Abstract
Background
Alternative mRNA splicing is an important mechanism for regulation of gene expression. Altered mRNA splicing occurs in association with several types of cancer, and a small number of disease-associated changes in splicing have been reported in heart disease. However, genome-wide approaches have not been used to study splicing changes in heart disease. We hypothesized that mRNA splicing is different in diseased hearts compared to control hearts.
Methods and Results
We used the Affymetrix exon array to globally evaluate mRNA splicing in LV myocardial RNA from control (n=15) and ischemic cardiomyopathy (ICM) patients. We observed a broad and significant decrease in RNA splicing efficiency in heart failure, which affected some introns to a greater extent than others. The profile of mRNA splicing separately clustered ICM and control samples, suggesting distinct changes in RNA splicing between groups. RTPCR validated 9 previously unreported alternative splicing events. Furthermore, we demonstrated that splicing of four key sarcomere genes, cardiac troponin T (TNNT2), cardiac troponin I (TNNI3), myosin heavy chain 7 (MYH7), and filamin C gamma (FLNC), was significantly altered in ICM, as well as in dilated cardiomyopathy and aortic stenosis (AS). In AS samples, these differences preceded the onset of heart failure. Remarkably, the ratio of minor to major splice variants of TNNT2, MYH7, and FLNC classified independent test samples as control or disease with greater than 98% accuracy.
Conclusions
Our data indicate that RNA splicing is broadly altered in human heart disease, and that patterns of aberrant RNA splicing accurately assign samples to control or disease classes.
Keywords: RNA Splicing, Cardiomyopathy, Heart Failure, Genes
Introduction
Nearly every human mRNA transcript is alternatively spliced1, and mRNA splicing is an important mechanism for generating transcriptional diversity and regulating gene expression. Tissue specific mRNA splicing confers specialized properties to organs such as the heart. A significant proportion of gene mutations that cause human disease disrupt mRNA splicing2. Perturbation of mRNA splicing may also influence the natural history of acquired diseases, as mRNA splicing is influenced by environmental signals and can become dysregulated in disease states3–5. Abnormal patterns of mRNA splicing are associated with diseases such as cancer, and splicing profiles have been shown to be effective in cancer classification5, 6.
Heart failure is a leading cause of morbidity and mortality. Biomechanical loads induce gene expression changes in the heart. Heart failure also influences mRNA splicing, as indicated by focused studies of a small number of mRNA transcripts documenting disease-associated changes in mRNA splicing7,8. For instance, human heart failure was associated with higher expression of a dominant negative variant of the key transcription factor Serum Response Factor (SRF)7. Ischemic cardiomyopathy was accompanied by higher expression of an alternatively spliced variant of the sarcomere gene titin, resulting in reduced titin-derived myofibrillar stiffness8. However, global, genome-wide studies of differences in RNA splicing between control and diseased myocardium have not been reported.
We hypothesized that mRNA splicing is more widely altered in human heart failure than has been appreciated based on anecdotal evidence. We used the Affymetrix Exon array to interrogate RNA splicing in ischemic cardiomyopathy (ICM; n=15) and non-failing control (n=15) myocardium, and we used RT-PCR to validate findings in an independent set of 60 samples. We found substantial differences in RNA splicing between control and diseased myocardium, including splicing of sarcomeric genes. Measures of RNA splicing assigned samples to disease or control categories with 98% accuracy. Our results indicate that mRNA splicing is broadly perturbed in heart disease.
Materials and methods
Patient samples
Left ventricular (LV) myocardial samples, remote from infarcted areas, were obtained from explanted cardiomyopathy hearts at the time of transplantation, from unused transplant donor hearts, and from hearts at the time of implant or explant of a LV assist device. Samples were frozen immediately in liquid nitrogen and stored at −80 °C. Collection and analysis of tissue samples was performed under protocols approved by Institutional Review Boards of participating institutions.
RNA extraction and transcript analysis
RNA was extracted with Trizol (Invitrogen) and further purified on RNeasy columns (Qiagen) with on column DNase digestion. Control and disease samples were of comparable high quality when analyzed using an Experion bio-analyzer (Bio-Rad). Probe generated from RNA was hybridized to Affymetrix GeneChip Human Exon 1.0 ST (HuEx) Arrays (Gene Expression Omnibus, accession number GSE16499).
RNA was converted to cDNA by random priming, using Superscript III reverse transcriptase (Invitrogen). PCR Primers are listed in Supplementary Table 1. RTPCR products were analyzed by gel electrophoresis, real time PCR, or capillary electrophoresis (see Supplementary Methods). Intergroup comparisons were performed using Welch's t-test. Values are expressed as mean ± SEM.
Bioinformatic analysis
We used several bioinformatic approaches to detect AS events and minimize false discoveries (Suppl Methods). We focused on genes with < 50 exons, because large exon numbers increase multiple simultaneous hypothesis testing. We defined alternative splicing events as novel if they were not present in the Alternative Splicing and Transcript Diversity database9. Assignment to diagnostic class was performed using a logistic regression-based classifier derived from biomarkers based on splicing state (Suppl Methods).
Results
Systematic alteration of RNA splicing in ischemic cardiomyopathy
We isolated total RNA from LV myocardium of patients with ischemic cardiomyopathy (ICM, n=15), obtained at the time of heart transplantation. Control samples (n=15) were obtained from transplant donor hearts not used for transplantation for non-cardiac reasons. Ischemic time in both groups was less than two hours. Groups were matched for sex (each 12 M and 3 F) and age (50.4 ± 1.9 yrs control vs 52.1 ± 1.1 yrs ICM). Clinical data are summarized in Suppl. Table 2.
