Abstract
In our recent paper, we demonstrated that the hypervirulence exhibited by a lineage of the fatal fungal pathogen Cryptococcus gattii is associated with its mitochondrial gene expression and an unusual mitochondrial morphology. As an importan organelle, the mitochondrion has been linked tovarious cellular activities, but its role in modulating virulence of pathogens remains unclear. In this addendum, the potential role of mitochondria in determining virulence in eukaryotic pathogens is discussed along with future experiments that may lead to an improved understanding of this topic.
Key words: Cryptococcus, virulence, mitochondrion, intracellular parasitism, mitochondrial fusion, Vancouver Island outbreak, macrophage
Cryptococcosis is a fatal fungal disease of humans and other animals, primarily caused by Cryptococcus neoformans infections in immunocompromised hosts. The related species Cryptococcus gattii can also cause disease, but this is generally restricted to very rare infections in tropical or subtropical areas. However, in 1999 this species was identified as the cause of an ongoing outbreak of cryptococcal disease in residents of Vancouver Island, Canada,1 an outbreak that has since spread to mainland Canada and the northwest region of the USA.2,3 This so-called Vancouver Island Outbreak (VIO) is remarkable for two reasons; firstly, because it represents a major expansion of C. gattii into a temperate area and, secondly, because most of the VIO infections have occurred in immunocompetent individuals.
Work by many groups over the last decade has clearly demonstrated that the VIO lineage of C. gattii is hypervirulent,4 but the underlying molecular reasons for this hypervirulence remain unclear. However, we have recently demonstrated a potential role for mitochondrial function in regulating virulence within this organism. Our study showed that C. gattii strains from within the VIO lineage, but not related control strains, exhibit enhanced intracellular proliferation within host macrophages.5 By comparing fungal gene expression whilst inside host macrophages between hypervirulent (VIO) and hypovirulent (non-VIO) strains using microarray approaches, we identified mitochondrial gene expression as the major hallmark of virulence within this species. Furthermore, we showed that VIO strains respond to the environment within the macrophage by producing long, tubular mitochondria (as opposed to the normal punctate mitochondria found in non-VIO strains during intracellular growth or in VIO strains that are grown in normal media).
In the past, the mitochondrion has been demonstrated to play a role in the fitness of microorganisms, as the organelle is essential for energy production and response to stress. For example, in S. cerevisiae, when mitochondria of wine yeasts were transferred to a laboratory strain, the latter showed increased viability and increased tolerance towards ethanol and high temperature.6 Furthermore, a fungal pathogen of plants, Heterobasidion annosum, exhibits differential virulence depending on the mitochondrial (but not nuclear) genotype.7 Our finding that the mitochondria of virulent C. gattii strains are more likely to fuse with each other during growth within host cells provides a potential explanation for the involvement of mitochondria in virulence regulation. Since mitochondrial fusion is generally thought to protect cells from the detrimental effect of mtDNA mutations and cell death,8–11 we proposed that the tubular morphology could be a protective response of the pathogen against the hostile intracellular environment.
Intriguingly, tubular mitochondrial formation inside host cells is very limited in less virulent strains within the same species, suggesting this trait is likely to have evolved very recently. Within the C. gattii species there are four genotypes, known as VGI, VGII, VGIII and VGIV, and most of the VIO strains belong to the VGII genotype. It has been demonstrated that the VGII C. gattii population has much lower within-lineage divergence in both the nuclear and mitochondrial genome in comparison with other groups, despite the fact that VGII is considered to be basal for the C. gattii species.12 This may point to a recent bottleneck event within the VGII population, during which both mitochondrially-regulated virulence and a generally fitter population may have been selected. It is tempting to speculate that the same-sex mating event that has been proposed as the source of the VIO lineage4 may provide just such a bottleneck during the recent evolution of this pathogen.
