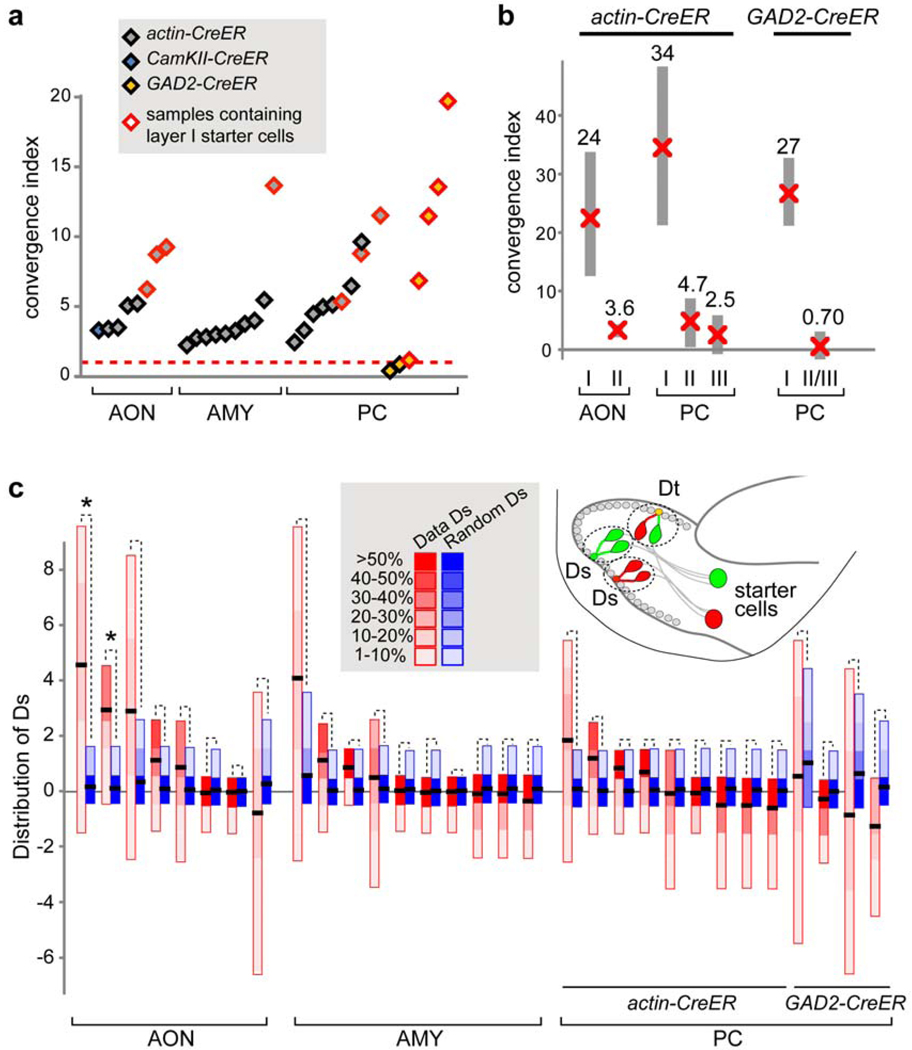
The convergence index, defined by the number of labeled mitral cells divided by the number of the starter cells in the cortex, exceeded 1 in all experiments using actin-CreER (Fig. 4a, Supplementary Table 1). This finding demonstrates that individual cortical neurons receive direct inputs from multiple mitral cells in vivo. As the vast majority of labeled mitral cells corresponded to different glomeruli (Supplementary Table 1), individual cortical neurons must receive direct inputs representing multiple glomeruli. This convergence index is likely an underestimate, as not all starter cells necessarily received direct mitral cell input (overestimation of the denominator), and not all cells presynaptic to starter cells were transsynaptically infected by the RV (underestimation of the numerator; see Supplementary Fig. 3).
Figure 4. Convergence and independence of mitral cell inputs.
a, Convergence index for each cortical injection experiment is represented by a diamond, with the type and layer of starter cells specified by the color code above. b, Multiple regression analysis to estimate the convergence indices of starter cells located in different layers of the AON and PC. Estimated mean convergence indices (“x”) and the corresponding 95% confidence intervals (grey bars) are shown. Data from actin-CreER and GAD2-CreER were analyzed separately. Injections into amygdala produced only one sample that contained layer I cells and were therefore excluded. c, Schematic of dually labeled glomeruli (D) resulting from two starter cells (Dt) or a single starter cell (Ds). Comparison of the distributions of Ds derived from experimentally observed frequency of D (Data Ds, red) and from simulated D based on the null hypothesis detailed in the Methods (Random Ds, blue). For each sample, the distributions of “Data Ds” and “Random Ds” are shown by colored heat maps. *, p<0.05.
