Abstract
The filamentous fungal pathogen Aspergillus fumigatus secretes hydrolytic enzymes to acquire nutrients from host tissues. The production of these enzymes exerts stress on the endoplasmic reticulum (ER), which is alleviated by two stress responses: the unfolded protein response (UPR), which adjusts the protein folding capacity of the ER, and ER-associated degradation (ERAD), which disposes of proteins that fail to fold correctly. In this study, we examined the contribution of these integrated pathways to the growth and virulence of A. fumigatus, focusing on the ERAD protein DerA and the master regulator of the UPR, HAcA. A ΔderA mutant grew normally and showed no increase in sensitivity to ER stress. However, expression of the UPR target gene bipA was constitutively elevated in this strain, suggesting that the UPR was compensating for the absence of DerA function. To test this, the UPR was disrupted by deleting the hacA gene. The combined loss of derA and hacA caused a more severe reduction in hyphal growth, antifungal drug resistance and protease secretion than the loss of either gene alone, suggesting that DerA and HacA cooperate to support these functions. Moreover, the ΔderA/ΔhacA mutant was avirulent in a mouse model of invasive aspergillosis, which contrasted the wild-type virulence of ΔderA and the reduced virulence of the ΔhacA mutant. Taken together, these data demonstrate that DerA cooperates with the UPR to support the expression of virulence-related attributes of A. fumigatus.
Key words: Aspergillus fumigatus, UPR, ERAD, virulence, hacA, derA
Introduction
Invasive aspergillosis is a life-threatening opportunistic infection caused by molds in the genus Aspergillus, most commonly Aspergillus fumigatus.1 The infection is acquired by the inhalation of aerosolized conidia that, if not effectively cleared by the immune system, germinate into hyphae that can rapidly spread to other tissues. Although the conidia of filamentous fungi contain sufficient ribosomes to initiate germination in the lung, they lack the protein synthetic capacity to sustain hyphal growth. Thus, the early stages of germination involve a burst of ribosome production to meet the unique demands of polarized hyphal growth.2–4 In addition, an expansion of the endoplasmic reticulum (ER) must also occur at this time to accommodate the ribosomes that are responsible for translocating proteins into the ER that are needed for filamentous growth, such as membrane proteins, enzyme complexes required for cell wall synthesis and secreted hydrolytic enzymes.5,6
The ER is the entry point for the secretory pathway and provides the necessary intracellular environment to fold proteins into a stable tertiary structure before delivery to the distal components of the secretory system. When the protein folding capacity of the ER becomes overwhelmed, either in response to adverse environmental conditions or a sudden increase in secretory capacity, misfolded protein can accumulate in the ER.7 Abnormally folded proteins illegitimately associate with other proteins in the ER lumen, creating toxic aggregates that disrupt ER homeostasis.8,9 Two of the major pathways responsible for mitigating the ensuing stress are the unfolded protein response (UPR)7 and ER-associated degradation (ERAD).10 The UPR restores ER homeostasis by increasing the expression of proteins involved in protein folding and secretion. The pathway is triggered by the ER-membrane protein Ire1, which constitutes the major sensor of the fungal UPR.7,11,12 The presence of misfolded proteins triggers the activation of Ire1, which in turn activates a transcription factor known as HacA in filamentous fungi.13–16 The HacA protein translocates to the nucleus, where it increases the transcription of genes that encode proteins with functions in protein folding and secretion, such as the ER-resident chaperone Bip.13,17,18 Polypeptides that ultimately fail to acquire the appropriate conformation are disposed of by ERAD, a multi-step process that begins with the selective recognition of misfolded proteins in the ER.19 The aberrant proteins are transferred across the ER membrane into the cytosol, where they are ubiquitinated on the cytosolic face of the ER and finally released for degradation by the proteasome.20 A central component of this pathway is a multi-protein complex in the ER membrane that is involved in the recognition, transport and ubiquitination functions of ERAD.21–23
Despite advances in our understanding of the molecular components of UPR and ERAD signaling, the involvement of these pathways in fungal virulence remains largely unexplored. In this study, we examined the contribution of ERAD to A. fumigatus virulence by deleting the derA gene, encoding the A. fumigatus homolog of Saccharomyces cerevisiae Der1p (Degraded in the ER), a transmembrane protein that is part of the multi-protein ER membrane complex required for ERAD.23 Although DerA was dispensable for most aspects of A. fumigatus physiology, it cooperated with the UPR to support growth on a complex substrate, cell wall homeostasis, antifungal drug resistance and virulence. These findings suggest that components of the UPR and ERAD pathways could represent promising new targets for novel antifungal drug development because of the central role that they play in sustaining A. fumigatus virulence.
Results
Disruption of the ERAD gene derA.
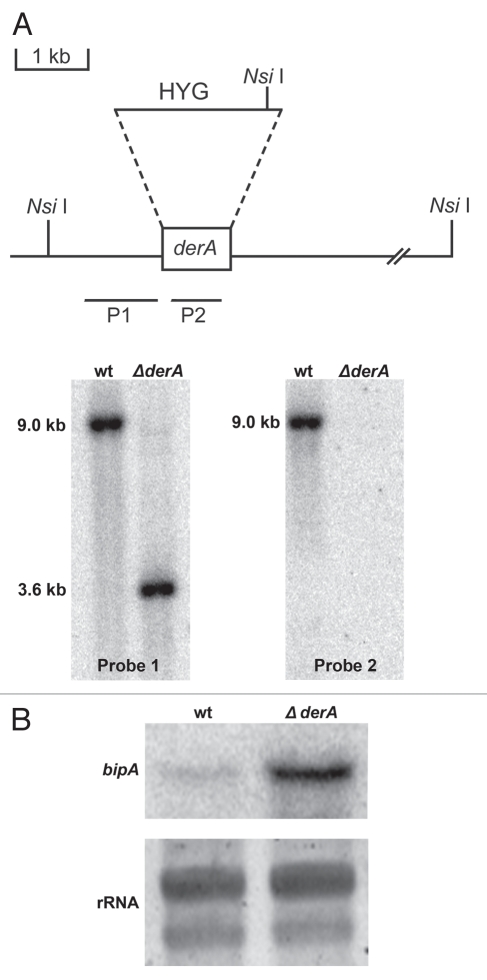
Der1p is a small ER-resident membrane protein that was initially identified in a screen for proteins necessary for ER-associated degradation in S. cerevisiae.23 A BLAST search of the A. fumigatus protein database using yeast Der1p as the query identified a 249 amino acid protein, annotated as DerA (Genbank accession XP_747611), with an E value of 1 × 10−11. The two proteins share 24% identity and 49% similarity and have the same predicted four transmembrane helices followed by a short cytosolic C-terminal domain.24 The presence of a single predicted intron in the A. fumigatus derA gene was confirmed by PCR amplification of the full-length cDNA (data not shown). The derA gene was deleted from A. fumigatus by replacing it with a hygromycin resistance cassette, resulting in truncation of a 9 kb NsiI fragment containing the derA gene (probe 1, Fig. 1A). A second probe located within the open reading frame confirmed the absence of the derA gene in the ΔderA mutant (probe 2, Fig. 1A).
