Abstract
The myocardium of the heart is composed of multiple highly specialized myocardial lineages, including those of the ventricular and atrial myocardium, and the specialized conduction system. Specification and maturation of each of these lineages during heart development is a highly ordered, ongoing process involving multiple signaling pathways and their intersection with transcriptional regulatory networks. Here, we attempt to summarize and compare much of what we know about specification and maturation of myocardial lineages from studies in several different vertebrate model systems. To date, most research has focused on early specification, and while there is still more to learn, less is known about factors that promote subsequent maturation of myocardial lineages required to build the functioning adult heart.
Keywords: myocardial lineages, specification, differentiation, transcriptional regulation, signaling, heart development
Introduction
The cardiomyocyte is the fundamental work unit of the heart. Although cardiomyocyte cell fate is a developmental endpoint, a diversity of cardiomyocyte subtypes exist within the heart. These include the outflow tract, right ventricle, left ventricle, atria, myocardial sleeves of cranial and caudal great vessels, and specialized conduction system tissue, including the SA node, AV node, and His-Purkinje tracts. The recent interest in production of cardiomyocytes for repair of the diseased heart has heightened the importance of a detailed understanding of cardiac lineage specification and differentiation.
To provide context for understanding the locations, contributions and patterning of progenitor cells, this review begins with a brief overview of the “basics” of heart development in mouse, chick, frog and fish, and then discusses aspects of myocardial development under the two broad headings of “specification”, dealing with patterning and growth of undifferentiated myocyte progenitors, and “differentiation”, dealing with subspecialization and growth of distinct myocyte lineages following differentiation. Buckingham and Vincent have recently published an excellent review of similar information and we refer the reader to the figures in that review for signaling and transcriptional network diagrams.1 A glossary of terms used in this review has been supplied as an online supplement.
I. Basic Mouse and Chick Heart Development (Figure 1, Table 1)
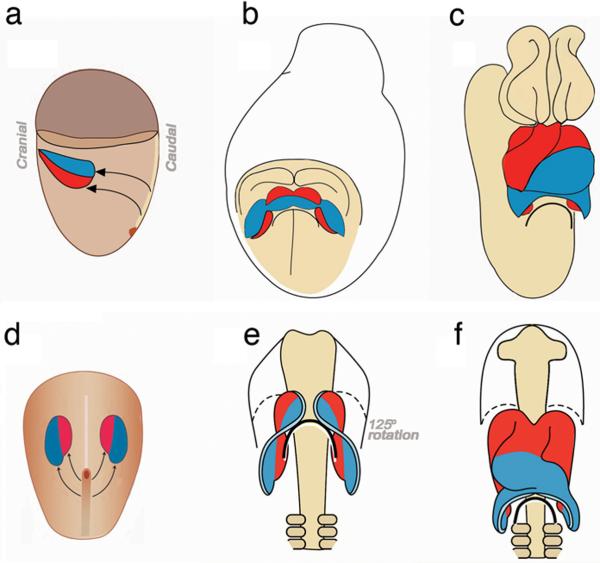
Figure 1.
Schematic representation of heart tube formation in mouse (a, b, c) and chick (d, e, f). First heart progenitors are shown in blue and second heart progenitors in red. (a,d) Fate mapping studies have defined the locations of chick and mouse cardiac progenitors in the heart fields. (a) Lateral view of an E6.5 mouse embryo. (d) Dorsal view of an HH5 chick embryo. (b,e) Fusion and differentiation of the heart fields is more rapid in the mouse leading to a cardiac crescent that is not seen in the chick. Second heart progenitors are located medially in both chick and mouse heart fields but quickly change positions to cranial as the heart fields converge on the midline. (b) Ventral view of an E8.5 mouse embryo showing the cardiac crescent and its relationship with the anterior intestinal portal (curved black line). (e) Ventral view of an HH8 chick embryo showing convergence of the heart fields in the ventral midline and how the first and second heart field progenitors have rotated their position from medial-lateral to craniocaudal. (c,f) Second heart progenitors are gradually added to the elongating cardiac tube. (c) Ventral view of an E9.5 mouse embryo. (f) Ventral view of an HH12 chick embryo. The caudal second heart progenitors are shifted by formation of the foregut pocket and anterior intestinal portal (curved black line) to cranial, thus putting them in place to contribute to the outflow pole. Some of the second heart field progenitors are also added to the venous pole: parts of the atrium and atrial septum but these are incorporated later than the stages shown, hence no red cells are seen at these stages in the venous pole. The proximal and distal outflow myocardium is added over an extended period of time (reprinted from 222 with permission).
Table 1.
Landmarks in heart development in the models discussed in this review
| Zebrafish (hpf) |
Frog (N&F) |
Chick (HH) |
Mouse (dpc) |
Human (days) |
|
|---|---|---|---|---|---|
| Gastrulation | 5.5 | 10 | 4 | 7 | 15-16 |
| Heart fields | 13 | 24 | 5 | 7 | 18 |
| Heart trough | 19 (cone) | 29/30 | 9 | 8 | 21-23 |
| Heart tube | 22 | 32/33 | 11 | 8.5 | 24 |
| Looping | 24-48 | 33-35 | 14-18 | 8.5-10.25 | 24-25 |
| Myocardial contraction |
22 | 32/33 | 10 | 8.5 | 24 |
| Anterograde circulation |
24 | 37 | 14 | 9 | 25 |
| Venous pole addition |
24-72 | ND | 14-18 | 8.5-10.25 | 25-40 |
| Proepicardium | 50 | 41 | 15 | 9.5 | 28-35 |
Before gastrulation, both chick and mouse embryos are composed of two cell layers, epiblast and hypoblast (chick) or primitive endoderm (mouse). The epiblast contributes all embryonic and some extra-embryonic tissues. Regional expression of genes and cell fate mapping suggest that embryonic anterior-posterior and left-right axes are established prior to gastrulation.2 Fate mapping experiments have demonstrated that heart precursors are located in the posterior/caudal epiblast and will be adjacent to the anterior/cranial two-thirds of the primitive streak when it forms, making heart progenitors among the earliest embryonic cells to gastrulate.3, 4
Formation of the three germ layers, ectoderm, mesoderm, and endoderm results from ingression of epiblast cells through the primitive streak at gastrulation.5 Early in gastrulation (embryonic day (E) 6.5, mouse); Hamilton Hamburger Stage 3(HH, chick), the primitive streak elongates cranially until mid-gastrulation, at which time most cardiac progenitors ingress.6, 7 Progenitor cells of pharyngeal and foregut endoderm are localized next to cardiogenic mesoderm progenitors in the epiblast, migrating through the early to mid-primitive streak to integrate with extraembryonic endoderm, progressively displacing the latter, coincident with the migration of the early cardiogenic mesoderm.8 Following ingression, cardiogenic mesoderm moves anterolaterally as a sheet of cells, and by late primitive streak stages is localized as identifiable bilateral “fields” in anterior lateral plate mesoderm (chick) or as a cardiac crescent (mouse).6, 9
During gastrulation, the craniocaudal arrangement of progenitors is shifted 90°: the most cranial cells in the epiblast become the most medial in the mesoderm and the most caudal cells in the epiblast become the most lateral in the mesoderm.9, 10 The chick embryo undergoes an extended period in which the bilateral heart fields remain as progenitors and do not show signs of differentiation until they move to the midline.11, 12 In contrast, in the mouse the cardiogenic fields move quickly to the midline, anterior to the forming neural plate, where they join to form a cardiac crescent of differentiating cardiomyocytes. In both chick and mouse, embryo folding to form the foregut pocket carries the cranial cells in the cardiogenic fields ventrocaudally resulting in an inversion of cardiogenic mesoderm.10 The cranial-lateral borders of the heart fields meet at E8.0 (HH9) to form the ventral seam of the early heart “tube” which is at this stage more accurately described as a myocardial trough open dorsally toward the ventral pharynx.10, 13 14
It is not until the tube closes dorsally and its attachment to the ventral pharynx, called the primary dorsal mesocardium, breaks down that an actual tube is formed. Partial rupture of the dorsal mesocardium allows the tube to be detached from the ventral pharynx except at its venous (caudal, inflow) and arterial (cranial, outflow) poles.15 The early myocardial tube comprises primarily cells destined to be the left ventricle with a little right ventricle and atrioventricular (AV) canal represented. The myocardium that will form the majority of the right ventricle, conus and truncus are all added via the arterial pole, while the AV canal, atria and atrial septum are added to the venous pole.10 Heart contractions can be seen as early as formation of the trough (E8.25, HH10) although anterograde circulation does not begin until after the tube has formed and begins to loop.16, 17
The heart tube loops to the right (E8.5, HH11), coincident with disappearance of the dorsal mesocardium. Initial looping is intrinsic to the myocardium followed by forces generated in the dorsal mesocardium just before it disappears.18-20 Much of the lengthening of the tube is due to addition of newly differentiated myocardium at the arterial and venous poles. With progressive growth of the tube and specification of the chambers, the atria, initially located caudal to the ventricles, become displaced cranial to the ventricles.
