Abstract
Histone proteins organize DNA into dynamic chromatin structures and regulate processes such as transcription, repair and replication. Control of chromatin function and structure is mediated in part by reversible posttranslational modifications (PTMs) on histones. The most N-terminal region of histone H3 contains a high density of modifiable residues. In this review, we focus on the dynamic interplay between histone modification states on the H3 N-terminus and the binding modules that recognize these states. Specifically, we will discuss the effect of auxiliary modifications to H3K4unmod/me3 binding modules (specifically H3R2 methylation, H3T3 phosphorylation and H3T6 phosphorylation). Emerging evidence suggests that histone PTMs behave less like a strict ‘code’, but rather like a ‘language’, which better illustrates the importance of context. Using androgen receptor-mediated gene activation as an example, we propose a model for how the combinatorial nature of PTMs on the H3 N-terminus and the complexes that recognize these epigenetic modifications control gene expression.
Keywords: Epigenetics, histone code, histone language, post translational modifications, histone H3
1. Introduction
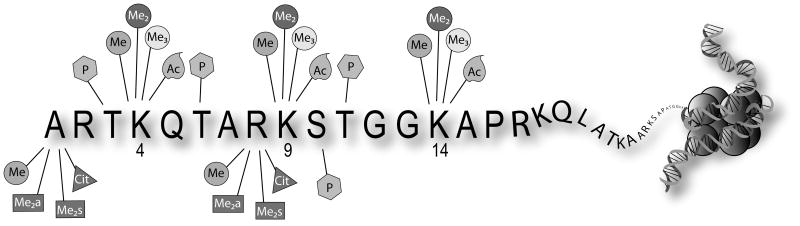
Histone proteins package eukaryotic DNA into chromatin to facilitate nuclear compaction and to modulate DNA expression and replication. The basic building block of chromatin, the nucleosome, is derived from an octamer of histone proteins: one H3/H4 tetramer and two H2A/H2B dimers. [1] Histone proteins are targets of post-translational modifications (PTMs) that include acetylation, methylation, phosphorylation and deimination. [2] The most N-terminal region of the histone (tail) is where the highest density of PTM possibilities exist (Figure 1).[3] These dynamic modifications regulate the structure and function of chromatin through two general mechanisms.
Figure 1.
Representation of possible modification states on the most N-terminal region of histone H3.
One mechanism involves the ability of a PTM to cause a change in chromatin dynamics (e.g., heterochromatin vs. euchromatin). For instance, acetylation of histone-4 lysine-16 (H4K16) inhibits the ability of chromatin to form a compact 30-nanometer-like structure and impedes formation of cross-fiber interactions.[4] The second model illustrates the ability of histone PTMs to serve as docking sites for non-histone proteins. In this case, the PTM indirectly affects chromatin organization and interpretation through recruitment and/or stabilization of module-associated proteins and complexes.[5] The associated complexes mediate diverse outcomes through further chromatin modification or recruitment of enzymes complexes that catalyze DNA-templated processes such as transcription, and DNA synthesis and repair.
The notion of a ‘histone-code’ is derived in part from the idea that modules associate with chromatin in a modification and position dependent manner. Unlike the genetic triplet code, one PTM mark does not necessarily correlate with one outcome (i.e. a triplet codon results in the same amino acid when translated, but the functional outcome of a histone PTM appears highly context dependent).[6] For an excellent review highlighting various chromatin binding modules with an emphasis on structural interactions see: Taverna et al.[2] The idea of a strict code is further complicated because combinations of histone modifications can affect the ability of modules to associate with chromatin (Figure 1).[7] In 2003, Fischle W., et al., hypothesized the existence of ‘binary switches’ and modification ‘cassettes’ on chromatin.[8] This idea expanded the histone-code hypothesis and highlighted a unique situation where phosphorylation of serine-10 on histone H3 (H3S10ph) antagonizes the ability of the HP1 chromodomain to recognize trimethylation at lysine-9 (H3K9me3). Though the authors focused mostly on this antagonistic mode of epigenetic regulation, several other classifications between histone modifications and proteins have been identified: cooperative (two or more marks act together to facilitate protein stabilization or recruitment), independent (modifications can exist together but do not interfere with each other's interacting proteins), or antagonistic (the presence of a modification will block interaction with an adjacent modified residue; binary switches).[9]
This review will focus on the interplay between several histone modification states on the H3 N-terminus (residues 1-15; Figure 1) with special attention to the cooperative, antagonistic and sequential categories of interaction. We will highlight the trimethylated and unmodified versions of histone H3 lysine 4 as distinct platforms for protein module recognition. The major concentration will be on the effect of auxiliary modifications to H3K4unmod/me3 binding modules (specifically H3R2 methylation, H3T3 phosphorylation and H3T6 phosphorylation), though some unique examples will be provided as well. The review will underscore the importance of considering context when interpreting the ‘language’ of these types of modifications. We have compiled a comprehensive table (Table 1) with measured dissociation constants (Kd values) for modules that have been evaluated for their ability to recognize histone H3 and to be affected by adjacent modifications. Finally we propose a simplified model for how the interplay between chromatin binding modules, enzymes, and histone modifications may help facilitate androgen receptor-mediated transcriptional activation.
Table 1. Dissociation constants for chromatin binding modules that recognize histone H3.