The convergence indices varied widely in different experiments, and did not differ substantially in the three cortical areas we examined. However, in experiments that contained starter cells located in layer I, which is mostly composed of GABAergic local interneurons36, the convergence indices were greater (Fig. 4a, red). Assuming all starter cells in a given layer contribute equally to mitral cell labeling, multiple regression analyses suggest that layer I neurons receive direct input from more mitral cells than layer II/III neurons (Fig. 4b).
To confirm the higher convergence index for layer I GABAergic neurons, we replaced the ubiquitous actin-CreER with GAD2-CreER, which is expressed only in GABAergic interneurons (Supplementary Fig. 13). We found that GABAergic neurons located in layer II or III of the piriform cortex received little direct mitral cell input, whereas those located in layer I showed a much greater convergence index (Fig. 4b, right; Supplemental Table 1). Thus, cortical GABAergic neurons are highly diverse with respect to mitral cell innervation. These observations agree with recent physiological studies30,34, and suggest different physiological roles for these GABAergic neurons: layer I and deeper layer GABAergic neurons provide global feedforward and feedback inhibition to cortical pyramidal neurons, respectively.
Sister Mitral Cells Connect Independently
Each glomerulus is innervated by the apical dendrites of ~25 electrically coupled mitral cells37. We refer to these cells as “sister” mitral cells. Are sister mitral cells more likely to connect to the same cortical postsynaptic target neurons compared to “non-sister” mitral cells that receive direct input from different glomeruli? Such an organization could increase the signal-to-noise ratio in information transmission from mitral cells to cortical neurons. Alternatively, sister mitral cells may connect to cortical neurons independently to deliver olfactory information widely across different cortical neurons.
We used the frequency of dually labeled glomeruli from our dataset and statistical simulation to distinguish between these possibilities. Dually labeled glomeruli (D) could result from a single starter cell (Ds) or two starter cells (Dt). Assuming that an individual starter cell can receive input from any of the 2000 glomeruli, we compared the distribution of Ds derived from our data and from a simulation according to the null hypothesis that sister mitral cells connect independently with postsynaptic targets. If sister mitral cells share significantly more postsynaptic targets than at random, then the “data Ds” distribution should be significantly higher than the simulated “random Ds” distribution. In all but two cases, these two distributions were not statistically different (Fig. 4c). Both exceptions came from transsynaptic labeling from the AON, which exhibited DV topography, so the original assumptions were not accurate. When we reduced the number of accessible glomeruli to 1500, no sample showed significant differences. Thus, our analysis suggests that individual mitral cells innervating the same glomerulus act independently in making connections with their cortical targets.
DISCUSSION
Our study revealed several general principles that define cortical representations of the olfactory bulb input. First, individual cortical neurons receive direct input from mitral cells originating from multiple glomeruli. On average, each excitatory neuron receives direct input from 4 mitral cells, but this number is likely to be an underestimate. Convergence of mitral cell inputs enables cortical neurons to integrate information from discrete olfactory channels. The lower bound of 4 already affords ~1012 glomerular combinations for 1000 olfactory channels, far exceeding the number of neurons in the mouse olfactory cortex. Thus, the olfactory cortical neuron repertoire samples only a small fraction of all possible combinations of direct OB inputs.
Second, neurons restricted to small olfactory cortical regions receive input from glomeruli that are broadly distributed in the OB. Although similar findings were reported previously16–19, our study provides a higher resolution analysis of direct connectivity between mitral cells and cortical neurons, rather than inferring connection from the presence of axons, which could be a major caveat of previous tracing studies (see Supplementary Fig. 5). At the same time, mitral cells representing the same glomerulus connect independently to postsynaptic cortical neurons, thus maximizing the spread of olfactory information originating from individual olfactory channels.
Third, different cortical areas receive differentially organized OB input (Supplementary Fig. 1c). The AON maintains a coarse topography along the DV axis, suggesting a pre-processing role for OB derived information before sending to other cortical areas. A lack of apparent spatial organization in the piriform cortex with regard to OB input provides an anatomical basis for recent physiological studies30,31, and suggests that the piriform cortex acts as an association cortex31,38. In the cortical amygdala, many neurons appear to receive strongly biased input from the dorsal OB. Mice lacking ORNs that project to the dorsal OB lose their innate avoidance of odors from predator urine and spoiled food, despite retaining the ability to sense these odors39. The cortical amygdala may preferentially process the olfactory information that directs innate behaviors. Our study agrees with similar findings using axon tracings from individual glomeruli40.
Interestingly, axonal arborization patterns of Drosophila olfactory projection neurons (PNs, equivalent to mitral cells) in higher olfactory centers exhibit a similar organizational principle. PN axon arborization patterns in the lateral horn, a processing center directing odor-mediated innate behavior, are highly stereotyped with respect to PN classes41,42, and are partitioned according to the biological significance of the odorants43. Arborization patterns of axon collaterals of the same PNs in the mushroom body, an olfactory memory center44, are much less stereotyped41,43, consistent with a physiological study suggesting non-stereotyped connections45. Therefore, from insects to mammals, a common theme emerges for the representations of olfactory information: more stereotyped and selective representation of odors is needed for directing innate behaviors while broader and less stereotyped sampling of the whole olfactory space is better suited for brain regions implicated in associative memories.
