Abstract
Objectives
To implement an elective course in pharmacogenomics designed to teach pharmacy students about the fundamentals of pharmacogenomics and the anticipated changes it will bring to the profession.
Design
The 8 sessions of the course covered the basics of pharmacogenomics, genomic biotechnology, implementation of pharmacogenetics in pharmacy, information security and privacy, ethical issues related to the use of genomic data, pharmacoepidemiology, and use and promotion of GeneScription, a software program designed to mimic the professional pharmacy environment.
Assessment
Student grades were based on completion of a patient education pamphlet, a 2-page paper on pharmacogenomics, and precourse and postcourse survey instruments. In the postcourse survey, all students strongly agreed that genomic data could be used to determine the optimal dose of a drug and genomic data for metabolizing enzymes could be stored in a safe place. Students also were more willing to submit deoxyribonucleic acid (DNA) data for genetic profiling and better understood how DNA analysis is performed after completing the course.
Conclusions
An elective course in pharmacogenomics equipped pharmacy students with the basic knowledge necessary to make clinical decisions based on pharmacogenomic data and to teach other healthcare professionals and patients about pharmacogenomics. For personalized medicine to become a reality, all pharmacists and pharmacy students must learn this knowledge and these skills.
Keywords: pharmacogenomics, personalized medicine, drug safety, instructional software
INTRODUCTION
The widespread application of genomic data to effectively influence health care outcomes is expected in the next decade as testing methods are improved and the public and health care professionals are educated about the potential benefits. The ability to identify single nucleotide polymorphisms (SNP) in drug metabolizing enzymes (eg, cytochrome P450) and targets represents an implementation model that can avoid some of the ethical issues previously associated with the identification of incurable genetic diseases. SNP identification has the potential to improve the cost-to-benefit ratio in the pharmacological management of patients with a wide variety of conditions.1 A potential barrier to the use of genomic information is the ability of pharmacists to interpret and communicate about genetic data and their importance in health care. We describe an elective course in pharmacogenomics designed to teach pharmacy students about the fundamentals of pharmacogenomics and the manner in which pharmacogenomics will change the practice of pharmacy in the future.
The Accreditation Council for Pharmacy Education (ACPE) recognizes the importance of pharmacogenomics to the future of pharmacy practice and has made strides in increasing the exposure pharmacy students have to pharmacogenomics in the curriculum.2 The elective course described in this paper has now been integrated into the core pharmacy curriculum and uses GeneScription, a software program that provides students with mock patient data to assist the student in making dosing decisions.3
GeneScription was developed by the authors under a grant from Microsoft Research specifically to provide hands-on instruction in applied personalized medicine and pharmacogenomics. GeneScription is designed to mimic the professional environment of the pharmacist, and therefore provides a practicum environment with a mock patient population, Food and Drug Administration (FDA) approved drugs, and algorithms that predict patient outcomes based on physiological parameters, genomic parameters, and drug and dose parameters. (A free online version of GeneScription is available at www.genescription.com).
This course was implemented to assist students in moving the basic science taught in the classroom to the actual practice site. In the near future, pharmacists will play a vital role in making personalized medicine a reality. To equip pharmacists to carry out this task, they need to be equipped with the fundamentals that will allow them to make clinical decisions based on pharmacogenomic data. Additionally, pharmacists will need to teach other healthcare professionals and patients about pharmacogenomics for personalized medicine to become a reality. This course was designed to prepare pharmacy students for this role.
Pharmacogenetics can be defined as inherited variations in drug effects as a result of single gene interactions with drugs. These single gene alterations or SNPs can alter drug disposition, safety, tolerability, and efficacy. Pharmacogenetic screening of drug metabolizing enzymes and pharmacologic targets can be used to improve patient outcomes. This course reviews the fundamentals of pharmacogenetics and genetic testing as a means to improve patient outcomes.
After completing this drug safety and pharmacogenomics course, pharmacy students should be able to:
(1) Compare and contrast pharmacogenetics and pharmacogenomics.
(2) Demonstrate an understanding of basic DNA terminology and genomic variations.
(3) Differentiate between the laboratory tests used in genetic testing for SNPs.