RNA samples were hybridized to HuEx arrays. Each exon is represented by a “probeset” consisting of four oligonucleotides. Probesets are grouped by the level of supportive evidence. The “Core Probeset” represents Refseq exons, while the “Complete Probeset” includes the Core Probeset plus probesets supported by expressed sequence tags (ESTs) and by computational prediction. There are 232,448 and 1,653,912 Core and Complete Probesets, respectively, on the HuEx Array.
Whole gene expression levels were calculated by combining probe intensities for exons that belong to the same gene. Consistent with prior reports10, 11, hierarchical clustering using genes with significant differential expression accurately segregated control and ICM samples (Suppl. Fig. 1a). We did not detect significantly differences in expression profiles between males and females within the control or ICM cohorts. 1036 genes were differentially expressed between control and ICM (nominal P < 0.01). The differentially expressed genes substantially overlapped with differentially expressed genes in an independent profiling study of ICM samples12 (Suppl. Fig 1b; Suppl. Table 3).
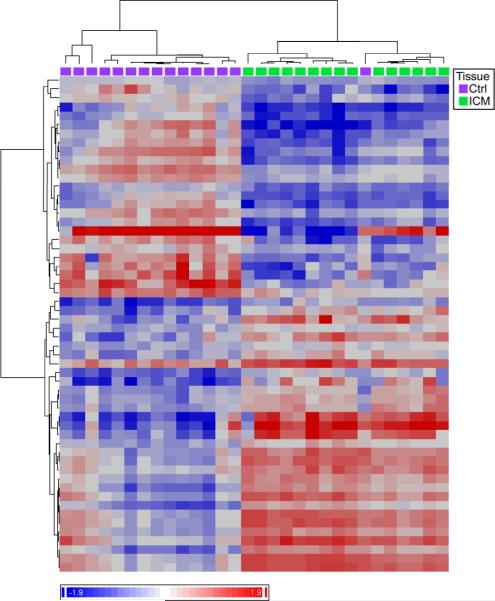
To determine if splicing profiles similarly segregate samples by diagnosis, we used the FIRMA method13 on Core Probesets to score each gene for alternative splicing. When we selected genes with FIRMA scores that differed significantly between groups (false discovery rate (FDR) < 1%), ICM and control samples clustered by diagnostic label (Fig. 1), with only one sample placed in the incorrect cluster. This analysis showed that splicing is systematically altered in ICM versus control samples.
Figure 1. Systematic differences of RNA splicing in ICM compared to control.
Genes were scored for likelihood of alternative splicing (FIRMA score). Two way clustering of samples and genes with significantly different FIRMA scores (FDR < 0.01) separated samples by diagnosis, with one misassignment. Heat map is color coded by FIRMA score (blue-red, −1.9 to +1.9 standard deviations from the mean). Genes with significantly different gene level expression (P < 0.05) were excluded.
Altered splicing of sarcomere genes in heart disease
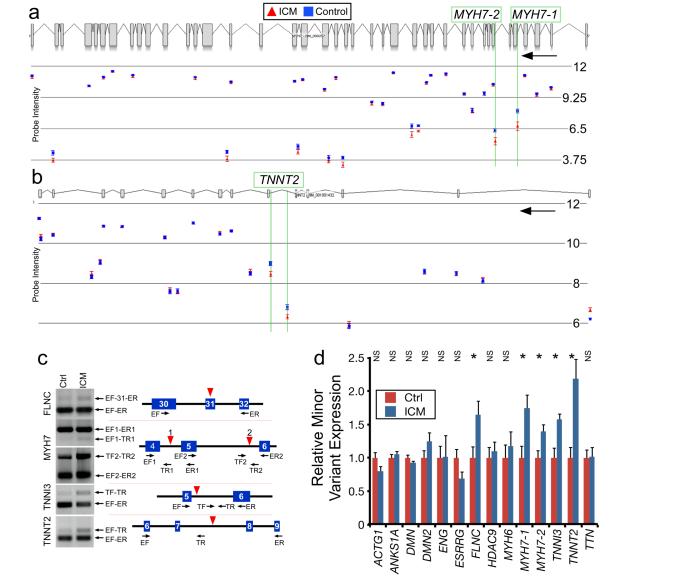
We next set out to identify genes that are differentially spliced between control and ICM groups. We analyzed Complete Probeset data using two different statistical approaches, AS-ANOVA and MADS14, to identify candidates likely to be differentially spliced between ICM and control groups. Those with significant scores by both methods (P < 0.001) are shown in Suppl. Table 4. A subset of genes identified by these methods was visually inspected to identify those with likely altered splicing between groups. Amongst these screened candidates were several genes already known to regulate cardiomyocyte contraction or hypertrophy, including multiple sarcomere genes. Subsequently, we visually inspected all cardiomyopathy-associated sarcomere genes for potential differential splicing between groups. Two of the candidates, MYH7 and TNNT2, are shown in Fig. 2a–b. The TNNT2 alternative splicing event discovered here is distinct from those previously reported15, 16.
Fig. 2. Differences in Alternative Splicing between ICM and control.
a–b. Examples of exon array data suggestive of differential alternative splicing between ICM and control. Expression values are in log2 scale. Black arrows indicate direction of transcription. n=15. c. Structure and representative RTPCR of major and minor isoforms in control and ICM heart samples. Blue boxes indicate RefSeq exons. Red triangles indicate probeset with differential signal detected by exon array. Primers are indicated as arrows beneath gene models. EF, exonic forward. ER, exonic reverse. TF, intronic forward. TR, intronic reverse. d. Percentage of minor variant by RTPCR. n=15. *, P < 0.05 by Welch's t-test. NS, not significant.
We selected 20 candidate alternative splicing events (in 17 genes) differentially regulated in ICM for validation by RTPCR (Table 1, Fig. 2, and Suppl. Fig. 2). Eleven involved intron retention, a relatively infrequent form of alternative splicing17. Additionally, eleven of these events were not previously reported in the Alternative Splicing Database9. To confirm these novel alternative splicing isoforms and define the mechanism of alternative splicing, we sequenced RTPCR products using primers predicted to span exon-exon boundaries. Of the eleven novel putative splicing events, we confirmed nine (Table 1 and Suppl. Fig. 2).
