Abstract
Peroxisomes are highly dynamic, multifunctional organelles that display remarkable changes in morphology, number and enzyme content. Peroxisomes multiply by growth and division of pre-existing organelles, but they can also form de novo from the ER. Growth and division of peroxisomes in mammalian cells involves elongation, membrane constriction and final fission and requires the peroxisome biogenesis Pex11 proteins as well as the recruitment of Dynamin-like protein DLP1/Drp1. We recently exploited the division-inhibiting properties of a unique Pex11pβ-YFP fusion protein to further dissect the process of peroxisomal growth and division. By applying life cell imaging and the HaloTag technology, our study revealed that Pex11pβ-mediated growth (elongation) and division of peroxisomes follows a multistep maturation pathway, which is initiated by the formation of an early peroxisomal membrane compartment from a pre-existing peroxisome and its stepwise conversion into a mature, metabolically active peroxisome compartment. Our observations support the view that peroxisomes formed by growth and division of pre-existing ones contain new membrane and matrix components. Peroxisome division is an asymmetric process, which is more complex than simple (symmetric) division of a preexisting organelle and equal distribution of the protein content. Our findings are in favor of Pex11pβ acting as a peroxisomal membrane shaping protein.
Key words: peroxisomes, organelle division, fission, peroxisome proliferation, Pex11p, membrane deformation
Peroxisomes are ubiquitous, multifunctional organelles, which adapt to different physiological conditions by changing their morphology, positioning, abundance and proteome in the cell.1,2 This dynamic behavior is essential for organellar and cellular function. Peroxisomes are able to proliferate by growth (e.g., elongation) of pre-existing organelles and subsequent division, but they can also form de novo from the ER via a maturation process.3–10 To what extend these alternative pathways contribute to peroxisome formation is controversial and might vary among organisms. In yeast, the bulk of the peroxisomes are formed by fission of pre-existing organelles,5 whereas in mammalian cells both mechanisms may operate simultaneously.11
We recently contributed to unveil the components of the peroxisomal division machinery in mammals.12,13 To divide peroxisomes house a set of proteins such as the dynamin-like large GTPase DLP1/Drp1, Fis1, a potential receptor for DLP1 at the peroxisomal membrane, or the putative membrane adaptor Mff (Mitochondrial fission factor),14 which are all shared with the fission machinery of mitochondria pointing to mechanistic parallels, coordinated biogenesis and organelle crosstalk.6,12,13,15,16 Interestingly, yeast species, filamentous fungi and plants use related dynamin-like GTPases and Fis homologues to divide peroxisomes, and share these components with mitochondria indicating that this is a general conserved principle among organisms.6,10,17 Recently, a novel lethal disorder affecting both mitochondria and peroxisomes based on a mutation in the human gene for DLP1 was discovered.18 Patient fibroblasts show peroxisomes arrested in a late stage of the fission process.
Members of the Pex11 family are unique PMPs and have been implicated in peroxisome proliferation and regulation of peroxisome abundance in all organisms studied.19 Whereas yeasts and fungi possess additional peroxins supposed to regulate peroxisome number,6,10 the three mammalian Pex11p isoforms (Pex11pα, Pex11pβ, Pex11pγ) are so far the only proteins known to mediate peroxisome proliferation and abundance in mammalian cells. Pex11pβ induces a prominent peroxisome proliferation via elongation, membrane constriction and final division,20 which can occur independently of peroxisome metabolism.21 Mice lacking PEX11β show neonatal lethality and other defects reminiscent of Zellweger syndrome,22 whereas mice lacking PEX11α develop normally.23 Nevertheless, the biochemical properties of Pex11p are still a matter of debate,19 and the growth and division process of peroxisomes is far from being fully understood.
