Abstract
Down syndrome (DS) or Trisomy 21 (Ts21) is caused by the presence of an extra copy of all or part of human chromosome 21 (Hsa21) and is the most frequent survivable congenital chromosomal abnormality. Bioinformatic annotation has established that Hsa21 harbors more than 400 genes, including five microRNA (miRNA) genes (miR-99a, let-7c, miR-125b-2, miR-155 and miR-802). MiRNAs are endogenous, single-stranded, small non-coding RNA molecules that regulate gene expression by interacting with specific recognition elements harbored within the 3′-untranslated region (3′-UTR) of mRNAs and subsequently target these mRNAs for translational repression or destabilization. MiRNA expression profiling, miRNA RT-PCR and miRNA in situ hybridization experiments have demonstrated that Hsa21-derived miRNAs were overexpressed in fetal brain and heart specimens isolated from individuals with DS. We now propose that Ts21 gene dosage overexpression of Hsa21-derived miRNAs in DS individuals result in the decreased expression of specific target proteins (i.e., haploinsufficiency) that contribute, in part, to the DS phenotype.
Key words: down syndrome, trisomy 21, microRNAs, haploinsufficiency
Introduction
DS or Ts21 usually results from the failure of chromosome pairs to separate properly during cell division (i.e., nondisjunction).1 In DS individuals nondisjunction usually occurs during meiotic division I and II (∼95% of the time), is frequently of maternal origin (∼90% of cases) and is often associated with advanced maternal age.2–4 Although Ts21 can be divided into four categories according to the size of the triplicated genomic region (i.e., complete trisomy, partial trisomy, microtrisomy and single-gene duplication), the majority of DS individuals (∼95%) have three complete copies of Hsa21.2–4 DS is the most frequent survivable congenital chromosomal abnormality occurring in approximately 750–1,000 live births with an estimated 5,000 infants born with DS each year in the USA.5,6
The clinical presentation of DS is complex and variable. A constellation of over 80 clinical features have been described in DS patients, but cognitive impairments are one of the most constant features with varying degrees of penetrance.7–9 In addition to cognitive dysfunction, the phenotypes of DS include congenital heart defects, craniofacial abnormalities, gastrointestinal anomalies, childhood leukemia, altered immune responses and early-onset Alzheimer's disease.7–9 Interestingly, even though DS individuals have a high incidence of leukemia, the risk of developing most solid tumors (e.g., neural tumors and breast cancer) is markedly reduced in DS patients across all age groups.10–14 Additionally, persons with DS have significantly lower systolic and diastolic blood pressure than do individuals with cognitive impairments due to other causes.15–19 Further, DS patients have a reduced risk of vascular anomalies compared with the general population.20 Finally, the prevalence of coronary artery disease is low in DS subjects and pathological studies revealed decreased levels of atherosclerosis in these; therefore, it has been suggested that DS individuals may represent an atherosclerosis-free model.21–25
While the genetic anomaly in DS has been identified to be a triplication of Hsa21, it is not known how this chromosomal anomaly causes DS characteristics. It is hypothesized that a “gene-dosage” effect resulting from the 50% increase in expression at the RNA level of the trisomic genes leads to an imbalance of critical genes, which in turn, initiates the DS phenotype.26–28 While the DS phenotype is most likely a multigenic condition, it is evident that only a specific subset of trisomic Hsa21 genes play a role in mediating the DS phenotype28–31 and, almost certainly, DS results from the interaction between Hsa21 genes and Hsa21 genes interacting with other disomic genes.
Hsa21-Derived miRNAs
MiRNAs are a family of small, ∼21-nucleotide long, nonprotein-coding RNAs that have emerged as key post-transcriptional regulators of gene expression.32,33 MiRNAs are processed from precursor molecules (pri-miRNAs), which are either transcribed from independent miRNA genes or are portions of introns of protein-coding RNA polymerase II transcripts. Pri-miRNAs fold into hairpin structures containing imperfectly base-paired stems and are subsequently endonucleolytically processed.32,33 In principle, the resulting miRNA duplex could give rise to two different mature miRNAs. However, in a manner similar to siRNA duplexes, only one strand is usually incorporated into miRNA-induced silencing complexes (miRISCs); the other strand is degraded (the complementary miRNA* strand).58 Still, recent data suggest that both arms of the pre-miRNA hairpin can give rise to mature miRNAs.32,33,38
Once the miRISC is assembled, the miRNA acts as an adaptor for this complex to specifically recognize and regulate particular mRNAs. Mature miRNAs recognize their target mRNAs by base-pairing interactions between nucleotides 2 and 8 of the miRNA (the seed region) and complementary nucleotides in the 3′-UTR of mRNAs. MiRISCs subsequently inhibit gene expression by targeting mRNAs for translational repression or destabilization.32,33
Bioinformatic annotation has established that Hsa21 harbors more than 400 genes,34,35 including five microRNA (miRNA) genes (miR-99a, let-7c, miR-125b-2, miR-155 and miR-802, http://www.mirbase.org/index.shtml).36 MiR-99a, let-7c and miR-125b-2 reside in the sense orientation within intron 6 of the C21orf34 gene (intron number designation changes because of alternative C21orf34 promoter utilization). The C21orf34 gene is located at the beginning of q21.1 and the protein encoded by this gene has not been characterized. MiR-99a and let-7c are only 659 bp apart, whereas miR-125b-2 is located just over 50,000 bp downstream of let-7c. It is currently not known whether this genomic cluster is transcribed and regulated independent of the host gene and/or one another. MiR-155 resides within exon 3 of the spliced and polyadenylated non-coding B-cell integration cluster (BIC) gene, which is frequently targeted for retroviral insertion in chickens.37 The BIC/miR-155 gene is located almost nine million base pairs downstream from the C21orf34 gene at Hsa21 genomic position q21.2. Finally, miR-802 is located over 10.1 million base pairs downstream from the BIC/miR-155 gene in the antisense orientation within intron 1 of the RUNX1 gene at position q22.11. Currently very little information is known regarding the expression distribution of this miRNA or the mechanisms by which it is regulated.