Table.
Selected Candidate Genes with Differential Alternative Splicing in ICM (Complete Probeset)
| Gene Symbol | Gene Name | Accesion Number | AS Type* | Changed in ICM^ |
|---|---|---|---|---|
| ACTG1 | Actin, gamma 1 | TRAN00000084206 | RI | N |
| ANKS1A | Ankyrin repeat and sterile alpha motif domain containing 1A | Novel | CE | N |
| DNM2 | Dynamin 2 | Novel | RI | N |
| ENG | Endoglin | ENST00000344849 | RI | N |
| ESRRG | Estrogen-related receptor gamma | ENST00000354407 | AFE | N |
| FLNC | Filamin C, gamma | ENST00000346177 | CE | Y |
| HDAC9 | Histone deacetylase 9 | Novel | RI | N |
| HOPX | HOP homeobox, event 1 | Novel | ND | ND |
| HOP homeobox, event 2 | ENST00000317745 | CE | U | |
| MKL2 | MKL/myocardin-like 2 | Novel (AK093577, AX748211) | ND | U |
| MYBPC3 | myosin binding protein C, cardiac | Novel | RI | U |
| MYH6 | myosin, heavy chain 6, event 1 | ENST00000356287 | CE | N |
| myosin, heavy chain 6, event 2 | Novel | RI | N | |
| MYH7 | myosin, heavy chain 7, event 1 | Novel | RI | Y |
| myosin, heavy chain 7, event 2 | Novel | RI | Y | |
| PARVB | parvin, beta | Novel | CE | U |
| TNNI3 | troponin I type 3 (cardiac) | Novel (AW027005) | RI | Y |
| TNNT2 | troponin T type 2 (cardiac) | TRAN00000060677 | RI | Y |
| TRABD | TraB domain containing | TRAN00000089822 | CE | N |
| TTN | Titin | ENST00000355565 | RI | N |
Database: If present in the alternative splicing database (ATSD), then a supporting accession number is provided. Otherwise the event was considered novel. If supporting ESTs were found, then up to 2 supporting accession numbers are provided.
Alternative splicing class for novel splicing events was validated by sequencing of RTPCR products.
Abbreviation:RI, retained intron. CE, cassette exon. ALE, alternative last exon. AFE, alternative first exon. ND, predicted minor isoform not detected by RTPCR. U, RTPCR unsuccessful for quantitating minor isoform abundance.
Motifs for the well-characterized splicing regulators muscleblind-like (MBNL) and hnRNP F were significantly overrepresented in sequences flanking differentially spliced exons (Suppl. Table 5; p < 10−6). Based on the exon array, hnRNP F expression was downregulated in ICM by 1.3-fold (nominal p-value 0.0012), while MBNL mRNA expression was not significantly changed. These data suggest that MBNL and hnRNP F participate in altered RNA splicing observed in human heart failure.
To validate differential alternative splicing between groups, we designed semi-quantitative RTPCR assays to measure the ratio of minor and major splice isoforms, and compared the ratio between groups. Out of the 20 splicing events, we were successful in designing RTPCR assays for 17. Of these 17 events, RTPCR showed that five splicing events involving four genes were enriched in ICM compared to control (P < 0.05; n=15; Figure 2c–d). Interestingly, these four genes are involved in sarcomere structure and function. One gene, Filamin C gamma (FLNC), a muscle-restricted actin binding protein localized to Z-discs and intercalated discs18, 19, exhibited higher inclusion of a previously described cassette exon (Fig. 2c–d). The other three genes, MYH7 (beta myosin heavy chain), TNNI3 (cardiac troponin I), and TNNT2 (cardiac troponin T), underwent greater intron retention in heart failure, resulting in incorporation of in-frame stop codons. (Figure 2c–d). Two separate intron retention events in MYH7, referred to as MYH7-1 and MYH7-2, were higher in ICM.
We performed several control experiments to verify that these intron retentions were not artifacts. First, all RTPCR experiments were performed with and without addition of reverse transcriptase, and we observed no RTPCR products in the absence of this enzyme (Suppl. Fig. 3a and data not shown). Second, sequencing of RTPCR products verified amplification of the intended fragment and demonstrated lack of splicing at the reference sequence exon-intron boundary in the minor isoform. Third, the intron retention products were also detected with oligo-dT-priming of RT reactions, indicating that the intron retention products are on polyadenylated transcripts.
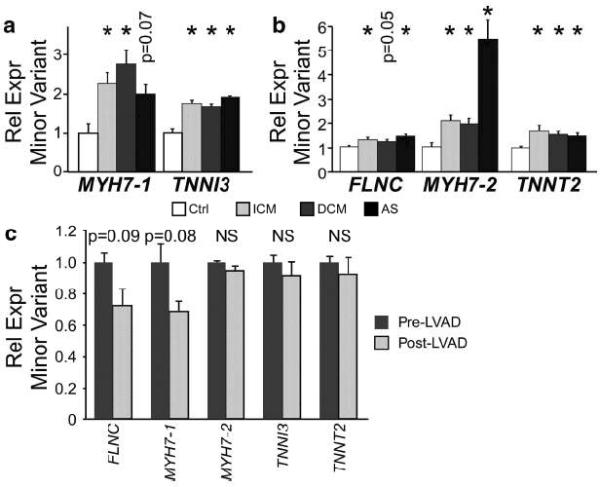
We next sought to validate and extend our findings in independent samples. We used quantitative RTPCR assays to measure minor to major isoform ratios of TNNI3, MYH7-1, MYH7-2, TNNT2, and FLNC in LV RNA from control, ICM, idiopathic dilated cardiomyopathy (DCM), and aortic stenosis (AS) patients, obtained at different centers from the 30 samples initially used for array analysis (Fig. 3a–b). These samples were described previously20. For ICM, we again found higher minor to major isoform ratios of MYH7-1, MYH7-2, TNNI3, TNNT2, and FLNC, indicating that these findings are reproducible in independent patient populations and in tissue collected at different centers (Fig. 3a). The minor to major isoform ratio of all four genes was also higher in dilated cardiomyopathy (DCM; P < 0.05) and aortic stenosis (AS; P < 0.05; Fig. 3a–b), distinct forms of heart disease.
