Abstract
Long noncoding RNAs (lncRNAs) are pervasively transcribed and critical regulators of the epigenome.1,2 These long, polyadenylated RNAs do not code for proteins, but function directly as RNAs, recruiting chromatin modifiers to mediate transcriptional changes in processes ranging from X-inactivation (XIST) to imprinting (H19).3 The recent discovery that lncRNA HOTAIR can link chromatin changes to cancer metastasis4 furthers the relevance of lncRNAs to human disease. Here, we discuss lncRNAs as regulatory modules and explore the implications for disease pathogenesis.
Key words: lncRNA, HOTAIR, chromatin, metastasis, PRC2
Although large-scale analyses of mammalian transcriptomes have revealed that more than 50% of transcripts have no protein coding potential,2,5,6 the functions of these putative transcripts are largely unknown. A subset of these noncoding transcripts are termed long noncoding RNAs (lncRNAs), based on an arbitrary minimum length of 200 nucleotides. LncRNAs are roughly classified based on their position relative to protein-coding genes: intergenic (between genes), intragenic/intronic (within genes) and antisense.2 Initial efforts to characterize these molecules demonstrated that they function in cis, regulating their immediate genomic neighbors. Examples include AIR, XIST and Kcnq1ot (reviewed in ref. 1, 7 and 8), which recruit chromatin modifying complexes to silence adjacent sites. The scope of lncRNAs in gene regulation was advanced with the finding that lncRNA HOTAIR exhibited trans regulatory capacities.
HOTAIR is transcribed at the intersection of opposing chromatin domains in the HOXC locus, but targets Polycomb Repressive Complex 2 (PRC2) to silence 40 kilobases of HOXD,9 a locus involved in developmental patterning. A subsequent study revealed that HOTAIR is overexpressed in approximately one quarter of human breast cancers, directing PRC2 to approximately 800 ectopic sites in the genome, which leads to histone H3 lysine 27 trimethylation and changes in gene expression.4 The impacts of lncRNA-mediated chromatin changes are noteworthy: not only did HOTAIR drive metastasis in a mouse model, but HOTAIR expression in human breast cancer was found to be an independent prognostic marker for death and metastasis.4 The fact that HOTAIR drives chromatin reprogramming genome-wide suggests that long-range regulation by lncRNAs may be a widespread mechanism. This is supported by a study showing that >20% of tested lncRNAs are bound by PRC2 and other chromatin modifiers.10 Furthermore, this is an underestimate of the total RNAs involved in chromatin modification, as PRC2 target genes also transcribe smaller 50–200 nt RNAs that interact with SUZ12 to mediate gene repression.11 These findings provoke questions regarding the initial triggers for HOTAIR overexpression and whether understanding of lncRNA mechanics may have clinical relevance.
Long Noncoding RNAs and Disease
The association of HOTAIR with cancer metastasis adds to a growing cohort of lncRNAs associated with disease.12 For example, although the blepharophimosis syndrome (BPES) is driven by dysregulation of the FOXL2 gene, numerous extragenic mutations have been reported in patients.13 One particular deletion occurring 283 kb away from FOXL2 disrupts a lncRNA, PISRT1, that was shown by chromatin confirmation capture to physically loop with FOXL2.14 Another example is the lncRNA EVF2, which recruits the transcription factor Dlx2 to activate the protein coding genes DLX5 and DLX6 that are associated with the Split Hand/Split Foot malformation disorder.15,16 Importantly, none of the known disease mutations reside within the protein coding loci, suggesting that disruption of noncoding transcripts may initiate pathogenesis. A third example is the BC200 RNA, a primate brain transcript that is reduced by 70% in Alzheimer's brain tissues.17 A role of BC200 in neurological disorders is further supported by its direct interaction with the fragile X mental retardation protein (FMRP),18 which is selectively lost in the majority of fragile X patients.19
Functional analysis of a noncoding locus within 9p21 that coincides with several exons of the 4 kb lncRNA ANRIL has bolstered a role for lncRNAs in disease. Despite a lack of protein coding genes, sequence polymorphisms in this 58 kb region are associated with coronary artery disease,20 including two SNPs within exons of ANRIL.21 Targeted deletion of the orthologous 70 kb region in a mouse model altered cardiac transcript levels of neighboring genes Cdkn2A and Cdkn2b and resulted in aberrant cell proliferation.22 Thus, an lncRNA locus may provide a mechanistic link between a disease polymorphism and its associated phenotype.