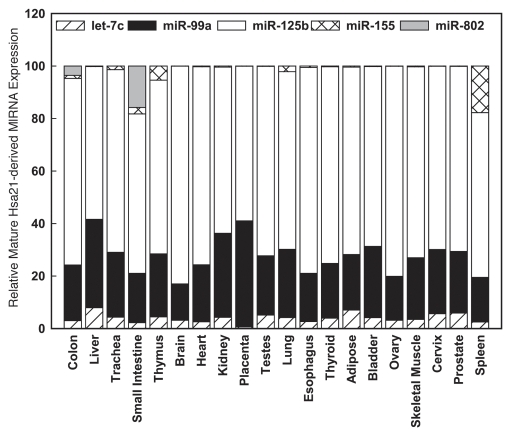
Experiments investigating the tissue specific distribution of the five Hsa21-derived miRNAs and their respective miRNA* species (probe sets are currently not available for miR-802*) demonstrated that miR-99a, let-7c and miR-125b-2 are abundantly expressed in a number of tissues including the DS relevant tissues, the brain and heart (Fig. 1). It is important to note that the expression levels of miR-125b-2 are an over-estimate due to cross-reactivity of qPCR probes with its miR-125b-1 ortholog.41 In contrast miR-155 and miR-802 are expressed at much lower levels with the highest expression being in the thymus, the colon and small intestine, respectively (Fig. 1). Although the Hsa21-derived miRNA* species were detectable in all the tissues investigated, they were usually at least 100-fold less abundant than their non-star miRNA complement (data not shown) suggesting that unlike some recent studies investigating the functionality of miRNA* strands,32,33,38 the Hsa21 miRNA* species may not be biologically important.
Figure 1.
Relative expression of Hsa21-derived miRNAs in human tissues. Mature human let-7c, miR-99a, miR-125b, miR-155 and miR-802 were quantified utilizing TaqMan microRNA assay kits specific for each Hsa21-derived miRNA (Applied Biosystems, Foster City, CA) as previously described.40,42 The expression values were normalized to RNU48 for each tissue. Relative gene expression was calculated as 2-(CTmiR-155-CT-RNU48). The relative abundance of each Hsa-21 derived miRNA is shown as a percentage of total combined expression of Hsa-21 derived miRNAs for each tissue investigated.
Hsa21-Derived miRNAs' Expression in DS Tissues
Given that bioinformatic annotation has established that Hsa21 harbors five miRNA genes, it was hypothesized that Ts21 may result in a gene dosage overexpression of these miRNAs. This hypothesis was first tested in fibroblasts isolated from an individual with DS and from their unaffected monozygotic twin.39 Twins were selected for study so that gene-expression differences could be attributed to only the supernumeracy of Hsa21 and not polymorphic variability in the rest of the genome. Importantly, these experiments demonstrated that miR-155 was more abundantly expressed in the fibroblasts isolated from the DS individual when compared with the unaffected twin;39 the expression levels of the other four Hsa21-derived miRNAs were not determined.
To extend these results miRNA expression profiling experiments were performed utilizing RNA isolated from control and DS fetal hippocampus samples.40 Of the 424 human mature miRNAs investigated, 10 were overexpressed and 22 were under-expressed by at least 50% in hippocampus specimens isolated from individuals with DS when compared with controls. Importantly, of the five Hsa21-derived miRNAs, miR-99a, let-7c and miR-155 were overexpressed in the profiling experiments. The values for miR-125b-2 were inconclusive and miR-802 was not profiled.40
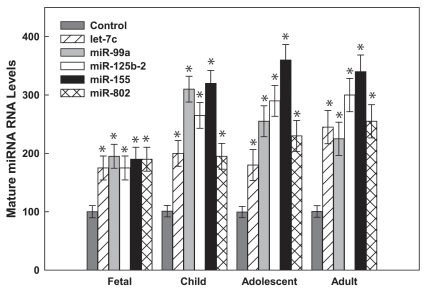
To validate the microarray studies, qPCR experiments were performed using primer sets specific for the five Hsa21-derived miRNAs. These experiments demonstrated that mature miR-99a, let-7c, miR-125b-2, miR-155 and miR-802 were overexpressed by at least 50% in fetal hippocampus and heart samples obtained from DS individuals when compared with age- and sex-matched control specimens.40 Hsa21-derived miRNA expression was also investigated in these samples by in situ hybridization experiments utilizing antisense miRNA locked nucleic acid (LNA) probes specific for each Hsa21-derived miRNA. Importantly, these experiments demonstrated that let-7c, miR-125b-2, miR-155 and miR-802 expressing neurons were increased by 10- to 15-fold and miR-99a expressing neurons were increased by ∼50-fold in DS samples.40 Finally, qPCR utilizing total RNA isolated from human prefrontal cortex specimens from brains of fetuses, infants/children, adolescents and adults with DS demonstrated that all five Hsa21-derived miRNAs were overexpressed by at least 50% in prefrontal cortex samples at all ages examined when compared with age- and sex-matched control brain specimens (Fig. 2).42
Figure 2.
Hsa21-derived miRNAs are overexpressed in human DS pre-frontal cortex brain specimens. Mature Hsa21-derived miRNAs were quantified utilizing RT-PCR as previously described by our laboratory40,42,85,86 using total RNA isolated from human fetal (18–22 weeks of gestation), child (1–8 years), adolescent (9–19 years old) and adult (20–50 years old) prefrontal cortex specimens from control and DS (age- and sex-matched, n = 3–5) samples. Gene expression was calculated relative to 18S rRNA and the data are expressed as percent over control (variation between control samples was never greater than 5%), which was assigned a value of 100%. The error bars represent the average ± S.E. of triplicate samples repeated in at least three independent experiments (*p < 0.01 DS vs. control).
Hsa21-Derived miRNA/mRNA Target Prediction
Given that Ts21 results in the gene dosage overexpression of Hsa21-derived miRNAs, we now hypothesize, as a consequence, the expression levels of specific target proteins would be decreased (i.e., haploinsufficiency), which in turn could contribute to the features of the DS phenotype (i.e., increased incidence of cognitive dysfunction, congenital heart defects, childhood leukemia, early-onset Alzheimer's disease (AD) and decreased incidence of solid tumors and cardiovascular disease). To test this hypothesis putative Hsa21-derived miRNA/mRNA targets must be identified. Computational miRNA/mRNA target programs remain the only source for rapid prediction of miRNA recognition sites harbored within the 3′-UTR of target mRNAs. For example, TargetScan analysis (TargetScan 5.0, http://www.targetscan.org),43 predicted that miR-99a could potentially interact with 40 putative mRNA targets; let-7c, 819 putative mRNA targets; miR-125b-2, 604 putative mRNA targets; miR-155, 281 putative mRNA targets and miR-802, 232 putative mRNA targets (Table 1). Although Hsa21-derived miRNAs may regulate a large number of mRNA targets (i.e., >1,695), it is still possible to prioritize and validate the importance of a predicted target in mediating the DS phenotype, given that target protein expression should be attenuated in DS individuals.