Figure 3. Disease-associated differences in splicing of sarcomere genes in diseased compared to control myocardium.
a. Minor variant fraction of TNNI3 and MYH7-1 were measured by RTPCR in control, ICM, DCM, and AS samples (5 per group). b. Minor variant fraction of TNNT2, MYH7-2, and FLNC sarcomere genes were measured by RTPCR in control, ICM, DCM, and AS samples (10 per group). c. LVAD mechanical unloading did not normalize splicing of the four sarcomere genes in heart failure. n=4. NS, not significant. *, P < 0.05, using Welch's t-test with Sidak correction.
The aortic stenosis samples gave us to opportunity to investigate if RNA splicing differences are evident prior to the onset of overt heart failure, as 5 AS samples were from patients with heart failure (EF < 50%), while 5 were from patients with compensated heart function (EF ≥ 50%). Importantly, the splicing ratios of these genes were the indistinguishable between compensated and failing AS myocardium. This suggests that splicing differences are present in diseased hearts with elevated mechanical load, prior to the onset of overt heart failure.
Conversely, we considered whether mechanical unloading of failing hearts, through placement of LV assist devices (LVADs), was sufficient to correct abnormal splicing patterns. Similar studies of gene expression have shown that mechanical unloading normalizes expression of some but not all genes21, 22. Therefore, we compared splicing of these genes before and after LVAD placement in patients with end stage heart disease (n = 4). RTPCR measurement of the minor to major isoform ratio showed that LVAD placement did not significantly change splicing of these genes (Fig. 3c). The splicing profile of these samples was consistent with the pattern observed in diseased hearts (see below).
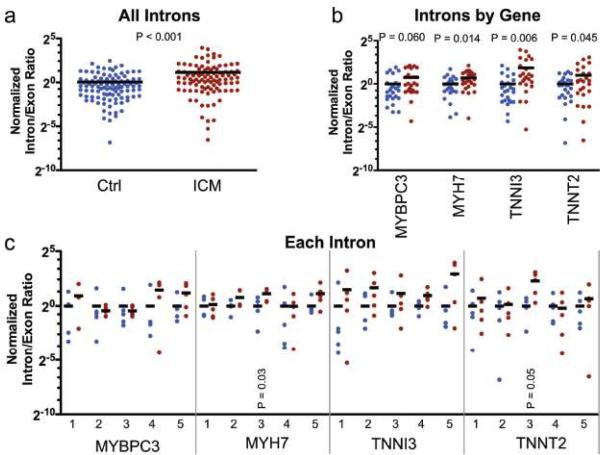
Exon array probes located in sarcomere gene introns identified several introns that were spliced less efficiently in heart failure. To determine if inefficient splicing was widespread in heart failure, we used quantitative real time RTPCR to measure 5 arbitrarily chosen introns and one exon in each of 4 sarcomere genes (TNNT2, TNNI3, MYH7, and MYBPC3). Comparison of ICM to control samples showed that introns were spliced out with significantly reduced efficiency in ICM. This was evident when the intron/exon ratio was compared between groups for all genes (Fig. 4a) or when intron/exon ratios were analyzed by gene (Fig. 4b). Analysis of each intron individually demonstrated considerable heterogeneity between samples. Despite reduced statistical power in this analysis, two additional introns showed statistically higher intron retention (Fig. 4c). Both of these intron retentions were significantly higher in an independent set of ICM and control samples (n = 5; P < 0.05; data not shown). These data indicate that ICM is associated with reduced splicing efficiency that affects some introns to a greater degree than others.
Figure 4. Lower RNA Splicing Efficiency in Heart Failure.
Expression of 5 introns and one exon for 4 genes was measured by qRTPCR. The intron/exon ratio was expressed relative to control (defined as 1). a. Comparison of all measured intron/exon ratios between control and ICM. b. Comparison of measured intron/exon ratios between control (blue) and ICM (red), analyzed by gene. c. Individual analysis of each intron/exon ratio. Black lines indicate the group mean. Each point represents the intron/exon ratio from a single sample, measured in duplicate. Controls lacking RT showed no detectable signal. P value indicates Welch's t-test between ICM and control.