(4) Explain “personalized medicine” from the standpoint of drug metabolism, bioactivation, and pharmacologic target screening.
(5) Describe the limitations to implementing pharmacogenetic screening in health care.
(6) Apply knowledge of pharmacogenetics to the initiation of warfarin therapy.
(7) Apply SNP data to patient dosing.
(8) Respond to drug-gene interactions in an appropriate fashion to improve patient outcomes.
(9) Explain the ethical issues associated with the use of pharmacogenomic data in health care.
(10) Interpret and apply pharmacoepidemiological data to the application of pharmacogenomic data.
DESIGN
The course in drug safety and pharmacogenomics was offered for the first time in spring 2008. The course was team taught with several departments on the Ohio Northern Unviversity (ONU) campus and with adjunct faculty members with expertise in the major areas of interest. The Department of Computer and Information Technology at Purdue University collaborated with ONU on the development of the GeneScription software. The participating faculty members’ areas of expertise included pharmacology, pharmacogenomics, information management systems, software development, pharmacoepidemology, ethics, and entrepreneurship. The class met for 2 hours a week for 10 weeks, and had a capped enrollment of 18 students. Capping enrollment at 18 students allowed for more of a discussion format and for easy use of computers for the software developed to apply the pharmacogenomic information.
The course began with an online survey designed to assess precourse knowledge and acceptance of pharmacogenomics (Table 1). The course was offered during the spring quarter of the fifth year of ONU's 6-year doctor of pharmacy (PharmD) program. Therefore, the course was offered at the end of the didactic curriculum, just prior to the students starting their advanced practice pharmacy experiences (APPEs). Entering the course, the students had experience in running polymerase chain reactions (PCRs) from their biomedical sciences laboratory and had received training in genetics from several courses, ranging from biochemistry to a biomedical sciences module that devoted 10 hours to genes and genetic diseases. The methods used in each course period are described below.
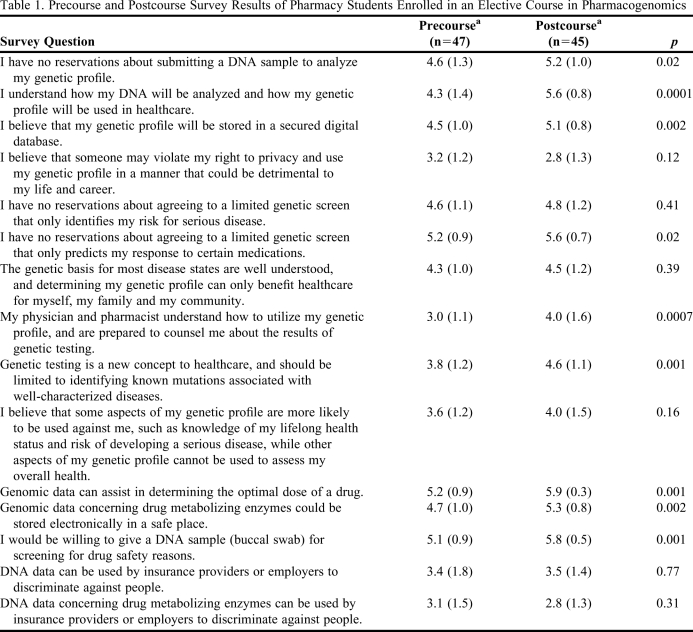
Table 1.
Precourse and Postcourse Survey Results of Pharmacy Students Enrolled in an Elective Course in Pharmacogenomics
a Rating scale: 1 = strongly disagree, 2 = moderately disagree, 3 = disagree, 4 = agree, 5 = moderately agree, and 6 = strongly agree.
Session 1 (2 hours) reviewed the basics of pharmacogenomics and known examples of SNPs including different genes/enzymes, known substrates, in vitro and in vivo effect of known SNPs on enzyme activity, gene expression, and substrate specificity. The topics covered in this session included: (1) comparison of pharmacogenomics and pharmacogenetics, (2) human genome overview, (3) DNA review (from chromosomes to transcription), (4) genomic variations with emphasis on SNPs, (5) goals of personalized medicine, (6) adverse drug reactions and cytochrome P450 application, (7) warfarin dosing example for applying pharmacogenetic data, and (8) discussion of the limitations on the implementation of personalized medicine.