DLP1 and its membrane adaptor Fis1 function later on in peroxisome division, whereas the membrane peroxin Pex11pβ appears to act upstream and early on in the process.24 In our recent study we discovered that a Pex11pβ-YFP fusion protein can be used as a specific tool to further dissect peroxisomal growth and division. In contrast to Pex11pβ-Myc, expression of Pex11pβ carrying a monomeric YFP tag at the C-terminus inhibited constriction (segmentation) and division of peroxisomes in mammalian cells, but instead resulted in the formation of elongated pre-peroxisomal membrane structures, so called TPAs (tubular peroxisomal accumulations) (Fig. 1). The alterations were specific for Pex11pβ-YFP (Fig. 1). As the division of these structures was blocked, single peroxisomes were apparently depleted from the cells. This observation challenges the current view that de novo formation from the ER and division of pre-existing peroxisomes may occur simultaneously in mammalian cells.11 As we do not observe the formation of new single peroxisomes, we might face a situation similar to yeast, where only the complete loss of peroxisomes triggers de novo formation.5 Alternatively, Pex11pβ may as well be required for de novo formation from the ER suggesting an overlap in the components involved. However, Pex11pβ has not been observed to localize to the ER in mammalian cells and appears to be directly inserted in the peroxisomal membrane in a Pex19p-dependent process. In cells lacking peroxisomes, Pex11pβ is instead targeted to mitochondria, and not to the ER. Interestingly, de novo formation of yeast peroxisomes from mitochondria was very recently induced upon mitochondrial targeting of Pex3p, a peroxin involved in peroxisomal membrane biogenesis and a key player in de novo formation from the ER.25 This indicates that natural or artificial targeting of Pex3p to any endomembrane may initiate peroxisome formation. Therefore, it will be important to elucidate whether the de novo synthesis of peroxisomes induced by the targeting of Pex3p to the ER is physiological or artificial.
Figure 1.
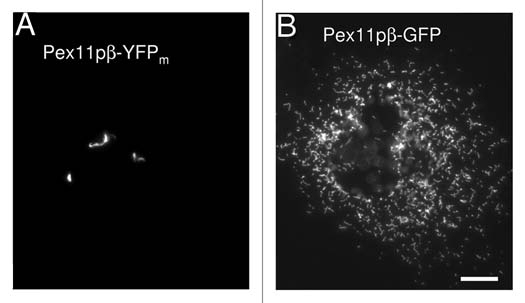
TPA formation is specific for Pex11pβ-YFP. Expression of Pex11pβ-YFP in COS-7 cells results in the formation of tubular peroxisomal membrane compartments (TPAs) (A) and inhibits peroxisome division and proliferation normally induced by untagged or Myc-tagged Pex11pβ. These alterations are specific for Pex11pβ-YFP. The expression of N-terminally tagged YFP-Pex11pβ or of Pex11pá-YFP, a homologue of Pex11pβ with similar membrane topology, does not induce TPAs. Interestingly, expression of a Pex11pβ-GFP construct does not induce TPAs (B) indicating that cloning-related differences in the linker region between the Pex11pβ and the (Y)GFP tag might have an effect on protein conformation. Potential oligomerization properties of non-monomeric GFP appear to be less crucial for TPA formation. Although the mechanistic reasons for the inhibitory effect of Pex11pβ-YFP are presently unclear, sterical hindrance during the formation of a functional constriction/division complex and/or some intrinsic “stacking” activity and tight trans-membrane interactions of Pex11pβ-YFP may explain the block in peroxisome division. Nevertheless, the Pex11pβ-YFP fusion protein represents a specific and useful novel tool to further dissect peroxisome proliferation and to investigate early events in peroxisomal growth and division. Bar, 10 µm.