Figure 1.
Deletion of A. fumigatus derA. (A) Deletion strategy for derA. The entire derA open reading frame was replaced with the hygromycin resistance cassette (HYG) as shown. Southern blot analysis of NsiI-digested genomic DNA using a probe located in the 5′ flanking region (P1) revealed the predicted truncation of a 9 kb wt NsiI fragment by the replacement of derA with HYG. Confirmation of derA deletion was obtained using a probe located within the derA open reading frame (P2). (B) Northern blot analysis of bipA expression in the absence of ER stress. Equal numbers of conidia from the indicated strains were inoculated into liquid YG medium (5% yeast extract/20% glucose) and shaken at 200 rpm for 16 h at 37°c. The steady-state levels of bipA mRNa in the two strains were then compared by northern blot analysis as described in Materials and Methods. Equal loading was confirmed by SYBR-green staining of the ribosomal RNA (rRNA) bands.
A. fumigatus derA is dispensable for growth.
The growth rate of the ΔderA mutant was indistinguishable from that of wild-type (wt) A. fumigatus on multiple types of medium (Fig. 2 and data not shown). Moreover, the ΔderA mutant showed normal sensitivity to conditions that stress the folding capacity of the ER, such as high temperature (50°C), treatment with the reducing agent dithiothreitol, disruption of ER-Golgi transport with brefeldin A or damaging the cell wall with calcofluor white (CFW) and Congo red (CR) (data not shown). Thus, derA is dispensable for the growth of A. fumigatus, even under situations that cause acute ER stress.
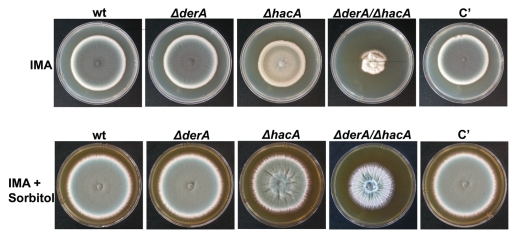
Figure 2.
DerA synergizes with the UPR to support radial growth. Colony morphology of ΔderA, ΔhacA, ΔderA/ΔhacA, and the complemented ΔderA/ΔhacA strain (C′). Equal numbers of conidia were spotted onto the center of a plate of rich medium (IMA) in the presence or absence of 1.2 M sorbitol and incubated for four days at 37°C.
The absence of a phenotype in the ΔderA mutant suggested that A. fumigatus can compensate for the absence of DerA function, possibly by increasing the activity of the UPR. To determine whether the UPR was induced by the loss of derA, we cultured the organism in the absence of ER stress and evaluated the steady-state levels of bipA mRNA by northern blot analysis. The bipA gene encodes an Hsp70 family chaperone that is a transcriptional target of the UPR in multiple species,15,17,25 including A. fumigatus,13 and thus provides a useful marker for UPR activity. The ΔderA mutant had higher levels of bipA mRNA relative to wt A. fumigatus grown under the same conditions (Fig. 1B). This suggests that loss of derA is sufficient to increase the level of ER stress in the fungus, which requires compensatory upregulation of the UPR.
DerA synergizes with the UPR to support radial growth and conidiation.
To determine whether the increased UPR activity in ΔderA was responsible for the normal growth of this mutant, the hacA gene was deleted in the ΔderA background. The absence of either derA or hacA alone had little-to-no effect on the radial growth rate of A. fumigatus on rich medium, but the growth of the ΔhacA/ΔderA mutant was severely reduced (Fig. 2). This indicates that A. fumigatus can tolerate the loss of either derA or hacA, but not both genes together. The impaired growth of the ΔhacA/ΔderA mutant could be partially rescued on medium containing sorbitol, an osmoprotectant that prevents swelling and lysis of strains that lack cell wall integrity.26 However, this protection was incomplete, suggesting that defects in processes other than cell wall homeostasis also contribute to the abnormal growth of this mutant (Fig. 2).
The ΔderA mutant conidiated normally, producing blue-green colonies with abundant conidia (Fig. 2). By contrast, conidiation was reduced in the ΔhacA mutant and was almost completely absent in ΔhacA/ΔderA colonies (evident by the absence of colony color, Fig. 2). The reduced conidiation of the ΔhacA mutant could be rescued by sorbitol, suggesting that the cell wall abnormalities previously reported in this strain interfere with asexual development.13 However, sorbitol was unable to fully restore the conidiation of the ΔhacA/ΔderA mutant, suggesting that cell wall defects are only partially responsible for the abnormal conidiation of this strain (Fig. 2).
Glucose and N-acetylglucosamine (GlcNAc) constitute the major cell wall monosaccharides in the A. fumigatus cell wall, forming interconnected polymers of glucan and chitin, respectively.27 The levels of these monosaccharides were unaffected by the ΔderA mutation (Table 1), which was consistent with the normal sensitivity of the ΔderA mutant to the cell wall damaging agents CFW and CR (data not shown). By contrast, the ΔhacA/ΔderA cell wall had reduced glucose levels and increased chitin, as well as slight differences in less abundant cell wall monosaccharides (Table 1). These differences could not be entirely attributed to the previously described effects of the hacA deletion alone,13 suggesting that derA deletion may partially influence cell wall composition when combined with loss of hacA.
Table 1.
Monosaccharide composition of wt, ΔderA, and ΔderA/ΔhacA cell walls
| Alkali insoluble | Alkali soluble | |||||
| wt | ΔderA | ΔderA/ΔhacA | wt | ΔderA | ΔderA/ΔhacA | |
| Mannose | 6.6 ± 04 | 9.6 ± 0.9* | 8.8 ± 0.6* | 1.3 ± 0.6 | 2.0 ± 0.4 | 2.3 ± 0.9 |
| Glucose | 45.9 ± 4.1 | 44.0 ± 1.7 | 34.8 ± 2.2* | 67.6 ± 3.9 | 76.6 ± 6.7 | 72.6 ± 5.4 |
| Galactose | 6.4 ± 1.0 | 8.7 ± 1.4 | 11.5 ± 2.1* | 2.1 ± 1.0 | 8.3 ± 3.9* | 8.8 ± 3.1* |
| GlcNAc | 24.6 ± 2.7 | 27.4 ± 5.4 | 32.1 ± 3.3* | 0.1 ± 0.2 | 0.3 ± 0.2 | 0.1 ± 0.1 |
| GalNAc | 0.6 ± 0.9 | 2.7 ± 3.1 | 0.0 ± 0.0 | 1.5 ± 1.0 | 4.2 ± 1.6* | 1.2 ± 1.0 |
Results are expressed as µg of individual sugars present in 100 µg of the cell wall alkali-insoluble and alkali-soluble fractions. Values represent the average of four replicates ± standard deviation.
denotes a significant difference relative to wt (p < 0 .05). The AI/AS ratio was 1.63 ± 0.3 for wt, 1.44 ± 0.2 for ΔderA and 1.94 ± 0.1 for ΔderA/ΔhacA.