Table 1.
Targetscan algorithm-predicted Hsa21-derived putative miRNA/mRNA targets
| Hsa-21-derived miRNA | Potential human targets* | Number of targets containing at least two potential binding sites | Number of targets containing at least three potential binding sites | Number of targets containing at least four potential binding sites | Number of targets containing at least five potential binding sites |
| let-7c | 819 | ||||
| miR-99a | 40 | ||||
| miR-125b-2 | 604 | ||||
| miR-155 | 281 | ||||
| miR-802 | 232 | ||||
| Total | 1695 | 541 | 194 | 33 | 0 |
Source: TargetScan (April, 2009). The number of potential targets is dramatically dependent upon the algorithm utilized.
Hsa21-Derived miRNAs and Cognitive Dysfunction
All people with DS have a mild-to-moderate learning disability.7–9 The cognitive deficiencies in DS caused by the triplication of Hsa21 genes, is mainly a consequence of developmental perturbations. Brains of DS individuals are characterized by neurological defects in cortex lamination and in cerebellar function (shape and volume).44 Morphological and functional defects in DS individuals have also been found at the cellular level determined by alteration in neurogenesis, neuronal differentiation, myelination, dendritogenesis and synaptogenesis.45–48 Given that Ts21 results in the gene dosage overexpression of all five Hsa21-derived miRNAs in the brain,40 these miRNAs may play a role in the developmental, morphological and functional defects in the brain and ultimately the cognitive impairments observed in DS individuals. Therefore, the capability of each of the individual 1,695 putative Hsa21-derived miRNA/mRNA targets in playing a role in neurodevelopment were prioritized based on the following criteria: first, is the mRNA target expressed in the brain?; second, does the protein encoded by this mRNA target play a role in some aspect of neurodevelopment?; third, are there any studies linking mutations in the gene of a given mRNA target to haploinsufficiency and problems in neurodevelopment (i.e., clinical relevance)?; and fourth, do any of these mRNA targets harbor multiple Hsa21-derived miRNA binding sites given that miRNA-mediated gene repression seems to be proportional to the number of miRNA recognition sites harbored in a given mRNA target (i.e., combinatorial miRNA inhibition)?49,50 Based on these criteria, we chose to investigate the transcription factor methyl-CpG-binding protein (MeCP2) mRNA as a potentially important Hsa21-derived miRNA target given that its 3′-UTR harbored two miR-155 and -802 and single miR-125b and let-7c, putative recognition sites.42 Additionally, MeCP2 was a potential miRNA target since it is highly expressed in neurons and has been shown to play a role in neurogenesis,51,52 a process that is abnormal in DS individuals.4,7 Finally, MeCP2 was a provocative clinical target since Rett syndrome (RS) is a neurodevelopmental disorder caused by mutations in the MECP2 gene.51,52
To validate that MeCP2 was indeed a true target of multiple Hsa21-derived miRNAs several independent experimental techniques were utilized. First, luciferase/target mRNA 3′-UTR reporter assays demonstrated that miR-155 and -802 could regulate MeCP2 protein expression levels by interacting with specific recognition sites harbored within its 3′-UTR.42 Second, miR-155 and -802 gain-of-function experiments demonstrated that MeCP2 mRNA and protein levels were attenuated and, conversely, miR-155 and -802 loss-of-function experiments demonstrated that MeCP2 mRNA and protein levels were augmented. These experiments suggested that MeCP2 mRNA was a target of both miR-155 and -802 and that these miRNAs markedly decreased MeCP2 expression by targeting MeCP2 mRNAs for degradation. Third, MeCP2 was under-expressed in DS brain specimens isolated from either humans or Ts65Dn mice (a widely used mouse model of DS53,54), suggesting that MeCP2 may also be a target of Hsa21-derived miRNAs in vivo. Fourth, by manipulating the concentration of MeCP2 in cell culture, transcriptionally-activated and -silenced MeCP2 target genes,55 Creb1 and Mef2c were also modulated.42 Fifth, CREB1 and MEF2C, were also aberrantly expressed in DS brain specimens isolated from either humans or Ts65Dn mice. Finally, in vivo silencing of miR-155 or -802 with antagomirs56,57 (chemically modified, cholesterol-conjugated, single-stranded RNA analogs complementary to miR-155 or -802) resulted in the normalization of the appropriate miRNA, MeCP2, CREB1 and MEF2C expression in Ts65Dn mice.42 Taken together these results suggest that Trisomy 21-induced, Hsa21-derived miR-155 and -802 overexpression directly inhibits MeCP2 expression which, in turn, leads to the aberrant expression of MeCP2-activated and -silenced target genes (e.g., Creb1 and Mef2c) in vivo.42 Importantly, these studies support the hypothesis that improper repression (haploinsufficiency) of MeCP2, secondary to trisomic overexpression of Hsa21-derived miRNAs, contribute in part, to the abnormalities in the neurochemistry observed in the brains of DS individuals.
Hsa21-Derived miRNAs and the DS Phenotype
Given that our results suggest that Ts21 gene dosage overexpression of Hsa21-derived miRNAs, play a role in mediating the cognitive dysfunction observed in individuals with DS, it is tempting to hypothesize that the dysregulation of these miRNAs may also contribute to the other observed DS phenotypes including increased incidence of congenital heart defects, childhood leukemia, early-onset AD and decreased incidence of solid tumors and cardiovascular disease. While this hypothesis has not been directly tested there are a number of studies that substantiate the view that Hsa21-derived miRNAs may play a role, in part, in most of the observed DS phenotypes.