By fluorescence and electron microscopy we further demonstrated that the pre-peroxisomal membrane compartment induced by Pex11pβ-YFP was composed out of globular and extended tubular membrane domains. Interestingly, peroxisomal matrix and PMPs were targeted to distinct regions of the peroxisomal structures suggesting a more complex architecture of the peroxisomal membrane. It will be important to study how the specific targeting of the PMPs to the distinct membrane domains within the TPAs is achieved. Are the PMPs directly inserted into specific membrane domains, or are pre-peroxisomal carriers (e.g., from the ER) involved? If different carriers with a distinct PMP composition exist, how do they distinguish between the globular and tubular membrane domains prior to fusion? If the carriers (or PMPs) fuse (or insert) over the whole of the globular and tubular domains or at a specific site (e.g., at the neck region), special, yet undefined sorting/retention mechanisms within the peroxisomal membrane must exist which require further investigation. Excitingly, lipid rafts essential for peroxisome biogenesis have very recently been identified in human HepG2 cells26 which might influence the mobility of certain PMPs.
By applying life cell imaging and the HaloTag technology, our study also revealed that Pex11pβ-mediated growth (elongation) and division follows a multistep maturation pathway, which is initiated by the formation of an early peroxisomal membrane compartment from a pre-existing peroxisome and its stepwise conversion into a mature, metabolically active peroxisome compartment (Fig. 2). Maturation is achieved by selective and stepwise import of certain PMPs, membrane lipids and matrix proteins. Thus, new peroxisomes formed by growth and division of pre-existing ones are likely to contain new membrane and matrix components. Our observations support the view that peroxisome division is an asymmetric process,27 which is far more complex than simple (symmetric) division of a pre-existing organelle and equal distribution of the matrix protein content. Furthermore, our findings support a role for Pex11pβ in shaping and deforming the peroxisomal membrane prior to division.
Figure 2.
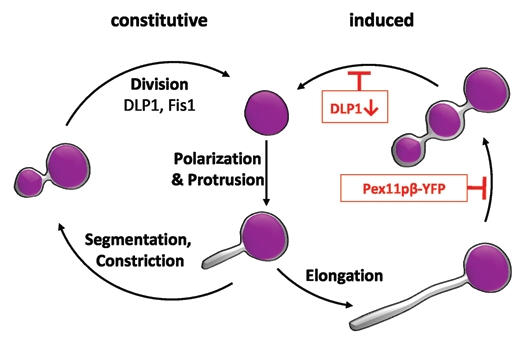
Pex11pβ-mediated peroxisome proliferation by growth and division follows a multistep maturation pathway. Pex11pβ localizes to the peroxisomal membrane and (upon activation) mediates the formation of a peroxisomal subdomain at one side of a pre-existing peroxisome (polarization28) as well as membrane protrusion. The membrane extension is growing (elongation by recruitment of lipids) and acquires a distinct set of PMPs, but contains no/few matrix proteins. Afterwards, the membrane extension segments and constricts. This step can be inhibited by expression of Pex11pβ-YFP, which keeps the peroxisomes in an elongated stage and results in TPA formation. The constricted peroxisomal tubule acquires other PMPs (e.g., components of the import machinery) and matrix proteins and is finally divided by hFis1 and DLP1 into several spherical peroxisomes. Inhibition of DLP1 function blocks fission resulting in the accumulation of constricted peroxisomal tubules positive for PMPs and matrix proteins.24 The process of fission may vary in different species and/or under non-inducing (constitutive) conditions, where peroxisomes may divide asymmetrically, with small daughter peroxisomes budding or pinching off from mature ones.7 The degree of membrane elongation may be influenced by the level of Pex11p expression and other regulatory events (adapted from ref. 24).
As organelle dynamics have been related to organelle functionality, developmental and physiological processes and as mutations in peroxisomal fission proteins result in severe diseases, the molecular characterization of peroxisomal dynamics and fission is of great cell biological and biomedical interest. The unique possibility to now generate and enrich division-arrested peroxisomes in cultured cells (Fig. 2) may allow the identification and further molecular characterization of peroxisomal division components in mammals.
Acknowledgements
In memory of Edgar F. Cruz e Silva, Director of the Centre for Cell Biology, Aveiro (1958–2010). This work was supported by grants from the DFG (SCHR 518/6-1, 2), FCT (REEQ/1023/BIO/2005; PTDC/BIABCM/71932/2006) and the University of Aveiro.