Although mitochondrial involvement in virulence of C. gattii species has not previously been proposed, indirect evidence from earlier studies suggest that the organelle might be involved in regulating virulence of another cryptococcal species, C. neoformans. Global in vivo transcriptional profiling of C. neoformans cells at the site of a central nervous system infection demonstrated that several respiratory genes were highly expressed by this yeast.13 Two other studies have shown the importance of mitochondria in responding to hypoxic conditions and oxidative stress,14,15 both of which occur during intracellular growth. However, a study conducted by creating stable AD hybrids to place serotype A and D mitochondria under different nuclear-genomic influences suggested that the mitochondrial genome itself is unlikely to have a significant influence on the differences between serotypes in their virulence composite in C. neoformans.16 This indicates that mitochondrial virulence regulation may involve complex interactions between nuclear and mitochondrial genes.
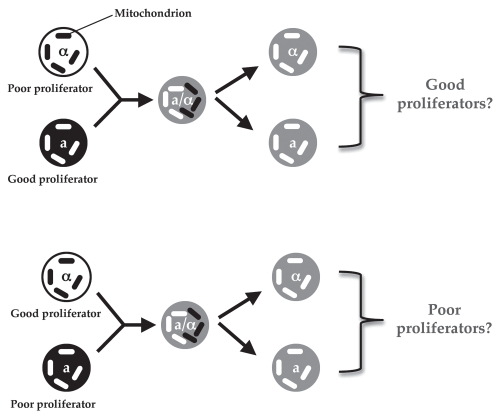
Based on our study, the ideal experiment to test the role of mitochondrial genotype in virulence of VIO strains would be to replace mitochondria of a poor intracellular proliferator (i.e., hypovirulent) strain with those from virulent (highly proliferative) strains or vice versa. However, such an experiment is technically challenging as, unlike S. cerevisiae, which can produce enough ATP by glycolysis (a pathway occurring in the cytoplasm that is independent of functional mitochondria17) the presence of mitochondria seems to be essential to cryptococcal viability.16 Fortunately, cryptococci exhibit a largely mating-type dependent uniparental mitochondrial inheritance: the offspring predominantly receive their mitochondria from the MATa parent, though a low level of leakage was also observed, during which biparental inheritance and mitochondrial recombination can occur.16,18–22 This means it is theoretically possible to cross two strains (one a good proliferator and one a poor proliferator) and generate F1 progeny that contain mitochondria only from their good or poor proliferator parent (see Fig. 1 for experimental design). In this case, the effect of mitochondrial genotype on virulence can be tested independently of nuclear genotype.
Figure 1.
A schematic illustration of the experimental design for two crosses to generate progeny with mitochondria from only their mating type a (mata) parent.
Attempts to explore this experimentally are currently ongoing in our group. Regrettably, both we and others have failed to produce viable progeny from crosses within the C. gattii group that contains the VIO isolates (the so-called VGII group), despite the fact the VGII strains are believed to be more fertile than the other C. gattii isolates.23–25 However, crosses between groups (e.g., VGII crossed with VGIII isolates) are feasible. It has been shown that such an inter-genotype mating can result in a loss of viability in the basidiospores (<5%) and the generation of many diploid and even aneuploid progeny,26 as the meiosis between the two genotypes is impaired because of their genomic divergence. This leads to the concern that progeny from such crosses may be generally less fit and, in addition, will contain part of the VGIII nuclear genome, which may lead to disruption of nuclear-mitochondrial crosstalk. Nonetheless, such crosses may still provide valuable information about the mitochondrial control of virulence.