| Module | Peptide | Kd ± s.d. (μM) |
Method | Reference |
|---|---|---|---|---|
| ING2-PHD | H3K4me3 | 1.5 ± 1 | TF | [44] |
| 0.98 ± 0.12 | ITC | [46] | ||
| H3R2me2s/K4me3 | 17.3 ± 2.3 | |||
| H3K4me3/T6ph | 19.9 ± 4.9 | |||
| H3K4me3/T3ph | N.D. | |||
| ING4-PHD | H3K4me3 | 3.0 ± 0.6 | NMR | [57] |
| 6.4 ± 0.8 | ITC | |||
| H3R2me2a/K4me3 | 19.2 ± 1.7 | NMR | ||
| RAG2-PHD | H3K4me3 | 33.8 ± 1.9 | FP | [28] |
| 17.9 ± 2.2 | ITC | [46] | ||
| H3R2me2a/K4me3 | 34.6 ± 5.0 | FP | [28] | |
| H3R2me2s/K4me3 | 25.1 ± 3.5 | FP | ||
| H3K4me3/T3ph | N.D. | ITC | [46] | |
| H3K4me3/T6ph | N.D. | |||
| BHC80-PHD | H3K4unmod | 19.6 ± 3.6 | ITC | [46] |
| H3R2me2s | 57.2 ± 5.4 | |||
| H3T3ph | N.D. | |||
| H3T6ph | N.D. | |||
| AIRE-PHD1 | H3K4me0 | 5.3 ± 1.2 | TF | [42] |
| 6.5 ± 0.2 | ITC | |||
| H3R2me1/K4me0 | 248.0 ± 20.0 | TF | ||
| 198.8 ± 12.4 | ITC | |||
| H3R2me2a/K4me0 | N.D. | TF | ||
| H3R2me2s/K4me0 | N.D. | |||
| H3K4me0/K9me3 | 30.4 ± 2.2 | |||
| 29.4 ± 0.5 | ITC | |||
| H3K4me0/K9ac | 39.2 ± 2.3 | TF | ||
| 47.6 ± 1.5 | ITC | |||
| H3K4me0/S10ph | 7.6 ± 1.2 | TF | ||
| 24.7 ± 0.2 | ITC | |||
| H3T3ph/K4me0 | N.D. | TF | ||
| N.D. | ITC | [46] | ||
| N.D. | ||||
| H3K4me0/T6ph | N.D. | |||
| CHD4-PHD2 | H3K4me0 | 18 ± 5 | TF | [98] |
| H3K4me0/K9me1 | 4 ± 1 | |||
| H3K4me0/K9me2 | 1.1 ± 1 | |||
| H3K4me0/K9me3 | 0.9 ± 0.4 | |||
| H3K4me0/K9ac | 0.6 ± 0.1 | |||
| CHD1-Chromo1,2 | H3K4me3 | 5.2 ± 0.6 | FP | [56] |
| H3K4me3/K9me3 | 6 ± 1 | |||
| H3T3ph/K4me3 | 140 ± 34 | |||
| H3K4me3/S10ph | 7.9 ± 0.6 | |||
| H3K4me3/K9ac/K14ac | 10 ± 2 | |||
| H3R2me2a/K4me3 | 24 ± 4 | |||
| TAF3-PHD | H3K4me3 | 0.31 ± 0.09 | TF | [95] |
| H3R2me2a/K4me3 | 2.5 ± 0.2 | |||
| BPTF-PHD | H3K4me3 | 3.6 ± 0.8 | ||
| H3R2me2a/K4me3 | 9.4 ± 0.3 | |||
| DNMT3A-ADD[a] | H3K4me0 | 0.26 | ITC | [99] |
| H3R2me2a/K4me0 | 0.74 | |||
| JMJD2A-DTD | H3K4me3 | 1.1 ± 0.0 | ITC | [46] |
| H3T3ph/K4me3 | N.D. | |||
| H3K4me3/T6ph | 1.8 ± 0.2 | |||
| 14-3-3 | H3S10ph | 240 ± 39 | FP | [69] |
| 24.07 ± 0.55[b] | ITC | [70] | ||
| 715 ±305[c] | ||||
| H3K9me2/S10ph | 200 ± 18 | FP | [69] | |
| H3K9ac/S10ph | 49 ± 18 | |||
| H3S10ph/K14ac | 26 ± 9 | |||
| 25.91 ± 0.74[b] | ITC | [70] | ||
| 281 ± 22[c] | ||||
| H3K9ac/S10ph/K14ac | 40 ±10 | FP | [69] | |
The ADD domain contains a PHD-finger
150 mM NaCl buffer
500 mM NaCl buffer.
N.D.: not detectable TF: tryptophan fluorescence ITC: isothermal titration calorimetry FP: fluorescence polarization.
References apply to all measurments below until a new reference number is cited.
2. Histone H3 N-terminal Methylation
2.1 Histone H3 Lysine 4 as a Recognition Platform
Lysine 4 on histone H3 is a well-documented site for post-translational modification.[5, 10] This position can be either mono-, di- or tri-methylated at the ε-amine. There is also evidence for an important role of acetylation at this site.[11] The most studied modification state on H3K4 is trimethylation (H3K4me3)[5], which is normally associated with start sites of transcriptionally active genes [12, 13] and has a strong positive correlation with active polymerase II occupancy, histone acetylation and transcription rates.[14-18]
Trimethylation of H3K4 falls into the category of histone modifications that serve to modulate binding interactions between non-histone proteins and histone tail domains. Chromatin-binding domains that specifically recognize trimethylation of H3K4 can be divided into two broad categories: the royal superfamily (including chromo and tudor domains) and the PHD-finger family. [19, 20]
The unmodified form of H3K4 (H3K4unmod) is also a platform for chromatin binding module recognition. Found in a multitude of diverse proteins, the majority of binding modules that recognize H3K4unmod are PHD fingers [19, 21], though other types of domains exist (i.e. the ADD-domain of the DNA-methyltransferase family).[2, 22]
2.2 Modification of Histone H3 at Arginine 2 (H3R2)
Histone arginine residues can exist in three distinct methylated states: monomethylation, asymmetric dimethylation, and symmetric dimethylation.[23] Type I protein methyl transferases (PRMTs) form monomethyl and asymmetric dimethyl arginine (Rme1 and Rme2a) by transferring methyl group(s) to the omega-nitrogen of the guanidinium side chain group. Type II PRMTs catalyze the formation of monomethyl and symmetric dimethyl arginine (Rme1 and Rme2s). Symmetric dimethylation differs slightly from asymmetric dimethylation where two methyl groups are transferred to a single omega-nitrogen of the guanidinium side chain. Type III PRMTs achieve specific catalysis of monomethyl arginine (Rme1) with no ability to create either dimethylated form.
The major type I PRMT responsible for mono- and asymmetric methylation of H3R2 is PRMT 6.[24-26] PRMT5 is a major type II PRMT that can preferentially catalyze mono- and symmetric dimethylation, however, H3R2 appears not to be a substrate for this enzyme.[27, 28] Whether symmetric dimethylation of H3R2 exists in vivo remains to be established.
Unlike histone lysine methylation, there is no known example where methylated arginine residues serve as an independent platform for recognition by a dedicated protein module. The modification status at H3R2, however, plays an important role in epigenetic regulation. For instance, asymmetric methylation at H3R2 antagonizes SET1a mediated trimethylation of H3K4 in Saccharomyces cerevisiae.[29] The Spp1 subunit of this complex is a critical component required for trimethylation to occur.[30] The PHD-finger domain of Spp1 arbitrates an interaction between H3K4me2/3 and the methyltransferase complex[31] helping to facilitate catalysis. Methylation at H3R2 regulates the activity of the Set1 complex towards H3K4 by antagonizing the ability of Spp1 to bind H3.[29]
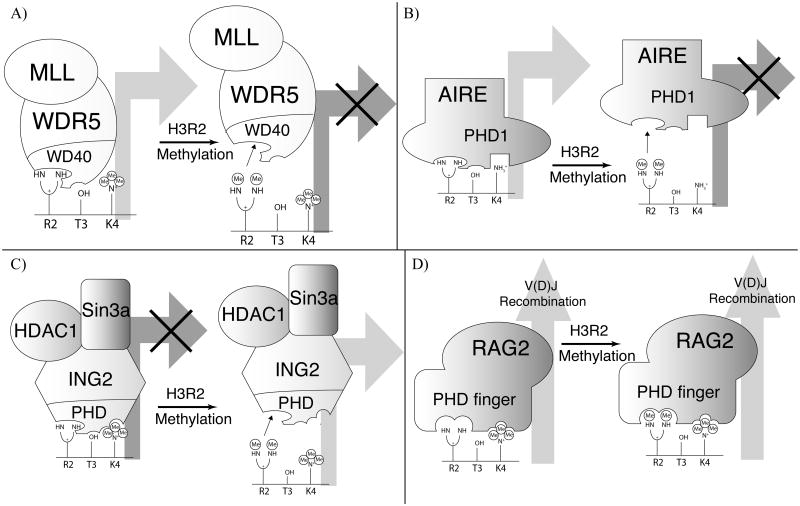
This regulatory mechanism is conserved to a similar extent in humans. The ability of an MLL-containing methyltransferase complex to methylate H3K4 is compromised in the presence of PRMT6-mediated methylation of H3R2 (asymmetric dimethylation).[25, 26] Interestingly, the complex contains several proteins with histone binding modules.[32] Though recent work suggests the primary function of WDR5 may be binding an arginine-bearing motif in MLL1, promoting complex assembly and activity[33], there is considerable evidence demonstrating the ability of the WD40 repeat domain of WDR5 to mediate catalysis at H3K4 by presenting this portion of the H3 tail for methylation.[34-37] Pulldowns with an H3 unmodified peptide were used to confirm that WDR5 could be recovered effectively from U937 nuclear lysates. The presence of H3K4me3 did not impinge upon the interaction, however, the existence of H3R2me2a reduced recovery to background levels[26] demonstrating the module's sensitivity to methylation at H3R2 (Figure 2a).