In the chick, myocardial progenitors proliferate at a very high rate with a cycle time of about 5.2 hrs.21 However, as soon as myocardial differentiation begins, the cells cease cycling for an extended period. Presumably, this allows differentiation and onset of function. Growth of the tubular heart ensues primarily from progressive addition of cells from the remaining progenitor pool.21 Retrospective clonal lineage analysis recognizes first and second lineages roughly corresponding to the first and second heart fields, based on their timing of differentiation and entry into the heart.22 The cells of the first heart field are the first to differentiate, and contribute to the left ventricle, AV canal and atria. The second heart field constitutes a highly proliferative progenitor population that contributes progressively to the remainder of the heart.21 These two populations of cells can be identified molecularly by expression of differentiation versus specification markers but since the process is progressive, there is not a definitive morphological boundary between them.
Two additional named subpopulations of the second heart field were identified using different techniques. Gene trapping showed an FGF10-expressing subpopulation of myocardium that contributed to the right ventricle and outflow tract. This subpopulation was called anterior heart field.23 A second subpopulation was identified by cell tracing techniques from the ventral splanchnic pharyngeal mesoderm that contributed myocardium and smooth muscle to the definitive arterial pole. This subpopulation was called secondary heart field.24, 25 Both subpopulations are second heart field progenitors. In addition, other subpopulations have been identified recently that contribute to the inflow pole of the heart including some of the atrial myocardial wall and the atrial septum via the dorsal mesenchymal protrusion (spina vestibulae) and the myocardial sleeves on the sinus venosus and pulmonary veins. Because all these subpopulations originate from the second heart field we will refer to them primarily by what they contribute to the heart and not use the more confusing subpopulation designations.
The progenitor population that contributes to the definitive arterial pole, comprised of truncal myocardium and smooth muscle that will form the tunica media at the base of the great arterial trunks, is located in the ventral pharynx behind the outflow tract.24-26 The myocardial cells are added in two spirals that become the myocardial underpinning of the aorta and pulmonary trunk.27, 28 The smooth muscle that forms the base of the arterial trunks is added after the myocardium and does not spiral.24 Ablation of chick arterial pole progenitors on the right leads to overriding aorta and pulmonary stenosis or atresia, which are key components of tetralogy of Fallot.27 Manipulations that interfere with proliferation of arterial pole progenitors cause the same phenotype 29, 30
Although initial differentiation of myocardial cells is accompanied by cessation of proliferation, chamber myocardial cells reinitiate a proliferative phase at about HH14 after anterograde circulation is established. This proliferation accounts for “ballooning” of the ventricles and establishment of the trabeculae.31, 32 Myocardium of the outflow tract and AV canal is relatively non-proliferative. The early trabecular ventricular myocardium of the embryonic heart undergoes growth and compaction to result in the final form of the ventricular myocardial wall.
At approximately E9.5 (HH15), the outflow tract and AV cushions form, which initially block retrograde blood flow and later contribute to septation of these regions. Formation of the cushions is succeeded by growth of the muscular ventricular septum and formation of the primary atrial septum by ingrowth of myocardializing mesoderm in the dorsal mesocardial protrusion from progenitors located dorsal to the common atrium.33 At E10 (HH24), cardiac neural crest cells migrate into the distal outflow cushions where they form the aorticopulmonary septation complex of condensed mesenchyme.34, 35 The neural crest-derived mesenchyme orchestrates septation of the aorta and pulmonary trunk and the distal outflow tract. Closure of the proximal outflow tract is by conal and AV cushion tissue after the distal septum has formed. Septation into four chambers is completed by E14.5 (HH34). Remodeling of outflow tract and AV cushions to form valves is an ongoing progress, which continues postnatally.36
Cells from the proepicardium give rise to the epicardial epithelium covering the heart. The proepicardium arises from septum transversum mesenchyme near the venous pole of the heart and is evident from approximately E9.5 (HH16) to E10.5.37, 38 Once formed the epicardial cells contribute a number of cell types to the heart, including smooth muscle cells of the coronary vessels, cardiac fibroblasts, and potentially, a subset of cardiomyocytes, although the latter is controversial.39-41
II. Frog and Fish Heart Development (Figure 2, Table 1)
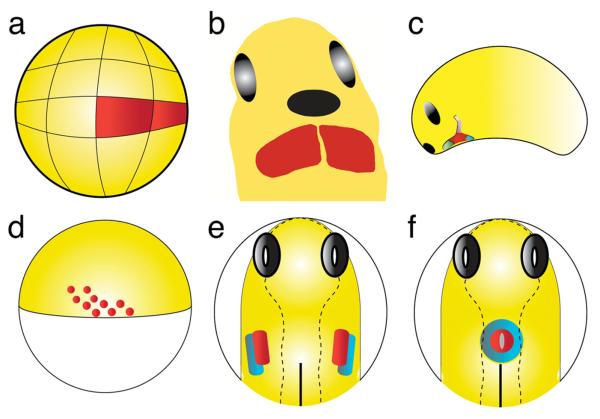
Figure 2.
Schematic representation of key stages in heart tube formation in frog (a, b, c) and fish (d, e, f). (a, d) Fate mapping studies have defined the locations of frog and fish cardiac progenitors prior to gastrulation. (a) Lateral view of a 32-cell stage Xenopus embryo, dorsal to the right, indicating the blastomeres (red) that will give rise to descendants in the adult heart. (d) Lateral view of a zebrafish embryo at the 40% epiboly stage, dorsal to the right, indicating the approximate locations of the blastomeres (red) that will give rise to the myocardium. (b, e) Fate maps and the expression patterns of molecular markers have demonstrated the locations of the bilateral heart fields in both frog and fish. (b) Anterior-ventral view of a Xenopus mid-tadpole, anterior to the top, just prior to the fusion of the heart fields (red) at the ventral midline. Cement gland is labeled in black and eyes are labeled in grey. Note: In Xenopus, unlike zebrafish, the atrial and ventricular precursor populations are not distinct populations but rather the precursors are intermingled until very late in heart development. (e) Dorsal view of a zebrafish embryo at the 7-somite stage, anterior to the top, indicating the locations of ventricular myocardial progenitors (red) and atrial myocardial progenitors (blue) prior to cardiac fusion. Notochord is labeled in black and eyes are labeled in grey. (c. f) Expression patterns of molecular markers have indicated the existence of discrete myocardial subpopulations as the heart tube forms. (c) Lateral view of a Xenopus tadpole at stage 25, anterior to the left, after fusion of the cardiac progenitors at the midline. Molecular markers distinguish cardiac populations expressing Tbx1 and Isl-1 (lavender) from those that do not (green). (f) Dorsal view of a zebrafish embryo at the 21-somite stage, anterior to the top, indicating the locations of ventricular cardiomyocytes (red) and atrial cardiomyocytes (blue) within the cardiac cone.
The early stages of frog and fish heart formation bear a strong resemblance to the early stages of mouse and chick heart development, despite the differences in embryonic anatomy between species. In this section, we highlight major stages of Xenopus and zebrafish heart development that correspond to the phases of mouse and chick development described above (Table 1). This correspondence guides our understanding of the conserved elements of the genetic pathways that regulate myocardial progenitor specification and differentiation.