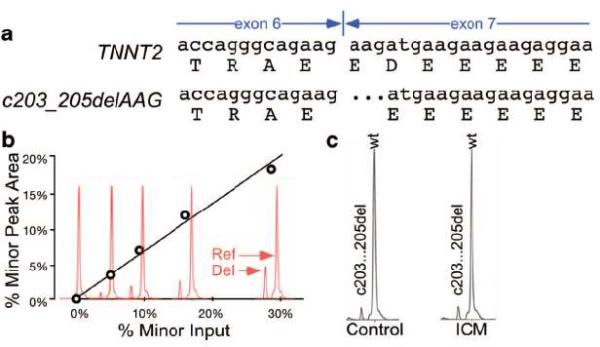
Lower splicing efficiency in heart failure led us to ask if splicing precision was also affected. To test this hypothesis, we developed a moderate throughput, coupled RTPCR-capillary electrophoresis assay to detect and measure minor variations at exon-exon junctions. For pilot studies, we examined a novel splice variant of TNNT2 that we discovered in the course of sequencing to validate TNNT2 intron retention. This variant arises from use of an alternative 5' splice acceptor on TNNT2 exon 7, resulting in a spliced transcript lacking three nucleotides and consequently resulting in an in-frame deletion of aspartate 41 (p.Asp41del; c.203_205delAAG [NM_001001431]; Fig. 5a). When known ratios of wild-type and c.203_205delAAG TNNT2 variants were mixed, we were able to detect the minor variant when it was present at 5% of the major isoform (Fig. 5b). Analysis of control and ICM samples (n=15) showed that the c.203_205delAAG variant comprised 5–10% of TNNT2 transcripts and did not significantly differ between groups (Fig. 5c and data not shown). We next used the RTPCR-capillary electrophoresis platform to anaylze 48 additional amplicons that encompass all coding exon-exon junctions of nine sarcomere genes that are known to genetically contribute to inherited cardiomyopathies (myosin binding protein c3 [MYBPC3], cardiac alpha actin [ACTC1], myosin light chain 2 [MYL2], myosin light chain 3 [MYL3], MYH7, cardiac troponin C [TNNC1], TNNI3, TNNT2, and alpha tropomyosin [TPM1]) for similar splice junction variation. This survey revealed only one other minor isoform involving fine differences of exon-exon junctions. This again occurred in TNNT2, in an amplicon (primers 3F/3R; Supplementary Table 1) that spans exons 12–16. The peak area of the minor isoform was < 5% of the major isoform and did not vary between control and ICM groups. These data indicate that the splicing of sarcomere genes remains precise in heart failure.
Fig 5. Analysis of sarcomere gene exon-exon junctions by capillary electrophoresis of RTPCR products.
a. TNNT2 transcript and alternative splicing variant due to differential exon 7 5' splice acceptor use. b. Detection of variation of exon-exon junctions by capillary electrophoresis. A standard curve was generated by mixing known proportions of cloned wild-type and c.203_205del TNNT2. The peak area ratios from capillary electrophoresis analysis were linearly related to the input ratio. c. Representative chromatograms of control and ICM samples analyzed using the capillary electrophoresis assay. No significant difference in the proportion of c203_205del variant was detected between groups (n=15).
Splicing Profiles Accurately Label Samples with Diagnostic Class
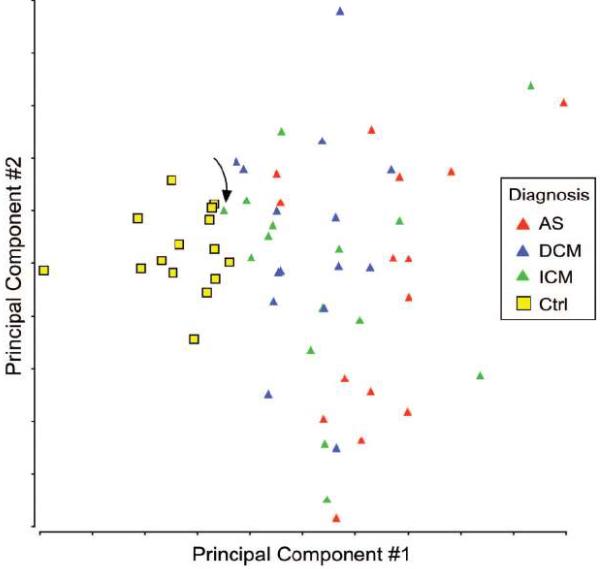
Splicing profiles are effective diagnostic classifiers in cancer5, 6. To begin to investigate the effectiveness of splicing profiles as biomarkers in heart disease, we asked whether the splicing pattern of MYH7-2, TNNT2, and FLNC was sufficient to classify samples as control or diseased. Pilot studies, in which we measured minor to major splicing ratios for MYH7-2, TNNT2, and FLNC in 20 samples (control, ICM, DCM, and AS, 5 per group) by qRTPCR, suggested that splicing profiles of these genes could accurately assign control and disease diagnostic labels. Therefore, we expanded the study to include an additional 40 samples (10 per group). Principal component analysis of the 60 samples indicated that splicing profiles of these genes effectively separated control and disease samples (Fig. 6). Consistent with this observation, a logistic regression-based classifier trained on the 40 samples to classify control versus disease samples assigned the remaining 20 samples to control or disease groups with 100% accuracy. Similarly, greater than 98% accuracy was attained when 20 samples were used as the training set and 40 samples were used as the validation set. This level of accuracy was also observed in a 5-fold cross validation study over 20 permutations of the combined 60 samples. One ICM sample, apparent on the Principal component analysis (arrow, Fig. 6), was frequently misclassfied as control. The classifier assigned both pre- and post- LVAD samples to the disease class.
Figure 6. Altered RNA Splicing Distinctly Clusters Control and Disease Samples.
MYH7-2, TNNT2, and FLNC splicing ratios measured in 60 samples were visualized by principal component analysis. The control samples were tightly clustered together and clearly distinguished from the disease samples. Arrow indicates sample prone to misclassification as control.
Discussion
We used the Affymetrix exon array to interrogate alternative splicing in human heart failure. Our data document for the first time global differences in RNA splicing that occur in diseased compared to control hearts. ICM and control samples clustered separately by their mRNA splicing profiles. We identified nine novel alternative splicing events, including six novel alternative splicing events in well-studied sarcomere genes (1 in TNNI3, TNNT2, MYH6, and MYBPC3, and 2 in MYH7). Moreover, we showed that splicing of four sarcomere genes, FLNC, TNNI3, TNNT2, and MYH7 was different in diseased compared to control hearts. These changes in splicing likely occur prior to the onset of overt heart failure, as they were also altered in AS patients with LV hypertrophy but preserved ventricular function. In addition, LVAD placement in failing hearts did not correct aberrant splicing of these genes, suggesting that molecular changes that lead to abnormal splicing of these genes are not readily reversible by mechanical unloading.