Session 2 (2 hours) provided students with a detailed description of genomic biotechnology. This session reviewed the basics of DNA detection, SNP discovery and detection, and biotechnologies, and concluded with a discussion on how the students felt genomic data derived from the application of these biotechnology methods could be used to support clinical decisions
Students were given a submitted grant proposal on the implementation of pharmacogenetics in pharmacy, which they were required to review and critique prior to session 3. For session 3, (2 hours) students met in a classroom designed for open discussion and each student identified 1 strength and 1 weakness of the proposal. As students presented their findings, the group routinely responded with additional comments.
Session 4 (2 hours) integrated the materials presented to this point into a usable model where information security, system usability, and related behaviors of a genomics-based drug safety that could impact healthcare. As the implementation of clinical genotyping requires healthcare providers to use software applications that correctly and efficiently manage patient genetic information, the purpose of this session was to provide students with a deeper understanding of information technology (IT) concepts and the impact of IT on the pharmacy profession, specifically in the context of clinical genotyping. This discussion included a brief background on IT and information systems as well as an introduction to user interfaces and their design. The session also contained material on pertinent information security concerns and data sensitivity, in addition to methods for implementing and improving privacy. Also discussed were the path to worldwide adoption of genomics-based drug safety systems, the IT-related obstacles to this adoption, and the tools and techniques for removing said obstacles.
Session 5 (2 hours) focused on the ethical issues related to the use of genomic data in health care. This session was led by an ethics faculty member, providing the students with a perspective outside of the health care team. The professor conducted an exercise in which he played the role of a patient and the students asked him questions regarding his perception of/feelings about the use of genomic data in health care. Students also were assigned 2 papers to read and then discuss in class.4,5
Session 6 (2 hours) focused on pharmacoepidemiology and began with 2 reading assignments, which were discussed in class.6,7 The outcomes for this session were: (1) describe the pharmacogenetic implications in the pharmacoepidemiology of drug response; (2) evaluate potential implications of pharmacogenetics information in risk minimization action plans; (3) describe how pharmacogenomic testing can be used to optimize clinical response; (4) discuss the availability of pharmacogenomic testing and the limitations of such testing.
Molecular pharmacoepidemiology is focused on understanding why groups of individuals respond differently to specific drug therapies based on underlying genotypic and phenotypic conditions. Specifically, it is the study of the manner in which molecular biomarkers alter the clinical effects of medications in populations. This session also focuses on the potential for pharmacogenetic tests to optimize clinical response, both in terms of beneficial and adverse effects. The class discussion provides a framework for bedside-to-population translational research through an introduction to statistics, research design, and methodology issues encountered in evaluating the efficacy of pharmacogenetic testing.
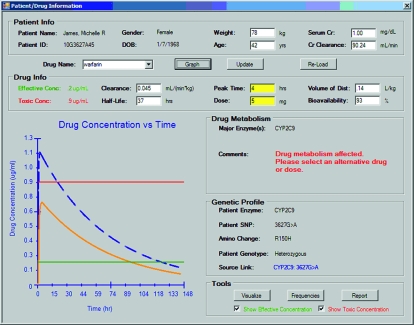
In Session 7 (4 hours) students used GeneScription, the software developed for applying pharmacogenomic information to pharmacy practice. The authors designed and developed this software system with a grant from Microsoft Research to create instructional software for teaching pharmacogenetics to healthcare professionals (Figure 1). During this session, each student was provided a laptop computer with the GeneScription system. Students were provided with a different fictitious patient and prescription. The student was then required to make decisions on altering the drug dose based on pharmacogenetic data provided by GeneScription.
Figure 1.
Effect of clinical genotyping data on warfarin clearance and ADR risk in GeneScription prototype.