Figure 2.
Effect of H3R2 methylation on chromatin binding modules. A) WD40-repeat of WDR5 displays sensitivity to H3R2 methylation and the transcriptional consequence is repression. B) The PHD-finger of ING2 recognizes H3K4me3 but is antagonized by H3R2 methylation, leading to transcriptional activation. C) AIRE-PHD1 recognizes H3K4unmod and is sensitive to H3R2 methylation, which leads to transcriptional repression. D) RAG2-PHD, which recognizes H3K4me3, is insensitive to methylation at H3R2 and V(D)J recombination is unaffected by this epigenetic modification.
In a study investigating the effect of H3R2me2a on a panel of chromatin binding modules, Iberg et al identified several isolated domains (PHD-fingers, tudor-domains, and WD40 repeats) that are sensitive to this modification.[38] Interestingly, it was discovered that PRMT6 could asymmetrically methylate H3R2 in the presence of H3K4me3 in vitro, though there is also evidence that H3K4me3 may actually antagonize PRMT6 activity.[25, 26]
2.3 H3R2 Methylation and Transcriptional Repression
Several studies demonstrated the repressive nature of H3R2me2a in the context of H3K4me3 at specific HoxA genes.[25, 38] By overexpressing PRMT6, MLL-WDR5 dependent transcription at these loci was dramatically decreased. This observation suggested that PRMT6-mediated H3R2 dimethylation influences recruitment of the methyltransferase complex and contributes to transcriptional repression.[25] (Figure 2a)
Another relevant example illustrating the repressive nature of H3R2me2a was reported with the human autoimmune regulator (AIRE) protein (Figure 2b). Through its first plant homeodomain (PHD1) finger, AIRE recognizes the H3K4unmod form of the H3 tail.[39] This interaction mediates organ-specific autoimmunity through transcriptional activation of tissue-specific genes with low methylation status at H3K4.[40, 41] Interestingly, H3R2 dimethylation decreases AIRE-PHD1 binding to H3 in vitro (Table 1) and reduces the in vivo activation of AIRE target genes in HEK293 cells. [42] This provides a unique example of modification crosstalk between H3K4unmod (an activation state in this context) and H3R2 dimethylation (a repressive effect in this context) (Figure 2b).
2.4 H3R2 Methylation and Transcriptional Activation
Asymmetric methylation at H3R2 may serve as a transcriptionally active modification in particularly contexts (Figure 2c). Expression of cylin D1 genes is negatively regulated by the ING2-HDAC1-mSin3a protein complex in response to DNA damage.[43] The interaction between the ING2-PHD finger and H3K4me3 is crucial for gene repression[43], and ING2 mutations in the PHD-finger affect the complex's ability to facilitate apoptosis in response to DNA damage.[44, 45] The presence of H3R2me2a results in decreased affinity of the ING2-PHD interaction for trimethylated H3K4 (Table 1).[38, 46] After doxorubicin treatment, repression of the cyclin D1 gene by the ING2-HDAC1 complex was robust in PRMT6 knockdown cells and weakened in PRMT6-overexpressing cells. Use of short hairpin RNA for PRMT6 (shPRMT6) resulted in increased physical association of ING2 and a significant decrease in cyclin D1 mRNA expression (Figure 2c).[38] This example illustrates the importance of considering context when analyzing the influence of H3R2 methylation at a transcriptional level.
2.5 H3R2 Methylation and V(D)J Recombination
Perhaps the most unique scenario with H3R2 methylation involves the PHD-finger of recombination activating gene 2 (RAG2). RAG2, in cooperation with RAG1, is essential for V(D)J-recombination, a process which mediates antigen-receptor gene assembly.[47, 48] The PHD-finger of RAG2 specifically recognizes H3K4me3. In contrast to many PHD-fingers (see Table 1), the RAG2-PHD finger is tolerant towards methylation at H3R2, and may even harbor a slight preference over the unmodified form (Figure 2d).[28] The ability of RAG2-PHD to recognize these dual-modifications offers a unique regulatory mechanism where V(D)J recombination is unaffected by H3R2 methylation, yet other processes (i.e. transcription) are inhibited by H3R2 methylation (Figure 2).
Methylation of H3R2 illustrates the inherent flexibility in histone PTM interpretation, and one could argue that such flexibility revokes the idea of a strict ‘histone code’. The word ‘code’ is conventionally used to describe histone PTMs, but may be misleading because of its implication in having a strict set of rules. The case here illustrates more of a histone ‘language’ where the meaning of a particular modification depends entirely on the context in which it exists. In support of this idea, H3R2 methylation status is a critical modulator of two unique circumstances where lack of H3K4me3 is involved in gene activation (AIRE), and where H3K4me3 is required for repression (ING2).
3. Histone H3 N-terminal Phosphorylation
3.1 Modification of Histone H3 at Threonine 3 (H3T3)
Histone 3 threonine-3 is phosphorylated by recently identified kinases. Mut9p phosphorylates histones H3, including H3T3, and H2A in Chlamydomonas reinhardtii.[49] Interestingly, phosphorylation of threonine-3 (H3T3ph) and monomethyl lysine 4 (H3K4me1) correlate with regions of repressed transcription and reduced Mut9p results in lower levels of both marks.[49, 50] MUT9p also appears to be involved in the inheritance of silent chromatin states. Haspin/Gsg2 also specifically phosphorylates H3T3.[51] RNAi-mediated depletion of haspin causes misalignment of metaphase chromosomes, while overexpression delays progression through early mitosis[52, 53] emphasizing the importance of this modification in these processes.
Much like methylation at H3R2, it is not known whether chromatin-binding modules exist that can specifically recognize H3T3ph. However, this modification appears to have a role in mediating interactions between histone H3 and the inhibitor of acetyltransferases (INHAT) complex. Two critical subunits of the INHAT complex, SET and pp32, can specifically bind to the H3 tail, but their interaction is decreased by phosphorylation at H3T3.[54]
In vitro, H3T3ph has a dramatic effect on a myriad of other modules that recognize both H3K4unmod[46, 55] and H3K4me3 (Table 1).[34, 46, 56] To date, there has not been a chromatin-binding module identified that recognizes either H3K4me3 or H3K4unmod and displays insensitivity to H3T3ph. It has been proposed, based on structural analysis, that the PHD-finger of ING4 might accommodate H3T3ph by a conformational change at the side chain of Lys232, in which a favorable electrostatic interaction could be established.[57] The ability of the ING4-PHD to successfully tolerate this mark, however, awaits experimental evidence. It is an intriguing possibility that H3T3ph functions as a general ‘off-switch’ for enzymes/complexes that recognize the H3 N-terminus, especially in the context of H3K4unmod or H3K4me3. The ability to transiently disrupt these transcriptional states may be an important component for the proper inheritance of silent chromatin states. Essentially, the modification status of H3K4 would be retained and protected from interacting partners that may interfere with the biological processes necessary for silent chromatin state inheritance.