Given the relatively well-conserved mitochondrial genome structure and gene synteny between cryptococci (Fig. 2), and the demonstration that mitochondrial genotype alone does not predict virulence in C. neoformans,16 one would also suspect that the observed mitochondrial differences between virulent and avirulent strains is at least partially due to changes in nuclear-encoded proteins that affect mitochondrial morphology and gene expression. In fact, our data identified several nuclear-encoded proteins that function in mitochondria and are upregulated in the VIO strains. The nuclear-encoded mitochondrial proteins are usually synthesised in the cytoplasm and then imported into mitochondria. They interact with mitochondrially encoded proteins (e.g., in the electron transport system), control mitochondrial biogenesis, regulate mtDNA copy number, influence mtDNA stability and alter mitochondrial morphology in a sophisticated manner.17,27 –29
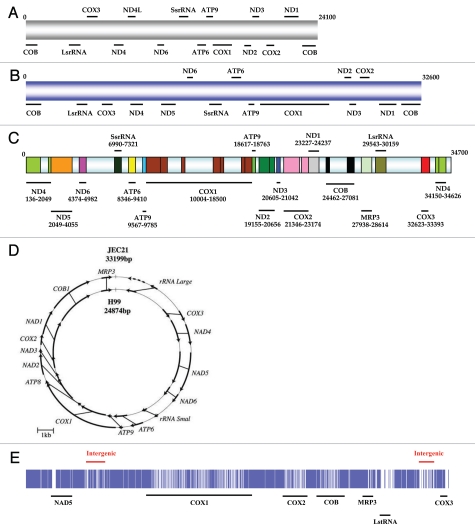
Figure 2.
mtDNA structure of Cryptococcus: all the mtDNAs show a conserved gene synteny but have different sizes. (A) IFM5844 (C. neoformans var. neoformans, serotype D) and (B) IFO410 (C. neoformans var. grubii, serotype A). These two mtDNA structures were drawn to scale based on information from litter et al.;37 (C) mtDNA structure of A1M-R265 (C. gattii, VGII). Sections with light blue colour are either introns or intergenic spaces;5 (D) Circular mtDNA structure of JEC21 (C. neoformans var. neoformans) and H99 (C. neoformans var. grubii) (taken from Toffaletti et al.16); (E) Simple alignment of A1M-R265 and WM276 (C. gattii, VGI) mtDNA using ClusterW. Before alignment, two repeat regions in both mtDNAs were removed (region one: 2434 nucleotides in COX1 gene; region two: 963 nucleotide at the end of the supercontig). Sections with white color stand for the variations between two mtDNA sequences.
Mitochondrial morphology can be affected by many genes as demonstrated by Ichishita et al.30 In yeast and mammals, several factors including Drp1/Dnm1 and Mfn/Fzo1 are known to regulate mitochondrial morphology by controlling membrane fission or fusion.28 Given our discovery that mitochondria in virulent (VIO) cryptococci adopt a tubular morphology during intracellular growth, such genes are prime candidates for regulators of virulence capacity in this pathogen. Interestingly, we find that FZO1 is upregulated in the VIO strains. Fzo (Fuzzy onions gene), first isolated from a screen for genes involved in Drosophila spermatogenesis, is the first molecule to be identified in regulating mitochondrial fusion.31 It is known as mitofusin and Fzo1p in mammals and yeast respectively.32,33 The protein contains a GTPase domain (exposed to the cytoplasm) at the N-terminus and a bipartite transmembrane domain (which spans the mitochondrial outer membrane twice) near the C-terminus.34,35 In S. cerevisiae, the mitochondrion of fzo1Δ mutants is highly fragmented due to ongoing fisson32,36 and overexpression of Fzo1p alters the fusion/fission protein ratio and thus inhibits cell apoptosis.11 Therefore, higher amounts of Fzo1p in VIO strains could be responsible for the tubular formation of mitochondria and also lead to a higher fusion/fission protein ratio, which is essential to increase the resistance of mitochondria and cells to apoptotic stimulation. The key unanswered questions are how nuclear-encoded proteins regulate mitochondrial activities and what external factors induce such regulation. To answer these questions, we are now subjecting the VIO strains to simple environmental stimuli commonly present in phagosomes in order to test whether these are sufficient to trigger mitochondrial morphology changes in vitro. In parallel, we are knocking out genes involved in mitochondrial fusion and fission (such as FZO1, MMM1, MDM10 and MDM12,28 to examine whether these proteins are the regulators of nuclear-mitochondrion communication and thus hypervirulence.