Classical fate mapping studies in Xenopus demonstrated that three hours after fertilization, a pair of blastomeres in the dorsal equatorial zone is fated to give rise to the adult heart (Figure 2a).42, 43 As with all mesolecithal embryos, Xenopus cells do not undergo extensive cell mixing during the early stages of embryogenesis; thus, by the onset of gastrulation (stage 10), the descendants of these two blastomeres give rise to a coherent bilateral population of cells in direct contact with the organizer and underlying endoderm. Fate mapping studies in the zebrafish early blastula found myocardial and endocardial progenitors at the ventrolateral margin of the embryo.44, 45 The distribution of progenitors at the embryonic margin shifts slightly over time, such that the myocardial progenitors are located in bilateral marginal zones at the onset of zebrafish gastrulation (Figure 2d).46 Following gastrulation, the myocardial progenitor population coalesces into distinct bilateral positions within the anterior lateral plate mesoderm (Figure 2e).47
Myocardial differentiation and heart tube formation begin contemporaneously in both frog and fish. In Xenopus, as shown by DiI labeling and single cell PCR, the heart field is comprised of an amalgamation of progenitors that include cells that will give rise to either the future atrium or ventricle. Just prior to fusion, the fields uniformly initiate expression of the first markers of cardiomyocyte differentiation, including troponin T, MLC2a and MHCα (stage 26/27)(Figure 2b).48, 49 As in chick, a marked decrease in the cardiac mitotic index accompanies the formation of myofibrils and the expression of markers of terminal cardiomyocyte differentiation.50, 51 However, differentiation of cardiomyocytes is not directly linked to cell cycle withdrawal since blocking the cell cycle from early stages (prior to stage 24) has no effect on timing of cardiomyocyte differentiation in whole embryos or in cardiac explants.51, 52 By early tadpole stages (stage 29/30), the midline heart field begins to round up at its lateral edges to form a myocardial trough that eventually covers the endocardial tube. The trough then gradually closes in a caudal-to-cranial fashion over the dorsal surface of the endocardium such that by mid-tadpole stage (stage 32), the heart comprises a bi-layered linear heart tube.53, 54
In zebrafish, myocardial differentiation initiates within the bilateral heart fields by 16 hours post-fertilization (hpf), as evidenced by the expression of genes encoding sarcomere components, such as cardiac myosin light chain 2.55 Time-lapse studies have tracked the migration of these bilateral sheets of cardiomyocytes as they travel toward the midline to create a ring of cells, referred to as the cardiac cone, through a process termed cardiac fusion (Figure 2f).56 The myocardial cone engulfs a centrally located sheet of endocardial precursors, and together these structures gradually elongate to transform from a shallow conical structure into a two-layered primitive heart tube.57, 58
The frog and fish heart tubes transform into looped, multi-chambered organs through a combination of morphogenesis and growth. By stage 35, the Xenopus heart begins to loop; shortly thereafter, its chambers begin to adopt distinct morphological features, including thickening and trabeculation of the ventricular myocardium. In the frog at stage 45/46, septation of the atrium completes formation of the mature three-chambered heart.48, 49 Although there is a continual decrease in hyperplastic growth during these stages, the number of cardiomyocytes continues to increase, implying that new cardiomyocytes are added through the recruitment of cells from extra-cardiac sources.49-51 Indeed, recent fate mapping studies in Xenopus support the existence of a second heart field that arises from a pool of cardiac progenitors coexpressing Isl-1 and Nkx2.5. A subset these cells later downregulate Isl-1 while maintaining Nkx2.5 expression thus, establishing the first and second heart field. In addition to Isl1, the Xenopus secondary heart field also expresses many markers associated with the second field in mouse and chick including Tbx1, Flk1 and Islet1 (Figure 2c).59, 60 However, the relative contribution of the second heart field to the poles of the Xenopus heart remains to be established.
Similarly, the zebrafish heart tube develops into a two-chambered organ between 24 and 48 hpf through looping, constriction of the AV canal, and the distinctive bulging that creates the characteristic curvatures of the ventricle and atrium.61 The number of cardiomyocytes increases during this transition although little proliferation occurs. The accretion of cardiomyocytes is a consequence of recruitment of new, late-differentiating cardiomyocytes to both the arterial and venous poles of the heart tube.62
Altogether, the similarities between the early steps of heart formation in mouse, chick, frog, and fish support the validity of all of these model organisms for investigation of the molecular mechanisms driving cardiac progenitor specification and differentiation.
III. Myocardial Specification
Regional patterning in the epiblast is likely to determine the time at which cells fated to become myocytes undergo gastrulation. Cells fated to become cardiomyocytes are among the first to ingress through the primitive streak.63 The ingressing cells are organized in a colinear alignment from inflow to outflow: atrial (venous pole) progenitors are located most caudally, while AV, left ventricular, right ventricular, and conus cardiomyocyte progenitors located successively more cranially.4, 9, 64, 65 Transplantation of cell clusters from the epiblast of mouse embryos demonstrated that descendents can contribute to myocardium, endocardium, and epicardium, suggesting progenitors of these lineages are in close proximity in the epiblast.3
Several experiments suggest that cells within the epiblast are not yet committed to specific lineages. Presumptive cardiac epiblast cells from mouse embryos transplanted into chick embryo somites do not undergo cardiomyocyte differentiation, but interaction with cardiogenic mesoderm in proximity to pharyngeal/foregut endoderm is sufficient to convert early primitive streak epiblast cells not normally destined for the heart to a myocardial cell fate.3,66 Ingression during gastrulation may result in restriction of lineage potency, as ingressed cells in the anterolateral plate mesoderm of a mid-streak mouse embryo, when transplanted to the epiblast, can undergo ingression, but do not colonize lateral plate mesoderm.3 Consistent with this, cardiogenic mesoderm from late streak (E7.5) explants transplanted into chick embryo somites differentiate into cardiomyocytes, suggesting that cell fate has become restricted at this stage.66 Retroviral lineage studies in chick embryos showed that myocardial and endocardial progenitors within early (HH3) primitive streak are already segregated.67, 68
In summary, regional signaling in the epiblast positions presumptive cardiac lineages within the posterior epiblast, where they undergo gastrulation from mid- to late- streak stages. The ordered arrangement of presumptive anterior-posterior progenitors may reflect the relative proximity of each progenitor to the node during ingression, as dictated by their original spatial orientation to each other. Specification and restriction of myocardial cell fate is likely to initiate during gastrulation, and also involves signaling within the cardiac crescent and adjacent pharyngeal endoderm. The timeframe for cardiac specification appears to be comparable in both frog and fish: in Xenopus, extirpation, explantation, and tissue recombination studies suggest that cardiac specification begins at the onset of gastrulation,69-71 and in zebrafish, experiments employing pharmacological antagonists suggest myocardial specification occurs during gastrulation.72-74
a. Early Cardiac Specification
As cells ingress through the cranial primitive streak they are exposed to signaling factors expressed in and around the node including retinoic acid and FGFs.75, 76, 81 FGFs produced by the node can induce expression of Nkx2.5 locally77 and an ectopic node can induce ventricular myosin heavy chain (vMHC) but not atrial myosin heavy chain (aMHC) expression. Because ventricular progenitors are closer to the node, this suggests that ventricular cardiomyocyte identity may be established in the primitive streak.78
The earliest identified indicator of cardiogenic mesoderm is the transcription factor Mesp1 expressed at the onset of gastrulation, E6.5-7.5.79 However Mesp1 is not specific for the cardiogenic mesoderm, as Mesp1Cre labeled lineages broadly contribute to multiple mesoderm-derived populations in the embryo. Mesp1 is downstream of Fgfr1 signaling, and both Mesp1 and Mesp2 are required for mesodermal ingression during gastrulation. Mutation of Mesp1 results in cardia bifida, and cells that are doubly mutant for Mesp1 and Mesp2 are selectively unable to contribute to the heart, suggesting a critical role in cardiac mesoderm specification. Mesp1 is upstream of a number of cardiac specification transcription factors, including Nkx2.5, Islet1 and myocardin, which begin to be expressed at the cardiac crescent stage.
After gastrulation, molecular markers begin to be useful indicators of the location of the heart fields in cranial lateral plate mesoderm. In Xenopus, the first markers of the cardiac lineage, including Nkx2.5, Tbx5, Tbx20, Gata4, Gata5, and Gata6, are not expressed until neurula stage, when the heart progenitors have moved anteroventrally. However, as in other vertebrate model systems, analysis of the cardiac lineage in Xenopus is confounded by the fact that most of these markers are expressed in neighboring non-cardiac tissue or in only a subset of cardiac progenitors.80 Thus, it is the combinatorial expression of these transcription factors that marks cardiac progenitor cells.59 The situation is similar in zebrafish, where the same transcription factors, with the addition of Hand2, combinatorially distinguish the locations of the heart fields.47
As cardiac progenitors move cranial-laterally during gastrulation to arrive at the cardiac crescent, they are exposed to factors produced by endoderm, including FGFs and BMPs.81, 82 In mouse, expression of Nkx2.5 in the cardiac crescent is dependent on BMP2 signaling.83, 84 However, in the second heart field, Nkx2.5 interacts directly with Smad to downregulate BMP2 signaling which limits the number of cardiomyocytes that differentiate to populate the initial heart tube.84 Signaling through the hedgehog receptor smoothened is also required for the early phase of Nkx2.5 expression in the cardiac crescent, although not for the later phase within the developing heart.85 In Xenopus embryos, inhibition of Wnt signaling in endoderm is required for expression of an as yet unidentified secreted factor required for cardiac specification.82
Key signals specifying myocardial progenitors are likely to be conserved among vertebrate species. Indeed, studies in zebrafish have indicated the importance of the BMP, FGF, and Nodal pathways. For example, swirl (bmp2b), acerebellar (fgf8), and one-eyed pinhead (oep) mutants all exhibit a decreased number of cardiomyocytes preceded by decreased expression of early markers of myocardial identity, such as nkx2.5.86, 87 Similarly, loss of Hedgehog signaling in zebrafish smoothened mutants reduces the number of myocardial progenitors.74 Furthermore, fate map analyses have suggested that loss of Nodal or Hedgehog signaling leads to a loss of cardiac progenitors via cell fate change, rather than through effects on progenitor proliferation.46, 74 Integration of BMP, FGF, Nodal, and Hedgehog signals within the zebrafish heart field is likely to trigger commitment to a myocardial fate.