3.2 Modification of Histone H3 at Threonine 6 (H3T6)
Only very recently has phosphorylation of H3T6 been identified. Garske et al. documented the existence of this modification in HeLa cells using both immunological and mass spectrometric techniques.[46] Shortly thereafter, protein kinase CβI (PKCβI) was identified as the protein kinase capable of phosphorylating H3T6 in vitro and in vivo.[58] Phosphorylation at T6 was shown to play an important role in LSD1 and JARID1B mediated demethylation of H3K4 in androgen receptor repression by preventing both enzymes from demethylating H3K4.[58]
Interestingly, H3T6ph has a varied effect on chromatin binding proteins that recognize either H3K4unmod or H3K4me3.[46] Similar to H3T3ph, phosphorylation at H3T6 antagonizes the ability of the BHC80 and AIRE PHD-fingers to bind H3K4unmod, and the ability of RAG2 PHD-finger to recognize H3K4me3 (Table 1). However, H3T6ph caused a modest decrease (∼10-fold) in the ability of the ING2 PHD-finger to recognize H3K4me3 and has no appreciable effect on the double-tudor domain of JMJD2A to bind H3K4me3 (Table 1).
3.3 Modification of Histone H3 at Serine 10 (H3S10)
Phosphorylation of serine-10 on histone H3 (H3S10ph) is a well-studied modification. First identified in 1968, AMP-dependent kinase was shown to phosphorylate H3S10 in vivo.[59] Since that time, additional kinases have been identified as bona fide histone H3S10 kinases.[60-62] Perhaps the most prominent example of histone modification crosstalk is that between H3S10ph and H3K9me3.[63, 64] Briefly, H3S10ph occurs at the onset of mitosis, interferes with a heterochromatin protein-1 (HP1) - H3K9me3 interaction, and ejects HP1 from its binding site. For a detailed perspective on this antagonistic mechanism see: [8, 65]
Although there are no reports of specific protein modules that solely bind threonine phosphorylation on the H3 histone tail, 14-3-3 family of proteins recognize H3S10ph[66-68], specifically the 14-3-3ζ isoform.[66] It appears that acetylation of the H3 tail, specifically at H3K14, enhances this interaction and demonstrates an interesting example where two marks cooperate to facilitate chromatin module binding (Table 1).[69, 70] In this scenario, lysine acetylation could function as an auxiliary modification that enhances the relatively weak interaction of 14-3-3 with H3S10ph. The dual modification contributes to localization of the 14-3-3 protein to target loci, ejection of HP1γ, and facilitating transcriptional activation.[69]
4. Androgen Receptor Activation and the Role of H3 N-terminus Modification
The androgen receptor (AR) is part of the steroid hormone receptor family of ligand-activated transcription factors. It shares a common structure with other nuclear receptors and consists of several domains that potentiate DNA binding, dimerization, ligand binding and transcriptional activity.[71] Upon hormone binding, the cytoplasmic androgen receptor dissociates from chaperones, dimerizes and translocates to the nucleus where it binds to androgen response elements (AREs) of target genes (such as prostate specific antigen (PSA) or kallikrein2).[72] The ARE-bound AR dimer can either interact directly with components of the transcription preinitiation complex or recruit other components that promote a similar interaction.[73-75] As a general definition, AR coregulators are proteins that are recruited by the AR and either enhance (i.e. coactivators) or reduce (i.e. corepressors) its transactivation, but they do not significantly alter the basal transcription rate and do not typically possess DNA binding ability.[76] Coregulators influence AR-mediated transcription by facilitating DNA occupancy, chromatin remodeling, and/or recruitment of general transcription factors associated with RNA polymerase II, or by assuring the competency of the AR to enhance gene expression directly.[73, 76] There are a myriad of putative coregulators for the AR that display a diverse array of functions and are involved in many different cellular pathways.[76] Our focus here will be on AR coactivators that modulate and interpret PTMs on chromatin. It is worth noting that further regulation of AR-dependent genes is achieved by both specific and general transcription factors, but the details of these interactions will not be discussed in this review. For a comprehensive overview on the multitude of AR coregulators with classification according to their intrinsic primary function see: [76].
Androgen-receptor mediated regulation is arbitrated in part by distal enhancer elements[77] which show distinct cell-type-specific histone modification patterns that strongly correlate to cell-type-specific gene expression programs on a global scale.[78] At AR-specific loci, the formation of an activation complex involves AR, coactivators, and RNA polymerase II recruitment to both the enhancer and promoter regions.[79] Below, we use AR-mediated gene activation as an example of how different combinations of histone H3 PTMs and enzyme complex (coactivator) binding events might intimately control chromatin structure and gene activity.
4.1 Demethylation of H3K9me3
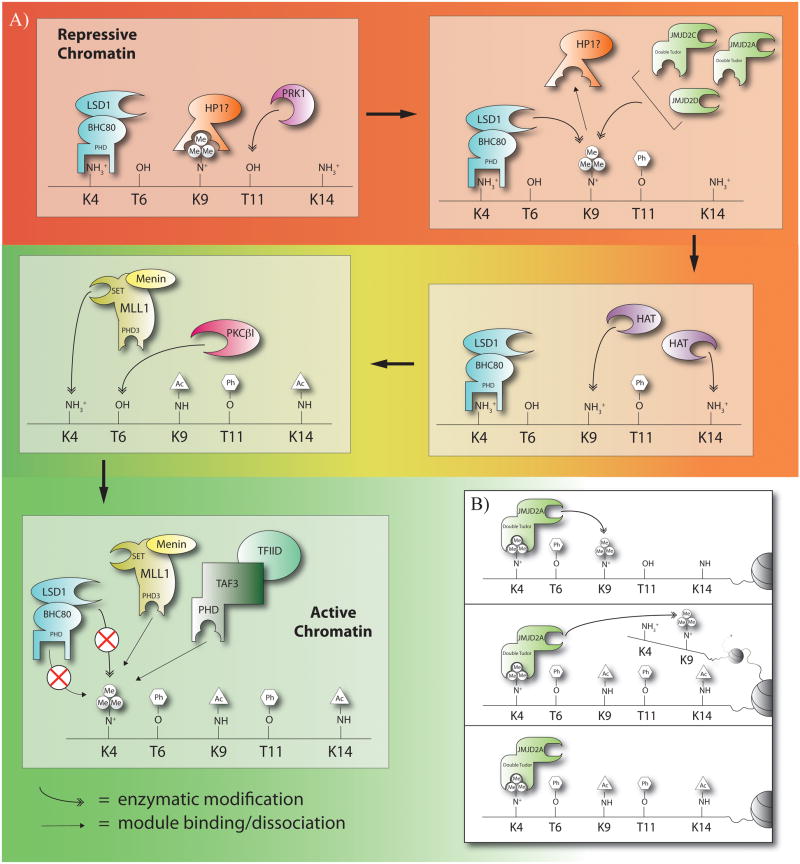
Activation requires proper PTM status of two lysine residues on H3: H3K9 and H3K4 (Figure 3a). An important first step in the transition from a repressed state to an activated state is the demethylation of H3K9me3, which is a repressive PTM in the context of AR-mediated transcription.[80] Removal of H3K9me3 is facilitated by cooperation of two histone demethylases, JMJD2C and LSD1 (Figure 3a).[81] The demethylase JMJD2C catalyzes demethylation of tri- and dimethylated H3K9 [82] while LSD1 facilitates removal of di- and monomethylated H3K9.[83] The actions of JMJD2C and LSD1 result in a shift from H3K9me3 to H3K9unmod. Presently, it is unclear to what extent JMJD2A and JMJD2D participate in AR activation through H3K9me3 demethylation (Figure 3a).[81, 84]
Figure 3.