Abbreviations
- ER
endoplasmic reticulum
- Pex
peroxin (peroxisomal protein required for peroxisome biogenesis)
- PMP
peroxisomal membrane protein
- TPA
tubular peroxisomal accumulation
References
- 1.Wanders RJA, Waterham HR. Biochemistry of mammalian peroxisomes revisited. Annu Rev Biochem. 2006;75:295–332. doi: 10.1146/annurev.biochem.74.082803.133329. [DOI] [PubMed] [Google Scholar]
- 2.Islinger M, Cardoso MJ, Schrader M. Be different—the diversity of peroxisomes in the animal kingdom. Biochim Biophys Acta. 2010;1803:881–897. doi: 10.1016/j.bbamcr.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 3.Hettema EH, Motley AM. How peroxisomes multiply. J Cell Sci. 2009;122:2331–2336. doi: 10.1242/jcs.034363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoepfner D, Schildknegt D, Braakman I, Philippsen P, Tabak HF. Contribution of the endoplasmic reticulum to peroxisome formation. Cell. 2005;122:85–95. doi: 10.1016/j.cell.2005.04.025. [DOI] [PubMed] [Google Scholar]
- 5.Motley AM, Hettema EH. Yeast peroxisomes multiply by growth and division. J Cell Biol. 2007;178:399–410. doi: 10.1083/jcb.200702167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nagotu S, Veenhuis M, van der Klei IJ. Divide et impera: the dictum of peroxisomes. Traffic. 2010;11:175–184. doi: 10.1111/j.1600-0854.2009.01019.x. [DOI] [PubMed] [Google Scholar]
- 7.Schrader M, Fahimi HD. Growth and division of peroxisomes. Int Rev Cytol. 2006;255:237–290. doi: 10.1016/S0074-7696(06)55005-3. [DOI] [PubMed] [Google Scholar]
- 8.Titorenko VI, Rachubinski RA. Mutants of the yeast Yarrowia lipolytica defective in protein exit from the endoplasmic reticulum are also defective in peroxisome biogenesis. Mol Cell Biol. 1998;18:2789–2803. doi: 10.1128/mcb.18.5.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Titorenko VI, Rachubinski RA. Dynamics of peroxisome assembly and function. Trends Cell Biol. 2001;11:22–29. doi: 10.1016/s0962-8924(00)01865-1. [DOI] [PubMed] [Google Scholar]
- 10.Saraya R, Veenhuis M, van der Klei IJ. Peroxisomes as dynamic organelles: peroxisome abundance in yeast. FEBS J. 2010;277:3279–3288. doi: 10.1111/j.1742-4658.2010.07740.x. [DOI] [PubMed] [Google Scholar]
- 11.Kim PK, Mullen RT, Schumann U, Lippincott-Schwartz J. The origin and maintenance of mammalian peroxisomes involves a de novo PEX16-dependent pathway from the ER. J Cell Biol. 2006;173:521–532. doi: 10.1083/jcb.200601036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delille HK, Alves R, Schrader M. Biogenesis of peroxisomes and mitochondria: linked by division. Histochem Cell Biol. 2009;131:441–446. doi: 10.1007/s00418-009-0561-9. [DOI] [PubMed] [Google Scholar]
- 13.Schrader M. Shared components of mitochondrial and peroxisomal division. Biochim Biophys Acta. 2006;1763:531–541. doi: 10.1016/j.bbamcr.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 14.Gandre-Babbe S, van der Bliek AM. The novel tail-anchored membrane protein Mff controls mitochondrial and peroxisomal fission in mammalian cells. Mol Biol Cell. 2008;19:2402–2412. doi: 10.1091/mbc.E07-12-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Camoes F, Bonekamp NA, Delille HK, Schrader M. Organelle dynamics and dysfunction: A closer link between peroxisomes and mitochondria. J Inherit Metab Dis. 2009;32:163–180. doi: 10.1007/s10545-008-1018-3. [DOI] [PubMed] [Google Scholar]