Comparable signaling pathways may also be active in Xenopus, where signals from the embryonic organizer and the endoderm act together to specify the cardiac lineage.69-71 Candidate molecules with either a passive or instructive role in Xenopus cardiac specification include members of the BMP, FGF, and Wnt families. In addition, an inhibitory role in cardiac specification has been proposed for the Notch pathway.88 However, neither activation nor inhibition of any components of these pathways has been shown to block Xenopus heart formation in vivo. Collectively, these studies suggest that cardiac specification in Xenopus depends on either a combination of redundant signal transduction pathways or an as yet unidentified signaling mechanism.
In all the animal models studied, the LIM homeodomain transcription factor Islet1 marks proliferating undifferentiated cardiogenic progenitors. Islet1 is required for proliferation and survival of second heart field progenitors, and may be required for their migration into the heart.89 Early expression of Islet1 is dependent on canonical Wnt signaling through beta-catenin and on Fgf8 signaling, both of which are required for proliferation of early cardiac progenitors.90-92 Wnt2a, 2b and 8 are expressed in appropriate regions and may serve as ligands in the mouse.93-95 Expression of a mesodermal enhancer of Islet1 is dependent on three conserved consensus Forkhead transcription factor binding sites.96 Islet is upstream of expression of BMP4, BMP7, and Fgf10, and regulates expression of Shh in pharyngeal endoderm, all of which may contribute to its role in driving proliferation. Mutants in which Fgf8 is ablated by Mesp1Cre exhibit arterial pole defects reflecting the required function of Fgf8 in arterial pole progenitors of the second heart field. By contrast Islet1 mutants have both arterial and venous pole defects.97, 98 In chick explants, FGF does not appear to enhance proliferation in outflow progenitors but is more likely to suppress myocardial differentiation and promote smooth muscle differentiation, suggesting perhaps an earlier role for Fgf8 in proliferation of mesodermal progenitors.99
Several experiments suggest a pivotal role for GATA4 or GATA5 in cardiomyocyte specification. In zebrafish embryos, ectopic expression of Gata5 is sufficient to specify non-cardiogenic mesoderm to a cardiogenic fate100 and ectopic Gata4 can induce myocardial differentiation of Xenopus ectodermal explants, in the absence of endoderm.101 Recent experiments in gastrulating mouse embryos demonstrated that ectopic expression of the chromatin remodeling factor Baf60c and GATA4 is sufficient to specify non-cardiogenic to cardiogenic mesoderm, and the addition of Tbx5 to these two factors is sufficient to promote cardiomyocyte differentiation.102
While some factors specify cardiogenic mesoderm, others restrict the cardiogenic field. Retinoic acid signaling imposes the posterior limit of the early cardiac crescent. In chick embryos, exposure to retinoic acid produces atrial expansion.103 Mutation of the retinoic acid synthesizing enzyme retinaldehyde dehydrogenase 2 (RALDH2) in mouse results in expansion of Islet1 and Fgf8 expression, and consequent enlargement of the heart.104, 105 Similarly, in zebrafish, mutation of raldh2 causes formation of large hearts composed of too many cardiomyocytes.106 Fate mapping demonstrated that this myocardial surplus results from a fate transformation that increases myocardial progenitor density within the forming heart field.106 Intriguingly, an increased number of cardiomyocytes is also observed in embryos lacking function of hoxb5b, a retinoic acid-responsive gene expressed within the forelimb field, which is located just posterior to the heart field.107 Thus, it seems that retinoic acid signaling restricts the developmental potential of the heart field at least in part through its influences on neighboring territories of lateral mesoderm. Wnt signaling also inhibits cardiac progenitor specification within the heart field, although it has also been shown to have a positive influence at earlier stages.108 Additional pathways that control endothelial and hematopoietic specification contribute to the restriction of myocardial progenitor specification. Ectopic cardiomyocytes and enlarged hearts form in zebrafish mutants that lack vessel and blood lineages, and fate mapping demonstrated that this phenotype is caused by transformation of a cranial region of lateral mesoderm that normally produces endothelial and hematopoietic progenitor cells.47 It is not yet clear how the network of multiple inductive and repressive pathways is interwoven to produce the proper number of myocardial progenitors.
b. Left Right Patterning
The heart exhibits left-right asymmetry, critical for a number of aspects of cardiogenesis.57 Humans with laterality defects exhibit abnormal venous and arterial connections of the heart. Evidence of left-right patterning in both chick and mouse can be observed in asymmetrical expression of a number of genes at late gastrulation.109, 110 Left-sided nodal expression in perinodal tissue is correlated with left sided expression of Nodal, Lefty, and Pitx2 in lateral mesoderm associated with cardiogenic mesoderm.111, 112
Selective contribution of left and right heart field progenitors to specific regions of the heart is determined by complex morphogenetic movements, regulated by differential cell proliferation and cell shape changes driven by distinct genetic programs within left and right progenitors. We are just beginning to understand the manner in which laterality cues drive these specific cell behaviors to shape organ morphogenesis and laterality. An interesting example of this is demonstrated by studies in zebrafish.57 An early manifestation of asymmetric morphogenesis in zebrafish is the leftward jogging of the heart tube, which is dependent on asymmetric BMP signaling, with greater signaling activity on the left. BMP signaling acts to direct the relative speed and direction of cardiomyocyte migration. Simultaneously with the onset of jogging, myocardial cells on the right side of the cardiac cone involute and move ventrally in a cranial and leftward direction. In this way, cardiomyocytes derived from the right heart field form the ventral part of the cardiac tube, while cardiomyocytes derived from the left heart field contribute to the dorsal roof of the tube. Perturbation of left-right signaling pathways affects the orientation of tissue involution and heart tube jogging.
Similar imposition of left-right patterning to dorsal and ventral domains within the heart occurs in other model organisms. Left and right heart field progenitors meet in the ventral midline. In the frog, cells that express a newly described gene called Castor populate the midline. This population is important for fusion of the two sides in Xenopus and may be important in maintaining a boundary between derivatives of the left and right heart field in the left ventricle.113 In chick and mouse, once the dorsal seam has closed and the dorsal mesocardium regressed, the looping tube rotates 90° to the right. This brings the left heart field derived myocardium on the left ventricle to the ventral surface of the looped tube while right heart field derived myocardium is turned dorsally.10 Even though the ventricles are from both heart fields the cells from left and right do not mix at the greater and lesser curvatures.10 It is not known if this arrangement is maintained by cells that are added to the tube to form the right ventricle and outflow tract myocardium.
Expression of Pitx2c in the heart delineates structures derived from left lateral plate mesoderm, which include the left AV canal, the inner curvature, the ventral part of the interventricular ring, the ventral portion of the right and left ventricles, and the left atrial appendage.114-116 Available data suggest that left and right atria are derived from left and right heart fields, respectively.10, 117 DiI labeling of the cardiogenic fields demonstrated that progenitor cells in the right or left caudal second heart field contribute to the right or left atrium, respectively.
Some atrial myocytes are contributed from the first heart field but the atria and atrial septum are substantially supplemented by addition of cells from the second heart field.33, 118, 119 The derivation of atrial cardiomyocytes is very diverse and encompasses at least four different lineages including atrioventricular myocardium which shifts into the atrium during later development, atrial myocardium derived from the first heart field, myocardium incorporated from the sinus venosus and pulmonary veins both of which are derived from second heart field.115, 120
Explant experiments with transgenic mouse embryos, in which a transgene marks the right atrium, show that atrial progenitor cells acquire right-left identity between the 4- and 6-somite stages, at the time when Pitx2c is first expressed. Pitx2 functions in second heart field myocardium to repress right atrial identity and proliferation of cells around the left sinus venosus.121 Myocardial specific ablation of Pitx2 results in abnormal persistence of BMP10 expression in fetal left atrium, resulting in an enlarged left atrium exhibiting right atrial morphology,122 consistent with right atrial isomerism previously observed in Pitx2 global null embryos.116 It is interesting to speculate that Pitx2 may also restrict growth of ventricular cells derived from progenitors that received left-sided cues earlier in development.
Shh is necessary for the production of asymmetry in the developing embryo. Fate mapping studies with an inducible Gli1-CreERT2 demonstrated that hedgehog-receiving progenitors migrate from the second heart field to populate the right-heart derived subpulmonary myocardium.119 Shh null embryos exhibit bilateral expression of Pitx2c at arterial and venous poles of the heart, suggesting that Shh signaling normally suppresses Pitx2c expression on the right.123 Consistent with this, Shh mutants also exhibit left atrial isomerism.