DerA cooperates with the UPR to resist antifungal stress.
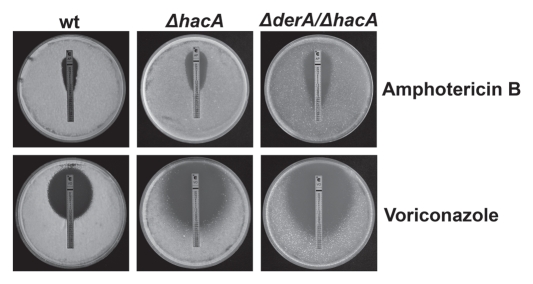
The susceptibility of the ΔderA mutant to caspofungin, amphotericin B and voriconazole was indistinguishable from that of wt (data not shown), indicating that derA is dispensable for resistance to the three major classes of antifungal drugs used for the treatment of aspergillosis. We have previously demonstrated that the ΔhacA mutant is hypersensitive to these.13 However, the ΔhacA/ΔderA mutant had increased sensitivity to both amphotericin B and voriconazole relative to the ΔhacA mutant (Fig. 3), suggesting that DerA contributes to, but is not required for, resistance to these two drugs. Interestingly, the caspofungin sensitivity of the ΔhacA/ΔderA mutant was no different from that of ΔhacA (data not shown), indicating that the combined loss of hacA and derA does not lead to a generalized increase in drug susceptibility.
Figure 3.
DerA cooperates with the UPR in antifungal resistance. Conidia from the indicated strains were spread evenly onto the surface of a plate of IMA and an Etest strip containing amphotericin B or voriconazole was applied to the inoculated agar surface. The plates were incubated at 37°C for 24 h.
DerA cooperates with the UPR to support protease secretion and growth on a complex substrate.
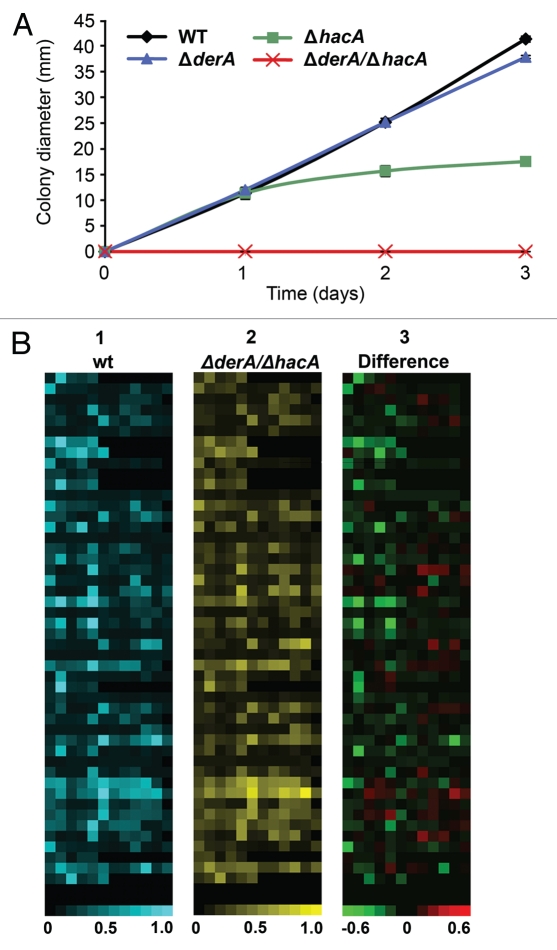
A. fumigatus relies on the secretion of extracellular hydrolytic enzymes to break down complex substrates into smaller molecules that are easier to absorb. To determine how loss of derA would affect growth on a substrate that requires degradation by secreted enzymes, the mutants were plated onto solid medium containing skim milk, a nutrient source that triggers the secretion of proteases by Aspergillus spp.28,29 The ΔderA mutant grew as well as wt on this medium (Fig. 4A). Although the growth of the ΔhacA mutant was indistinguishable from wt during the first 24 h of incubation on skim milk agar, subsequent radial expansion was severely impaired. By contrast, the ΔderA/ΔhacA mutant was unable to form colonies (Fig. 4A). Microscopic analysis revealed that the ΔderA/ΔhacA conidia were able to germinate and undergo limited hyphal growth on this medium. However, there was insufficient hyphal growth to generate a macroscopically visible colony, even after 6 d of incubation, indicating that skim milk is a poor substrate for this mutant. The ΔderA/ΔhacA was also unable to grow on explants of mouse lung tissue after 4 d of incubation (Fig. 3, Sup. Data). Since the ΔderA/ΔhacA mutant was able to form colonies after 4 d on medium containing pre-digested proteins (Fig. 2), the inability to utilize undigested milk proteins or mouse lung tissue as a substrate suggests that derA and hacA are required under conditions where secretion is needed for nutrient breakdown.
Figure 4.
DerA cooperates with the UPR to support protease secretion and growth on a complex substrate. (A) Growth on skim milk agar. Conidia from the indicated strains were inoculated onto the center of a plate of skim milk agar and colony diameter was monitored for 3 d at 37°C. (B) Analysis of secreted protease activity using a library of fluorescence resonance energy transfer (FRET) labeled peptide substrates. Equimolar mixtures of up to 8 individual FRET peptides in each well of a microtiter plate were incubated with culture supernatants from the indicated strains and heat maps were generated from the fluorescent signals generated by substrate cleavage, with each square corresponding to a single assay well. For part 1 (wt; blue) and part 2 (ΔderA/ΔhacA; yellow), the brightness of each square corresponds to the fluorescence intensity, quantified as fluorescence fold change, which indicates the extent of cleavage of the FRET peptides in that well. For part 3, the brightness of a given well indicates the extent to which specific protease activities are disproportionally decreased (green) or increased (red) in the ΔderA/ΔhacA mutant as compared to wt.
The A. fumigatus genome encodes numerous secreted proteases.30 To determine how loss of derA and hacA would impact the secretion of these enzymes, we used activity-based protease substrate profiling to compare the in vitro secreted proteolytic signatures of ΔderA/ΔhacA and wt A. fumigatus. Substrate profiling was accomplished using a library of fluorescence resonance energy transfer (FRET) labeled peptide substrates.31 The FRET peptide library is comprised of a microtiter plate, with each well containing up to eight individual peptides. When these substrates are cleaved, a fluorophore is liberated from a quenching moiety, resulting in a fluorescence signal that is proportional to the extent of cleavage. A. fumigatus culture supernatants were used to screen the FRET library and heat maps were generated from the resulting data (Fig. 4B). We found that secreted proteolytic activity was reduced in the ΔderA/ΔhacA mutant, requiring a 20-fold reduction in supernatant dilution to achieve comparable levels of activity seen in the wt supernatants. The overall proteolytic signature was similar, but not identical, between wt and the ΔderA/ΔhacA mutant (Fig. 4B, parts 1 and 2). Part 3 shows the relative change in the substrate specificity profile, expressed as the difference of normalized values in Parts A and B (ΔderA/ΔhacA-wt). Some of the substrates showed increased cleavage in the ΔderA/ΔhacA mutant (part 3, red), whereas others showed decreased cleavage (part 3, green), suggesting that the loss of both derA and hacA caused a dysregulation of protease secretion rather than a uniform decrease.