- 16.Schrader M, Yoon Y. Mitochondria and peroxisomes: Are the ‘Big Brother’ and the ‘Little Sister’ closer than assumed? Bioessays. 2007;29:1105–1114. doi: 10.1002/bies.20659. [DOI] [PubMed] [Google Scholar]
- 17.Kaur N, Hu J. Dynamics of peroxisome abundance: a tale of division and proliferation. Curr Opin Plant Biol. 2009;12:781–788. doi: 10.1016/j.pbi.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 18.Waterham HR, Koster J, van Roermund CW, Mooyer PA, Wanders RJ, Leonard JV. A lethal defect of mitochondrial and peroxisomal fission. N Engl J Med. 2007;356:1736–1741. doi: 10.1056/NEJMoa064436. [DOI] [PubMed] [Google Scholar]
- 19.Thoms S, Erdmann R. Dynamin-related proteins and Pex11 proteins in peroxisome division and proliferation. FEBS J. 2005;272:5169–5181. doi: 10.1111/j.1742-4658.2005.04939.x. [DOI] [PubMed] [Google Scholar]
- 20.Schrader M, Reuber BE, Morrell JC, Jimenez-Sanchez G, Obie C, Stroh TA, et al. Expression of PEX11beta mediates peroxisome proliferation in the absence of extracellular stimuli. J Biol Chem. 1998;273:29607–29614. doi: 10.1074/jbc.273.45.29607. [DOI] [PubMed] [Google Scholar]
- 21.Li X, Gould SJ. PEX11 promotes peroxisome division independently of peroxisome metabolism. J Cell Biol. 2002;156:643–651. doi: 10.1083/jcb.200112028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li X, Baumgart E, Morrell JC, Jimenez-Sanchez G, Valle D, Gould SJ. PEX11beta deficiency is lethal and impairs neuronal migration but does not abrogate peroxisome function. Mol Cell Biol. 2002;22:4358–4365. doi: 10.1128/MCB.22.12.4358-4365.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li X, Baumgart E, Dong GX, Morrell JC, Jimenez-Sanchez G, Valle D, et al. PEX11alpha is required for peroxisome proliferation in response to 4-phenylbutyrate but is dispensable for peroxisome proliferator-activated receptor alpha-mediated peroxisome proliferation. Mol Cell Biol. 2002;22:8226–8240. doi: 10.1128/MCB.22.23.8226-8240.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koch A, Schneider G, Luers GH, Schrader M. Peroxisome elongation and constriction but not fission can occur independently of dynamin-like protein 1. J Cell Sci. 2004;117:3995–4006. doi: 10.1242/jcs.01268. [DOI] [PubMed] [Google Scholar]
- 25.Rucktaschel R, Halbach A, Girzalsky W, Rottensteiner H, Erdmann R. De novo synthesis of peroxisomes upon mitochondrial targeting of Pex3p. Eur J Cell Biol. 2010;89:947–954. doi: 10.1016/j.ejcb.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 26.Woudenberg J, Rembacz KP, Hoekstra M, Pellicoro A, van den Heuvel FA, Heegsma J, et al. Lipid rafts are essential for peroxisome biogenesis in HepG2 cells. Hepatology. 2010;52:623–633. doi: 10.1002/hep.23684. [DOI] [PubMed] [Google Scholar]
- 27.Huybrechts SJ, Van Veldhoven PP, Brees C, Mannaerts GP, Los GV, Fransen M. Peroxisome dynamics in cultured mammalian cells. Traffic. 2009;10:1722–1733. doi: 10.1111/j.1600-0854.2009.00970.x. [DOI] [PubMed] [Google Scholar]
- 28.Koch J, Pranjic K, Huber A, Ellinger A, Hartig A, Kragler F, et al. PEX11 family members are membrane elongation factors that coordinate peroxisome proliferation and maintenance. J Cell Sci. 2010;123:3389–3400. doi: 10.1242/jcs.064907. [DOI] [PubMed] [Google Scholar]