Consistent with left-right information being important for normal patterning of the outflow tract, Pitx2 is upstream of a genetic cascade, including Wnt11 and TGFbeta2, which is required for correct alignment of the great arteries with their respective ventricles.124 Pitx2 interacts with beta-catenin to activate expression of Wnt11, which then acts through a non-canonical Wnt signaling pathway to activate transcription of TGFbeta2. Vangl2, a component of the non-canonical planar cell polarity pathway (PCP), is also required. Loop-tail (Lp) mice, which have mutations in Vangl2, exhibit double outlet right ventricle and aortic arch defects.125 As with Wnt11 and TGFbeta2, Vangl2 is required for the polarization and movement of myocardializing cells into the outflow tract cushions, and RhoA and ROCK1 are downstream mediators of the PCP signaling pathway in the developing outflow tract.125
Tbx1 is co-expressed with Pitx2 in second heart field cells, and Pitx2 expression is downregulated in Tbx1 mutants, suggesting a role for Tbx1 in asymmetric cardiac morphogenesis even though Tbx1 expression is not asymmetric.126 Asymmetric expression of Pitx2 is regulated by a Tbx1-Nkx2.5 binding enhancer. Semaphorin3C-expressing subpulmonary myocardium is reduced and malpositioned in Tbx1 mutant hearts, consistent with perturbation of left-right patterning.127
c. Craniocaudal Patterning
Although some of the cues involved in craniocaudal patterning during myocardial specification have been elucidated, we know relatively little mechanistically as to how craniocaudal patterning is integrated by developing myocardial cells. A key regulator of craniocaudal patterning in the embryo generally, and for myocardial cells specifically is retinoic acid. As discussed above, at early stages of cardiac specification, retinoic acid signaling restricts the heart field. At later stages, a posterior to anterior gradient of retinoic acid signaling is important for normal patterning of both posterior and anterior segments of the heart.
Tbx5 expression is downstream of retinoic acid signaling, and is selectively expressed within the left ventricle and atria. Higher levels of expression of Tbx5 in atria than in left ventricle suggest that levels of Tbx5 reflect a retinoic acid gradient important for chamber specialization.128, 129 Ectopic expression of Tbx5 utilizing a ventricular specific promoter results in ectopic expression of atrial genes. Tbx5 may also define a boundary between right and left ventricles, which is important for placement of the ventricular septum.130, 131
At the arterial pole of the heart, Crkl1 and Tbx1 positively regulate expression of retinoic acid metabolizing enzymes, and negatively regulate RALDH2, reducing local concentrations of retinoic acid in second heart field progenitors that will contribute to anterior segments of the heart.132 Increased retinoic acid in Crkl1 and Tbx1 mutants affects pharyngeal segmentation and patterning along the craniocaudal axis.132, 133 In turn, retinoic acid represses Tbx1 expression in second heart field progenitors that will contribute to posterior segments of the heart. It is interesting to note that Tbx1 appears at the intersection between craniocaudal and left-right patterning.
IV. Cardiomyocyte Differentiation
Transitioning from proliferative progenitors to differentiated cardiomyocytes requires both downregulation of expansion pathways and upregulation of differentiation pathways. Several pathways that promote proliferation of cardiac progenitors also actively prevent differentiation, likely by inhibition of pathways required for differentiation.
BMP signaling is required for cardiomyocyte differentiation. Expression of a dominant negative ALK3 receptor in Xenopus embryos prevented expression of differentiation markers in cardiogenic mesoderm, and BMP signaling is required for maintenance of Tbx20 expression.134, 135,136, 137
Nkx2.5 represses BMP2 activated Smad1 signaling to inhibit differentiation.84 In addition to positively regulating second heart field proliferation, Tbx1 negatively regulates differentiation in the caudal pharyngeal region by inhibition of BMP signaling.138 Tbx1 binds Smad1 and suppresses BMP4/Smad1 signaling, and expression of Tbx1 in transgenic mice generates phenotypes similar to those associated with loss of a BMP receptor. In Tbx1 null mutants, premature differentiation occurs within second heart field progenitors.138 Islet1, Nkx2.6, and Hod are downregulated in Tbx1 mutants, while myocardial differentiation genes including Raldh2, Gata4, Tbx5, and a subset of muscle contractile genes are ectopically expressed. Tbx20 is a positive effector of differentiation downstream of BMP signaling within the heart, and also represses expression of Islet1 as progenitors enter the heart.134, 139
Genetic analysis and protein depletion studies demonstrate that embryos lacking Tbx20 show a progressive loss of cardiomyocytes, a failure of the heart to undergo looping and chamber formation, as well as defects in cardiomyocyte maturation.140-143 Despite the early cardiac expression of Tbx20 in vertebrate development and the associated downregulation of early heart markers such as Nkx2.5 and Tbx5, TBX20 depleted embryos still form a primary heart tube which appears to be correctly patterned along the anterior-posterior axis, since polarized expression of numerous cardiac molecular markers, including a-MyHC, b-MyHC, Tbx5, and Gata4 are unaltered in Tbx20 mutants. Instead, the cardiac abnormalities in Tbx20-depleted and mutant embryos appear to be the outcome of a fundamental requirement for Tbx20 in proliferation of cardiomyocytes as demonstrated by a dramatic decrease in the number of terminally differentiated cardiomyocytes in Tbx20 mutants.140-143
Hedgehog signaling is required for proliferation of second heart field progenitors,29 and is required to prevent differentiation of second heart field cells as evidenced by premature differentiation of second heart field progenitors following ablation of the smoothened receptor with a second heart field Cre.144 Downregulation of another pathway involved in proliferation of second heart field progenitors, Wnt/beta catenin, is required for differentiation of myocardial progenitors, as evidenced by lack of differentiation when beta-catenin is constitutively activated selectively in cardiac progenitors.91-93, 145
Although beta-catenin is initially required for proliferation of undifferentiated progenitors, beta-catenin also directly regulates expression of Wnt11, which stimulates a non-canonical Wnt pathway to effect critical aspects of cell polarity as myocardial cells differentiate.124, 125 It is interesting to speculate that activation of non-canonical signaling through Wnt11 may in turn downregulate beta-catenin, to further promote differentiation in a feedback loop, as there are many examples where non-canonical Wnt pathways downregulate canonical Wnt signaling.
SRF, a MADS box transcription factor, is required to regulate sarcomeric protein genes and excitation-contraction coupling proteins. Ablation of SRF in the Nkx2-5 expression domain results in failure to initiate cardiac beating.146 SRF regulates expression of microRNAs miR-1 and miR-133, which are derived from a common precursor transcript. SRF interacts with GATA4 and Nkx2.5 to regulate cardiac genes147,148. Multiple loci encode miR-1 and miR-133, including miR-1-1, miR-1-2, miR-133a-1, miR-133a-2, and miR133b. MicroRNAs are evolutionarily conserved small, noncoding RNA molecules that function posttranscriptionally to negatively mRNA translation and stability thus playing critical roles in heart development.149, 150 In keeping with the role of SRF in promoting differentiation, MiR-1 and MiR-133 may promote cell cycle withdrawal. Overexpression of MiR-1 under the control of the betaMHC promoter induces premature exit from cell cycle, in part by inhibiting translation of Hand2.147 Hand2 null mutants die at E10.5 with right ventricular hypoplasia and decreased trabeculation in the left ventricle.151 Loss of miR-1-2 in mice, resulting in a 50% decrease in total miR-1, results in partial lethality between E15.5 and birth, with ventricular septal defects and cardiac dysfunction.152 Loss of miR-133a-1 and miR-133a-2 results in excessive cardiac proliferation, partial lethality and ventricular septal defects, similar to miR-1-2 mutants.153
A hypoxia driven pathway stimulates myofibrillogenesis. Ablation of Hypoxia-Inducible-Factor (HIF) 1 alpha in differentiating myocytes results in lack of differentiation, owing to decreased titin expression. The contractile apparatus cannot be assembled in its absence.154
In summary, downregulation of beta-catenin and Shh, BMP, and non-canonical Wnt signaling are required to promote cardiomyocyte differentiation. Regulation of myofibrillar gene expression is driven by SRF in concert with Nkx2.5 and GATA factors. SRF also regulates expression of micro RNAs that promote cell cycle withdrawal. Hypoxia pathways may also contribute to myocyte differentiation.
a. Chamber versus Non-chamber Myocardium
As morphogenesis of the heart proceeds from the early phases of differentiation, the myocardium becomes subdivided into either chamber (ventricles, atria) or non-chamber (outflow tract, AV canal, inner curvature, central conduction system) myocardium.155 Non-chamber myocardium, similar to early-differentiated cardiomyocytes, is relatively non-proliferative, whereas chamber myocardium undergoes a “ballooning” phase of growth, beginning at about E9.5 that is driven by signals from both endocardium and epicardium.
Delineation of chamber and non-chamber myocardium is achieved in part by expression of a T-box transcription factor code, including Tbx2, Tbx3, Tbx20, Tbx5, and Tbx18 which imparts regional myocyte identity and coordinately regulates regionally appropriate levels of proliferation.156 T-box genes confer regional myocyte identity both by activating expression of identity-appropriate genes, and by repressing expression of identity-inappropriate genes.