Epigenetic control of androgen receptor activation. A) The transition from repressive to active chromatin. B) Three different models for JMJD2A/C-DTD recognition of H3K4me3in the presence of H3T6ph. Top panel, JMJD2A/C facilitates demethylation of H3K9me3 at sites where H3T6ph is already present (“intra-tail” activation). Middle panel, JMJD2A/C interacts with active chromatin containing H3K4me3/T6ph and facilitates the start of activation (“inter-tail” activation). Bottom panel, JMJD2A/C remains localized to the active chromatin via the DTD in order to maintain demethylated H3K9.
Interestingly, the ability of JMJD2C to demethylate H3K9me3 is augmented by phosphorylation at H3T11.[85] Blocking protein-kinase-C-related kinase 1's (PRK1) ability to phosphorylate H3T11 inhibits androgen-induced demethylation of histone H3. However, more work is needed to understand the complete molecular details for how H3T11ph accelerates demethylation by the Jumonji C (JmjC)-domain-containing protein JMJD2C. Knockdown of PRK1 also affects acetylation of both H3K9/K14 (modifications corresponding to transcriptional activation) at several AR-receptor dependent genes.[85] Synergistic coupling of histone H3 phosphorylation and acetylation has been observed in response to epidermal growth factor stimulation[86], and it is interesting to speculate a similar mechanism for AR-mediated activation. Furthermore, hyperacetylation of histones is common in nuclear-hormone receptor mediated transcriptional activation.[87]
4.2 Trimethylation of H3K4
The next step in AR-mediated activation involves methylation of H3K4. In a study by Kim et al, substantial increases in H3K4me2/3 were observed in the coding region of the PSA gene correlating with expression.[88] Interestingly, it is reported that the histone methyltransferase complex MLL1/MLL2, in concert with menin, is a transcriptional coactivator of the nuclear receptors for estrogen and vitamin D.[89] Menin interacts with MLL1/MLL2 histone methyltransferases[90, 91] and is crucial for trimethylation of H3K4 (Figure 3a). The menin-MLL1/MLL2 complex could function similarly for AR-dependent gene activation, though more work will be needed to explore this possibility. The presence of H3K9ac contributes to the ability of the MLL1-SET domain to catalyze trimethylation of H3K4 and provides a possible role for H3 acetylation in activation. Enhanced H3K4 methylation by the MLL1-SET domain of an H3K9 acetylated substrate may arise from increased binding affinity, as charge neutralization by acetylation favors a hydrophobic interaction.[92]
To facilitate activation, it is important that newly trimethylated H3K4me3 is maintained, and this is largely accomplished by phosphorylation of H3T6ph. Phosphorylation at H3T6 was recently discovered and is catalyzed by protein kinase C beta I (PKCβ1).[46, 58] Phosphorylation eliminates the ability of LSD1 to demethylate H3K4me2/1 and prevents JARID1B from demethylating H3K4me3/2.[93] PKCβ1 acts in an androgen- and PRK1-dependent manner (downstream of T11 phosphorylation) shedding light onto the modification chronology of AR-activated transcription.[58]
Trimethylation of H3K4 recruits the basal transcription factor TFIID in a TAF3 PHD-finger dependent manner.[94] This interaction results in enhanced recruitment or stability of the RNA polymerase II preinitiation complex [94], and provides the basis for its role as a transcriptional coactivator. Interestingly, acetylation of H3K9/14 augments the ability of the PHD-finger to recognize H3K4me3[94] and provides evidence for a further role of these acetyl marks in transcriptional activation. It would be interesting to test the effect of H3T6 phosphorylation on the ability of the TAF3 PHD-finger to recognize H3K4me3. Because recognition is based primarily on interactions with the first 4 amino acids of the H3 tail, phosphorylation at H3T6 may have a very small effect. This is corroborated by structural similarity to the ING2-PHD finger[95] and the observation that peptides containing H3K4me3T6ph yield a relatively small decrease in binding affinity compared to the singly modified H3K4me3 peptide for the ING2-PHD finger (Table 1).[46]
Interestingly, the double tudor domain (DTD) of JMJD2A recognizes H3K4me3 and its binding affinity is not appreciably affected by H3T6ph (Table 1).[46] Because of structural similarities, the DTD of JMJD2C may harbor similar insensitivity to this modification, but this claim awaits experimental evidence. The significance of this interaction for AR-mediated genes falls into three general models (Figure 3b). Firstly, H3K4me3T6ph protects lysine-4 trimethylation but allows JMJD2A/C to demethylate H3K9me3/2. This may be an initial activation step if H3K4 trimethylation and H3T6 phophorylation occur before H3K9 demethylation (“intra-tail” demethylation). The second possible function of this interaction could be a platform for “inter-tail” activation by keeping the H3K9 demethylases JMJD2A/C localized to chromatin containing both H3K4me3 and H3T6ph. In this scenario, the demethylases would facilitate demethylation of neighboring H3-tails. Finally, recognition of H3K4me3T6ph by the DTD may serve as a platform for continued maintenance of the H3 epigenetic state. This would ensure that H3K9 remains demethylated even in the presence of methyl transferases that remain in close proximity.
5. Outlook
Since the proposal of a ‘histone code’[96] there has been great progress in understanding the molecular underpinnings of PTM-based modulation of chromatin structure and function. Here we have outlined several pertinent examples that demonstrate the importance of considering histone PTMs in the context of other neighboring modifications. The highlighted examples underscore the difficulty in generalizing transcriptional outcome based solely on the correlation mapping of single post-translational modifications. A good example is H3R2 methylation. The downstream biological effect of this modification is reliant on a.) whether the appropriate protein module is expressed and b.) how R2 methylation affects that modules ability to bind the H3 tail. Although there are no reports of chromatin-binding modules that specifically recognize methylated versions of H3R2, recent structural evidence demonstrated the ability of a tudor domain to bind Rme2s from a non-histone protein [97] It seems highly likely, based on this new discovery, that H3R2me/me2-binding domains exist.
The recent discoveries of additional phosphorylation sites (T3 and T6) on the H3 N-terminus highlight the combinatorial complexity of PTMs in close proximity, especially those that occur within tail ‘hotspots’. An important piece of the epigenetic puzzle is understanding the effect of histone modifications on modules that recognize neighboring modifications (for example, the effect of H3T6ph on H3K4me3 binders). So far phosphorylation at T3 or T6 have either a neutral or antagonistic effect on module binding, but it remains to be discovered whether phosphorylation at these sites might enhance some protein:histone interactions, or whether there exists dedicated protein modules that specifically recognize these threonine phosphorylations.