In addition to a user interface for patient data input, drug/dose selection, and a population of fictitious patients that harbor clinically relevant genetic and physiological traits, the GeneScription system has a data management component that includes FDA-approved drugs that are known to be metabolized through p450 enzymes, and known SNPs within drug metabolism genes that have an effect in vivo (ie, published clinical studies). This database is extensive and includes all relevant details about the drug (such as half-life and bioavailability). Although there are thousands of known SNPs in genes relevant to drug metabolism, the system does not include SNP data that has not been linked yet to altered drug metabolism capabilities through clinical testing; doing so would complicate the learning environment without adding additional value to the training process. Instead, GeneScription makes simple single-dose predictions as a method of unambiguous education and training. Importantly, the GeneScription system was developed to include patients who are both heterozygous (1 of the 2 gene copies is “altered”) and homozygous (both of the gene copies are “altered”), and make adverse drug predictions based on results from clinical studies that have investigated the impact on heterozygous and homozygous alleles. Hyperlinks to the associated published studies are incorporated into the Genescription system, allowing users to gain more information about these studies. For example, Genescription includes the case of a 45-year-old female patient who was prescribed 10 mg warfarin orally and is homozygous for the variant CYP2C9*3, and provides hyperlinks to the articles from which the information was derived.8,9
In GeneScription, after the student selects a patient and then chooses the drug to be prescribed to that patient and the dose (in this case, a single oral dosing regimen), the system accesses the patient's genetic information to determine whether the patient harbors an SNP in the metabolic enzyme that is known to metabolize the prescribed drug. If the selected patient harbors an SNP capable of altering the pharmacokinetics of the prescribed drug, the GeneScription system provides this information, as well as an interactive graph that displays the area under the curve, minimum effective concentration, and minimum toxic concentration. If the patient does not harbor a clinically relevant SNP, then the system simply notifies the user that the prescribed drug and dose are within normal limits. Figure 1 is a screen shot demonstration of the clinical genotyping data on warfarin clearance and adverse drug reactions (ADRs). In Figure 1, a normal (“mock”) patient's drug concentration vs. time curve (solid line) following a standard warfarin oral dose shows the changes in drug plasma concentration over time. Warfarin is metabolized to 7-hydroxywarfarin by the oxidative metabolism enzyme 2C9, which is the primary mechanism for warfarin clearance in humans. There are 2 variant alleles that have a reduced capability for metabolizing warfarin, with 11% and 7% frequency in the Caucasian population for variants CYP2C9*2 and CYP2C9*3, respectively. Patients who are homozygous for these variant alleles (ie, having 2 variant copies of the 2C9 gene) experience a 65% decrease in drug clearance rate (dashed line). The presence of a variant allele leads to increased drug plasma concentrations above the minimum toxic concentration and markedly increases the risk of an ADR. GeneScription's applied learning environment also provides basic genetic and biochemical information including details about the relevant SNP and metabolism enzyme, its location in the human genome, the codon and amino acid changes that are involved, other SNPs in the gene, and the frequency of the SNPs in the human population. GeneScription represents a hands-on learning environment that allows students to determine the manner in which dosing, genetic, and physiological variations (relevant to drug clearance) impact therapeutics using a real-time predictive model of drug plasma concentrations.
Session 8 (2 hours) was led by a faculty member from the business college and explored the challenges of adoption of pharmacogenomics by consumers, physicians, support staff members, administrators, and healthcare companies; the interdependence between distinct professional domains; the business opportunities created by this system; and the benefits to the practice of pharmacy. This was an open discussion with no lecture handouts and the faculty member serving as the facilitator.
As a part of the course, students worked in teams of 3 to develop announcements and a variety of educational materials to be used in patient education. The student programs were evaluated by the instructor and 5 randomly selected individuals representing the lay public. The team receiving the highest assessment from the lay public received a gift certificate. Students also were required to write a 2-page perspective paper on the future use of pharmacogenomics in the practice of pharmacy and use a minimum of 5 references. There were no examinations in this first offering of the course. Grades were based on the quality of the students’ patient education materials and pharmacogenomics paper and completion of the pre- and post-course survey instruments described below. Multiple-choice and essay type examinations were used once the course was integrated into the core curriculum.