Lastly, the genetically-controlled mono-transsynaptic tracing described here should be widely applicable for mapping neuronal circuitry throughout the mouse brain. It is currently unknown how RV crosses synapses, and whether the efficiency and specificity vary with cell type, connection strength and activity3,5,46. Further applications of these transsynaptic methods to other neurons and circuits47 will be necessary to address these questions. Nevertheless, the control experiments (Fig. 1, Supplementary Fig. 2–6) confirmed that our strategy labels neurons that are directly presynaptic to starter cells but not neurons whose axons pass through the injection sites without making synapses. Our method will be especially valuable for analyzing long distance connections that are usually refractory to physiological mapping strategies2. This method can be further extended to genetic manipulation of starter cells to combine circuit tracing with genetic loss- or gain-of-function experiments. These approaches will facilitate the investigation of not only the organization of information flow within neural circuits, but also the molecular basis of neuronal connections at a single cell resolution in vivo.
METHODS SUMMARY
Detailed methods on the generation of CAG-stop-tTA2 mice, viral preparations, animal surgery, tissue processing, 3D reconstruction, and quantitative analyses can be found in Methods in the Supplementary Information.
METHODS
Generation of CAG-stop-tTA2 Mice
The tTA2 transactivator gene50 was placed after the CAG promoter of plasmid pCA-HZ2 (Ref. 51) using a PCR-based cloning method. A neomycin resistance (neor) gene and a transcriptional stop signal52 were flanked by loxP sites to create loxP-neor-stop-loxP cassette. This cassette was then introduced between the CAG promoter and tTA2 using PCR-based cloning. An EcoRI fragment obtained from ETLpA-/LTNL13, which contains the IRES-tau-lacZ cassette, was introduced after the tTA2 coding sequence. The resulting cassette (CAG-stop-tTA2-IRES-tau-lacZ) was cloned into pBT264 to flank the cassette with two copies of ~250 bp beta-globin HS4 insulator sequence53 on each side. pBT264 (pii-TRE-tdTomato-3Myc-ii) was generated by inserting PCR-amplified copies of ~250 bp-long core insulator fragment (i) from the chicken beta-globin HS4 insulator on each side of TRE-tdTomato-3Myc in pBT239. The insulator fragments were amplified from pJC13-1 (ref. 53). The final construct pKM1 (pii-CAG-stop-tTA2-IRES-tau-lacZii) was tested by transient co-transfection with pBT264 into cultured HEK293 cells. When a Cre-encoding plasmid pBT140 (CMV promoter driving nuclear localization signal-Cre) was further introduced into the same cell, strong tdTomato fluorescence was detected 72 hours post-transfection. pKM2 was digested with restriction enzymes SwaI and AscI, the insert was gel-purified using Qiagen gel extraction kit and eluted into 10mM Tris-HCl, pH 7.4, 0.1 mM EDTA. The purified and linearized DNA devoid of plasmid backbone was used for mouse transgenesis via standard pronuclear injection procedure. Founders were screened by PCR primers to detect the neor gene. Four independent transgenic lines were established. They were crossed with mice containing β-actin-CreER7 and TRE-Bi-SG-T reporter54 to screen for functional CAG-stop-tTA2 transgenes. Mice containing all three transgenes were injected with 1 mg of tamoxifen in corn oil at postnatal day (PD) 10, and brains were collected at PD21 for the analysis. Two lines showed broad tdTomato fluorescence throughout the brain. One line (containing 2–3 copies of the transgene based on Southern) was used exclusively in this study.
Virus Preparations
All viral procedures followed the Biosafety Guidelines approved by the Stanford University Administrative Panel on Laboratory Animal Care (A-PLAC) and Administrative Panel of Biosafety (APB). To make the AAV containing the TRE-HTG cassette, which encodes histone-GFP, TVA and G linked by the 2A “self-cleaving” peptides, the HTG cassette obtained from pBOB-synP-HTB (I.W. & E.M.C., unpublished plasmid) was placed after the TRE-Tight promoter in pTRE-Tight (Clontech), and then the entire construct was subcloned into the pAAV vector (Stratagene). Recombinant AAV serotype 2 was produced using the pAAV helper free kit (Stratagene) according to the manufacturer’s instructions. AAV was also produced commercially by the Gene Therapy Center of the University of North Carolina. The AAV titer was estimated to be ~4×1012 viral particles/ml based on serial dilution and blot hybridization analysis. Pseudotyped ΔG RV was prepared as previously described3,55. The pseudotyped RV titer was estimated to be ~5×108 infectious particles/ml based on the infections of cell line 293-TVA800 by serially diluted virus stocks.