Localized expression of Tbx2 confers AV canal identity to the region separating the ventricle from the atrium, repressing expression of chamber specific genes ANF (Nppa), chisel, connexin 40 (Cx40), and connexin 43 (Cx43), and repressing proliferation.155 Tbx2 represses regulatory fragments of Cx40, Cx43, and Nppa, interacting with Nkx2.5 to repress expression of the latter.157 Ectopic expression of Tbx2 in chamber myocardium results in repression of chamber specific genes.155 Tbx2 is also required for development of the outflow tract.158
Although active expression of Tbx2 within the heart is restricted to the AV canal and outflow tract, recent genetic lineage tracing with a Tbx2Cre has revealed that Tbx2 lineages make an unexpectedly large contribution to Tbx2-negative ventricles, with the embryonic left ventricle only forming the left part of the definitive ventricular septum and the apex.159 Tbx2 lineages also contribute to the AV node, but not the AV bundle. In the outflow tract, Tbx2 lineages formed the right ventricle and right part of the ventricular septum.
Tbx3 is co-expressed with Tbx2 in outflow tract progenitors and AV canal, and is selectively expressed in the central conduction system. Tbx3 is required for normal alignment of the outflow tract and normal ventricular septation.160 Tbx3 is also required for specification of the AV conduction system, at least in part by repressing expression of chamber specific genes.161
Tbx20, which is expressed throughout the developing heart, is required for proliferation and expression of chamber myocardial genes and represses expression of Tbx2 in ventricular chamber myocardium.140, 142, 162 Tbx5, expressed in atria and left ventricle, is required to maintain expression of Nppa and Cx40.163 Tbx18 is expressed in sinus venosus myocardium and in a subset of SA nodal cells and is required in both lineages for their normal development.164
b. Patterning of the AV Canal
The non-chamber myocardium of the AV canal performs several specialized functions during heart development, serving as a slow conducting “sphincter” in the early heart tube, and later actively participating in signaling to form cardiac cushions and partition the heart. Patterning of AV myocardium is also likely to affect development of the central conduction system, the AV node and associated structures.
Many details of the molecular pathway that regulates specification of AV canal myocardium have been worked out. Normal AV patterning requires myocardial BMP4 and BMP2.165, 166 BMP2 An important role of BMP4 may be to activate transcription of the forkhead transcription factor Foxn4 as studies in zebrafish embryos have demonstrated that Foxn4 directly regulates expression of Tbx2 in the AV canal.167 BMP2 also signals directly to cushion-forming endocardium to induce EMT at least in part by activating expression of Tbx2.166, 168 This has been demonstrated genetically with mice that are compound heterozygous null for Bmp2 and Bmp4 are embryonic lethal, exhibiting outflow tract malalignment, and defective septation of atria, ventricles, and AV canal revealing novel functions of BMP2 in concert with BMP4.169 Msx1 genes are not only a target of BMP2, but both regulate and cooperate with Tbx2 to pattern AV myocardium and to effect AV cushion formation.170 Msx1 and Msx2 can directly interact with Tbx2 and Tbx3 to suppress Cx43 promoter activity.171
BMP2 expression in mouse, and BMP4 expression in zebrafish is restricted to the developing AV canal and inner curvature (IC) at least in part through the activity of Notch2 and Hey proteins.172 For example, in mice, embryos mutant for Hey2 exhibit an expanded AVC domain of BMP2 and zebrafish mutants in Hey2 (aka gridlock) show ectopic expression of BMP4. Conversely, in developing chick heart, ectopic activation of Notch2 signaling, or misexpression of either Hey1 or Hey2, represses BMP2. Moreover, ectopic expression of Tbx2 inhibits expression of Hey1 and Hey2. Likewise, mouse embryos ectopically expressing Hey1 throughout the heart display poorly defined boundaries of the AV canal and expression levels of AV myocardial markers BMP2 and Tbx2 are weak or undetectable though in this case, independent of Notch2 signaling. 173 Taken together these studies suggest that Tbx2 expression may sharpen the border of developing AV canal and IC by restricting Hey gene expression and hence BMP2 and BMP4 to prospective chamber regions.
Myocardial identity can also regulate regionally vy MicroRNAs. For example, MicroRNA-138 is expressed in AV canal in zebrafish heart and plays a role in establishing the AV canal myocardium.174 Disruption of miR-138 function leads to ventricular expansion of gene expression normally restricted to the AV canal and disrupted ventricular cardiomyocyte morphology and function.
In summary, data are consistent with a model in which Hey1 and Hey2 expression bordering the AV canal restricts BMP2 to AV canal myocardium. BMP signaling in turn activates expression of Tbx2, Msx1 and Msx2, which cooperate to regulate the AV myocardial phenotype. How Foxn4 interacts with BMP signaling remains to be determined.
c. Cardiac Conduction System
The specialized cardiac conduction system is comprised of central slow-conducting components, which include the SA node, AV node, His bundle and bundle branches, and fast-conducting components, including internodal tracts, and Purkinje fibers.175-177 A number of connexins are specifically expressed within certain components of the cardiac conduction system, and confer fast or slow conducting properties.178-180 Cx45 and Cx30.2 are expressed within nodal tissues, and Cx40 within specialized fast-conducting tissues, with some Cx40 expression within the AV node. High conductance channels Cx43 and Cx40 are expressed within the working myocardium of the atria and ventricles. Recently a number of transcription factors important for conduction system formation have been described, including Nkx2.5, Shox2, Hop, Irx4/Irx5, Tbx2, Tbx3, Tbx18, Tbx5, and Id2. Their role in conduction system formation has been recently reviewed.181, 182
Viral lineage tracing studies in chick embryos showed that conduction cells are progressively recruited from cardiomyogenic cells with recruitment to elements of the central conduction system (e.g. the His bundle) preceding recruitment to the peripheral components of the network (i.e. subendocardial and periarterial Purkinje fibres).183-185 Birth dating studies in rodents suggest an analogous recruitment process occurs.186
Factors secreted by endothelial cells appear to be important for recruitment of myocardial cells into Purkinje fiber lineages, and, at least in zebrafish, for AV conduction system formation. In chick embryos, endothelin secreted by coronary vascular cells and endocardium can influence recruitment of myocardial cells into the Purkinje fiber lineage.187 Intriguingly, stretch influences the processing and secretion of ET-1 and stretch/flow activates the expression of endothelin converting enzyme-1 (ECE-1). This in turn regulates recruitment of myocardial cells into the Purkinje fiber lineage.188 In mouse embryos, neuregulin, which is secreted by endocardium, can recruit myocardial cells into the Purkinje fiber lineages.189 Endocardial signals are also important for formation of the AV conduction ring in zebrafish heart, and Neuregulin and Notch1b are required for formation of this central conduction tissue.190
Although a number of transcription factors are known to pattern the conduction system, we know relatively little about the signaling pathways required for conduction system formation, or the manner in which these signaling pathways cross talk with the transcription factors that are important for conduction system specification.
d. Chamber Regionalization/Specialization
As discussed above, patterning, in large part by T-box genes, differentiates chamber myocardium from non-chamber myocardium. However, within chamber myocardium, sub-regionalization is also important to confer atrial or ventricular identity, or to discriminate between left and right atria or ventricles. A number of early studies suggested that both selective transcriptional activation and repression are important for conferring regional chamber identity. Early studies of myofibrillar protein mRNA expression in developing mouse heart demonstrated that many myofibrillar protein isoforms are first expressed throughout early differentiating myocardium and then become selectively downregulated in either atrial or ventricular chambers, within a time frame specific to each isoform.191, 192 Three myosin genes which become transcriptionally restricted to the mouse atria between embryonic day E12.5 and birth are coordinately downregulated in compact myocardium of the left ventricle before that of the right ventricle.115 Other isoforms exhibit more restricted domain specific expression from the beginning, including MLC2v in mouse, and ventricular myosin heavy chain in zebrafish.55, 192 Analysis of transgene expression patterns has also revealed regional transcriptional regulation.193 A myosin light chain (MLC) 3F-nlacZ-2E transgene is expressed in embryonic right atrium, AV canal, and left ventricle, whereas an MLC3F-nlacZ-9 transgene is expressed in atria, ventricles and AV canal, but not in inflow and outflow tracts.194
In keeping with these observations, a number of transcription factors have been described that regulate activation or repression of region-specific gene expression. Tbx5 plays an important role in conferring atrial identity and establishing the boundary between left and right ventricles. Irx4 is a ventricle-specific transcription factor that confers ventricular identity by activating ventricular specific gene expression, and by repressing atrial specific gene expression.195, 196 The basic helix-loop-helix transcriptional repressor Hey2 is expressed specifically in ventricular myocytes, and confers ventricular identity.197 Cardiomyocyte-specific ablation of Hey2 results in ectopic activation of atrial genes in ventricular myocardium, with an associated impairment of cardiac contractility and a unique distortion in morphology of the right ventricular chamber. Forced expression of Hey2 in atrial cardiomyocytes is sufficient to repress atrial cardiac genes.