In support of the finding that Hsa21-derived miRNAs may play a role in the leukemias observed in DS children,66 a subset of these miRNAs have been implicated in normal and pathologic hematopoiesis. For example, two of the most highly expressed miRNAs in pediatric AML (non-DS) were miR-125b and miR-99a.67 Notably, miR-155 was also detected at elevated levels in the bone marrow of some patients suffering acute myeloid leukemia (AML).68 Additionally, overexpression of miR-155 in hematopoietic stem cells in the mouse resulted in gross expansion of myeloid lineages in the bone marrow and peripheral blood at the expense of erythroid and lymphoid populations.68 Moreover, transduction of miR-155 in the mouse macrophage RAW264.7 cell line induces dysregulation of a large number of genes, including 89 genes that were predicted to be targets of miR-155 on the basis of in silico analysis. Importantly, ten of the predicted miR-155 target genes (e.g., PU.1 and C/EBPβ) are involved in hematopoietic and myeloproliferative disorders.69
To further corroborate our hypothesis that Hsa21-derived miRNAs may play a role in the early-onset Alzheimer's disease observed in DS individuals,7–9 miR-125b has been consistently reported to be upregulated in the cortex, hippocampus, cerebellum and the temporal lobe neocortex of AD (non-DS) affected tissue.70–72 The dysregulation of miR-125b in AD may be particularly important as this miRNA is abundantly expressed in the normal brain and is involved in brain development.73 Although experimentally validated miR-125b/mRNA targets that may play a role in mediating AD have not yet been identified, it is important to note that synapsin II, a neuron-specific phosphoprotein that selectively binds to small synaptic vesicles in the presynaptic nerve terminal, is a miR-125b predicted target (www.targetscan.org). Thus, the small increases in miR-125b observed in AD may have a bearing on the synaptic protein deficits previously observed in Alzheimer affected brains.74,75
In further support of our hypothesis that Hsa21-derived miRNAs may play a role in the reduced incidence of solid tumors observed in DS individuals,10–14 other investigators have demonstrated that miR-99a, let-7c and miR-125b may act as tumor suppressors given that homozygous deletion (HD) of the miR-99a, let-7c and miR-125b-2 cluster is frequently associated with lung cancers.76,77 Additional evidence demonstrated that members of the let-7 family (which includes let-7c) were poorly expressed or deleted in human lung tumors and, importantly, one of the experimentally validated targets that was silenced by let-7 was the RAS oncogene.78 Furthermore, many genes involved in cell proliferation were also silenced by let-7, including cyclins B1, E2, F, G2 and cell division cycle 2 (CDC2), CDC25 and CDC34.79 Interestingly, the exogenous delivery of let-7 prevented the formation of lung tumors from premalignant lesions and reduced tumors with activating RAS mutations.80 Additionally, the under-expression of miR-125b-2 also seems to play an important role in lung carcinogenesis, and one of its genetic targets includes the ERBB2 proto-oncogene that encodes the epidermal growth factor (EGF) receptor, which is highly expressed in carcinomas.81,82 Overexpression of miR-125a/b in an ERBB2-positive breast cancer cell line impaired cell growth and mobility capability,82 and frequent overexpression or amplification of ERBB2 was reported in ∼20% of serous ovarian cancers.83 Lastly, it has also been demonstrated that miR-99a, let-7c and miR-125b-2 expression is attenuated in prostate cancer.84
Finally, to substantiate the finding that Hsa21-derived miRNAs may play a role in the reduced incidence of cardiovascular disease observed in DS individuals,15–25 our laboratory has demonstrated that the human angiotensin II type 1 receptor (AT1R) mRNA is a miR-155 target.85,86 AT1R activation initiates a cascade of pathological events, including altered vascular tone, endothelial dysfunction, structural remodeling and vascular inflammation and it has been speculated that overproduction of the AT1R contributes to the development of cardiovascular disease.87 Interestingly, a silent single nucleotide polymorphism (SNP, +1166 A/C) in the human AT1R gene has been associated with essential hypertension,88,89 cardiac hypertrophy,90 aortic stiffness,91 myocardial infarction92 and increased oxidative stress levels in human heart failure.93 Additionally, it has been demonstrated that patients with the human AT1R CC genotype have enhanced vascular reactivity94–96 and increased renovascular sensitivity to Ang II.97 Initial miRNA experimental studies suggested that miR-155 could bind to the 3′-UTR of human AT1R mRNAs and translationally repress the expression of this protein in vivo.85 Computer alignment of the human AT1R 3′-UTR sequence with the miR-155 sequence demonstrated that the +1166 A/C SNP occurs within the miR-155 cis-regulatory binding site (Fig. 3). The interaction between miR-155 and the human AT1R 3′-UTR harboring the A-allele fulfills the seed sequence rule for miRNA/mRNA target interaction given that there is a 7 bp region of complementarity between the 5′ end of miR-155 and the human AT1R mRNA target site. In contrast, if a hAT1R mRNA that harbors the +1166 C-allele is expressed, the complementary seed site is interrupted and the thermodynamics of the miRNA:mRNA duplex would be significantly altered.86 Thus, the presence of the +1166 C-allele would decrease the ability of miR-155 to interact with the cis-regulatory site located in the hAT1R 3′-UTR and, as a consequence, it would be expected that aberrantly high levels of the human AT1R would be synthesized.
Figure 3.
The human AT1R +1166 A/C SNP occurs in the miR-155-binding site. (A) Complementarity between miR-155 and the hAT1R 3′-UTR site targeted (70–90 bp downstream from the human AT1R stop codon). The +1166 A/C SNP corresponds to the nucleotide 86 bp downstream from the human AT1R stop codon (shown in bold print). The binding of miR-155 to the hAT1R 3′-UTR target site fulfills the requirement of a 7 bp seed sequence of complementarity at the miRNA 5′ end when the +1166 A-allele is expressed. (B) Complementarity between miR-155 and the human AT1R 3′-UTR harboring the +1166 C-allele. If the +1166 C-allele is expressed, the seed sequence requirement would not be met and, as a consequence, it would be expected that human AT1R expression would be elevated.
Given that the human AT1R is an experimentally validated target of Hsa21-derived miR-155, it has been hypothesized that the AT1R expression would be attenuated in individuals with DS due to the Ts21-induced gene dosage overexpression of miR-155. In support of this hypothesis, Ts21-isolated fibroblasts (which express higher levels of miR-155), showed reduced protein levels of the human AT1R when compared with that of euploid fibroblasts, which express lower levels of miR-155.39 Taken together, these studies suggest that one possible explanation for the observed lack of cardiovascular disease in DS individuals may be due, in part, to the Ts21 gene dosage overexpression of miR-155 and the subsequent under-expression of the AT1R.
Conclusion
Dysfunctional miRNA-mediated regulation has been implicated in the pathogenesis of many disease states. Given that Hsa21-derived miRNAs (miR-99a, let-7c, miR-125b-2, miR-155 and miR-802) may be involved in regulating either a single gene or sets of genes in common and divergent genetic networks, bioinformatics and systems biology tools will have to be employed to assist in the delineation of regulatory networks and feedback loops, by integrating signaling and transcription factors and identifying the presence of regulatory elements and links with other miRNAs. This line of investigation may result in the development of therapeutics that would benefit individuals with DS as well as individuals in the general population that suffer from aberrant brain development, congenital heart disease, childhood leukemia, Alzheimer's disease, cardiovascular disease and solid tumors.