To summarise, our study reveals an interesting link between the mitochondrion and virulence of C. gattii. Since mitochondria are found in all eukaryotic pathogens, and have been implicated in virulence in at least one other fungus (H. annosum),7 mitochondrial regulation of pathogenicity may be a widespread phenomenon. More detailed experiments are therefore urgently needed to provide a clearer understanding of how mitochondria fulfil such a role.
Acknowledgements
This work was made possible with financial support from the Medical Research Council (G0601171) and the Wellcome Trust (WT088148MF). We also would like to thank Wenjun Li from the Heitman lab at the Duke University for the help on the A1M-R265 and WM276 mtDNA alignment.
Addendum to: Ma H, Hagen F, Stekel DJ, Johnston SA, Sionov E, Falk R, et al. The fatal fungal outbreak on Vancouver Island is characterized by enhanced intracellular parasitism driven by mitochondrial regulation. Proc Natl Acad Sci USA. 2009;106:12980–12985.
Footnotes
Previously published online: www.landesbioscience.com/journals/virulence/article/11053
References
- 1.Kidd SE, Hagen F, Tscharke RL, Huynh M, Bartlett KH, Fyfe M, et al. A rare genotype of Cryptococcus gattii caused the cryptococcosis outbreak on Vancouver Island (British Columbia, Canada) Proc Natl Acad Sci USA. 2004;101:23–25. doi: 10.1073/pnas.0402981101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Byrnes EJ, III, Bildfell RJ, Frank SA, Mitchell TG, Marr KA, Heitman J. Molecular Evidence that the Range of the Vancouver Island Outbreak of Cryptococcus gattii Infection Has Expanded into the Pacific Northwest in the United States. J Infect Dis. 2009;199:1081–1086. doi: 10.1086/597306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.MacDougall L, Kidd SE, Galanis E, Mak S, Leslie MJ, Cieslak PR, et al. Spread of Cryptococcus gattii in British Columbia, Canada, and detection in the Pacific Northwest USA. Emerg Infect Dis. 2007;13:42–50. doi: 10.3201/eid1301.060827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fraser JA, Giles SS, Wenink EC, Geunes-Boyer SG, Wright JR, Diezmann S, et al. Same-sex mating and the origin of the Vancouver Island Cryptococcus gattii outbreak. Nature. 2005;437:1360–1364. doi: 10.1038/nature04220. [DOI] [PubMed] [Google Scholar]
- 5.Ma H, Hagen F, Stekel DJ, Johnston SA, Sionov E, Falk R, et al. The fatal fungal outbreak on Vancouver Island is characterized by enhanced intracellular parasitism driven by mitochondrial regulation. Proc Natl Acad Sci USA. 2009;106:12980–12985. doi: 10.1073/pnas.0902963106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jimenez J, Benitez T. Yeast cell viability under conditions of high temperature and ethanol concentrations depends on the mitochondrial genome. Curr Genet. 1988;13:461–469. doi: 10.1007/BF02427751. [DOI] [PubMed] [Google Scholar]
- 7.Olson A, Stenlid J. Plant pathogens. Mitochondrial control of fungal hybrid virulence. Nature. 2001;411:438. doi: 10.1038/35078147. [DOI] [PubMed] [Google Scholar]
- 8.Chan DC. Mitochondrial fusion and fission in mammals. Annu Rev Cell Dev Biol. 2006;22:79–99. doi: 10.1146/annurev.cellbio.22.010305.104638. [DOI] [PubMed] [Google Scholar]