Altogether, the long range regulation of mRNAs by noncoding sequences appears to be a reoccurring theme in disease development. Yet undiscovered lncRNAs may underlie the functional significance of unexplained disease polymorphisms and expands the catalogue of potential “first-hits” in pathogenesis. For example, one avenue to explore would be whether the gross overexpression of HOTAIR in metastastic tumors can be explained by mutations of the noncoding gene.
Mechanisms for Targeting of Long Noncoding RNAs
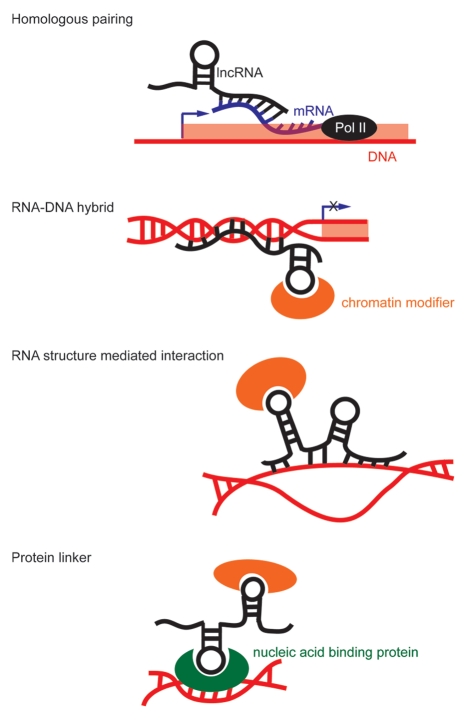
The role of lncRNAs in disease processes creates an urgency to understand the mechanisms by which these RNAs seek their targets. The earliest lncRNAs suggested a simplistic model where the RNA remains tethered to the site of origin to regulate transcriptional changes in cis. One example is an lncRNA upstream of the CCND1 promoter that recruits the RNA binding protein TLS to mediate heterochromatin formation.23 However, with trans acting RNAs such as HOTAIR affecting genome wide chromatin changes, it is clear that additional targeting mechanisms must be involved. The extensive sequence space available to lncRNAs provide plausible strategies for highly discriminative binding to the genome in an allele- or gene-specific fashion (reviewed in ref. 3). Possible RNA targeting schemes include the following (see fig. 1):
Figure 1.
Possible lncRNA targeting mechanisms.
Sequence-specific recognition: RNA-RNA.
Global RNA targeting may occur through direct sequence homology, a mechanism that is common for antisense lncRNAs such as p15AS24 and an RNA antisense to CDKN1A.25 A skewed equilibrium between sense and antisense transcripts can lead to disease, as seen in vascular anomalies tissues with altered ratios of TIE-1 mRNA to TIE1-AS lncRNA.26 Since as many as 70% of transcripts have antisense partners,27 antisense regulation is likely to be a widespread phenomenon.
RNA-DNA hybrids.
Sequence complementarity can also be employed in more complex configurations such as RNA-DNA duplexes and triplexes. An example occurs at the DHFR locus, where an lncRNA forms a triplex with the promoter to mediate sequence-specific transcriptional repression.28 In the case of XIST, a lncRNA that spreads over 150 Mb of the inactive X chromosome to mediate gene silencing, there is also evidence that genomic sequence plays a role in the silencing function of the RNA, albeit in a non homologous fashion. By identifying DNA features unique to genes that undergo or escape inactivation, the inactivation status of 80% of the genes could be predicted based on sequence alone.29 This example suggests that in a situation where lncRNA-mediated silencing is nearly compulsory, genomic sequence factors can still confer specificity to the targets. However, despite having identical genomic sequences, there is heterogeneity in the inactivation profile of different fibroblast cell lines,30 suggesting that DNA sequence alone is not sufficient to guide the lncRNA complexes.
Structure-mediated interactions.
The ability of RNA molecules to form secondary and tertiary structures enables more complex schemes for lncRNA targeting. First, base-pairing and looping within an RNA molecule may connect distant sequences to create a binding module that is not evident by primary sequence. Secondly, lncRNAs appear to form secondary structure configurations that mediate their functions. For example, repA RNA forms a duplex of four double hairpin repeats that mediates binding to PRC2.31,32 Similarly, Gas5 lncRNA serves as a decoy for the Glucocorticoid Receptor to titrate it away from its DNA binding sites,33 and a mutation disrupting a hairpin structure inhibits this activity. The importance of secondary structure can be seen when comparing two RNAs, human Alu and mouse B2. Although they have no obvious sequence homology, both are transacting RNAs that sequester RNA polymerase II from initiating transcription.34,35 Indeed, thousands of mouse and human transcripts with no primary sequence conservation share commonalities in RNA structure.36 Thus, it is possible that a structure-mediated mechanism may underlie the specificity of lncRNA targeting.