In the mouse, the basic helix-loop-helix transcription factor Hand2 is expressed in cardiac precursors throughout the cardiac crescent and the linear heart tube, before becoming restricted to the right ventricular chamber at the onset of looping morphogenesis. A Hand2 enhancer that directs lacZ expression in this pattern requires two conserved GATA factor consensus sites for expression in right ventricle but since GATAs are expressed universally by myocardial cells this does not explain why Hand2 expression is limited to the right ventricle.198 Hand1 is selectively expressed in the left ventricle, and, as for Tbx5, the boundary between Hand1 expressing and non-expressing myocardium may affect formation of the ventricular septum.199 Hand1 and Hand2 are critical for ventricular chamber expansion, as ablation of both transcription factors in heart severely reduces ventricular tissue.200
Thus, regional chamber identity is regulated by transcription factors that both activate and restrict gene expression to confer chamber identity. Epigenetic mechanisms are also likely to be involved in maintaining patterning of the mammalian myocardium.201 Maintenance of Mlc3f transgene expression is regulated by a cell-autonomous mechanism that is independent of DNA methylation, trichostatin A-sensitive histone deacetylation, or misexpression of transcription factors that are expressed in left or right ventricles of embryonic heart. This recalcitrance to ectopic activation is suggestive of a “locked” epigenetic configuration, similar to silencing mediated by polycomb in Drosophila.
f. Epicardium and Myocardial Lineages
As discussed briefly above, cells from the proepicardium migrate onto the early looping heart to give rise to the epicardium. One of the roles of the developing epicardium is to promote proliferation of underlying myocardium. Lineage studies in avians demonstrated that epicardial cells undergo epithelial-mesenchymal transformation and delaminate from the epicardium to enter the myocardium to give rise to cells of the coronary vasculature and interstitial fibroblasts.202, 203 It is likely that each of these cell types is involved in crosstalk with myocardial cells, and in fact embryonic cardiac fibroblasts have recently been shown to promote myocardial proliferation.204 Myocardial signaling is reciprocally important for development of coronary vessels, which arise in part from epicardial lineages. Crosstalk between the epicardium and myocardium has recently been reviewed in depth in an article in the same series, and will not be discussed further here.205 Here, we will focus on recent studies that have suggested that epicardial lineages have the potential to adopt cardiomyocyte cell fates.
Proepicardial progenitors are contiguous with progenitors that will give rise to sinus venosus myocardium, and both sets of progenitors express the T-box transcription factor Tbx18.206, 207 Studies in chick embryos have suggested that the ability of Tbx18 progenitors in this domain to adopt either cardiomyocyte or proepicardial fate is based upon differential BMP or FGF signaling respectively.208-210 Proepicardial cells can adopt a cardiomyocyte cell fate when BMP-Smad signaling is activated in the context of inhibition of FGF signaling via mitogen-activated protein kinase kinase 1/2.
Lineage studies in mouse embryos utilizing two distinct epicardial-Cre mouse lines, Tbx18-Cre and Wt1-Cre, and fluorescent tracker dye labeling of epicardial cells in heart organ explant cultures, have suggested that, in addition to cardiac fibroblasts and coronary vascular support cells, epicardial lineages may contribute to a subset of cardiomyocyte lineages.40 Clonal analysis of proepicardial progenitors demonstrated that single clones were able to give rise to both smooth muscle cells and cardiomyocytes in vitro. In the same culture conditions, however, adult epicardial cells were able to adopt smooth muscle, but not cardiomyocyte cell fate.40 As Cre lineage studies are subject to uncertainties concerning expression domains of the Cre-driver,41 the question as to whether epicardial lineages contribute to myocardial lineages in mouse embryos awaits “bona fide” in vivo lineage tracing studies. In this regard, Kispert and colleagues have recently shown at the Weinstein meeting that Tbx18 is expressed in the left ventricle and ventricular septum as well as in the epicardium, thus leaving open the question of an epicardium-derived cardiomyocyte lineage.211
The foregoing suggests that proepicardial lineages have the capacity to be directed toward a myocardial cell fate, at least in vitro, whereas later stage epicardial cells may have lost this ability. Understanding genetic differences between proepicardial and adult epicardial cells that are responsible for differences in ability to adopt cardiomyocyte cell fates may have important implications for utilization of epicardial cells for cardiac repair.
g. Growth Signals from Endocardium
As with the epicardium, growth factor signaling from endocardium to myocardium is responsible for stimulation of myocardial proliferation following cardiac looping. The early ventricular myocardium is composed of trabeculae, fingerlike projections of myocardium that are lined by endocardium, the specialized vascular endothelium lining the heart. This structure provides for a high endocardial cell to myocardial cell ratio.
Neuregulin 1 (NRG1) secreted by endocardial cells activates ErbB2 and ErbB4 receptors in myocardial cells to promote formation of trabecular myocardium.212-214 Serotonin signaling through 5-HT(2B) receptors is required for expression of ErbB2 in ventricular myocardium, and mutants of 5-HT(2B) receptors exhibit impaired trabeculation.215 Notch signaling within endocardium functions upstream of NRG1 with mutants for Notch 1 and the transcriptional effector downstream of Notch, RbpJ, exhibiting impaired trabeculation, and decreased myocardial proliferation.216 Thus, Notch signaling in endocardium is required for expression of EphrinB2, which in turn activates expression of NRG1.
Endocardial Notch signaling also activates expression of BMP10 in myocardium, which is required for trabecular proliferation. BMP10 null mice display ectopic and elevated expression of p57(kip2) and a dramatic reduction in proliferative activity in cardiomyocytes at E9.5. BMP10 is also required for maintaining normal expression levels of Nkx2.5 and Mef2C in developing myocardium at mid-gestation.217 Myocardial specific ablation of Nkx2.5 results in increased expression of BMP10, resulting in over-proliferation of trabeculae,218 suggesting a negative feedback loop from BMP10 to Nkx2.5 which is required for regulation of normal trabecular growth. It is interesting that this echoes an earlier BMP-Nkx2.5 feedback loop required during early cardiac specification.84
FGF signaling from either epicardium or endocardium stimulates myocardial proliferation. FGF9, FGF16, and FGF20 are expressed in endocardium as well as epicardium, and endocardially-derived FGF signals regulate myocardial proliferation during midgestation heart development.219 Ablation of FGF16 results in embryonic lethality with poor trabeculation and thin-walled myocardium.220
Modulation of extracellular matrix by proteases secreted by endocardium can also modulate myocardial growth. For example, ADAMTS1 is a metalloprotease normally expressed in the matrix assocaited with the endocardium at later stages of trabecular development, when trabecular growth is slowing221. Ablation of the chromatin remodeling protein Brg1 results in precocious expression of ADAMTS1 and thus, inhibits early trabecular growth.
In summary, Notch signaling within endocardium activates mid-gestation myocardial growth through two pathways. Endocardial Notch signaling upregulates EphrinB2 required to upregulate endocardial Neuregulin, which binds to myocardial ErbB2/ErbB4 receptors to stimulate growth. Notch signaling within endocardium also upregulates BMP10 in myocardium, through as yet unknown mediators. Serotonin, FGF, and protease pathways are also critical to the crosstalk between endocardium and myocardium at mid-gestation to regulate proliferation of myocardium. Although FGF and BMP signaling are also important for proliferation of cardiac progenitors prior to differentiation, ErbB2/ErbB4 signaling appears to be specific for proliferation of differentiated cardiomyocytes. It will be of future interest to investigate commonalities and differences between endocardial and epicardial pathways which crosstalk to cardiomyocytes. It is interesting to speculate that differential stimulation of pathways from either source may influence endocardial to epicardial gradients of gene expression.
Summary
The journey of a prospective myocardial epiblast cell to a highly specialized cardiomyocyte is a “long and winding road”. Proximity to signals emanating from the node (retinoic acid, nodal) may impart initial craniocaudal and left-right axis information to the presumptive cardiomyocyte, which will be reinforced later by signals from lateral plate mesoderm and genetic pathways within cardiogenic progenitors and cardiomyocytes themselves. Axial patterning programs are likely to drive individual cell migratory and proliferative behavior. Reliance on and proximity to endodermal signals during development is another theme for presumptive cardiomyocytes, including early position in the epiblast, during initial ingression at gastrulation, and later when the cardiac crescent forms adjacent to definitive anterior endoderm. As cardiogenic progenitors enter the heart to become myocytes, they are in proximity to other cardiac cell types, including endocardial and epicardial lineages, which are also involved in signaling and crosstalk to influence myocardial development.
Becoming a specialized cardiomyocyte is a complex ordered process involving specification, differentiation and specialization. This process involves an ordered activation and repression of growth factor signaling pathways, regulated by transcription factor regulation of ligands, receptors, transcription factors, and microRNAs. Activation of a particular pathway has several outcomes, the most immediate goal of stimulating proliferation, cell motility/polarity, differentiation, or further specialization, and at the same time the initiation of the next phase in the cascade, which in turn downregulates activation of the initial pathway, ensuring progression. During and after chamber formation, signaling to establish and maintain specialization and proliferation of the myocardium to form trabeculae and other specializations falls to the endocardium and epicardium. Maintenance of the specialized state is also likely to require active signaling processes.
Much of our work to date has focused on earliest stages of heart development, while less is known as to signaling pathways required for maturation of cardiomyocytes from late fetal stages onward. Understanding the complexities of building cardiac tissue from these stages will be of great future interest and will inform research aimed at cardiac tissue engineering.