The observation that auxiliary modifications affect histone-binding module recognition is also important when interpreting experiments that use an antibody developed against a single histone PTM. A case in point, chromatin immunoprecipitation (ChIP) experiments that utilize these antibodies to characterize the genome-wide or loci-specific existence of individual histone PTMs might be misleading in genome regions where no signal is detected. The ability of an antibody to recognize a particular modification depends on the local structural environment, which can affect cross-linking efficiency, and the lack of neighboring modifications that might otherwise occlude antibody binding. Therefore, ChIP-type experiments that suggest an absence of a particular PTM might reflect alterations in chromatin structure and neighboring PTMs that preclude antibody binding to these regions of chromatin. In order to determine their specificity in the context of multiple PTMs, an important future endeavor will be the detailed characterization of antibodies used for such experiments. Particularly useful in characterizing such antibodies would be the utilization of PTM-based peptide arrays that include adjacent PTMs. For ChIP-type experiments, the use of non-antibody based detection, or the use of antibodies raised against peptides having multiple PTMs might avoid some of these potential issues and provide vital missing information that is contained within this complex PTM language.
Using androgen receptor-mediated gene activation, we have constructed a model that describes how combinatorial PTMs contribute to the different chromatin states. The exact chronology of these events is not entirely clear and more work is needed. Further investigations are required to provide a complete understanding of how combinations of modifications affect histone modifying enzymes and chromatin binding modules. For instance, how does H3T11ph augment JMJD2C demethylation of H3K9me3? Is H3K9me3T11ph a better enzyme substrate or is there a module that specifically recognizes H3T11ph and localizes JMJD2C for demethylation of H3K9me3? It is important to appreciate that histone modifications in the context of neighboring modifications reveal a deeper understanding of transcriptional control at the epigenetic level.
Supplementary Material
Acknowledgments
This work was supported by NIH (GM059785 to JMD and T32GM008505 to SSO). We would like to thank J.P. Svaren for his helpful comments on the manuscript.
References
- 1.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Nature. 1997;389:251. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. Nat Struct Mol Biol. 2007;14:1025. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kouzarides T. Cell. 2007;128:693. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 4.Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Science. 2006;311:844. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 5.Ruthenburg AJ, Allis CD, Wysocka J. Mol Cell. 2007;25:15. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 6.Turner BM. Nat Cell Biol. 2007;9:2. doi: 10.1038/ncb0107-2. [DOI] [PubMed] [Google Scholar]
- 7.Ruthenburg AJ, Li H, Patel DJ, Allis CD. Nat Rev Mol Cell Biol. 2007;8:983. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fischle W, Wang Y, Allis CD. Nature. 2003;425:475. doi: 10.1038/nature02017. [DOI] [PubMed] [Google Scholar]
- 9.Seet BT, Dikic I, Zhou MM, Pawson T. Nat Rev Mol Cell Biol. 2006;7:473. doi: 10.1038/nrm1960. [DOI] [PubMed] [Google Scholar]
- 10.Shilatifard A. Curr Opin Cell Biol. 2008;20:341. doi: 10.1016/j.ceb.2008.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xhemalce B, Kouzarides T. Genes Dev. 24:647. doi: 10.1101/gad.1881710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin C, Zhang Y. Nat Rev Mol Cell Biol. 2005;6:838. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- 13.Dehe PM, Geli V. Biochem Cell Biol. 2006;84:536. doi: 10.1139/o06-081. [DOI] [PubMed] [Google Scholar]
- 14.Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T. Nature. 2002;419:407. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 15.Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, Zeitlinger J, Lewitter F, Gifford DK, Young RA. Cell. 2005;122:517. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 16.Schubeler D, MacAlpine DM, Scalzo D, Wirbelauer C, Kooperberg C, van Leeuwen F, Gottschling DE, O'Neill LP, Turner BM, Delrow J, Bell SP, Groudine M. Genes Dev. 2004;18:1263. doi: 10.1101/gad.1198204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schneider R, Bannister AJ, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T. Nat Cell Biol. 2004;6:73. doi: 10.1038/ncb1076. [DOI] [PubMed] [Google Scholar]
- 18.Ng HH, Robert F, Young RA, Struhl K. Mol Cell. 2003;11:709. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 19.Bienz M. Trends Biochem Sci. 2006;31:35. doi: 10.1016/j.tibs.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 20.Maurer-Stroh S, Dickens NJ, Hughes-Davies L, Kouzarides T, Eisenhaber F, Ponting CP. Trends Biochem Sci. 2003;28:69. doi: 10.1016/S0968-0004(03)00004-5. [DOI] [PubMed] [Google Scholar]
- 21.Mellor J. Cell. 2006;126:22. doi: 10.1016/j.cell.2006.06.028. [DOI] [PubMed] [Google Scholar]
- 22.Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, Cheng X, Bestor TH. Nature. 2007;448:714. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bedford MT, Clarke SG. Mol Cell. 2009;33:1. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lakowski TM, Frankel A. J Biol Chem. 2008;283:10015. doi: 10.1074/jbc.M710176200. [DOI] [PubMed] [Google Scholar]
- 25.Hyllus D, Stein C, Schnabel K, Schiltz E, Imhof A, Dou Y, Hsieh J, Bauer UM. Genes Dev. 2007;21:3369. doi: 10.1101/gad.447007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guccione E, Bassi C, Casadio F, Martinato F, Cesaroni M, Schuchlautz H, Luscher B, Amati B. Nature. 2007;449:933. doi: 10.1038/nature06166. [DOI] [PubMed] [Google Scholar]