Acknowledgements
T.S.E. acknowledges financial support from the NIH (HL48848, HD058997) and the Fondation Jerome Lejeune. We extend our apologies to any colleagues whose contributions to DS and miRNA studies might have been inadvertently overlooked in this review.
Abbreviations
- DS
down syndrome
- Ts21
trisomy 21
- Hsa21
human chromosome 21
- miRNA
microRNA
- 3′-UTR
3′-untranslated region
- AD
alzheimer's disease
- miRISCs
miRNA-induced silencing complexes
- LNA
locked nucleic acid
- MeCP2
methyl-CpG-binding protein
- RS
rett syndrome
- AML
acute myeloid leukemia
- HD
homozygous deletion
- CDC2
cell division cycle 2
- EGF
epidermal growth factor
- AT1R
angiotensin II type 1 receptor
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/12685
References
- 1.LeJeune J, Gautier M, Turpin R. Study of somatic chromosomes from mongoloid children. Comptes Renus de l'Academic les Sciences. 1959;248:1721–1722. [PubMed] [Google Scholar]
- 2.Muller F, Rebiffe M, Taillandier A, Oury JF, Mornet E. Parental origin of the extra chromosome in prenatally diagnosed fetal trisomy 21. Hum Genet. 2000;106:340–344. doi: 10.1007/s004390051047. [DOI] [PubMed] [Google Scholar]
- 3.Mutton D, Alberman E, Hook EB. Cytogenetic and epidemiological findings in Down syndrome. England and Wales 1989–1993. National Down Syndrome Cytogenetic Register and the Association of Clinical Cytogeneticists. J Med Genet. 1996;33:387–394. doi: 10.1136/jmg.33.5.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hassold T, Hunt P. To err (meiotically) is human: The genesis of human aneuploidy. Nat Rev Genet. 2001;2:280–291. doi: 10.1038/35066065. [DOI] [PubMed] [Google Scholar]
- 5.Canfield MA, Honein MA, Yuskiv N, Xing J, Mai CT, Collins JS, et al. National estimates and race/ethnic-specific variation of selected birth defects in the United States 1999–2001. Birth Defects Res A Clin Mol Teratol. 2006;76:747–756. doi: 10.1002/bdra.20294. [DOI] [PubMed] [Google Scholar]
- 6.Allen EG, Freeman SB, Druschel C, Hobbs CA, O'Leary LA, Romitti PA, et al. Maternal age and risk for trisomy 21 assessed by the origin of chromosome nondisjunction: a report from the Atlanta and National Down Syndrome Projects. Hum Genet. 2009;125:41–52. doi: 10.1007/s00439-008-0603-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Epstein CJ. The Consequences of Chromosome Imbalance: Principles, Mechanism and Models. New York, NY: Cambridge University Press; 1986. pp. 253–259. [Google Scholar]
- 8.Korenberg J. Down syndrome phenotype mapping. In: Epstein C, editor. Progress in Clinical and Biological Research. New York NY: Wiley-Liss; 1991. pp. 43–52. [Google Scholar]
- 9.Roizen NJ, Patterson D. Down's syndrome. Lancet. 2003;361:1281–1289. doi: 10.1016/S0140-6736(03)12987-X. [DOI] [PubMed] [Google Scholar]
- 10.Hasle H, Clemmensen IH, Mikkelsen M. Risks of leukaemia and solid tumours in individuals with Down's syndrome. Lancet. 2000;355:165–169. doi: 10.1016/S0140-6736(99)05264-2. [DOI] [PubMed] [Google Scholar]
- 11.Yang Q, Rasmussen SA, Friedman JM. Mortality associated with Down's syndrome in the USA from 1983 to 1997: a population-based study. Lancet. 2002;359:1019–1025. doi: 10.1016/s0140-6736(02)08092-3. [DOI] [PubMed] [Google Scholar]
- 12.Satge D. A decreased incidence of neuroblastomas in Down syndrome and over-production of S-100 b protein: a hypothesis. Med Hypotheses. 1996;46:393–399. doi: 10.1016/s0306-9877(96)90193-0. [DOI] [PubMed] [Google Scholar]
- 13.Satge D, Sasco AJ, Azema B, Culine S. Mental retardation research focus. New York NY: Nova Sciences Publisher; 2007. Cancers in persons with intellectual deficiency: current data; pp. 47–84. [Google Scholar]
- 14.Benard J, Beron-Gaillard N, Satge D. Down's syndrome protects against breast cancer: is a constitutional cell microenvironment the key? Int J Cancer. 2005;113:168–170. doi: 10.1002/ijc.20532. [DOI] [PubMed] [Google Scholar]
- 15.Murdoch JC, Rodger JC, Rao SS, Fletcher CD, Dunnigan MG. Down's syndrome: an atheroma-free model? Br Med J. 1977;2:226–228. doi: 10.1136/bmj.2.6081.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yla-Herttuala S, Luoma J, Nikkari T, Kivimaki T. Down's syndrome and atherosclerosis. Atherosclerosis. 1989;76:269–272. doi: 10.1016/0021-9150(89)90110-x. [DOI] [PubMed] [Google Scholar]
- 17.Morrison RA, McGrath A, Davidson G, Brown JJ, Murray GD, Lever AF. Low blood pressure in Down's syndrome, a link with Alzheimer's disease? Hypertension. 1996;28:569–575. doi: 10.1161/01.hyp.28.4.569. [DOI] [PubMed] [Google Scholar]
- 18.Kapell D, Nightingale B, Rodriguez A, Lee JH, Zigman WB, Schupf N. Prevalence of chronic medical conditions in adults with mental retardation: Comparison with the general population. Mental Retardation. 1998;36:269–279. doi: 10.1352/0047-6765(1998)036<0269:POCMCI>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 19.Draheim CC, McCubbin JA, Williams DP. Differences in cardiovascular disease risk between nondiabetic adults with mental retardation with and without Down syndrome. Am J Ment Retard. 2002;107:201–211. doi: 10.1352/0895-8017(2002)107<0201:DICDRB>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 20.Greene AK, Kim S, Rogers GF, Fishman SJ, Olsen BR, Mulliken JB. Risk of vascular anomalies with Down syndrome. Pediatrics. 2008;121:135–140. doi: 10.1542/peds.2007-1316. [DOI] [PubMed] [Google Scholar]
- 21.Baird PA, Sadovnick AD. Causes of death to age 30 in Down syndrome. Am J Hum Genet. 1988;43:239–238. [PMC free article] [PubMed] [Google Scholar]
- 22.Brattstrom L, England E, Brun A. Does Down syndrome support homocysteine theory of arteriosclerosis? Lancet. 1987;1:391–392. doi: 10.1016/s0140-6736(87)91772-7. (1987) [DOI] [PubMed] [Google Scholar]