Protein-mediated interactions.
Nearly a fourth of known human proteins have nucleic acid binding domains,37 so it is possible that proteins may link lncRNAs to target loci. The telomere complex is a prime model for proteins serving as adapters between RNAs and DNAs:38,39 the ribonucleoprotein hnRNP A2 binds both telomerase RNA40 and telomeric DNA repeats.41 Likewise, the telomere repeat factor TRF2 forms a stable complex with telomere-repeat-encoding RNA (TERRA) and telomere DNA repeats.42 The strategies used to probe these interactions should be applied to the lncRNA field to determine whether these formations occur beyond the telomere.
Altogether, the diversity and abundance of noncoding transcripts suggests that several permutations of the aforementioned mechanisms may exist. In the early examples of cis regulatory RNAs, it was difficult to distinguish lncRNA targeting from mere diffusion of the RNAs. For example, the mechanism by which the XIST RNA spreads across the entire X chromosome is still undefined. The recent analysis of HOTAIR thus provides a unique avenue to explore requirements for lncRNA targeting. Specifically, analysis of sequence elements associated with the 800+ genes affected by HOTAIR overexpression may identify features that guide these RNAs. Future analysis of ectopic protein localization mediated by other disease-associated RNAs would further enhance our understanding of ncRNAs in disease. As the mechanics of lncRNA localization become elucidated, we may eventually develop strategies to interfere with their targeting, thus blocking the epigenetic reprogramming that contributes to diseases such as cancer.
Acknowledgements
We thank members of the Chang lab for discussion. Supported by National Science Foundation, American Society for Engineering Education (T.H.), National Institutes of Health (H.Y.C.) and California Institute for Regenerative Medicine (H.Y.C.). H.Y.C. is an Early Career Scientist of the Howard Hughes Medical Institute.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/13216
References
- 1.Bernstein E, Allis CD. RNA meets chromatin. Genes Dev. 2005;19:1635–1655. doi: 10.1101/gad.1324305. [DOI] [PubMed] [Google Scholar]
- 2.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 3.Lee JT. Lessons from X-chromosome inactivation: long ncRNA as guides and tethers to the epigenome. Genes Dev. 2009;23:1831–1842. doi: 10.1101/gad.1811209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 6.Claverie JM. Fewer genes, more noncoding RNA. Science. 2005;309:1529–1530. doi: 10.1126/science.1116800. [DOI] [PubMed] [Google Scholar]
- 7.Umlauf D, Fraser P, Nagano T. The role of long non-coding RNAs in chromatin structure and gene regulation: variations on a theme. Biol Chem. 2008;389:323–331. doi: 10.1515/BC.2008.047. [DOI] [PubMed] [Google Scholar]
- 8.Hekimoglu B, Ringrose L. Non-coding RNAs in polycomb/trithorax regulation. RNA Biol. 2009;6:129–137. doi: 10.4161/rna.6.2.8178. [DOI] [PubMed] [Google Scholar]
- 9.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kanhere A, Viiri K, Araújo CC, Rasaiyaah J, Bouwman RD, Whyte WA, et al. Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Mol Cell. 38:675–688. doi: 10.1016/j.molcel.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prasanth KV, Spector DL. Eukaryotic regulatory RNAs: an answer to the ‘genome complexity’ conundrum. Genes Dev. 2007;21:11–42. doi: 10.1101/gad.1484207. [DOI] [PubMed] [Google Scholar]
- 13.Beysen D, Raes J, Leroy BP, Lucassen A, Yates JR, Clayton-Smith J, et al. Deletions involving long-range conserved nongenic sequences upstream and downstream of FOXL2 as a novel disease-causing mechanism in blepharophimosis syndrome. Am J Hum Genet. 2005;77:205–218. doi: 10.1086/432083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D'haene B, Attanasio C, Beysen D, Dostie J, Lemire E, Bouchard P, et al. Disease-causing 7.4 kb cis-regulatory deletion disrupting conserved non-coding sequences and their interaction with the FOXL2 promotor: implications for mutation screening. PLoS Genet. 2009;5:1000522. doi: 10.1371/journal.pgen.1000522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crackower MA, Scherer SW, Rommens JM, Hui CC, Poorkaj P, Soder S, et al. Characterization of the split hand/split foot malformation locus SHFM1 at 7q21.3-q22.1 and analysis of a candidate gene for its expression during limb development. Hum Mol Genet. 1996;5:571–579. doi: 10.1093/hmg/5.5.571. [DOI] [PubMed] [Google Scholar]