EVALUATION AND ASSESSMENT
The evaluation and assessment of the teaching methodology used to facilitate the learning of pharmacogenomics followed the IDEAS format (introduction, design, evaluation, assessment, and summary).10 The IDEAS model uses 3 types of evaluative data: (1) attitudinal scales, eg, precourse and postcourse surveys; (2) observational data, eg, students’ care plans; and (3) performance assessment, eg, test scores.
Attitudinal assessment was performed using an online survey instrument designed to assess precourse knowledge and acceptance of pharmacogenomics. Survey results are shown in Table 1. Precourse and postcourse survey responses were compared using a paired t test. Significant differences were found in all but 5 of the 15 areas assessed. After completing the course, all students strongly agreed that genomic data could be used to determine the optimal dose of a drug and that genomic data for metabolizing enzymes could be stored in a safe place. Students also were more willing to submit DNA data for genetic profiling (p = 0.02) and better understood how DNA analysis is performed (p = 0.0001) after completing the course.
Observational assessment was performed using the student-developed patient education pamphlets and an evaluation of the 2-page perspective paper on the future use of pharmacogenomics. This observational assessment was expanded to include essay-style examinations, which required the student to apply the knowledge gained in class to a case-based situation in subsequent offerings of the course. A sample case and essay style examination is provided in Appendix 1.
Once the course was integrated into the core curriculum, student performance was assessed based on traditional multiple-choice examinations and application-based essay questions. Student performance on the formal examinations has been excellent (mean grade = 89.4 ± 8.5).
Additionally, 5 to 7 students were randomly selected from the class to serve on focus groups. The focus group met after receiving an introduction to the GeneScription program. We asked each student the following questions: (1) To date, what aspects of the program helped facilitate your learning in this course? (2) To date, what aspects of the program hindered your learning in this course? (3) To date, what would you like to see changed?
Students stated that they appreciated the ability to visualize plasma concentrations, frequency data, and enzyme searches. Students provided suggestions for improving the warning mechanism, bioavailability graphing, and some pharmacokinetic parameter monitoring. The information gleaned from the focus group discussions was used to improve the GeneScription system. These changes have been made to the current version of the program.
DISCUSSION
Drug safety and personalized medicine are clearly the future of pharmacy practice. This course was designed in such a way that students could learn how pharmacists can use pharmacogenetics data to improve drug safety and therapeutic outcomes. Furthermore, pharmacy students were given the opportunity to develop strategies to improve patient acceptance of genetic data. The GeneScription system gave the students the opportunity for hands-on experience in filling prescriptions with a computer system that could screen for drug-gene interactions as readily as systems currently screen for drug-drug interactions. We predict that, in the future, patients will be able to enter any hospital or community pharmacy practice setting and obtain a buccal swab for DNA analysis identifying relevant P450 polymorphisms. This information then will be seamlessly integrated into prescription filling systems. During the prescription filling process, the pharmacist will be alerted if there is a drug-genomic interaction. The pharmacist then will be provided with therapeutic and genomic data (examples can be found in the GeneScription system) that will assist them in their consultation with the physician about tailoring the patient's drug therapy. The genescription system is a unique tool to address ethical and educational hindrances by (1) supporting a key component of individualized medicine that does not rely on using DNA variance linked to a particular disease state(s) and therefore minimizes patient privacy concerns, and (2) can be used by healthcare professionals as an education and training tool to better understand the role of DNA and genotyping in patient care. As with session 1, the GeneScription system also has been used in a live continuing education program for pharmacists. Results similar to this study were obtained from practicing pharmacists after attending a 4-part continuing education program consisting of sessions 1, 2, and 7. This demonstrates the practicality of the course as well as versatility.
With the didactic information provided in this manuscript, the course would be transferable to other colleges and schools of pharmacy. Furthermore, the GeneScription software is available for other institutions to use in training students in the area of pharmacogenomics.
Because most practicing pharmacists (and other healthcare providers for that matter) were trained long before the use of human genomic information was included in pharmacy curricula, educating pharmacists and future pharmacists is paramount to the adoption of clinical genotyping in patient care. Furthermore, patient acceptance only can be gained through their understanding of genomic data use in health care. The course in drug safety and pharmacogenomics was designed to assist in addressing these gaps.