Differences in minor to major isoform ratios of specific genes were highly characteristic of heart disease, such that the ratio of three splice variants of MYH7, TNNT2, and FLNC assigned samples into normal and disease classes with greater than 98% accuracy. The classification accuracy of splicing profiles suggests that splicing ratios may be developed as useful diagnostic and prognostic biomarkers in heart failure. mRNA expression profiling of myocardial samples has been investigated as a diagnostic and prognostic tool in heart disease, and initial studies indicate that transcriptional profiles predict prognosis and disease severity in ICM23, 24. Our results suggest that measurement of RNA splicing may be a useful adjunct to gene level measurements of transcript abundance. Further proof of concept will involve identifying splicing events that accurately label samples with disease class, which has been used as further proof of concept of prognostic utility of gene expression profiles25 Although the three splicing events at MYH7, TNNT2, and FLNC were not able to discriminator among different subtypes of heart disease, this is not unexpected because they were discovered in a screen of splicing events differentially regulated in control versus ICM. Screening for splicing events differentially regulated between disease classes may identify splicing events that distinguish between disease classes.
A surprising finding of the exon array, validated by RTPCR, was the high frequency of intron retention events among splicing events differentially regulated between ICM and control hearts. Consequently, we surveyed 20 unselected introns of sarcomere genes and found significantly reduced excision of multiple introns in ICM. This suggests that ICM is accompanied by widespread reduction of splicing efficiency. This reduction in intron splicing affected some introns to a greater degree than others. While RNA splicing efficiency was lower in heart failure, the precision of splicing was not significantly affected, as demonstrated by our survey of all coding exon-exon junctions of eight sarcomere genes for variation in splice donor or acceptor sites.
We validated differential splicing of four sarcomere genes, MYH7, TNNI3, TNNT2, and FLNC, in diseased versus normal myocardium. FLNC is a large, rod-like actin binding protein expressed in striated muscles and localized to Z-disks and intercalated discs18, 19. Mutation of FLNC caused dilated cardiomyopathy. FLNC contains a cassette exon encoding a flexible hinge region18. In normal muscle, this cassette exon is largely spliced out to yield the FLNC (−H1) variant26. We show that the FLNC variant that includes this flexible hinge region, FLNC (+H1), increases in heart disease, potentially altering structure or function of Z-disks, key signaling centers in cardiomyocytes.
The sarcomere genes MYH7, TNNI3, and TNNT2 exhibited increased intron retention in diseased myocardium. The fraction of total transcripts with retained introns was low (by quantitative PCR for MYH7-2, 0.1% and 0.3% in control and ICM hearts, respectively), but the fold increase in transcripts with retained introns was substantial (up to 15-fold; Fig. 4). The functional significance of increased abundance of incompletely spliced transcripts will require further investigation. Intron retention also results in transcripts containing premature stop codons, which may reduce effective translation, cause nonsense-mediated decay, produce incomplete proteins that have dominant negative activity27, or contribute to protein aggregates seen in some forms of heart disease.
In addition to alternative splicing of TNNT2 involving retention of the intron following exon 7, we also found a novel TNNT2 isoform generated by alternative splice acceptor usage at the 5' boundary of exon 7. This confirmed prior a suggestion there may be two closely space splice acceptors at this location28. Variable 5' splice acceptor usage resulted in 5–10% of adult TNNT2 transcripts encoding a protein lacking Asp41. Expression of this variant was not different in ICM. The N-terminus of TNNT2 encoded by exons 2–7 is less well conserved than the C-terminus and undergoes developmentally regulated alternative splicing29 that influences differential sensitivity of the fetal heart to hypoxia and acidosis30. The importance of this region of TNNT is underscored by an intron 3 polymorphism that alters splicing of exon 4 and predisposes to LV hypertrophy31.
Several limitations of this study should be noted. First, while the exon array revealed interesting aspects of RNA splicing in heart disease, the it has limited resolution and sensitivity. Perhaps as a result of this limitations, previously reported alternative splicing of titin8 and TNNT2 exons 4–616, 32 were not detected by our exon array analysis. Future studies might take advantage of the recently reported application of massively parallel sequencing strategies to sequence the entire mRNA transcriptome (mRNA-seq)1, 17, which permits quantitative delineation of alternative splicing at nucleotide resolution. Second, our global analysis compared ICM to control myocardium, and therefore it is not surprising that the splicing events arising from this screen were not effective in differentiating between disease states. Additional studies of mRNA splicing in different heart disease classes will be required to provide further proof of principle that myocardial splicing-related biomarkers contain this type of clinically important information.
In summary, we showed that RNA splicing is altered in human heart failure. We identified novel splicing events and heart failure associated differences in RNA splicing of several key sarcomere genes. Our data demonstrate reduction of RNA splicing efficiency in heart failure. These changes in RNA splicing may contribute to pathogenesis of heart disease. In addition, RNA splicing patterns accurately classified samples by diagnostic label, providing proof of concept that RNA splicing patterns may have utility as diagnostic or prognostic markers in heart disease.
Changes in gene expression accompany and contribute to heart failure. These gene expression changes have been documented at the whole gene level using microarray based, genome-wide expression profiling approaches. However, global changes in mRNA splicing in heart failure have not been previously studied. Here, we use a microarray that individually measures expression of each known or putative exon and reverse transcription PCR assays to examine splicing changes that accompany heart failure. We found that broad changes in RNA splicing occur in the failing heart. Many of these changes were indicative of decreased efficiency of RNA splicing, and among the affected genes were several sarcomere genes. These splicing changes were also observed in hypertrophied myocardium with preserved systolic function, suggesting that the splicing changes may occur prior to the onset of overt heart failure. The splicing changes were characteristic of diseased myocardium. A prediction model using the splicing of three sarcomere genes discriminated failing from non-failing hearts with 98% accuracy, providing proof of principle that splicing-based biomarkers may provide diagnostic and prognostic information on patients with heart disease. Future studies will be needed to prospectively evaluate the value of splicing-based myocardial biomarkers in individualizing therapy in heart failure. Future work will also be needed to determine whether changes in RNA splicing are functionally important in the pathogenesis of heart disease, and whether targeted modulation of splicing is a useful therapeutic strategy for heart failure.