- 23.Pueschel SM, Craig WY, Haddow JE. Lipids and lipoproteins in persons with Down's syndrome. J Intellect Disabil Res. 1992;36:365–369. doi: 10.1111/j.1365-2788.1992.tb00535.x. [DOI] [PubMed] [Google Scholar]
- 24.Licastro F, Marocchi A, Penco S, Porcellini E, Lio D, Dogliotti G, et al. Does Down's syndrome support the homocysteine theory of atherogenesis? Experience in elderly subjects with trisomy 21. Arch Gerontol Geriatr. 2006;43:381–387. doi: 10.1016/j.archger.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 25.Goi G, Baquero-Herrera C, Licastro F, Dogliotti G, Corsi MM. Advanced oxidation protein products (AOPP) and high-sensitive C-reactive protein (hs-CRP) in an ‘atheroma-free model’: Down's syndrome. Intern J Cardiol. 2006;113:427–429. doi: 10.1016/j.ijcard.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 26.Delabar JM, Theophile D, Rahmani Z, Chettouh Z, Blouin JL, Prieur M, et al. Molecular mapping of twenty-four features of Down syndrome on chromosome 21. Eur J Hum Genet. 1993;1:114–124. doi: 10.1159/000472398. [DOI] [PubMed] [Google Scholar]
- 27.Korenberg JR, Chen XN, Schipper R, Sun Z, Gonsky R, Gerwehr S, et al. Down syndrome phenotypes: the consequences of chromosomal imbalance. Proc Natl Acad Sci USA. 1994;91:4997–5001. doi: 10.1073/pnas.91.11.4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ait-Yahya-Graison E, Aubert J, Dauphinot L, Rivals I, Prieur M, Golfier G, et al. Classification of human chromosome 21 gene-expression variations in Down syndrome: impact on disease phenotypes. Am J Hum Genet. 2007;81:475–491. doi: 10.1086/520000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deutsch S, Lyle R, Dermitzakis ET, Attar H, Subrahmanyan L, Gehrig C, et al. Gene expression variation and expression quantitative trait mapping of human chromosome 21 genes. Human Mol Genet. 2005;14:3741–3749. doi: 10.1093/hmg/ddi404. [DOI] [PubMed] [Google Scholar]
- 30.Prandini P, Deutsch S, Lyle R, Gagnebin M, Delucinge VC, Delorenzi M, et al. Natural gene-expression variation in Down syndrome modulates the outcome of gene—dosage imbalance. Am J Hum Genet. 2007;81:252–263. doi: 10.1086/519248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sultan M, Piccini I, Balzereit D, Herwig R, Saran NG, Lehrach H, et al. Gene expression variation in Down's syndrome mice allows prioritization of candidate genes. Genome Biol. 2007;8:91. doi: 10.1186/gb-2007-8-5-r91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bushati N, Cohen SM. microRNA functions. Annu. Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 33.Bartel DP. MicroRNAs: Target Recognition and Regulatory Functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gardiner G, Costa ACS. The proteins of human chromosome 21. Am J Med Genet. 2006;142:196–205. doi: 10.1002/ajmg.c.30098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kahlem P. Gene-dosage effect on chromosome 21 transcriptome in trisomy 21: implication in Down Syndrome cognitive disorders. Behav Genet. 2006;36:416–428. doi: 10.1007/s10519-006-9053-z. [DOI] [PubMed] [Google Scholar]
- 36.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:154–158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eis PS, Tam W, Sun L, Chadburn A, Li Z, Gomez MF, et al. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci USA. 2005;102:3627–3632. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okamura K, Phillips MD, Tyler DM, Duan H, Chou YT, Lai EC. The regulatory activity of microRNA* species has substantial influence on microRNA and 3′ UTR evolution. Nat Struct Mol Biol. 2008;15:354–363. doi: 10.1038/nsmb.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sethupathy P, Borel C, Gagnebin M, Grant GR, Deutsch S, Elton TS, et al. Human microRNA-155 on chromosome 21 differentially interacts with its polymorphic target in the AGTR1 3′ untranslated region: A mechanism for functional single-nucleotide polymorphisms related to phenotypes. Am J Hum Genet. 2007;81:405–413. doi: 10.1086/519979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuhn DE, Nuovo GJ, Martin MM, Malana GE, Pleister AP, Jiang J, et al. Human chromosome 21-derived miRNAs are overexpressed in down syndrome brains and hearts. Biochem Biophys Res Commun. 2008;370:473–477. doi: 10.1016/j.bbrc.2008.03.120. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41.Yamada H, Yanagisawa K, Tokumaru S, Taguchi A, Nimura Y, Osada H, et al. Detailed characterization of a homozygously deleted region corresponding to a candidate tumor suppressor locus at 21q11-21 in human lung cancer. Genes Chromosomes Cancer. 2008;47:810–818. doi: 10.1002/gcc.20582. [DOI] [PubMed] [Google Scholar]
- 42.Kuhn DE, Nuovo GJ, Terry A, Jr, Martin MM, Malana GE, Sansom SE, et al. Chromosome 21-derived mirnas provide an etiological basis for aberrant protein expression in human down syndrome brains. J Biol Chem. 2010;285:1529–1543. doi: 10.1074/jbc.M109.033407. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of.microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dierssen M, Herault Y, Estivill X. Aneuploidy: from a physiological mechanism of variance to Down syndrome. Physiol Rev. 2009;89:887–920. doi: 10.1152/physrev.00032.2007. [DOI] [PubMed] [Google Scholar]
- 45.White NS, Alkire MT, Haier RJ. A voxel-based morphometric study of nondemented adults with Down Syndrome. Neuroimage. 2003;20:393–403. doi: 10.1016/s1053-8119(03)00273-8. [DOI] [PubMed] [Google Scholar]
- 46.Teipel SJ, Schapiro MB, Alexander GE, Krasuski JS, Horwitz B, Hoehne C, et al. Relation of corpus callosum and hippocampal size to age in nondemented adults with Down's Syndrome. Am J Psychiatry. 2003;160:1870–1878. doi: 10.1176/appi.ajp.160.10.1870. [DOI] [PubMed] [Google Scholar]