Gilkey and colleagues11 outlined 3 important areas for developing patient education materials on patient safety: risk perception, community participation, and social marketing. When the students were given the task to develop the patient education pamphlets, we also discussed the importance of these 3 areas in developing patient educational campaigns.
Although many aspects of the course were successful in advancing the students’ knowledgebase in the area of pharmacogenomics, students stated that the GeneScription software system was the most beneficial aspect of the course and helped them visualize the manner in which pharmacogenomic data could be integrated into the practice of pharmacy. Furthermore, GeneScription strengthened the foundational information provided in class by demonstrating the pharmacokinetic effects of SNPs and providing potential therapeutic alternatives and links to scientific literature relevant to the SNPs.
Spurred by the interest and value expressed by the students in relationship to the GeneScription software, a Web-based version is now available online at http://www.genescription.com. At this site, visitors also can view a multimedia introduction to pharmacogenomics. Upon entering the Web version of GeneScription, the user first selects a patient and then a drug. In the event that we detect an interaction, GeneScription warns the user, and then the user has the opportunity to view the current dose curve and select an alternate dose. After selecting an alternative, the user then sees the dose curve that is associated with the alternate dose. This Web version also uses a case-based approach in addition to providing the user the full capabilities of the GeneScription system.
The use of GeneScription also assisted us in finding one area of weakness in the overall design of the course. We learned as we walked through the computer program with the students that drug-drug interactions coupled with drug-gene interactions needed to be addressed. For example, if a patient were heterozygotic for CYP2C9*2, they may have a slightly reduced metabolism of warfarin. However, if the patient also were taking a CYP2C9 inhibitor, they would now have an even greater reduction in warfarin metabolism. As a result of these discussions with the students, we expanded the course and added a section on pharmacogenomics and pharmacokinetic implications.
This course now has been integrated into the core curriculum. Additionally, continuing education programs for pharmacists and physicians have been developed as a result of the course. This general introductory session on pharmacogenomics also was converted into a continuing education publication for licensed pharmacists of the Ohio Pharmacists Association. The live continuing education programs have been evaluated in a similar fashion to the course and the findings from the precourse and postcourse surveys have been very similar. Our overarching goal is to improve therapeutic outcomes and reduce adverse drug reactions through the advancement of personalized medicine. This course is the first step in that direction. We offer our findings and methods to our colleagues in pharmacy education to assist them in training pharmacists to be mediators of change in personalized medicine.
SUMMARY
An elective course in pharmacogenomics was implemented to assist students in moving the basic science knowledge learned in the classroom to the actual practice site. In the near future, pharmacists will play a vital role in making personalized medicine a reality. To do this, pharmacists need to be equipped with the fundamentals that will allow them to make clinical decisions based on pharmacogenomic data. Additionally, pharmacists will need to teach other healthcare professionals and patients about pharmacogenomics for personalized medicine to become a reality. This course was designed to prepare pharmacy students for these tasks.
The elective course described in this paper has now been integrated into the core pharmacy curriculum and uses the GeneScription software. The student learning measures employed in this course indicate that the goals and objectives of the course were met. Students’ responses and feedback to the GeneScription software suggest that it is an effective tool to support accomplishment of the stated course objectives.
ACKNOWLEDGMENTS
This work was supported by a generous award from Microsoft Research's Computational Challenges of Genome Wide Association Studies (GWAS) program.
Appendix 1. Sample Case Essay Examination Question
LL is a 68-year-old female presenting with recent stent placement, recent stroke, and pulmonary embolism. She has a 20-year history of hyperlipidemia, osteoarthritis × 3 years, and is currently being treated with warfarin therapy for the pulmonary embolism she experienced 2 weeks ago. She came into your pharmacy today for a medication review and to get her prescriptions filled. LL's family history and social history are non-contributory. Her current medications include atorvastatin 10 mg po QD, melatonin 3 mg po QHS for sleep, acetaminophen 500 mg TID for osteoarthritis pain, and she was recently started on warfarin 5 mg po QD. Your pharmacy has the newest pharmacogenetic testing technology so you test her genetic information while you are reviewing LL's medications. Genetic testing reveals LL has a polymorphism at CYP2C19 and a CYP2C9*2/*3 mutation.