Animal Surgery
All animal procedures followed animal care guidelines approved by Stanford University’s Administrative Panel on Laboratory Animal Care (A-PLAC). To activate Cre in animals carrying a CreER transgene, we injected intraperitoneally 0.1 mg - 1 mg of tamoxifen (Sigma) dissolved in corn oil into mice around PD10. For transsynaptic labeling, 0.1–0.3 µl of AAV-TRE-HTG was injected into brain at PD21 by using a stereotactic apparatus (KOPF). During surgery, animals were anesthetized with 65mg/kg ketamine and 13mg/kg xylazine (Ben Venue Laboratories). For motor cortex injections, the needle was placed 1.5 mm anterior and 1.5 mm lateral from the Bregma, and 0.4 mm from the brain surface. For olfactory cortex injections, see Supplementary Fig. 1b for the stereotactic parameters. After recovery, animals were housed in regular 12 hour-dark/light cycle with food and water ad libitum. Two weeks later, 0.3 µl of pseudotyped RV (ΔG-mCherry+EnvA) was injected into the same brain location under anesthesia. After recovery, animals were housed in a biosafety room for 7 days to allow RV to infect, transsynaptically spread and express sufficient amount of mCherry to label pre-synaptic cells. All animals were healthy and their brain structures were normal 7 days after RV infection, confirming non-pathogenicity of ΔG-mutant RV.
Tissue Processing
Brain tissue was processed according to previously described procedures56. To set the common coronal plane among different animals, the cerebellum was cut off, and the brain was embedded in the Optimum Cutting Temperature (O.C.T.) compound (Tissue-Tek) with the cut surface facing the bottom of the mold. The brain was adjusted to ensure that the left-right axis was parallel to the section plane. Neither mCherry nor histone-GFP required immunostaining for visualization. In some cases, brain sections were immunostained for better signal preservation according to the published methods56 using the following antibodies: chicken anti-GFP (1:500; Aves Labs), rabbit anti-DsRed (1:1000, Clontech), donkey anti-chicken FITC and donkey anti-rabbit Cy3 (1:200; Jackson ImmunoResearch). In most transsynaptic labeling experiments starting from the olfactory cortex, every one of four sections of the OB was immunostained by the free-floating method with goat anti-OCAM (1:100, R&D Systems) and donkey anti-Goat Alexa488 (Invitrogen) to label OCAM(+) ORN axons. For immunostaining against GABA, 60 µm free-floating coronal sections were treated with rabbit anti-GABA (Sigma, 1:2000 in PBS with 0.3% Triton-X100) for 48 hours. GABA(+) cells were visualized with donkey anti-rabbit Cy3 (1:200, Jackson ImmunoResearch). Sections were imaged with a Nikon CCD camera by using a 10x objective or by 1-µm optical sectioning using confocal microscopy (Zeiss 510).
3D Reconstruction
To compare distribution of labeled glomeruli (OB) and starter cells (AON) across different samples, we needed to map them in a common 3D reference frame. To do this, we first saved manual annotations carried out in Adobe Illustrator in a Scalable Vector Graphics (SVG) format. The SVG file saved all the annotations as an Extensible Markup Language (XML) file describing the ellipses and contours (defined below), making it feasible to accurately parse the information by MATLAB scripts. In the OB, we represented all glomeruli as ellipses. We used the center of mass for each ellipse to define a single point, and calculated the center of mass of all the points to define the center of each slice. For the AON, we defined the contour as the boundary between layer I and layer II, which can be clearly distinguished by differences in the density of DAPI staining. To define the center of mass for each contour, we replaced it with a dense series of points and used these points to calculate the center of mass. Now, each slice is represented by a series of points and the center of mass contained within an SVG file. To assemble the slices represented by SVG files into a 3D shape, we first aligned the center of mass for each slice to that of the previous slice to form the cylindrical (Z) axis. Then, we refined the alignment by sequentially applying the Iterative Closest Points (ICP) algorithm57, which can identify the local rotation and translation parameters for each slice to maximize the overlap with the previous slice. Once we have aligned all the slices in a sample to generate a 3D shape, we needed to identify an orientation for the polar axis that can be most reliably identified in different 3D reconstructions. As the OB is ellipsoidal, the Principle Component Analysis (PCA) can reliably find a plane that contains the Z axis and intersects the 3D shape to maximize the surface of the intersection (plane m). We then defined the polar axis to be contained within the plane m, perpendicular to the Z-axis, and pointing in the dorsal direction. For the AON, we approximated the contours of the most posterior slide of the AON as a triangle and calculated the rotation around the Z-axis that minimizes the distance of the three vertices to those of a standard AON sample. We applied the same rotation to the whole 3D shape. To define the orientation of the polar axis, we used the side of the triangle that connects two of its medial vertices and points in the dorsal direction. Then we defined the polar axis as the line that is parallel to it and that intersects the Z-axis. Finally, we calculated the volume occupied by each shape and applied a uniform scaling factor to account for different sizes of the anatomical structures in different animals.