DerA cooperates with the UPR to support virulence.
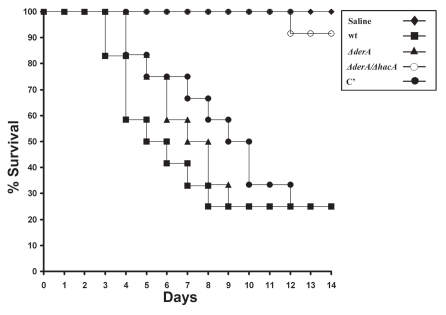
The ΔhacA mutant was shown in a previous study to have attenuated virulence, although it still retained the ability to cause 30–60% mortality, depending on the infection model that was used.13 To determine whether derA has a role during infection, we compared the virulence of ΔderA to that of ΔderA/ΔhacA in a mouse infection model. The mortality curve of the ΔderA mutant was indistinguishable from wt A. fumigatus (Fig. 5). By contrast, the virulence of the ΔderA/ΔhacA mutant was not statistically different from the mock-infected controls (Fig. 5), indicating a requirement for both derA and hacA in A. fumigatus virulence. Moreover, no viable fungus could be recovered from any of the ΔderA/ΔhacA-inoculated mice after two weeks, indicating that the mice were able to clear this infection.
Figure 5.
DerA cooperates with the UPR to support virulence. Groups of 12 mice were immunosuppressed with a single dose of triamcinolone acetonide (40 mgkg−1 of body weight) injected subcutaneously on day-1 and infected intranasally with conidia from the indicated strains on day 0. Percent survival was monitored for 14 days. The Statistical significance was assessed by Kruskal-Wallis ANOVA. ΔderA/ΔhacA is attenuated in virulence relative to wt (p < 0.001), whereas ΔderA is indistinguishable from wt.
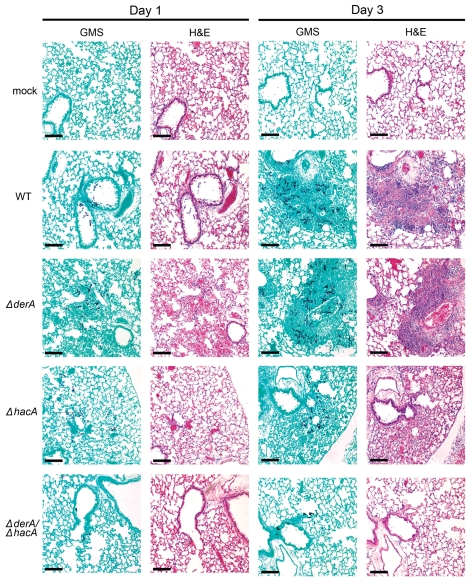
Histopathologic analysis of infected lungs was consistent with these mortality data. The ΔderA-infected lung tissue was indistinguishable from wt-infected lung tissue; multiple germlings were visible in the bronchioles and alveolar spaces by day 1, and by day 3 there was increased hyphal growth with extensive tissue inflammation and necrosis (Fig. 6). The extent of fungal growth and tissue damage was relatively lower in the ΔhacA-infected lungs, consistent with the known attenuated virulence of this strain.13 By contrast, the ΔderA/ΔhacA-infected lungs revealed very limited fungal growth and pulmonary inflammation, even by day 3. Some swollen conidia were evident in the ΔderA/ΔhacA-infected lungs, a few of which had started to form small germlings (Fig. 2, Sup. Data). However, no hyphae were visible on day 3, and minimal inflammation was evident in the lung parenchyma. These observations demonstrate that the ΔderA/ΔhacA conidia can initiate growth in the lung, but are unable to sustain the infection in the presence of a progressively reconstituting immune system. This correlates with the results in Figure 4A, demonstrating that the ΔderA/ΔhacA mutant can initiate, but not sustain, growth on a complex substrate in vitro, suggesting that impaired growth in the host may also involve a reduced capacity for nutrient acquisition.
Figure 6.
Histopathology of infected lung tissue. Mice were infected as described in Figure 5 and sacrificed on day 1 and 3 post-infection. The lungs were sectioned at 5 µm and stained with hematoxylin and eosin (H&E) or Grocott methenamine silver (GMS). Microscopic examinations were performed on a Zeiss Axioscope 2-plus microscope using Zeiss Axiovision version 4.4 software. Scale bar represents 100 µm.
Discussion
The attenuated virulence caused by disruption of the UPR in A. fumigatus suggests that this fungus needs the UPR to support ER homeostasis in the host environment.13 The purpose of this study was to determine the extent to which A. fumigatus also relies upon ERAD, a related pathway that works in conjunction with the UPR to maintain protein quality control in the ER.10 The loss of the ERAD protein DerA had no detectable effect on the growth of the fungus, even under conditions of severe ER stress, similar to what has been described for the corresponding mutant in S. cerevisiae.23 The ability of the ΔderA mutant to grow normally in the presence of ER stress contrasts the ΔhacA mutant, which is unable to survive when ER stress levels are high.13 This likely reflects the dominant role that HacA plays as the master regulator of the UPR,32 as opposed to the more specialized role of DerA as part of a multi-protein ER membrane complex.22 No homologs of DerA could be clearly identified in the A. fumigatus genome, although it is difficult to rule out the existence of divergent proteins that could provide functional redundancy. The combined loss of DerA and HacA caused a more severe reduction in growth capacity than the loss of either gene alone, suggesting that the functions provided by these two proteins act synergistically to support the growth of A. fumigatus hyphae. Since precursor molecules that are needed at the growing hyphal tips are transported in secretory vesicles,33 we speculate that the support functions provided by DerA and HacA to the secretory network are necessary for optimal filamentous growth.
The cell wall of A. fumigatus is divided biochemically into two fractions: an alkali-insoluble (AI) fraction containing predominantly β-(1,3) glucan and chitin, which is thought to provide the major structural rigidity to the wall, and an alkali-soluble (AS) fraction comprised of α-(1,3) glucan and galactomannan.27 The ΔderA mutant showed no difference in either glucose or GlcNAc composition, indicating that the major structural components of the cell wall are unaffected by the absence of DerA function. The ΔderA/ΔhacA cell wall contained the same reduction in AI glucose [β-(1, 3) glucan] that was previously reported in the ΔhacA mutant,13 which was consistent with the comparable sensitivity of these two strains to the β-(1, 3) glucan synthesis inhibitor caspofungin (data not shown). An increase in chitin levels was also seen in ΔderA/ΔhacA, which may reflect compensatory cell wall changes caused by the loss of both hacA and derA. Interestingly, the ΔderA/ΔhacA mutant was slightly more resistant to CFW and CR than the ΔhacA mutant (Fig. 1, Sup. Data). Since these compounds bind to nascent chitin chains and inhibit enzymes that connect chitin to glucan,34 the lower levels of chitin in the ΔhacA mutant relative to ΔderA/ΔhacA may account for the difference in CFW and CR sensitivity between the two strains.