-
1. LL is wondering how this affects her treatment, why you need this information, and who has access to it (3 points)
a. How would you explain the importance of pharmacogenetics/genomics and how this information is being used, to her as a patient, and put her concerns regarding genetic testing at ease?
-
2. LL's physician has given her a prescription for clopidogrel 75mg po QD.
a. Based on her CYP2C19 polymorphism should she have the clopidogrel prescription filled? (2 points)
b. Why should or why shouldn't LL have this prescription filled (will this polymorphism affect clopidogrel treatment and if so, how)? (4 points)
-
3. The physician initiated LL on warfarin 5mg po QD for anticoagulant therapy. (9 points-3 points each)
a. Is this dose appropriate based on the patient's INR goal of 2-3 and their genetic information? Why or why not?
b. If not, what dose would you recommend based on LL's polymorphism? ****Must show work to receive credit for question!
c. Is the patient at an increased risk for ADRs based on the dose recommended by the physician? If yes, what risks?
REFERENCES
- 1.Veenstra D, Higashi M, Phillips K. Assessing the cost-effectiveness of pharmacogenomics. AAPS J. 2000;2(3):e29. doi: 10.1208/ps020329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Accreditation Standards and Guidelines for the Professional Program in Pharmacy Leading to the Doctor of Pharmacy Degree. The Acceditation Council of Pharmacy Education, Chicago, IL 2007. http://www.acpe-accedit.org/standards/default.asp. Accessed August 5, 2010.
- 3.Kane MD, Springer JA, Sprague JE. Drug safety assurance through clinical genotyping: near-term considerations for a systems-wide implementation of personalized medicine. Person Med. 2008;5(4):387–397. doi: 10.2217/17410541.5.4.387. [DOI] [PubMed] [Google Scholar]
- 4.Hedgecoe A. Context, Ethics and pharmacogenetics. Stud Hist Philos Biol Biomed Sci. 2006;37(3):566–582. doi: 10.1016/j.shpsc.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 5.Van Delden J, Bolt I, Kalis A, Derijks J, Leufkens H. Tailor-made pharmacotherapy: future developments and ethical challenges in the field of pharmacogenomics. Bioethics. 2004;18(4):303–321. doi: 10.1111/j.1467-8519.2004.00399.x. [DOI] [PubMed] [Google Scholar]
- 6.Caraco Y, Blotnick S, Muszkat M. CYP2C9 genotype-guided warfarin prescribing enhances the efficacy and safety of anticoagulation: a prospective randomized controlled study. Clin Pharmacol Ther. 2008;83(3):460–470. doi: 10.1038/sj.clpt.6100316. [DOI] [PubMed] [Google Scholar]
- 7.Anderson J, Horne B, Stevens S, et al. Randomized trial of genotype-guided versus standard warfarin dosing in patients initiating oral anticoagulation. Circulation. 2007;116(22):2563–2570. doi: 10.1161/CIRCULATIONAHA.107.737312. [DOI] [PubMed] [Google Scholar]
- 8.Sullivan-Klose T, Ghanayem B, Bell D, et al. The role of the CYP2C9-Leu359 allelic variant in the tolbutamide polymorphism. Pharmacogenetics. 1996;6(4):341–349. doi: 10.1097/00008571-199608000-00007. [DOI] [PubMed] [Google Scholar]
- 9.Shintani M, Ieiri I, Inoue K, et al. Genetic polymorphisms and functional characterization of the 5'-flanking region of the human CYP2C9 gene: in vitro and in vivo studies. Clin Pharmacol Ther. 2001;70(2):175–182. doi: 10.1067/mcp.2001.117367. [DOI] [PubMed] [Google Scholar]
- 10.Poirier T, Crouch M, Mackinnon G, et al. Updated guidelines for manuscripts describing instructional design and assessment: the IDEAS format. Am J Pharm Educ. 2009;73(3) doi: 10.5688/aj730355. Article 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilkey M, Earp J, French E. Applying health education theory to patient safety programs: three case studies. Health Promot Pract. 2008;9(2):123–129. doi: 10.1177/1524839907312703. [DOI] [PubMed] [Google Scholar]