- 9.Karbowski M, Arnoult D, Chen H, Chan DC, Smith CL, Youle RJ. Quantitation of mitochondrial dynamics by photolabeling of individual organelles shows that mitochondrial fusion is blocked during the Bax activation phase of apoptosis. J Cell Biol. 2004;164:493–499. doi: 10.1083/jcb.200309082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sato A, Nakada K, Hayashi J. Mitochondrial dynamics and aging: Mitochondrial interaction preventing individuals from expression of respiratory deficiency caused by mutant mtDNA. Biochim Biophys Acta. 2006;1763:473–481. doi: 10.1016/j.bbamcr.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 11.Sugioka R, Shimizu S, Tsujimoto Y. Fzo1, a protein involved in mitochondrial fusion, inhibits apoptosis. J Biol Chem. 2004;279:52726–52734. doi: 10.1074/jbc.M408910200. [DOI] [PubMed] [Google Scholar]
- 12.Xu J, Yan Z, Guo H. Divergence, hybridization and recombination in the mitochondrial genome of the human pathogenic yeast Cryptococcus gattii. Mol Ecol. 2009;18:2628–2642. doi: 10.1111/j.1365-294X.2009.04227.x. [DOI] [PubMed] [Google Scholar]
- 13.Steen BR, Zuyderduyn S, Toffaletti DL, Marra M, Jones SJ, Perfect JR, et al. Cryptococcus neoformans gene expression during experimental cryptococcal meningitis. Eukaryot Cell. 2003;2:1336–1349. doi: 10.1128/EC.2.6.1336-1349.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ingavale SS, Chang YC, Lee H, McClelland CM, Leong ML, Kwon-Chung KJ. Importance of mitochondria in survival of Cryptococcus neoformans under low oxygen conditions and tolerance to cobalt chloride. PLoS Pathog. 2008;4:1000155. doi: 10.1371/journal.ppat.1000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Narasipura SD, Chaturvedi V, Chaturvedi S. Characterization of Cryptococcus neoformans variety gattii SOD2 reveals distinct roles of the two superoxide dismutases in fungal biology and virulence. Mol Microbiol. 2005;55:1782–1800. doi: 10.1111/j.1365-2958.2005.04503.x. [DOI] [PubMed] [Google Scholar]
- 16.Toffaletti DL, Nielsen K, Dietrich F, Heitman J, Perfect JR. Cryptococcus neoformans mitochondrial genomes from serotype A and D strains do not influence virulence. Curr Genet. 2004;46:193–204. doi: 10.1007/s00294-004-0521-9. [DOI] [PubMed] [Google Scholar]
- 17.Osiewacz HD, Kimpel E. Mitochondrial-nuclear interactions and lifespan control in fungi. Exp Gerontol. 1999;34:901–909. doi: 10.1016/s0531-5565(99)00063-7. [DOI] [PubMed] [Google Scholar]
- 18.Xu J. Mitochondrial DNA polymorphisms in the human pathogenic fungus Cryptococcus neoformans. Curr Genet. 2002;41:43–47. doi: 10.1007/s00294-002-0282-2. [DOI] [PubMed] [Google Scholar]
- 19.Xu J. The inheritance of organelle genes and genomes: patterns and mechanisms. Genome. 2005;48:951–958. doi: 10.1139/g05-082. [DOI] [PubMed] [Google Scholar]
- 20.Xu J, Luo G, Vilgalys RJ, Brandt ME, Mitchell TG. Multiple origins of hybrid strains of Cryptococcus neoformans with serotype AD. Microbiology. 2002;148:203–212. doi: 10.1099/00221287-148-1-203. [DOI] [PubMed] [Google Scholar]
- 21.Yan Z, Sun S, Shahid M, Xu J. Environment factors can influence mitochondrial inheritance in the fungus Cryptococcus neoformans. Fungal Genet Biol. 2007;44:315–322. doi: 10.1016/j.fgb.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 22.Yan Z, Xu J. Mitochondria are inherited from the MATa parent in crosses of the basidiomycete fungus Cryptococcus neoformans. Genetics. 2003;163:1315–1325. doi: 10.1093/genetics/163.4.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fraser JA, Subaran RL, Nichols CB, Heitman J. Recapitulation of the sexual cycle of the primary fungal pathogen Cryptococcus neoformans var. gattii: implications for an outbreak on Vancouver Island, Canada. Eukaryot Cell. 2003;2:1036–1045. doi: 10.1128/EC.2.5.1036-1045.