- 47.Aylward EH, Li Q, Honeycutt NA, Warren AC, Pulsifer MB, Barta PE, et al. MRI volumes of the hippocampus and amygdala in adults with Down's syndrome with and without dementia. Am J Psychiatry. 1999;156:564–568. doi: 10.1176/ajp.156.4.564. [DOI] [PubMed] [Google Scholar]
- 48.Pinter JD, Brown WE, Eliez S, Schmitt JE, Capone GT, Reiss AL. Amygdala and hippocampal volumes in children with Down syndrome: a high-resolution MRI study. Neurology. 2001;56:972–974. doi: 10.1212/wnl.56.7.972. [DOI] [PubMed] [Google Scholar]
- 49.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 50.Hon LS, Zhang A. The roles of binding site arrangement and combinatorial targeting in microRNA repression of gene expression. Genome Biology. 2007;8:166. doi: 10.1186/gb-2007-8-8-r166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kaufmann WE, Johnston MV, Blue ME. MeCP2 expression and function during brain development: implications for Rett syndrome's pathogenesis and clinical evolution. Brain & Develop. 2005;27:77–87. doi: 10.1016/j.braindev.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 52.Bienvenu T, Chelly J. Molecular genetics of Rett syndrome: when DNA methylation goes unrecognized. Nature Rev Genet. 2006;7:415–426. doi: 10.1038/nrg1878. [DOI] [PubMed] [Google Scholar]
- 53.Reeves R, Irvin N, Moran T, Wohn A, Kitt C, Sisodia S, et al. A mouse model for Down syndrome exhibits learning and behaviour deficits. Nat Genet. 1995;11:177–183. doi: 10.1038/ng1095-177. [DOI] [PubMed] [Google Scholar]
- 54.Reeves RH. Down syndrome mouse models are looking up. Trends Mol Med. 2006;12:237–240. doi: 10.1016/j.molmed.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 55.Chahrour M, Jung SY, Shaw C, Zhou X, Wong ST, Qin J, et al. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 57.Krützfeldt J, Kuwajima S, Braich R, Rajeev KG, Pena J, Tuschl T, et al. Specificity, duplex degradation and subcellular localization of antagomirs. Nucleic Acids Res. 2007;35:2885–2828. doi: 10.1093/nar/gkm024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schwarz DS, Hutvágner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 59.Freeman SB, Taft LF, Dooley KJ, Allran K, Sherman SL, Hassold TJ, et al. Population-based study of congenital heart defects in Down syndrome. Am J Med Gen. 1998;80:213–217. [PubMed] [Google Scholar]
- 60.Abbag FI. Congenital heart diseases and other major anomalies in patients with Down syndrome. Saudi Med J. 2006;27:219–222. [PubMed] [Google Scholar]
- 61.Vis JC, Duffels MG, Winter MM, Weijerman ME, Cobben JM, Huisman SA, et al. Down syndrome: a cardiovascular perspective. J Intellect Disabil Res. 2009;53:419–425. doi: 10.1111/j.1365-2788.2009.01158.x. [DOI] [PubMed] [Google Scholar]
- 62.Bruneau BG. The developmental genetics of congenital heart disease. Nature. 2008;451:943–948. doi: 10.1038/nature06801. [DOI] [PubMed] [Google Scholar]
- 63.Sayed D, Hong C, Chen IY, Lypowy J, Abdellatif M. MicroRNAs play an essential role in the development of cardiac hypertrophy. Circ Res. 2007;100:416–424. doi: 10.1161/01.RES.0000257913.42552.23. [DOI] [PubMed] [Google Scholar]
- 64.Tatsuguchi M, Seok HY, Callis TE, Thomson JM, Chen JF, Newman M, et al. Expression of microRNAs is dynamically regulated during cardiomyocyte hypertrophy. J Mol Cell Cardiol. 2007;42:1137–1141. doi: 10.1016/j.yjmcc.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thum T, Galuppo P, Wolf C, Fiedler J, Kneitz S, van Laake LW, et al. MicroRNAs in the human heart: a clue to fetal gene reprogramming in heart failure. Circulation. 2007;116:258–267. doi: 10.1161/CIRCULATIONAHA.107.687947. [DOI] [PubMed] [Google Scholar]
- 66.Taub JW. Relationship of chromosome 21 and acute leukemia in children with Down syndrome. J Pediatr Hematol Oncol. 2001;23:175–178. doi: 10.1097/00043426-200103000-00012. [DOI] [PubMed] [Google Scholar]
- 67.Zhang H, Luo XQ, Zhang P, Huang LB, Zheng YS, Wu J, et al. MicroRNA patterns associated with clinical prognostic parameters and CNS relapse prediction in pediatric acute leukemia. PLoS One. 2009;4:7826. doi: 10.1371/journal.pone.0007826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.O'Connell RM, Rao DS, Chaudhuri AA, Boldin MP, Taganov KD, Nicoll J, et al. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J Exp Med. 2008;205:585–594. doi: 10.1084/jem.20072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Metcalf D, Dakic A, Mifsud S, Di Rago L, Wu L, Nutt S. Inactivation of PU.1 in adult mice leads to the development of myeloid leukaemia. Proc Natl Acad Sci USA. 2006;103:1486–1491. doi: 10.1073/pnas.0510616103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lukiw WJ. Micro-RNA speciation in fetal, adult and Alzheimer's disease hippocampus. Neuroreport. 2007;18:297–300. doi: 10.1097/WNR.0b013e3280148e8b. [DOI] [PubMed] [Google Scholar]
- 71.Cogswell JP, Ward J, Taylor IA, Waters M, Shi YL, Cannon B, et al. Identification of miRNA changes in Alzheimer's disease brain and CSF yields putative biomarkers and insights into disease pathways. J Alzheimers Disease. 2008;14:27–41. doi: 10.3233/jad-2008-14103. [DOI] [PubMed] [Google Scholar]
- 72.Sethi P, Lukiw WJ. Micro-RNA abundance and stability in human brain: specific alterations in Alzheimer's disease temporal lobe neocortex. Neurosci Lett. 2009;459:100–104. doi: 10.1016/j.neulet.2009.04.052. [DOI] [PubMed] [Google Scholar]