All the steps explained above were implemented in MATLAB that ran automatically without human intervention to avoid biasing the registration results. Once we have registered each shape, we used a standard algorithm to extract surfaces from 2D contours58 in order to transform the point cloud into a triangulated mesh that can be saved in the Visualization Toolkit (VTK)59 format for visualization and analysis purposes.
We used the following landmarks to map starter cells in the amygdala and piriform cortex (Fig. 3): (I) appearance of the olfactory tubercle, (II) end of the olfactory tubercle; (III) appearance of hippocampus; (IV) appearance of dentate gyrus of hippocampus in the ventral edge of the cortex.
Quantitative Analyses
For each tracing experiment where we analyzed the distribution of labeled glomeruli along the DV axis using mean θ’OB (Fig. 3e), we generated a corresponding random distribution of simulated mean θ’OB (mean simθ’OB) from M glomeruli, where M is the number of labeled glomeruli in the injection. To generate this random distribution for each experiment, we randomly selected M glomeruli from a given 3D reconstruction model (generated from that injection) and calculated the mean simθ’OB value for those randomly selected M glomeruli to get the mean simθ’1. We then repeated the same simulation 50,000 times to obtain mean simθ’2,…, mean simθ’50,000, and therefore to obtain the range of mean simθ’ for M glomeruli that are randomly distributed throughout the OB. Once we obtained distributions for mean simθ’OB that corresponded to each injection, we compared the mean simθ’OB distribution with the experimental mean θ’OB. If the value for the experimental mean θ’OB was outside of the 95% of the mean simθ’OB distribution, we considered the glomerular distribution to be non-random for that sample.
Multiple regression analysis (Fig. 4b) was conducted by using Excel (Microsoft). Data from every experiment in Table S1 (n=8 for AON, n=10 PC using actin-CreER) was used for the left part of the Fig. 4b. Data from 7 experiments obtained from GAD2-CreER in the APC were used in the right part of the Fig. 4b. The number of labeled mitral cells in the OB was set as a dependent variable, Y, and the number of starter cells in layer i was set as an independent variable, Xi. The constant was set to zero. Excel then calculated the values of coefficients Ai (shown in red crosses in Fig. 4b) and 95% confidence intervals of Ai based on the student’s t statistics (shown in gray bars in Fig. 4b). R2 for these multiple regression assays were: 0.98 for the AON, 0.96 for the APC (actin-CreER), and 0.97 for the APC (GAD2-CreER) datasets.
To estimate the number of dually labeled glomeruli originating from single starter cells (data Ds in Fig. 4c) in our experimental data, we first simulated hypothetical number of dually labeled glomeruli originating from single starter cells (Ds) and two independent starter cells (Dt) according to the null hypothesis that mitral cells connect randomly with postsynaptic targets. This situation can be modeled by “balls and bins”: there are 2,000 bins (a bin represents a single glomerulus) and N balls (a ball represents a single transsynaptic labeling event). N balls were randomly thrown into 2,000 bins, and the number of bins that received more than one ball (i.e., glomeruli labeled more than once) was counted. To distinguish Ds vs. Dt, we further introduced n different colors to the balls, where each color represented an individual starter cell in the cortex. We assumed that an equal number of balls (N/n) was labeled with n different colors. These balls were randomly thrown into one of 2,000 bins, and the number of bins containing more than one ball was counted. We separately counted the bins with more than one ball of an identical color (representing Ds) and the bins with more than one ball of different colors (representing Dt). We fixed the number of bins (glomeruli) to be 2,000, while N and n corresponded to the number of labeled mitral cells and the number of starter cells, respectively, in each experiment. We repeated this simulation 100,000 times for each set of N and n to obtain the simulated distribution of Ds and Dt (we call these ‘random Ds’ and ‘random Dt’). To estimate the Ds components in experimental data (data Ds), we assumed that individual starter cells contributed independently to the labeling (random Dt = data Dt). Based on the equation: D (number of observed dually labeled glomeruli) = Ds + Dt, we estimated the data Ds distribution by subtracting the random Dt from observed D (Fig. 4c). Then we determined if there was a significant difference in the distribution of data Ds and random Ds. We considered two distributions to be significantly different if the probability of data Ds > random Ds or Data Ds < random Ds exceed 0.95 (shown by asterisks in Fig. 4c). To accurately count dually labeled glomeruli, samples with more than 200 labeled mitral cells were excluded from this analysis.