- 27.Lacroix M, Messaoudi SE, Rodier G, Le Cam A, Sardet C, Fabbrizio E. EMBO Rep. 2008;9:452. doi: 10.1038/embor.2008.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramon-Maiques S, Kuo AJ, Carney D, Matthews AG, Oettinger MA, Gozani O, Yang W. Proc Natl Acad Sci U S A. 2007;104:18993. doi: 10.1073/pnas.0709170104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kirmizis A, Santos-Rosa H, Penkett CJ, Singer MA, Vermeulen M, Mann M, Bahler J, Green RD, Kouzarides T. Nature. 2007;449:928. doi: 10.1038/nature06160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schneider J, Wood A, Lee JS, Schuster R, Dueker J, Maguire C, Swanson SK, Florens L, Washburn MP, Shilatifard A. Mol Cell. 2005;19:849. doi: 10.1016/j.molcel.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 31.Shi X, Kachirskaia I, Walter KL, Kuo JH, Lake A, Davrazou F, Chan SM, Martin DG, Fingerman IM, Briggs SD, Howe L, Utz PJ, Kutateladze TG, Lugovskoy AA, Bedford MT, Gozani O. J Biol Chem. 2007;282:2450. doi: 10.1074/jbc.C600286200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dou Y, Milne TA, Ruthenburg AJ, Lee S, Lee JW, Verdine GL, Allis CD, Roeder RG. Nat Struct Mol Biol. 2006;13:713. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- 33.Trievel RC, Shilatifard A. Nat Struct Mol Biol. 2009;16:678. doi: 10.1038/nsmb0709-678. [DOI] [PubMed] [Google Scholar]
- 34.Couture JF, Collazo E, Trievel RC. Nat Struct Mol Biol. 2006;13:698. doi: 10.1038/nsmb1116. [DOI] [PubMed] [Google Scholar]
- 35.Han Z, Guo L, Wang H, Shen Y, Deng XW, Chai J. Mol Cell. 2006;22:137. doi: 10.1016/j.molcel.2006.03.018. [DOI] [PubMed] [Google Scholar]
- 36.Ruthenburg AJ, Wang W, Graybosch DM, Li H, Allis CD, Patel DJ, Verdine GL. Nat Struct Mol Biol. 2006;13:704. doi: 10.1038/nsmb1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schuetz A, Allali-Hassani A, Martin F, Loppnau P, Vedadi M, Bochkarev A, Plotnikov AN, Arrowsmith CH, Min J. Embo J. 2006;25:4245. doi: 10.1038/sj.emboj.7601316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iberg AN, Espejo A, Cheng D, Kim D, Michaud-Levesque J, Richard S, Bedford MT. J Biol Chem. 2008;283:3006. doi: 10.1074/jbc.C700192200. [DOI] [PubMed] [Google Scholar]
- 39.Org T, Chignola F, Hetenyi C, Gaetani M, Rebane A, Liiv I, Maran U, Mollica L, Bottomley MJ, Musco G, Peterson P. EMBO Rep. 2008;9:370. doi: 10.1038/embor.2008.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Musco G, Peterson P. Epigenetics. 2008;3:310. doi: 10.4161/epi.3.6.7182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koh AS, Kuo AJ, Park SY, Cheung P, Abramson J, Bua D, Carney D, Shoelson SE, Gozani O, Kingston RE, Benoist C, Mathis D. Proc Natl Acad Sci U S A. 2008;105:15878. doi: 10.1073/pnas.0808470105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chignola F, Gaetani M, Rebane A, Org T, Mollica L, Zucchelli C, Spitaleri A, Mannella V, Peterson P, Musco G. Nucleic Acids Res. 2009;37:2951. doi: 10.1093/nar/gkp166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, Carney D, Pena P, Lan F, Kaadige MR, Lacoste N, Cayrou C, Davrazou F, Saha A, Cairns BR, Ayer DE, Kutateladze TG, Shi Y, Cote J, Chua KF, Gozani O. Nature. 2006;442:96. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pena PV, Davrazou F, Shi X, Walter KL, Verkhusha VV, Gozani O, Zhao R, Kutateladze TG. Nature. 2006;442:100. doi: 10.1038/nature04814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nagashima M, Shiseki M, Miura K, Hagiwara K, Linke SP, Pedeux R, Wang XW, Yokota J, Riabowol K, Harris CC. Proc Natl Acad Sci U S A. 2001;98:9671. doi: 10.1073/pnas.161151798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garske AL, Oliver SS, Wagner EK, Musselman CA, LeRoy G, Garcia BA, Kutateladze TG, Denu JM. Nat Chem Biol. 6:283. doi: 10.1038/nchembio.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jones JM, Simkus C. Arch Immunol Ther Exp (Warsz) 2009;57:105. doi: 10.1007/s00005-009-0011-3. [DOI] [PubMed] [Google Scholar]
- 48.Liu Y, Subrahmanyam R, Chakraborty T, Sen R, Desiderio S. Immunity. 2007;27:561. doi: 10.1016/j.immuni.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Casas-Mollano JA, Jeong BR, Xu J, Moriyama H, Cerutti H. Proc Natl Acad Sci U S A. 2008;105:6486. doi: 10.1073/pnas.0711310105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Dijk K, Marley KE, Jeong BR, Xu J, Hesson J, Cerny RL, Waterborg JH, Cerutti H. Plant Cell. 2005;17:2439. doi: 10.1105/tpc.105.034165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eswaran J, Patnaik D, Filippakopoulos P, Wang F, Stein RL, Murray JW, Higgins JM, Knapp S. Proc Natl Acad Sci U S A. 2009;106:20198. doi: 10.1073/pnas.0901989106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dai J, Higgins JM. Cell Cycle. 2005;4:665. doi: 10.4161/cc.4.5.1683. [DOI] [PubMed] [Google Scholar]
- 53.Dai J, Sultan S, Taylor SS, Higgins JM. Genes Dev. 2005;19:472. doi: 10.1101/gad.1267105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schneider R, Bannister AJ, Weise C, Kouzarides T. J Biol Chem. 2004;279:23859. doi: 10.1074/jbc.C400151200. [DOI] [PubMed] [Google Scholar]
- 55.Nady N, Min J, Kareta MS, Chedin F, Arrowsmith CH. Trends Biochem Sci. 2008;33:305. doi: 10.1016/j.tibs.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 56.Flanagan JF, Mi LZ, Chruszcz M, Cymborowski M, Clines KL, Kim Y, Minor W, Rastinejad F, Khorasanizadeh S. Nature. 2005;438:1181. doi: 10.1038/nature04290. [DOI] [PubMed] [Google Scholar]
- 57.Palacios A, Munoz IG, Pantoja-Uceda D, Marcaida MJ, Torres D, Martin-Garcia JM, Luque I, Montoya G, Blanco FJ. J Biol Chem. 2008;283:15956. doi: 10.1074/jbc.M710020200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Metzger E, Imhof A, Patel D, Kahl P, Hoffmeyer K, Friedrichs N, Muller JM, Greschik H, Kirfel J, Ji S, Kunowska N, Beisenherz-Huss C, Gunther T, Buettner R, Schule R. Nature. 464:792. doi: 10.1038/nature08839. [DOI] [PubMed] [Google Scholar]
- 59.Langan TA. Science. 1968;162:579. doi: 10.1126/science.162.3853.579. [DOI] [PubMed] [Google Scholar]
- 60.Prigent C, Dimitrov S. J Cell Sci. 2003;116:3677. doi: 10.1242/jcs.00735. [DOI] [PubMed] [Google Scholar]
- 61.DeManno DA, Cottom JE, Kline MP, Peters CA, Maizels ET, Hunzicker-Dunn M. Mol Endocrinol. 1999;13:91. doi: 10.1210/mend.13.1.0222. [DOI] [PubMed] [Google Scholar]