- 16.Feng J, Bi C, Clark BS, Mady R, Shah P, Kohtz JD. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 2006;20:1470–1484. doi: 10.1101/gad.1416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lukiw WJ, Handley P, Wong L, Crapper McLachlan DR. BC200 RNA in normal human neocortex, non-Alzheimer dementia (NAD) and senile dementia of the Alzheimer type (AD) Neurochem Res. 1992;17:591–597. doi: 10.1007/BF00968788. [DOI] [PubMed] [Google Scholar]
- 18.Zalfa F, Adinolfi S, Napoli I, Kühn-Hölsken E, Urlaub H, Achsel T, et al. Fragile X mental retardation protein (FMRP) binds specifically to the brain cytoplasmic RNAs BC1/BC200 via a novel RNA-binding motif. J Biol Chem. 2005;280:33403–33410. doi: 10.1074/jbc.M504286200. [DOI] [PubMed] [Google Scholar]
- 19.Jin P, Warren ST. Understanding the molecular basis of fragile X syndrome. Hum Mol Genet. 2000;9:901–908. doi: 10.1093/hmg/9.6.901. [DOI] [PubMed] [Google Scholar]
- 20.McPherson R, Pertsemlidis A, Kavaslar N, Stewart A, Roberts R, Cox D, et al. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Samani NJ, et al. Genomewide association analysis of coronary artery disease. N Engl J Med. 2007;357:443–453. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Visel A, et al. Targeted deletion of the 9p21 noncoding coronary artery disease risk interval in mice. Nature. 464:409–412. doi: 10.1038/nature08801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X, et al. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454:126–130. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu W, et al. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morris KV, et al. Bidirectional transcription directs both transcriptional gene activation and suppression in human cells. PLoS Genet. 2008;4:1000258. doi: 10.1371/journal.pgen.1000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li K, et al. A noncoding antisense RNA in tie-1 locus regulates tie-1 function in vivo. Blood. 115:133–139. doi: 10.1182/blood-2009-09-242180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katayama S, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 28.Martianov I, et al. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007;445:666–670. doi: 10.1038/nature05519. [DOI] [PubMed] [Google Scholar]
- 29.Wang Z, et al. Evidence of influence of genomic DNA sequence on human X chromosome inactivation. PLoS Comput Biol. 2006;2:113. doi: 10.1371/journal.pcbi.0020113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carrel L, Willard HF. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434:400–404. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- 31.Maenner S, et al. 2-D structure of the A region of Xist RNA and its implication for PRC2 association. PLoS Biol. 8:1000276. doi: 10.1371/journal.pbio.1000276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao J, et al. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kino T, et al. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal. 3:8. doi: 10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mariner PD, et al. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol Cell. 2008;29:499–509. doi: 10.1016/j.molcel.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 35.Espinoza CA, et al. B2 RNA binds directly to RNA polymerase II to repress transcript synthesis. Nat Struct Mol Biol. 2004;11:822–829. doi: 10.1038/nsmb812. [DOI] [PubMed] [Google Scholar]
- 36.Torarinsson E, et al. Thousands of corresponding human and mouse genomic regions unalignable in primary sequence contain common RNA structure. Genome Res. 2006;16:885–889. doi: 10.1101/gr.5226606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Venter JC, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 38.Zappulla DC, Cech TR. RNA as a flexible scaffold for proteins: yeast telomerase and beyond. Cold Spring Harb Symp Quant Biol. 2006;71:217–224. doi: 10.1101/sqb.2006.71.011. [DOI] [PubMed] [Google Scholar]
- 39.Cech TR. Life at the End of the Chromosome: Telomeres and Telomerase. Angew Chem Int Ed Engl. 2000;39:34–43. doi: 10.1002/(sici)1521-3773(20000103)39:1<34::aid-anie34>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 40.Dreyfuss G, et al. hnRNP proteins and the biogenesis of mRNA. Annu Rev Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- 41.Kamma H, et al. Interaction of hnRNP A2/B1 isoforms with telomeric ssDNA and the in vitro function. Biochem Biophys Res Commun. 2001;280:625–630. doi: 10.1006/bbrc.2000.4173. [DOI] [PubMed] [Google Scholar]
- 42.Bilaud T, et al. Telomeric localization of TRF2, a novel human telobox protein. Nat Genet. 1997;17:236–239. doi: 10.1038/ng1097-236. [DOI] [PubMed] [Google Scholar]