Supplementary Material
Acknowledgments
Funding sources: This work was funded a charitable donation from Edward Marram and Karen Carpenter (WTP) and grants from NIH (WTP, R21HL89417), the Charles H. Hood Foundation (SWK), and the University of Sydney (JWKH). The Molecular Genetics Core Facility at Children's Hospital performed microarray experiments (NIH-P50-NS40828 and NIH-P30-HD18655).
Footnotes
Conflict of Interest Disclosures: Jennifer Hall is a consultant for Catholic Health Care West.
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blencowe BJ. Alternative splicing: new insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 3.Stamm S. Signals and their transduction pathways regulating alternative splicing: a new dimension of the human genome. Hum Mol Genet. 2002;11:2409–2416. doi: 10.1093/hmg/11.20.2409. [DOI] [PubMed] [Google Scholar]
- 4.Li C, Kato M, Shiue L, Shively JE, Ares M, Jr., Lin RJ. Cell type and culture condition-dependent alternative splicing in human breast cancer cells revealed by splicing-sensitive microarrays. Cancer Res. 2006;66:1990–1999. doi: 10.1158/0008-5472.CAN-05-2593. [DOI] [PubMed] [Google Scholar]
- 5.Zhang C, Li HR, Fan JB, Wang-Rodriguez J, Downs T, Fu XD, Zhang MQ. Profiling alternatively spliced mRNA isoforms for prostate cancer classification. BMC Bioinformatics. 2006;7:202. doi: 10.1186/1471-2105-7-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Venables JP, Klinck R, Bramard A, Inkel L, Dufresne-Martin G, Koh C, Gervais-Bird J, Lapointe E, Froehlich U, Durand M, Gendron D, Brosseau JP, Thibault P, Lucier JF, Tremblay K, Prinos P, Wellinger RJ, Chabot B, Rancourt C, Elela SA. Identification of alternative splicing markers for breast cancer. Cancer Res. 2008;68:9525–9531. doi: 10.1158/0008-5472.CAN-08-1769. [DOI] [PubMed] [Google Scholar]
- 7.Davis FJ, Gupta M, Pogwizd SM, Bacha E, Jeevanandam V, Gupta MP. Increased expression of alternatively spliced dominant-negative isoform of SRF in human failing hearts. Am J Physiol Heart Circ Physiol. 2002;282:H1521–33. doi: 10.1152/ajpheart.00844.2001. [DOI] [PubMed] [Google Scholar]
- 8.Neagoe C, Kulke M, del Monte F, Gwathmey JK, de Tombe PP, Hajjar RJ, Linke WA. Titin isoform switch in ischemic human heart disease. Circulation. 2002;106:1333–1341. doi: 10.1161/01.cir.0000029803.93022.93. [DOI] [PubMed] [Google Scholar]
- 9.Koscielny G, Le Texier V, Gopalakrishnan C, Kumanduri V, Riethoven JJ, Nardone F, Stanley E, Fallsehr C, Hofmann O, Kull M, Harrington E, Boue S, Eyras E, Plass M, Lopez F, Ritchie W, Moucadel V, Ara T, Pospisil H, Herrmann A, G Reich J, Guigo R, Bork P, Doeberitz MK, Vilo J, Hide W, Apweiler R, Thanaraj TA, Gautheret D. ASTD: The Alternative Splicing and Transcript Diversity database. Genomics. 2009;93:213–220. doi: 10.1016/j.ygeno.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 10.Hwang JJ, Allen PD, Tseng GC, Lam CW, Fananapazir L, Dzau VJ, Liew CC. Microarray gene expression profiles in dilated and hypertrophic cardiomyopathic end-stage heart failure. Physiol Genomics. 2002;10:31–44. doi: 10.1152/physiolgenomics.00122.2001. [DOI] [PubMed] [Google Scholar]
- 11.Kittleson MM, Minhas KM, Irizarry RA, Ye SQ, Edness G, Breton E, Conte JV, Tomaselli G, Garcia JG, Hare JM. Gene expression analysis of ischemic and nonischemic cardiomyopathy: shared and distinct genes in the development of heart failure. Physiol Genomics. 2005;21:299–307. doi: 10.1152/physiolgenomics.00255.2004. [DOI] [PubMed] [Google Scholar]
- 12.Hannenhalli S, Putt ME, Gilmore JM, Wang J, Parmacek MS, Epstein JA, Morrisey EE, Margulies KB, Cappola TP. Transcriptional genomics associates FOX transcription factors with human heart failure. Circulation. 2006;114:1269–1276. doi: 10.1161/CIRCULATIONAHA.106.632430. [DOI] [PubMed] [Google Scholar]
- 13.Purdom E, Simpson KM, Robinson MD, Conboy JG, Lapuk AV, Speed TP. FIRMA: a method for detection of alternative splicing from exon array data. Bioinformatics. 2008;24:1707–1714. doi: 10.1093/bioinformatics/btn284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xing Y, Stoilov P, Kapur K, Han A, Jiang H, Shen S, Black DL, Wong WH. MADS: a new and improved method for analysis of differential alternative splicing by exon-tiling microarrays. RNA. 2008;14:1470–1479. doi: 10.1261/rna.1070208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anderson PA, Malouf NN, Oakeley AE, Pagani ED, Allen PD. Troponin T isoform expression in humans. A comparison among normal and failing adult heart, fetal heart, and adult and fetal skeletal muscle. Circ Res. 1991;69:1226–1233. doi: 10.1161/01.res.69.5.1226. [DOI] [PubMed] [Google Scholar]