- 62.Schmitt A, Gutierrez GJ, Lenart P, Ellenberg J, Nebreda AR. FEBS Lett. 2002;518:23. doi: 10.1016/s0014-5793(02)02630-3. [DOI] [PubMed] [Google Scholar]
- 63.Fischle W, Tseng BS, Dormann HL, Ueberheide BM, Garcia BA, Shabanowitz J, Hunt DF, Funabiki H, Allis CD. Nature. 2005;438:1116. doi: 10.1038/nature04219. [DOI] [PubMed] [Google Scholar]
- 64.Hirota T, Lipp JJ, Toh BH, Peters JM. Nature. 2005;438:1176. doi: 10.1038/nature04254. [DOI] [PubMed] [Google Scholar]
- 65.Dormann HL, Tseng BS, Allis CD, Funabiki H, Fischle W. Cell Cycle. 2006;5:2842. doi: 10.4161/cc.5.24.3540. [DOI] [PubMed] [Google Scholar]
- 66.Macdonald N, Welburn JP, Noble ME, Nguyen A, Yaffe MB, Clynes D, Moggs JG, Orphanides G, Thomson S, Edmunds JW, Clayton AL, Endicott JA, Mahadevan LC. Mol Cell. 2005;20:199. doi: 10.1016/j.molcel.2005.08.032. [DOI] [PubMed] [Google Scholar]
- 67.Dougherty MK, Morrison DK. J Cell Sci. 2004;117:1875. doi: 10.1242/jcs.01171. [DOI] [PubMed] [Google Scholar]
- 68.Mackintosh C. Biochem J. 2004;381:329. doi: 10.1042/BJ20031332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Winter S, Simboeck E, Fischle W, Zupkovitz G, Dohnal I, Mechtler K, Ammerer G, Seiser C. Embo J. 2008;27:88. doi: 10.1038/sj.emboj.7601954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Walter W, Clynes D, Tang Y, Marmorstein R, Mellor J, Berger SL. Mol Cell Biol. 2008;28:2840. doi: 10.1128/MCB.01457-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mangelsdorf DJ, Evans RM. Cell. 1995;83:841. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 72.Metzger E, Wissmann M, Schule R. Curr Opin Genet Dev. 2006;16:513. doi: 10.1016/j.gde.2006.08.013. [DOI] [PubMed] [Google Scholar]
- 73.Heinlein CA, Chang C. Endocr Rev. 2002;23:175. doi: 10.1210/edrv.23.2.0460. [DOI] [PubMed] [Google Scholar]
- 74.Lee DK, Chang C. J Steroid Biochem Mol Biol. 2003;84:41. doi: 10.1016/s0960-0760(03)00005-0. [DOI] [PubMed] [Google Scholar]
- 75.Wang L, Hsu CL, Chang C. Prostate. 2005;63:117. doi: 10.1002/pros.20170. [DOI] [PubMed] [Google Scholar]
- 76.Heemers HV, Tindall DJ. Endocr Rev. 2007;28:778. doi: 10.1210/er.2007-0019. [DOI] [PubMed] [Google Scholar]
- 77.Magee JA, Chang LW, Stormo GD, Milbrandt J. Endocrinology. 2006;147:590. doi: 10.1210/en.2005-1001. [DOI] [PubMed] [Google Scholar]
- 78.Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, Ching KA, Antosiewicz-Bourget JE, Liu H, Zhang X, Green RD, Lobanenkov VV, Stewart R, Thomson JA, Crawford GE, Kellis M, Ren B. Nature. 2009;459:108. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shang Y, Myers M, Brown M. Mol Cell. 2002;9:601. doi: 10.1016/s1097-2765(02)00471-9. [DOI] [PubMed] [Google Scholar]
- 80.Hublitz P, Albert M, Peters AH. Int J Dev Biol. 2009;53:335. doi: 10.1387/ijdb.082717ph. [DOI] [PubMed] [Google Scholar]
- 81.Wissmann M, Yin N, Muller JM, Greschik H, Fodor BD, Jenuwein T, Vogler C, Schneider R, Gunther T, Buettner R, Metzger E, Schule R. Nat Cell Biol. 2007;9:347. doi: 10.1038/ncb1546. [DOI] [PubMed] [Google Scholar]
- 82.Shin S, Janknecht R. Biochem Biophys Res Commun. 2007;353:973. doi: 10.1016/j.bbrc.2006.12.147. [DOI] [PubMed] [Google Scholar]
- 83.Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH, Gunther T, Buettner R, Schule R. Nature. 2005;437:436. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 84.Shin S, Janknecht R. Biochem Biophys Res Commun. 2007;359:742. doi: 10.1016/j.bbrc.2007.05.179. [DOI] [PubMed] [Google Scholar]
- 85.Metzger E, Yin N, Wissmann M, Kunowska N, Fischer K, Friedrichs N, Patnaik D, Higgins JM, Potier N, Scheidtmann KH, Buettner R, Schule R. Nat Cell Biol. 2008;10:53. doi: 10.1038/ncb1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cheung P, Tanner KG, Cheung WL, Sassone-Corsi P, Denu JM, Allis CD. Mol Cell. 2000;5:905. doi: 10.1016/s1097-2765(00)80256-7. [DOI] [PubMed] [Google Scholar]
- 87.Chen H, Lin RJ, Xie W, Wilpitz D, Evans RM. Cell. 1999;98:675. doi: 10.1016/s0092-8674(00)80054-9. [DOI] [PubMed] [Google Scholar]
- 88.Kim J, Jia L, Tilley WD, Coetzee GA. Nucleic Acids Res. 2003;31:6741. doi: 10.1093/nar/gkg909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dreijerink KM, Mulder KW, Winkler GS, Hoppener JW, Lips CJ, Timmers HT. Cancer Res. 2006;66:4929. doi: 10.1158/0008-5472.CAN-05-4461. [DOI] [PubMed] [Google Scholar]
- 90.Yokoyama A, Wang Z, Wysocka J, Sanyal M, Aufiero DJ, Kitabayashi I, Herr W, Cleary ML. Mol Cell Biol. 2004;24:5639. doi: 10.1128/MCB.24.13.5639-5649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hughes CM, Rozenblatt-Rosen O, Milne TA, Copeland TD, Levine SS, Lee JC, Hayes DN, Shanmugam KS, Bhattacharjee A, Biondi CA, Kay GF, Hayward NK, Hess JL, Meyerson M. Mol Cell. 2004;13:587. doi: 10.1016/s1097-2765(04)00081-4. [DOI] [PubMed] [Google Scholar]
- 92.Southall SM, Wong PS, Odho Z, Roe SM, Wilson JR. Mol Cell. 2009;33:181. doi: 10.1016/j.molcel.2008.12.029. [DOI] [PubMed] [Google Scholar]
- 93.Xiang Y, Zhu Z, Han G, Ye X, Xu B, Peng Z, Ma Y, Yu Y, Lin H, Chen AP, Chen CD. Proc Natl Acad Sci U S A. 2007;104:19226. doi: 10.1073/pnas.0700735104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Vermeulen M, Mulder KW, Denissov S, Pijnappel WW, van Schaik FM, Varier RA, Baltissen MP, Stunnenberg HG, Mann M, Timmers HT. Cell. 2007;131:58. doi: 10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 95.van Ingen H, van Schaik FM, Wienk H, Ballering J, Rehmann H, Dechesne AC, Kruijzer JA, Liskamp RM, Timmers HT, Boelens R. Structure. 2008;16:1245. doi: 10.1016/j.str.2008.04.015. [DOI] [PubMed] [Google Scholar]
- 96.Jenuwein T, Allis CD. Science. 2001;293:1074. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 97.Liu H, Wang JY, Huang Y, Li Z, Gong W, Lehmann R, Xu RM. Genes Dev. 24:1876. doi: 10.1101/gad.1956010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Musselman CA, Mansfield RE, Garske AL, Davrazou F, Kwan AH, Oliver SS, O'Leary H, Denu JM, Mackay JP, Kutateladze TG. Biochem J. 2009;423:179. doi: 10.1042/BJ20090870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Otani J, Nankumo T, Arita K, Inamoto S, Ariyoshi M, Shirakawa M. EMBO Rep. 2009;10:1235. doi: 10.1038/embor.2009.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.