- 73.Wu L, Belasco JG. Micro-RNA regulation of the mammalian lin-28 gene during neuronal differentiation of embryonal carcinoma cells. Mol Cell Biol. 2005;25:9198–9208. doi: 10.1128/MCB.25.21.9198-9208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lukiw WJ, Rogaev EI, Bazan NG. Synaptic and cytoskeletal RNA message levels in sporadic Alzheimer neocortex. Alzheimer's Res. 1996;2:221–227. [Google Scholar]
- 75.Lukiw WJ. Gene expression profiling in fetal, aged and Alzheimer hippocampus: a continuum of stress-related signaling. Neurochem Res. 2004;29:1287–1297. doi: 10.1023/b:nere.0000023615.89699.63. [DOI] [PubMed] [Google Scholar]
- 76.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancer. Proc Natl Acad Sci USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nagayama K, Kohno T, Sato M, Arai Y, Minna JD, Yokota J. Homozygous deletion scanning of the lung cancer genome at a 100-kb resolution. Genes Chromosomes Cancer. 2007;46:1000–1010. doi: 10.1002/gcc.20485. [DOI] [PubMed] [Google Scholar]
- 78.Johnson CD, Esquela-Kerscher A, Stefani G, Byrom M, Kelnar K, Ovcharenko D, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67:7713–7722. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- 79.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 80.Esquela-Kerscher A, Slack FJ. Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 81.Fujimoto N, Wislez M, Zhang J, Iwanaga K, Dackor J, Hanna AE, et al. High expression of ErbB family members and their ligands in lung adenocarcinomas that are sensitive to inhibition of epidermal growth factor receptor. Cancer Res. 2005;65:11478–11485. doi: 10.1158/0008-5472.CAN-05-1977. [DOI] [PubMed] [Google Scholar]
- 82.Scott GK, Goga A, Bhaumik D, Berger CE, Sullivan CS, Benz CC. Coordinate suppression of ERBB2 and ERBB3 by enforced expression of micro-RNA miR-125a or miR-125b. J Biol Chem. 2007;282:1479–1486. doi: 10.1074/jbc.M609383200. [DOI] [PubMed] [Google Scholar]
- 83.Lassus H, Leminen A, Vayrynen A, Cheng G, Gustafsson JA, Isola J, et al. ERBB2 amplification is superior to protein expression status in predicting patient outcome in serous ovarian carcinoma. Gynecol Oncol. 2004;92:31–39. doi: 10.1016/j.ygyno.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 84.Ozen M, Creighton CJ, Ozdemir M, Ittmann M. Widespread deregulation of microRNA expression in human prostate cancer. Oncogene. 2008;27:1788–1793. doi: 10.1038/sj.onc.1210809. [DOI] [PubMed] [Google Scholar]
- 85.Martin MM, Lee EJ, Buckenberger JA, Schmittgen TD, Elton TS. MicroRNA-155 regulates human angiotensin II type 1 receptor expression in fibroblasts. J Biol Chem. 2006;281:18277–84. doi: 10.1074/jbc.M601496200. [DOI] [PubMed] [Google Scholar]
- 86.Martin MM, Buckenberger JA, Jiang J, Malana GE, Nuovo GJ, Chotani M, et al. The human angiotensin II type 1 receptor +1166 A/C polymorphism attenuates microrna-155 binding. J Biol Chem. 2007;282:24262–24269. doi: 10.1074/jbc.M701050200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 87.Billet S, Aguilar F, Baudry C, Clauser E. Role of angiotensin II AT1 receptor activation in cardiovascular diseases. Kidney Int. 2008;74:1379–1384. doi: 10.1038/ki.2008.358. [DOI] [PubMed] [Google Scholar]
- 88.Bonnardeaux A, Davies E, Jeunemaitre X, Fery I, Charru A, Clauser E, et al. Angiotensin II type 1 receptor gene polymorphisms in human essential hypertension. Hypertension. 1994;24:63–69. doi: 10.1161/01.hyp.24.1.63. [DOI] [PubMed] [Google Scholar]
- 89.Wang WYS, Zee RYL, Morris BJ. Association of angiotensin II type 1 receptor gene polymorphism with essential hypertension. Clin Genet. 1997;51:31–34. doi: 10.1111/j.1399-0004.1997.tb02410.x. [DOI] [PubMed] [Google Scholar]
- 90.Osterop APRM, Kofflard MJM, Sandkuiji LA, ten Cate FJ, Krams R, Schalekamp MADH, et al. AT1 receptor A/C1166 polymorphism contributes to cardiac hypertrophy in subjects with hypertrophic cardiomyopathy. Hypertension. 1998;32:825–830. doi: 10.1161/01.hyp.32.5.825. [DOI] [PubMed] [Google Scholar]
- 91.Benetos A, Gautier S, Ricard S, Topouchian J, Asmar R, Poirier O, et al. Influence of angiotensin-converting enzyme and angiotensin II type 1 receptor gene polymorphisms on aortic stiffness in normotensive and hypertensive patients. Circulation. 1996;94:698–703. doi: 10.1161/01.cir.94.4.698. [DOI] [PubMed] [Google Scholar]
- 92.Tiret L, Bonnardeax A, Pourier O, Ricard S, Marques-Vidal P, Evans A, et al. Synergistic effects of angiotensin-converting enzyme and angiotensin-II type 1 receptor gene polymorphisms on risk of myocardial infarction. Lancet. 1994;344:910–913. doi: 10.1016/s0140-6736(94)92268-3. [DOI] [PubMed] [Google Scholar]
- 93.Cameron VA, Mocatta TJ, Pilbrow AP, Frampton CM, Troughton RW, Richards AM, et al. Angiotensin type-1 receptor A1166C gene polymorphism correlates with oxidative stress levels in human heart failure. Hypertension. 2006;47:1155–1161. doi: 10.1161/01.HYP.0000222893.85662.cd. [DOI] [PubMed] [Google Scholar]
- 94.Amant C, Hamon M, Bauters C, Richard F, Helbecque N, McFadden EP, et al. The angiotensin II type 1 receptor gene polymorphism is associated with coronary artery vasoconstriction. J Am Coll Cardiol. 1997;29:486–490. doi: 10.1016/s0735-1097(96)00535-9. [DOI] [PubMed] [Google Scholar]
- 95.Henrion D, Amant C, Benessiano J, Philip I, Plantefeve G, Chatel D, et al. Angiotensin II type 1 receptor gene polymorphism is associated with an increased vascular reactivity in the human mammary artery in vitro. J Vasc Res. 1998;35:356–362. doi: 10.1159/000025605. [DOI] [PubMed] [Google Scholar]
- 96.van Geel PP, Pinto YM, Voors AA, Buikema H, Oosterga M, Crijns HJGM, et al. Angiotensin II type 1 receptor A1166C gene polymorphism is associated with an increased response to angiotensin II in human arteries. Hypertension. 2000;35:717–721. doi: 10.1161/01.hyp.35.3.717. [DOI] [PubMed] [Google Scholar]