The sensitivity of the ΔderA/ΔhacA mutant to amphotericin B and voriconazole was also greater than that of ΔhacA. The exact mechanism for this heightened susceptibility is unclear. Both of these drugs share the characteristic of disrupting membrane ergosterol, suggesting that DerA and HacA have synergistic functions that protect against attack on membrane homeostasis. It is possible that the cell wall changes in the ΔderA/ΔhacA mutant increase access to the membrane and cytosol, thereby potentiating the effects of these drugs. Alternatively, the more severe growth defect of the ΔderA/ΔhacA mutant could increase the amphotericin B and voriconazole sensitivity of this strain, although slow growth is unlikely to be the sole explanation since the caspofungin sensitivity of ΔderA/ΔhacA was no different from that of ΔhacA (data not shown). Regardless, the increased antifungal susceptibility of both ΔhacA and ΔderA/ΔhacA raises the possibility that disrupting these pathways with novel antifungal therapy could be exploited as a strategy to enhance the potency of other antifungal drugs.
The ΔderA/ΔhacA mutant was unable to form colonies on medium containing undigested milk proteins as a nutrient source. Activity-based substrate profiling of in vitro culture supernatants revealed a reduction in overall secreted protease capacity that may contribute to the inability of this strain to grow well on skim milk agar. However, some activities were actually increased in the ΔderA/ΔhacA mutant on a relative basis. This suggests that the ER stress caused by loss of derA and hacA selectively regulates the expression of a subset of proteolytic activities that are essential to the fungus, rather than simply downregulating all proteolytic activities together. These differentially regulated activities may provide new insight into how the fungus balances the need for nutrients with protection against secretion stress, and efforts to uncover their identity are in progress.
The three mutants examined in this study showed a graded virulence phenotype: the ΔderA mutant was fully virulent, the ΔhacA mutant was partially virulent13 and the ΔderA/ΔhacA mutant was avirulent (Fig. 5). These observations suggest that HacA and DerA act synergistically to support aspects of A. fumigatus physiology that have important roles in virulence, the most obvious of which would be rapid invasive growth. The inability of the ΔderA/ΔhacA mutant to efficiently deliver plasma membrane and cell wall material to the hyphal tips would severely limit the invasive growth properties of the mutant hyphae, a problem that would be compounded by the reduced ability to secrete sufficient hydrolytic enzymes for nutrient acquisition. In the transient immunosuppression model used in this study, the progressively reconstituting immune system would encounter a much lower fungal burden in animals inoculated with the ΔderA/ΔhacA mutant relative to those inoculated with the ΔderA or wt strains, which could account for the observed clearance of the ΔderA/ΔhacA mutant in surviving mice. The loss of derA and hacA could also reduce the overall viability of the fungus in the host environment, which would also be expected to reduce fungal burden. Finally, the cell wall changes associated with this mutant could expose carbohydrate epitopes on the cell wall that increase recognition and clearance by the reconstituting immune system. Regardless of the exact mechanism involved, the synergism between DerA and HacA in the maintenance of protein quality control in the ER may represent a vulnerability of A. fumigatus that could be exploited to increase the efficacy of antifungal therapy.
Materials and Methods
Strains and culture conditions.
The strains used in this study are listed in Table 3. Conidia were harvested from cultures maintained on Aspergillus minimal medium (AMM) osmotically stabilized with 1.2 M sorbitol because this medium best supported the conidiation of the mutant strains used in this study. Growth rate analysis was determined by spotting 5,000 conidia onto the center of a 100 mm plate containing 40 mL of inhibitory mold agar (IMA) or IMA supplemented with 1.2 M sorbitol (Sigma) where indicated, and monitoring colony diameter over time. IMA is a rich medium that best supported the growth of the mutant strains used in this study. Growth on skim milk was performed by inoculating 5,000 conidia onto plates containing 0.5% skim milk and incubating at 37°C for 3 d.
Table 3.
Strains used in this study
Sensitivity to ER stress was determined by monitoring growth in the presence of dithiothreitol (DTT) or the ER-Golgi transport inhibitor brefeldin A (BFA). Sensitivity to cell wall stress was determined by evaluating growth in the presence of calcofluor white (CFW) or Congo red (CR). All compounds were obtained from Sigma. DTT sensitivity was performed by inoculating 20,000 conidia into each well of a 24-well plate containing liquid AMM supplemented with the indicated concentrations of DTT. For analysis of BFA, CFW and CR sensitivity, 2,000 conidia were spotted in the center of each well of a 24-well plate containing solid IMA medium supplemented with the indicated concentrations of each compound. Plates were photographed after 2–3 d of incubation at 37°C.
Antifungal susceptibility.
Antifungal susceptibility was determined using the Etest diffusion assay (AB BIODISK) according to the manufacturer's instructions, with slight modification. Briefly, conidial suspensions were prepared in sterile distilled water and adjusted to 1 × 106 conidia/mL. One milliliter of the conidial suspension was then spread evenly onto the surface of a 100 mm plate of IMA medium using a glass spreader. The inoculated agar surface was allowed to dry for 20 min before Etest strips containing amphotericin B, caspofungin or voriconazole, were applied. The plates were incubated at 37°C for 24 h before being photographed.
Deletion and reconstitution of the A. fumigatus derA gene.
All primers used in this study are shown in Table 2. The annotated A. fumigatus derA gene (Genbank accession XP_747611) is predicted to contain a single intron and this was confirmed by PCR amplification of reverse-transcribed RNA isolated from vegetative cultures using a 5′ primer 16 nt upstream of the ATG (5-CTC GAT CTC CGT CGT AAT G) and a 3′ primer located 126 nt downstream of the predicted stop codon (5-TAA GCC GCT TCC TTC TCA G). The ΔderA gene was disrupted using the split-marker deletion strategy.35 The left arm of derA was PCR-amplified from wt genomic DNA using iProof polymerase (Bio-Rad) with primers 579 and 580 creating PCR product #1. The first two thirds of the hygromycin resistance cassette were PCR-amplified from plasmid pAN7-1 using primers 395 and 398 to create PCR product #3. PCR products #1 and #3 were then combined in an overlap PCR reaction with primers 395 and 579 to generate PCR product #5. PCR product #5 was cloned into pCR-Blunt II-TOPO (Invitrogen) to create p540. The right arm of the derA gene was PCR-amplified from genomic DNA using primers 581 and 582 to generate PCR product #2. The second two-thirds of the hygromycin resistance cassette was amplified from pAN7-1 with primers 396 and 399 to make PCR product #4, and PCR products #2 and #4 were combined in an overlap PCR reaction with primers 396 and 582 to generate PCR product #6. PCR product #6 was then cloned into pCR-Blunt II-TOPO to create p541.