Supplementary Material
Acknowledgements
We thank Stanford Transgenic Facility for help in generating transgenic mice, F. Zhang and K. Deisseroth for teaching us stereotactic injection, Y. Yoshihara and H. Zeng for reagents, C. Manalac and M. Shu for technical assistance, R. Axel, L. Buck, L. Stryer, R. Yuste and members of the Luo Lab for discussion and critical reading of the manuscript. K.M. was supported by JSPS program for Research Abroad and Human Frontier Science Program. K.M. is currently a research associate, and L.L. is an investigator, of the Howard Hughes Medical Institute. This work was also supported by an NIH grant.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author contributions K.M. planned and performed all the experiments. F.A. and F.M. developed the computer programs for 3D reconstructions and statistical simulation under the supervision of M.A.H. C.W. and Z.H. performed the initial AAV production. I.W., N.R.W. and E.M.C. provided the modified RV and the construct to make the AAV vector. H.T. and Z.J.H. provided the GAD2-CreER mice. B.T. provided DNA constructs. L.L. supervised the project. L.L. and K.M. wrote the manuscript, with contribution from B.T.
Author Information Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
References
- 1.Cowan WM. The emergence of modern neuroanatomy and developmental neurobiology. Neuron. 1998;20:413–426. doi: 10.1016/s0896-6273(00)80985-x. [DOI] [PubMed] [Google Scholar]
- 2.Luo L, Callaway EM, Svoboda K. Genetic dissection of neural circuits. Neuron. 2008;57(5):634–660. doi: 10.1016/j.neuron.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wickersham IR, et al. Monosynaptic restriction of transsynaptic tracing from single, genetically targeted neurons. Neuron. 2007;53(5):639–647. doi: 10.1016/j.neuron.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ugolini G. Specificity of rabies virus as a transneuronal tracer of motor networks: transfer from hypoglossal motoneurons to connected second-order and higher order central nervous system cell groups. J Comp Neurol. 1995;356(3):457–480. doi: 10.1002/cne.903560312. [DOI] [PubMed] [Google Scholar]
- 5.Ugolini G. Advances in viral transneuronal tracing. J Neurosci Methods. 2010 doi: 10.1016/j.jneumeth.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 6.Wickersham IR, Finke S, Conzelmann KK, Callaway EM. Retrograde neuronal tracing with a deletion-mutant rabies virus. Nat Methods. 2007;4(1):47–49. doi: 10.1038/NMETH999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo C, Yang W, Lobe CG. A Cre recombinase transgene with mosaic, widespread tamoxifen-inducible action. Genesis. 2002;32(1):8–18. doi: 10.1002/gene.10021. [DOI] [PubMed] [Google Scholar]
- 8.Wise SP, Jones EG. The organization and postnatal development of the commissural projection of the rat somatic sensory cortex. J Comp Neurol. 1976;168(3):313–343. doi: 10.1002/cne.901680302. [DOI] [PubMed] [Google Scholar]
- 9.Iriki A, Pavlides C, Keller A, Asanuma H. Long-term potentiation in the motor cortex. Science. 1989;245(4924):1385–1387. doi: 10.1126/science.2551038. [DOI] [PubMed] [Google Scholar]
- 10.Taymans JM, et al. Comparative analysis of adeno-associated viral vector serotypes 1, 2, 5, 7, and 8 in mouse brain. Hum Gene Ther. 2007;18(3):195–206. doi: 10.1089/hum.2006.178. [DOI] [PubMed] [Google Scholar]
- 11.Vassar R, et al. Topographic organization of sensory projections to the olfactory bulb. Cell. 1994;79(6):981–991. doi: 10.1016/0092-8674(94)90029-9. [DOI] [PubMed] [Google Scholar]
- 12.Ressler KJ, Sullivan SL, Buck LB. Information coding in the olfactory system: evidence for a stereotyped and highly organized epitope map in the olfactory bulb. Cell. 1994;79(7):1245–1255. doi: 10.1016/0092-8674(94)90015-9. [DOI] [PubMed] [Google Scholar]
- 13.Mombaerts P, et al. Visualizing an olfactory sensory map. Cell. 1996;87(4):675–686. doi: 10.1016/s0092-8674(00)81387-2. [DOI] [PubMed] [Google Scholar]
- 14.Malnic B, Hirono J, Sato T, Buck LB. Combinatorial receptor codes for odors. Cell. 1999;96(5):713–723. doi: 10.1016/s0092-8674(00)80581-4. [DOI] [PubMed] [Google Scholar]
- 15.Wilson RI, Mainen ZF. Early events in olfactory processing. Annu Rev Neurosci. 2006;29:163–201. doi: 10.1146/annurev.neuro.29.051605.112950. [DOI] [PubMed] [Google Scholar]
- 16.Luskin MB, Price JL. The distribution of axon collaterals from the olfactory bulb and the nucleus of the horizontal limb of the diagonal band to the olfactory cortex, demonstrated by double retrograde labeling techniques. J Comp Neurol. 1982;209(3):249–263. doi: 10.1002/cne.902090304. [DOI] [PubMed] [Google Scholar]