- 16.Mesnard-Rouiller L, Mercadier JJ, Butler-Browne G, Heimburger M, Logeart D, Allen PD, Samson F. Troponin T mRNA and protein isoforms in the human left ventricle: pattern of expression in failing and control hearts. J Mol Cell Cardiol. 1997;29:3043–3055. doi: 10.1006/jmcc.1997.0519. [DOI] [PubMed] [Google Scholar]
- 17.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van der Flier A, Sonnenberg A. Structural and functional aspects of filamins. Biochim Biophys Acta. 2001;1538:99–117. doi: 10.1016/s0167-4889(01)00072-6. [DOI] [PubMed] [Google Scholar]
- 19.Estigoy CB, Pontén F, Odeberg J, Herbert B, Guilhaus M, Charleston M, Ho JWK, Cameron D, dos Remedios CG. Intercalated discs: multiple proteins perform multiple functions in non-failing and failing human hearts. Biophysical Reviews. :43–49. doi: 10.1007/s12551-008-0007-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ikeda S, Kong SW, Lu J, Bisping E, Zhang H, Allen PD, Golub TR, Pieske B, Pu WT. Altered microRNA expression in human heart disease. Physiol Genomics. 2007;31:367–373. doi: 10.1152/physiolgenomics.00144.2007. [DOI] [PubMed] [Google Scholar]
- 21.Hall JL, Grindle S, Han X, Fermin D, Park S, Chen Y, Bache RJ, Mariash A, Guan Z, Ormaza S, Thompson J, Graziano J, de Sam Lazaro SE, Pan S, Simari RD, Miller LW. Genomic profiling of the human heart before and after mechanical support with a ventricular assist device reveals alterations in vascular signaling networks. Physiol Genomics. 2004;17:283–291. doi: 10.1152/physiolgenomics.00004.2004. [DOI] [PubMed] [Google Scholar]
- 22.Margulies KB, Matiwala S, Cornejo C, Olsen H, Craven WA, Bednarik D. Mixed messages: transcription patterns in failing and recovering human myocardium. Circ Res. 2005;96:592–599. doi: 10.1161/01.RES.0000159390.03503.c3. [DOI] [PubMed] [Google Scholar]
- 23.Heidecker B, Kasper EK, Wittstein IS, Champion HC, Breton E, Russell SD, Kittleson MM, Baughman KL, Hare JM. Transcriptomic biomarkers for individual risk assessment in new-onset heart failure. Circulation. 2008;118:238–246. doi: 10.1161/CIRCULATIONAHA.107.756544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Margulies KB, Bednarik DP, Dries DL. Genomics, transcriptional profiling, and heart failure. J Am Coll Cardiol. 2009;53:1752–1759. doi: 10.1016/j.jacc.2008.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kittleson MM, Ye SQ, Irizarry RA, Minhas KM, Edness G, Conte JV, Parmigiani G, Miller LW, Chen Y, Hall JL, Garcia JG, Hare JM. Identification of a gene expression profile that differentiates between ischemic and nonischemic cardiomyopathy. Circulation. 2004;110:3444–3451. doi: 10.1161/01.CIR.0000148178.19465.11. [DOI] [PubMed] [Google Scholar]
- 26.Xie Z, Xu W, Davie EW, Chung DW. Molecular cloning of human ABPL, an actin-binding protein homologue. Biochem Biophys Res Commun. 1998;251:914–919. doi: 10.1006/bbrc.1998.9506. [DOI] [PubMed] [Google Scholar]
- 27.Watkins H, Seidman CE, Seidman JG, Feng HS, Sweeney HL. Expression and functional assessment of a truncated cardiac troponin T that causes hypertrophic cardiomyopathy. Evidence for a dominant negative action. J Clin Invest. 1996;98:2456–2461. doi: 10.1172/JCI119063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Farza H, Townsend PJ, Carrier L, Barton PJ, Mesnard L, Bahrend E, Forissier JF, Fiszman M, Yacoub MH, Schwartz K. Genomic organisation, alternative splicing and polymorphisms of the human cardiac troponin T gene. J Mol Cell Cardiol. 1998;30:1247–1253. doi: 10.1006/jmcc.1998.0698. [DOI] [PubMed] [Google Scholar]
- 29.Townsend PJ, Farza H, MacGeoch C, Spurr NK, Wade R, Gahlmann R, Yacoub MH, Barton PJ. Human cardiac troponin T: identification of fetal isoforms and assignment of the TNNT2 locus to chromosome 1q. Genomics. 1994;21:311–316. doi: 10.1006/geno.1994.1271. [DOI] [PubMed] [Google Scholar]
- 30.Solaro RJ, Kumar P, Blanchard EM, Martin AF. Differential effects of pH on calcium activation of myofilaments of adult and perinatal dog hearts. Evidence for developmental differences in thin filament regulation. Circ Res. 1986;58:721–729. doi: 10.1161/01.res.58.5.721. [DOI] [PubMed] [Google Scholar]
- 31.Komamura K, Iwai N, Kokame K, Yasumura Y, Kim J, Yamagishi M, Morisaki T, Kimura A, Tomoike H, Kitakaze M, Miyatake K. The role of a common TNNT2 polymorphism in cardiac hypertrophy. J Hum Genet. 2004;49:129–133. doi: 10.1007/s10038-003-0121-4. [DOI] [PubMed] [Google Scholar]
- 32.Anderson PA, Greig A, Mark TM, Malouf NN, Oakeley AE, Ungerleider RM, Allen PD, Kay BK. Molecular basis of human cardiac troponin T isoforms expressed in the developing, adult, and failing heart. Circ Res. 1995;76:681–686. doi: 10.1161/01.res.76.4.681. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.