Table 2.
PCR primers used in this study
| Primer | Gene | Sequence (5′-3′) | Purpose |
| 395 | hph | CTC CAT ACA AGC CAA CCA CGG | deletion of derA |
| 396 | hph | CGT TGC AAG ACC TGC CTG AA | deletion of derA |
| 398 | hph/ble | CGC CAG GGT TTT CCC AGT CAC GAC AAG TGG AAA GGC TGG TGT GC | deletion of derA |
| 399 | hph | AGC GGA TAA CAA TTT CAC ACA GGA TCG CGT GGA GCC AAG AGC GG | deletion of derA |
| 579 | derA | CTG GTA TCT GCG GAT CTC TT | deletion of derA/probe1 |
| 580 | derA | GTC GTG ACT GGG AAA ACC CTG GCG GAC AGT GAG TGA GTG ATA TG | deletion of derA/probe1 |
| 581 | derA | TCC TGT GTG AAA TTG TTA TCC GCT TCC GAT GAC TTG CTG GCA TT | deletion of derA |
| 582 | derA | TCG AAC AGC TCT TTT CTG CC | deletion of derA |
| 584 | derA | GCG ACC GTG GCT ACT TCA GT | probe 2 |
| 585 | derA | GTT CAC ATT ATC CGT GTC TG | probe 2 |
| 586 | derA | GCC ACT TGA CAT AGA GCC CT | ΔderA complementation |
| 587 | derA | GAG TAT CCC AAA CAG CTG AC | ΔderA complementation |
| 408 | ble | TGC TCG CCG ATC TCG GTC AT | deletion of derA |
| 409 | ble | AGC GGA TAA CAA TTT CAC ACA GGA TTA AAG CCT TCG AGC GTC C | deletion of derA |
| 410 | ble | GAC AAG GTC GTT GCG TCA GTC | deletion of derA |
| 494 | bipA | GTC TGA TTG GAC GCA AGT TC | creation of bipA probe |
| 495 | bipA | ATC TGG GAA GAC AGA GTA CG | creation of bipA probe |
M13-derived sequences used for overlap PCR are underlined.
The insert from p540 was excised with HindIII and XbaI and the insert from p541 was excised with EcoRI. Both inserts were gel-purified and 10 µg of each was used to transform wt-ΔakuA36 protoplasts as previously described.37 Loss of the derA gene was confirmed by Southern blot analysis of genomic DNA isolated from hygromycin resistant monoconidial isolates. Probe 1 (Fig. 1) was PCR-amplified from wt genomic DNA using primers 579 and 580, whereas Probe 2 was PCR-amplified using primers 584 and 585.
To construct the derA complementation plasmid, the derA gene including 511 bp upstream of the ATG was PCR-amplified from wt genomic DNA using primers 586 and 587 and cloned into pCR-Blunt II-TOPO (Invitrogen) to create p546. A phleomycin resistance cassette flanked by HindIII sites was then inserted into p546 to create p547. Ten micrograms of the derA complementation plasmid was linearized with NsiI and transformed into ΔderA mutant protoplasts. Ectopic reconstitution of the derA gene was demonstrated by Southern blot analysis of genomic DNA from phleomycin-resistant monoconidial isolates using the internal probe Probe 2 (data not shown).
To create the ΔderA/ΔhacA mutant, the derA gene was deleted in the ΔhacA mutant background using the same split-marker strategy described above, except that phleomycin was used as the selectable marker. The first two-thirds of the phleomycin-resistance cassette were PCR-amplified from plasmid pBC-phleo using primers 398 and 408 to generate PCR product #7. PCR products #1 (derA left arm) and #7 where then combined in an overlap PCR reaction with primers 579 and 408 to create PCR product #8. PCR product #8 was then cloned into pCR-Blunt II-TOPO to create p543. The second two-thirds of the phleomycin-resistance cassette were PCR-amplified from plasmid pBC-phleo using primers 409 and 410 to create PCR product #9. PCR products #2 (derA right arm) and #9 were then combined in an overlap PCR reaction with primers 410 and 582 to generate PCR product #10. PCR product #10 was cloned into pCR-Blunt II-TOPO (Invitrogen) to create p544. The inserts from p543 and p544 were excised with EcoRI and 10 µg of each was used to transform ΔhacA protoplasts. Loss of the derA gene was confirmed by Southern blot analysis of genomic DNA isolated from phleomycin resistant monoconidial isolates (data not shown).
The ΔderA/ΔhacA mutant was complemented by simultaneously introducing hacA and derA complementing plasmids into the ΔderA/ΔhacA mutant and selecting for restoration of thermotolerant growth. Briefly, ten µg of a previously published ΔhacA complementation plasmid13 was linearized with ApaI and transformed into ΔderA/ΔhacA mutant protoplasts, together with 50 µg of the derA complementation plasmid p547 digested with NsiI.13 Since all three commonly used dominant selectable markers were used for the construction of the ΔderA/ΔhacA mutant, selection was accomplished by screening for restoration of growth at 45°C. Since the thermosensitive phenotype is primarily attributed to loss of hacA,13 any transformant that was able to grow at 45°C would be expected to have at least the return of hacA. Thus, a molar excess of the derA complementation plasmid was used in the transformation mix to enhance the probability of reconstituting both genes in the ΔderA/ΔhacA mutant. Following transformation, the plates were incubated overnight at room temperature to allow for cell wall regeneration. The temperature was then increased stepwise to avoid heat-shock stress: 4 h at 30°C, 4 h at 37°C and then 45°C for 3 d. Conidia were isolated from 17 colonies that were able to grow within 3 d at 45°C and spread for isolation on a fresh IMA plate at 45°C. An equivalent number of ΔderA/ΔhacA protoplasts that were not transformed with DNA did not yield any colonies at 45°C. Southern blot analysis identified a transformant that had greater than one ectopic integration of both hacA and derA and this strain was used as the complemented strain.
Northern blot analysis.
A total of 5 × 106 conidia were inoculated into a 50 mL conical tube containing 5 mL of YG (5% yeast extract/20% glucose) and shaken at 200 rpm for 16 h at 37°C. Total RNA was extracted by crushing the mycelium in liquid nitrogen and resuspending in TRI reagent (Sigma). The RNA was fractionated by formaldehyde gel electrophoresis and equal loading was confirmed by visualization of ribosomal RNA using SYBR-Green II (Invitrogen). The RNA was transferred to BioBond nylon membrane (Sigma), and hybridized with a 32P-labeled DNA fragment derived from the A. fumigatus bipA gene. The bipA probe was PCR-amplified from genomic DNA using primers 494 and 495. Hybridization intensity and rRNA loading was determined using a STORM Phosphorimager (Molecular Dynamics).