- 17.Scott JW, McBride RL, Schneider SP. The organization of projections from the olfactory bulb to the piriform cortex and olfactory tubercle in the rat. J Comp Neurol. 1980;194(3):519–534. doi: 10.1002/cne.901940304. [DOI] [PubMed] [Google Scholar]
- 18.Shipley MT, Adamek GD. The connections of the mouse olfactory bulb: a study using orthograde and retrograde transport of wheat germ agglutinin conjugated to horseradish peroxidase. Brain Res Bull. 1984;12(6):669–688. doi: 10.1016/0361-9230(84)90148-5. [DOI] [PubMed] [Google Scholar]
- 19.Nevelle KR, Haberly LB. Olfactory Cortex. In: Shepherd GM, editor. The synaptic organization of the brain. Oxford: Oxford University Press; 2004. [Google Scholar]
- 20.Feinstein P, Mombaerts P. A contextual model for axonal sorting into glomeruli in the mouse olfactory system. Cell. 2004;117(6):817–831. doi: 10.1016/j.cell.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 21.Schaefer ML, Finger TE, Restrepo D. Variability of position of the P2 glomerulus within a map of the mouse olfactory bulb. J Comp Neurol. 2001;436(3):351–362. [PubMed] [Google Scholar]
- 22.Brunjes PC, Illig KR, Meyer EA. A field guide to the anterior olfactory nucleus (cortex) Brain Res Brain Res Rev. 2005;50(2):305–335. doi: 10.1016/j.brainresrev.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 23.Yan Z, et al. Precise circuitry links bilaterally symmetric olfactory maps. Neuron. 2008;58(4):613–624. doi: 10.1016/j.neuron.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 24.Schoenfeld TA, Macrides F. Topographic organization of connections between the main olfactory bulb and pars externa of the anterior olfactory nucleus in the hamster. J Comp Neurol. 1984;227(1):121–135. doi: 10.1002/cne.902270113. [DOI] [PubMed] [Google Scholar]
- 25.Scott JW, Ranier EC, Pemberton JL, Orona E, Mouradian LE. Pattern of rat olfactory bulb mitral and tufted cell connections to the anterior olfactory nucleus pars externa. J Comp Neurol. 1985;242(3):415–424. doi: 10.1002/cne.902420309. [DOI] [PubMed] [Google Scholar]
- 26.Miyamichi K, Serizawa S, Kimura HM, Sakano H. Continuous and overlapping expression domains of odorant receptor genes in the olfactory epithelium determine the dorsal/ventral positioning of glomeruli in the olfactory bulb. J Neurosci. 2005;25(14):3586–3592. doi: 10.1523/JNEUROSCI.0324-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoshihara Y, et al. OCAM: A new member of the neural cell adhesion molecule family related to zone-to-zone projection of olfactory and vomeronasal axons. J Neurosci. 1997;17(15):5830–5842. doi: 10.1523/JNEUROSCI.17-15-05830.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Swanson LW, Petrovich GD. What is the amygdala? Trends Neurosci. 1998;21(8):323–331. doi: 10.1016/s0166-2236(98)01265-x. [DOI] [PubMed] [Google Scholar]
- 29.Scalia F, Winans SS. The differential projections of the olfactory bulb and accessory olfactory bulb in mammals. J Comp Neurol. 1975;161(1):31–55. doi: 10.1002/cne.901610105. [DOI] [PubMed] [Google Scholar]
- 30.Poo C, Isaacson JS. Odor representations in olfactory cortex: "sparse" coding, global inhibition, and oscillations. Neuron. 2009;62(6):850–861. doi: 10.1016/j.neuron.2009.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stettler DD, Axel R. Representations of odor in the piriform cortex. Neuron. 2009;63(6):854–864. doi: 10.1016/j.neuron.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 32.Lei H, Mooney R, Katz LC. Synaptic integration of olfactory information in mouse anterior olfactory nucleus. J Neurosci. 2006;26(46):12023–12032. doi: 10.1523/JNEUROSCI.2598-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Franks KM, Isaacson JS. Strong single-fiber sensory inputs to olfactory cortex: implications for olfactory coding. Neuron. 2006;49(3):357–363. doi: 10.1016/j.neuron.2005.12.026. [DOI] [PubMed] [Google Scholar]
- 34.Stokes CC, Isaacson JS. From Dendrite to Soma: Dynamic Routing of Inhibition by Complementary Interneuron Microcircuits in Olfactory Cortex. Neuron. 2010;67(3):452–465. doi: 10.1016/j.neuron.2010.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Apicella A, Yuan Q, Scanziani M, Isaacson JS. Pyramidal cells in piriform cortex receive convergent input from distinct olfactory bulb glomeruli. J Neurosci. 2010;30(42):14255–14260. doi: 10.1523/JNEUROSCI.2747-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suzuki N, Bekkers JM. Inhibitory neurons in the anterior piriform cortex of the mouse: classification using molecular markers. J Comp Neurol. 2010;518(10):1670–1687. doi: 10.1002/cne.22295. [DOI] [PubMed] [Google Scholar]