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Halliday CL, Carter DA. Clonal reproduction and limited dispersal in an environmental population of Cryptococcus neoformans var gattii isolates from Australia. J Clin Microbiol. 2003;41:703–711. doi: 10.1128/JCM.41.2.703-711.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ngamskulrungroj P, Sorrell TC, Chindamporn A, Chaiprasert A, Poonwan N, Meyer W. Association between fertility and molecular sub-type of global isolates of Cryptococcus gattii molecular type VGII. Med Mycol. 2008:1–9. doi: 10.1080/13693780802210734. [DOI] [PubMed] [Google Scholar]
- 26.Lengeler KB, Cox GM, Heitman J. Serotype AD strains of Cryptococcus neoformans are diploid or aneuploid and are heterozygous at the mating-type locus. Infect Immun. 2001;69:115–122. doi: 10.1128/IAI.69.1.115-122.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cannino G, Di Liegro CM, Rinaldi AM. Nuclear-mitochondrial interaction. Mitochondrion. 2007;7:359–366. doi: 10.1016/j.mito.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 28.Okamoto K, Shaw JM. Mitochondrial morphology and dynamics in yeast and multicellular eukaryotes. Annu Rev Genet. 2005;39:503–536. doi: 10.1146/annurev.genet.38.072902.093019. [DOI] [PubMed] [Google Scholar]
- 29.Poyton RO, McEwen JE. Crosstalk between nuclear and mitochondrial genomes. Annu Rev Biochem. 1996;65:563–607. doi: 10.1146/annurev.bi.65.070196.003023. [DOI] [PubMed] [Google Scholar]
- 30.Ichishita R, Tanaka K, Sugiura Y, Sayano T, Mihara K, Oka T. An RNAi screen for mitochondrial proteins required to maintain the morphology of the organelle in Caenorhabditis elegans. J Biochem. 2008;143:449–454. doi: 10.1093/jb/mvm245. [DOI] [PubMed] [Google Scholar]
- 31.Hales KG, Fuller MT. Developmentally regulated mitochondrial fusion mediated by a conserved, novel, predicted GTPase. Cell. 1997;90:121–129. doi: 10.1016/s0092-8674(00)80319-0. [DOI] [PubMed] [Google Scholar]
- 32.Hermann GJ, et al. Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p. J Cell Biol. 1998;143:359–373. doi: 10.1083/jcb.143.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Santel A, Fuller MT. Control of mitochondrial morphology by a human mitofusin. J Cell Sci. 2001;114:867–874. doi: 10.1242/jcs.114.5.867. [DOI] [PubMed] [Google Scholar]
- 34.Fritz S, Rapaport D, Klanner E, Neupert W, Westermann B. Connection of the mitochondrial outer and inner membranes by Fzo1 is critical for organellar fusion. J Cell Biol. 2001;152:683–692. doi: 10.1083/jcb.152.4.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rojo M, Legros F, Chateau D, Lombes A. Membrane topology and mitochondrial targeting of mitofusins, ubiquitous mammalian homologs of the transmembrane GTPase Fzo. J Cell Sci. 2002;115:1663–1674. doi: 10.1242/jcs.115.8.1663. [DOI] [PubMed] [Google Scholar]
- 36.Rapaport D, Brunner M, Neupert W, Westermann B. Fzo1p is a mitochondrial outer membrane protein essential for the biogenesis of functional mitochondria in Saccharomyces cerevisiae. J Biol Chem. 1998;273:20150–20155. doi: 10.1074/jbc.273.32.20150. [DOI] [PubMed] [Google Scholar]
- 37.Litter J, Keszthelyi A, Hamari Z, Pfeiffer I, Kucsera J. Differences in mitochondrial genome organization of Cryptococcus neoformans strains. Antonie Van Leeuwenhoek. 2005;88:249–255. doi: 10.1007/s10482-005-8544-x. [DOI] [PubMed] [Google Scholar]