Analysis of cell wall polysaccharide composition.
Fractionation of the mycelial cell wall was performed according to Fontaine et al.38 with slight modification. Briefly, the wt, ΔderA and ΔderA/ΔhacA strains were grown in a 1.2 L fermenter in liquid Sabouraud medium. After 24 h of cultivation (linear growth phase), the mycelia were collected by filtration, washed extensively with water and disrupted in 50 mL Falcon tubes using the FastPrep-24 instrument (MP Biomedicals, Solon, United States). Disruptions were performed at 4°C in two intervals of one minute at 6 m/s using 1 mm glass beads. The disrupted mycelial suspensions were centrifuged (3,000 g for 10 min), and the cell wall fractions (pellet) obtained were washed three times with water. Subsequent removal of proteins using SDS and β-mercaptoethanol, alkali-fractionation and estimation of the hexose composition using gas-liquid chromatography were performed as reported previously.13
Mouse model of invasive aspergillosis.
Conidia were harvested from plates of AMM-glucose supplemented with 1.2 M sorbitol and resuspended in sterile saline. Groups of 12 CF-1 outbred female mice (22–32 g) were immunosuppressed with a single dose of triamcinolone acetonide (40 mgkg−1 of body weight) injected subcutaneously on day −1. The mice were anesthetized with 3.5% isofluorane and inoculated intranasally with 2 × 106 conidia in a 20 µl suspension of saline. Survival was monitored for 14 d. Statistical significance was assessed by Kruskal-Wallis ANOVA using Sigma Stat 3.5 (p < 0.001).
For histopathology, mice were infected as described above and sacrificed on days 1 and 3 post-infection. The lungs were fixed in 4% phosphate-buffered paraformaldehyde, dehydrated and embedded in paraffin, sectioned at 5 µm, and stained with hematoxylin and eosin (H&E) or Grocott methenamine silver (GMS). Microscopic examinations were performed on an Olympus BH-2 microscope and imaging system using Spotsoftware version 4.6.
Analysis of protease secretion.
Conidia were inoculated at a concentration of 1 × 105/mL in 50 mL of AMM containing 10% heat-inactivated human AB serum (Innovative Research, Novi, Michigan). After incubating at 37°C for 72 h at 150 rpm, the mycelium was removed by centrifugation and the culture supernatant was diluted 1:100 (wild-type) or 1:5 (ΔderA/ΔhacA) in sterile-filtered HEPES buffer (50 mM HEPES, 100 mM NaCl, 10 mM CaCl2, pH 8.0). These dilutions were found to give comparable fluorescence intensity values in preliminary experiments.
A FRET peptide library comprised of 512 microtiter plate wells, each containing an equimolar mixture of up to eight individual peptides, was obtained (Mimotopes, Clayton, Australia).31 Each well (5 nmol of peptide) was dissolved in 5 µL 50% acetonitrile in ultrapure water and further diluted in 45 µL HEPES buffer. This solution (25 µL per well) was transferred to low volume black microtiter plates (Molecular Devices, Sunnyvale, CA). Diluted fungal culture supernatant (20 µL per well) was added to each well. Time-resolved fluorescence data were obtained on an Analyst HT instrument (Molecular Devices) using excitation and emission filters of 320 nm and 420 nm, respectively. The fluorescence intensity fold change after 5 h at room temperature was calculated as Ffinal/Finitial and each data set was normalized to the highest global signal intensity. No fluorescence was observed in control culture medium lacking fungal supernatant. Heatmaps were generated from these data in which each square corresponds to a single assay well (Heatmap Builder, Ashley Lab, Stanford).39
Acknowledgements
This work was supported by National Institutes of Health grant R01AI072297 and a Cystic Fibrosis Research Foundation Grant to D.S.A. D.L.R. was supported by an NIH F31 predoctoral fellowship (AI064121). The authors thank Jay Card for photography and illustration assistance.
Supplementary Material
References
- 1.Segal BH. Aspergillosis. N Engl J Med. 2009;360:1870–1884. doi: 10.1056/NEJMra0808853. [DOI] [PubMed] [Google Scholar]
- 2.Mirkes PE. Polysomes, ribonucleic acid and protein synthesis during germination of Neurospora crassa conidia. J Bacteriol. 1974;117:196–202. doi: 10.1128/jb.117.1.196-202.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Winther MD, Stevens L. RNA synthesis during the germination of conidia of Aspergillus nidulans. Microbios. 1981;30:153–162. [PubMed] [Google Scholar]
- 4.Brogden KA, Phillips M, Thurston JR, Richard JL. Electron microscopic examination of ribosome preparations from germinated spores of Aspergillus fumigatus. Mycopathologia. 1984;86:59–64. doi: 10.1007/BF00437231. [DOI] [PubMed] [Google Scholar]
- 5.Harris SD. Cell polarity in filamentous fungi: shaping the mold. Int Rev Cytol. 2006;251:41–77. doi: 10.1016/S0074-7696(06)51002-2. [DOI] [PubMed] [Google Scholar]
- 6.Steinberg G. Hyphal growth: a tale of motors, lipids and the Spitzenkorper. Eukaryot Cell. 2007;6:351–360. doi: 10.1128/EC.00381-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kohno K. Stress-sensing mechanisms in the unfolded protein response: similarities and differences between yeast and mammals. J Biochem. 2010;147:27–33. doi: 10.1093/jb/mvp196. [DOI] [PubMed] [Google Scholar]
- 8.Wobus U, Schubert I. People must be judged in the context of their time. Nature. 2000;404:330. doi: 10.1038/35006259. [DOI] [PubMed] [Google Scholar]
- 9.Dobson CM. Protein folding and misfolding. Nature. 2003;426:884–890. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- 10.Hoseki J, Ushioda R, Nagata K. Mechanism and components of endoplasmic reticulum-associated degradation. J Biochem. 2010;147:19–25. doi: 10.1093/jb/mvp194. [DOI] [PubMed] [Google Scholar]
- 11.Shamu CE, Walter P. Oligomerization and phosphorylation of the Ire1p kinase during intracellular signaling from the endoplasmic reticulum to the nucleus. EMBO J. 1996;15:3028–3039. [PMC free article] [PubMed] [Google Scholar]