- 37.Christie JM, et al. Connexin36 mediates spike synchrony in olfactory bulb glomeruli. Neuron. 2005;46(5):761–772. doi: 10.1016/j.neuron.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 38.Haberly LB. Parallel-distributed processing in olfactory cortex: new insights from morphological and physiological analysis of neuronal circuitry. Chem Senses. 2001;26(5):551–576. doi: 10.1093/chemse/26.5.551. [DOI] [PubMed] [Google Scholar]
- 39.Kobayakawa K, et al. Innate versus learned odour processing in the mouse olfactory bulb. Nature. 2007;450(7169):503–508. doi: 10.1038/nature06281. [DOI] [PubMed] [Google Scholar]
- 40.Sosulski DL, Lissitsyna MV, Curforth T, Axel R, Datta SR. Distinct representations of olfactory information in the cortex. doi: 10.1038/nature09868. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marin EC, Jefferis GS, Komiyama T, Zhu H, Luo L. Representation of the glomerular olfactory map in the Drosophila brain. Cell. 2002;109(2):243–255. doi: 10.1016/s0092-8674(02)00700-6. [DOI] [PubMed] [Google Scholar]
- 42.Wong AM, Wang JW, Axel R. Spatial representation of the glomerular map in the Drosophila protocerebrum. Cell. 2002;109(2):229–241. doi: 10.1016/s0092-8674(02)00707-9. [DOI] [PubMed] [Google Scholar]
- 43.Jefferis GS, et al. Comprehensive maps of Drosophila higher olfactory centers: spatially segregated fruit and pheromone representation. Cell. 2007;128(6):1187–1203. doi: 10.1016/j.cell.2007.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Davis RL. Olfactory memory formation in Drosophila: from molecular to systems neuroscience. Annu Rev Neurosci. 2005;28:275–302. doi: 10.1146/annurev.neuro.28.061604.135651. [DOI] [PubMed] [Google Scholar]
- 45.Murthy M, Fiete I, Laurent G. Testing odor response stereotypy in the Drosophila mushroom body. Neuron. 2008;59(6):1009–1023. doi: 10.1016/j.neuron.2008.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marshel JH, Mori T, Nielsen KJ, Callaway EM. Targeting single neuronal networks for gene expression and cell labeling in vivo. Neuron. 2010;67(4):562–574. doi: 10.1016/j.neuron.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stepien AE, Tripodi M, Arber S. Monosynaptic rabies virus reveals premotor network organization and synaptic specificity of cholinergic partition cells. Neuron. 2010;68(3):456–472. doi: 10.1016/j.neuron.2010.10.019. [DOI] [PubMed] [Google Scholar]
- 48.Madisen L, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13(1):133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lein ES, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445(7124):168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 50.Urlinger S, et al. Exploring the sequence space for tetracycline-dependent transcriptional activators: novel mutations yield expanded range and sensitivity. Proc Natl Acad Sci U S A. 2000;97(14):7963–7968. doi: 10.1073/pnas.130192197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zong H, Espinosa JS, Su HH, Muzumdar MD, Luo L. Mosaic analysis with double markers in mice. Cell. 2005;121(3):479–492. doi: 10.1016/j.cell.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 52.Muzumdar MD, Luo L, Zong H. Modeling sporadic loss of heterozygosity in mice by using mosaic analysis with double markers (MADM) Proc Natl Acad Sci U S A. 2007;104(11):4495–4500. doi: 10.1073/pnas.0606491104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chung JH, Whiteley M, Felsenfeld G. A 5' element of the chicken beta-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell. 1993;74(3):505–514. doi: 10.1016/0092-8674(93)80052-g. [DOI] [PubMed] [Google Scholar]
- 54.Li L, et al. Visualizing the distribution of synapses from individual neurons in the mouse brain. PLoS One. 2010;5(7):e11503. doi: 10.1371/journal.pone.0011503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wickersham IR, Sullivan HA, Seung HS. Production of glycoprotein-deleted rabies viruses for monosynaptic tracing and high-level gene expression in neurons. Nat Protoc. 2010;5:595–606. doi: 10.1038/nprot.2009.248. [DOI] [PubMed] [Google Scholar]
- 56.Espinosa JS, Wheeler DG, Tsien RW, Luo L. Uncoupling dendrite growth and patterning: single-cell knockout analysis of NMDA receptor 2B. Neuron. 2009;62(2):205–217. doi: 10.1016/j.neuron.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang Z. Iterative point matching for registration of free-form curves and surfaces. Int J Comput Vision. 1994;13(2):119–152. [Google Scholar]
- 58.Cong G, Parvin B. Surface Reconstruction from Sparse Fringe Contours. Proceedings of the 4th IEEE Workshop on Applications of Computer Vision. 1998;140 [Google Scholar]
- 59.Schroeder W, Martin K, Lorensen B. The Visualization Toolkit: An Object-Oriented Approach to 3-D Graphics. Prentice Hall; 1997. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.