- 12.Lee KP, Dey M, Neculai D, Cao C, Dever TE, Sicheri F. Structure of the dual enzyme Ire1 reveals the basis for catalysis and regulation in nonconventional RNA splicing. Cell. 2008;132:89–100. doi: 10.1016/j.cell.2007.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richie DL, Hartl L, Aimanianda V, Winters MS, Fuller KK, Miley MD, et al. A role for the unfolded protein response (UPR) in virulence and antifungal susceptibility in Aspergillus fumigatus. PLoS Pathog. 2009;5:1000258. doi: 10.1371/journal.ppat.1000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dave A, Jeenes DJ, Mackenzie DA, Archer DB. HacA-independent induction of chaperone-encoding gene bipA in Aspergillus niger strains overproducing membrane proteins. Appl Environ Microbiol. 2006;72:953–955. doi: 10.1128/AEM.72.1.953-955.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mulder HJ, Saloheimo M, Penttila M, Madrid SM. The transcription factor HACA mediates the unfolded protein response in Aspergillus niger, and upregulates its own transcription. Mol Genet Genomics. 2004;271:130–140. doi: 10.1007/s00438-003-0965-5. [DOI] [PubMed] [Google Scholar]
- 16.Valkonen M, Ward M, Wang H, Penttila M, Saloheimo M. Improvement of foreign-protein production in Aspergillus niger var. awamori by constitutive induction of the unfolded-protein response. Appl Environ Microbiol. 2003;69:6979–6986. doi: 10.1128/AEM.69.12.6979-6986.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Back SH, Schroder M, Lee K, Zhang K, Kaufman RJ. ER stress signaling by regulated splicing: IRE1/HAC1/XBP1. Methods. 2005;35:395–416. doi: 10.1016/j.ymeth.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 18.Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- 19.Vembar SS, Brodsky JL. One step at a time: endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirsch C, Gauss R, Horn SC, Neuber O, Sommer T. The ubiquitylation machinery of the endoplasmic reticulum. Nature. 2009;458:453–460. doi: 10.1038/nature07962. [DOI] [PubMed] [Google Scholar]
- 21.Horn SC, Hanna J, Hirsch C, Volkwein C, Schutz A, Heinemann U, et al. Usa1 functions as a scaffold of the HRD-ubiquitin ligase. Mol Cell. 2009;36:782–793. doi: 10.1016/j.molcel.2009.10.015. [DOI] [PubMed] [Google Scholar]
- 22.Carvalho P, Goder V, Rapoport TA. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 2006;126:361–373. doi: 10.1016/j.cell.2006.05.043. [DOI] [PubMed] [Google Scholar]
- 23.Knop M, Finger A, Braun T, Hellmuth K, Wolf DH. Der1, a novel protein specifically required for endoplasmic reticulum degradation in yeast. EMBO J. 1996;15:753–763. [PMC free article] [PubMed] [Google Scholar]
- 24.Hitt R, Wolf DH. Der1p, a protein required for degradation of malfolded soluble proteins of the endoplasmic reticulum: topology and Der1-like proteins. FEMS Yeast Res. 2004;4:721–729. doi: 10.1016/j.femsyr.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 25.Mulder HJ, Nikolaev I, Madrid S. HACA, the transcriptional activator of the unfolded protein response (UPR) in Aspergillus niger, binds to partly palindromic UPR elements of the consensus sequence 5′-CAN(G/A)NTGT/GCCT-3′. Fungal Genet Biol. 2006;43:560–572. doi: 10.1016/j.fgb.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 26.Pinchai N, Juvvadi PR, Fortwendel JR, Perfect BZ, Rogg LE, Asfaw YG, et al. The Aspergillus fumigatus P-type Golgi apparatus Ca2+/Mn2+ ATPase PmrA is involved in cation homeostasis and cell wall integrity but is not essential for pathogenesis. Eukaryot Cell. 2010;9:472–476. doi: 10.1128/EC.00378-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gastebois A, Clavaud C, Aimanianda V, Latge JP. Aspergillus fumigatus: cell wall polysaccharides, their biosynthesis and organization. Future Microbiol. 2009;4:583–595. doi: 10.2217/fmb.09.29. [DOI] [PubMed] [Google Scholar]
- 28.Sharon H, Hagag S, Osherov N. Transcription factor PrtT controls expression of multiple secreted proteases in the human pathogenic mold Aspergillus fumigatus. Infect Immun. 2009;77:4051–4060. doi: 10.1128/IAI.00426-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mattern IE, van Noort JM, van den Berg P, Archer DB, Roberts IN, van den Hondel C. Isolation and characterization of mutants of Aspergillus niger deficient in extracellular proteases. Mol Gen Genet. 1992;234:332–336. doi: 10.1007/BF00283855. [DOI] [PubMed] [Google Scholar]
- 30.Nierman WC, Pain A, Anderson MJ, Wortman JR, Kim HS, Arroyo J, et al. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature. 2005;438:1151–1156. doi: 10.1038/nature04332. [DOI] [PubMed] [Google Scholar]
- 31.Thomas DA, Francis P, Smith C, Ratcliffe S, Ede NJ, Kay C, et al. A broad-spectrum fluorescence-based peptide library for the rapid identification of protease substrates. Proteomics. 2006;6:2112–2120. doi: 10.1002/pmic.200500153. [DOI] [PubMed] [Google Scholar]
- 32.Malhotra JD, Kaufman RJ. The endoplasmic reticulum and the unfolded protein response. Semin Cell Dev Biol. 2007;18:716–731. doi: 10.1016/j.semcdb.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malavazi I, Semighini CP, Kress MR, Harris SD, Goldman GH. Regulation of hyphal morphogenesis and the DNA damage response by the Aspergillus nidulans ATM homolog AtmA. Genetics. 2006;173:99–109. doi: 10.1534/genetics.105.052704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ram AF, Klis FM. Identification of fungal cell wall mutants using susceptibility assays based on Calcofluor white and Congo red. Nat Protoc. 2006;1:2253–2256. doi: 10.1038/nprot.2006.397. [DOI] [PubMed] [Google Scholar]
- 35.Catlett NL, Lee BN, Yoder OC, Turgeon BG. Splitmarker recombination for efficient targeted deletion of fungal genes. Fungal Genet News. 2002;50:9–11. [Google Scholar]
- 36.Krappmann S, Sasse C, Braus GH. Gene targeting in Aspergillus fumigatus by homologous recombination is facilitated in a nonhomologous end-joining-deficient genetic background. Eukaryot Cell. 2006;5:212–215. doi: 10.1128/EC.5.1.212-215.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bhabhra R, Miley MD, Mylonakis E, Boettner D, Fortwendel J, Panepinto JC, et al. Disruption of the Aspergillus fumigatus gene encoding nucleolar protein CgrA impairs thermotolerant growth and reduces virulence. Infect Immun. 2004;72:4731–4740. doi: 10.1128/IAI.72.8.4731-4740.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fontaine T, Simenel C, Dubreucq G, Adam O, Delepierre M, Lemoine J, et al. Molecular organization of the alkali-insoluble fraction of Aspergillus fumigatus cell wall. J Biol Chem. 2000;275:41528. [PubMed] [Google Scholar]
- 39.King JY, Ferrara R, Tabibiazar R, Spin JM, Chen MM, Kuchinsky A, et al. Pathway analysis of coronary atherosclerosis. Physiol Genomics. 2005;23:103–118. doi: 10.1152/physiolgenomics.00101.2005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.