Supplementary Material
Acknowledgements
In revision of this review, the authors were required to eliminate references in order to comply with the word limit. We apologize to those colleagues who are not appropriately cited.
Sources of Funding
Supported by NIH (SE), HL069594 (DY), American Heart Association (DY) and the March of Dimes (DY), DE018825 (FLC), HL089641 (FLC), American Heart Association (FLC), and HL036059 (MLK), HL070140 (MLK), HL083240 (MLK) and the George and Jean Brumley, Jr. Neonatal-Perinatal Research Institute at Duke University (MLK).
Non-standard Abbreviations and Acronyms
- E
Embryonic day
- HH
Hamburger-Hamilton staging procedure for chick
- AV
atrioventricular
- MLC
myosin light chain
- MHC
myosin heavy chain
- Isl1
islet 1
- Tbx
T-box transcription factor
- Flk
fetal liver kinase 1
- Nkx
NK homeodomain factor
- FGF
fibroblast growth factor
- aMHC
atrial myosin heavy chain
- vMHC
ventricular myosin heavy chain
- Mesp1
mesoderm posterior 1 homolog
- Cre
cyclization recombination
- Fgfr
fibroblast growth factor receptor
- BMP
bone morphogenetic protein
- Smad
drosophila protein mothers against decapentaplegic (MAD) and C. elegans protein SMA
- Oep
one-eyed pinhead
- Wnt
Wg (wingless) and Int
- LIM
LIN-11, Islet-1, and MEC-3
- Shh
Sonic hedgehog
- RALDH
retinoic acid dehydrogenase
- Hox
homeobox gene
- DiI
1,1′-dioctadecyl-3,3,3′3′-tetramethylindocarbocyanine perchlorate
- TGF
transforming growth factor
- PCP
planar cell polarity
- Vangl
vang-like
- NRG
neuregulin
- Lp
Loop tail
- RhoA
Ras homolog gene family, member A
- ROCK
Rho-associated, coiled-coil containing protein kinase
- Gli
glioblastoma
- ERT
estrogen replacement therapy
- Crkl
Crk-like protein
- ALK
activin like kinase
- Hod
homeodomain-only protein
- Mef
myocyte enhancer factor
- SRF
serum response factor
- MADS
mothers against decapentaplegic
- miR
microRNA
- HIF
hypoxia inducible factor
- RNA
ribonucleic acid
- DNA
deoxynucleic acid
- ANF
atrial natriuretic factor
- Nppa
atrial natriuretic peptide precursor
- Cx
connexin
- Foxn
ForkheadN
- EMT
epithelial-mesenchymal transformation
- Hey
Hairy and Enhancer-of-split-related basic helix-loop-helix (bHLH) transcription factor
- IC
inner curvature
- AVC
atrioventricular canal
- Shox
Short stature homeobox gene
- Irx
Iroquois
- ET
endothelin
- ECE
endothelin converting enzyme
- ErbB
erythroblastic leukemia viral oncogene homolog
- 5HT
5-hydroxytryptamine
- ADAMTS
a disintegrin and metalloproteinase with a thrombospondin type 1 motif
Footnotes
Disclosures: None
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Vincent SD, Buckingham ME. How to make a heart: the origin and regulation of cardiac progenitor cells. Curr Top Dev Biol. 2010;90:1–41. doi: 10.1016/S0070-2153(10)90001-X. [DOI] [PubMed] [Google Scholar]
- 2.Thomas PQ, Brown A, Beddington RS. Hex: a homeobox gene revealing peri-implantation asymmetry in the mouse embryo and an early transient marker of endothelial cell precursors. Development. 1998;125:85–94. doi: 10.1242/dev.125.1.85. [DOI] [PubMed] [Google Scholar]
- 3.Tam PPL, Parameswaran M, Kinder SJ, Weinberger RP. The allocation of epiblast cells to the embryonic heart and other mesodermal lineages: The role of ingression and tissue movement during gastrulation. Development. 1997;124:1631–1642. doi: 10.1242/dev.124.9.1631. [DOI] [PubMed] [Google Scholar]
- 4.Garcia-Martinez V, Schoenwolf GC. Primitive-streak origin of the cardiovascular system in avian embryos. Dev. Biol. 1993;159:706–719. doi: 10.1006/dbio.1993.1276. [DOI] [PubMed] [Google Scholar]
- 5.Bellairs R. The primitive streak. Anat Embryol (Berl) 1986;174:1–14. doi: 10.1007/BF00318331. [DOI] [PubMed] [Google Scholar]
- 6.Parameswaran M, Tam PP. Regionalisation of cell fate and morphogenetic movement of the mesoderm during mouse gastrulation. Dev Genet. 1995;17:16–28. doi: 10.1002/dvg.1020170104. [DOI] [PubMed] [Google Scholar]
- 7.Lopez-Sanchez C, Garcia-Martinez V, Schoenwolf GC. Localization of cells of the prospective neural plate, heart and somites within the primitive streak and epiblast of avian embryos at intermediate primitive-streak stages. Cells Tiss. Organs. 2001;169:334–346. doi: 10.1159/000047900. [DOI] [PubMed] [Google Scholar]
- 8.Tam PP, Beddington RS. Establishment and organization of germ layers in the gastrulating mouse embryo. Ciba Found Symp. 1992;165:27–41. doi: 10.1002/9780470514221.ch3. discussion 42-29. [DOI] [PubMed] [Google Scholar]
- 9.Stalsberg H, DeHaan RL. The precardiac areas and formation of the tubular heart in the chick embryo. Dev. Biol. 1969;19:128. doi: 10.1016/0012-1606(69)90052-9. [DOI] [PubMed] [Google Scholar]
- 10.Abu-Issa R, Kirby ML. Patterning of the heart field in the chick. Dev Biol. 2008;319:223–233. doi: 10.1016/j.ydbio.2008.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colas JF, Lawson A, Schoenwolf GC. Evidence that translation of smooth muscle alpha-actin mRNA is delayed in the chick promyocardium until fusion of the bilateral heart-forming regions. Dev. Dyn. 2000;218:316–330. doi: 10.1002/(SICI)1097-0177(200006)218:2<316::AID-DVDY6>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 12.Yuan S, Schoenwolf GC. Islet-1 marks the early heart rudiments and is asymmetrically expressed during early rotation of the foregut in the chick embryo. Anat Rec. 2000;260:204–207. doi: 10.1002/1097-0185(20001001)260:2<204::AID-AR90>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 13.de la Cruz MV, Markwald RR. Living Morphogenesis of the Heart. 1998 [Google Scholar]
- 14.Christoffels VM, Gabets PE, Franco D, Campione M, de Jong F, Lamers WH, Bao ZZ, Palmer S, Biben C, Harvey RP, Moorman AF. Chamber formation and morphogenesis in the developing mammalian heart. Dev. Biol. 2000;223:266–278. doi: 10.1006/dbio.2000.9753. [DOI] [PubMed] [Google Scholar]
- 15.Kirby ML. Cardiac Development. Oxford; New York: 2007. [Google Scholar]
- 16.Van Mierop LHS, Bertuch CJJ. Development of arterial blood pressure in the chick embryo. Am. J. Physiol. 1967;212:43–48. doi: 10.1152/ajplegacy.1967.212.1.43. [DOI] [PubMed] [Google Scholar]
- 17.Hirota A, Sakai T, Fujii S, Kamino K. Initial development of conduction pattern of spontaneous action potential in early embryonic precontractile chick heart. Dev. Biol. 1983;99:517–523. doi: 10.1016/0012-1606(83)90301-9. [DOI] [PubMed] [Google Scholar]
- 18.Latacha KS, Remond MC, Ramasubramanian A, Chen AY, Elson EL, Taber LA. Role of actin polymerization in bending of the early heart tube. Dev. Dyn. 2005;233:1272–1286. doi: 10.1002/dvdy.20488. [DOI] [PubMed] [Google Scholar]
- 19.Itasaki N, Nakamura H, Sumida H, Yasuda M. Actin bundles on the right side in the caudal part of the heart tube play a role in dextro-looping in the embryonic chick heart. Anat. Embryol. (Berl) 1991;183:29–39. doi: 10.1007/BF00185832. [DOI] [PubMed] [Google Scholar]
- 20.Voronov DA, Alford PW, Xu G, Taber LA. The role of mechanical forces in dextral rotation during cardiac looping in the chick embryo. Dev. Biol. 2004;171:339–350. doi: 10.1016/j.ydbio.2004.04.033. [DOI] [PubMed] [Google Scholar]
- 21.van den Berg G, Abu-Issa R, de Boer BA, Hutson MR, de Boer PA, Soufan AT, Ruijter JM, Kirby ML, van den Hoff MJ, Moorman AF. A caudal proliferating growth center contributes to both poles of the forming heart tube. Circ Res. 2009;104:179–188. doi: 10.1161/CIRCRESAHA.108.185843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buckingham M, Meilhac S, Zaffran S. Building the mammalian heart from two sources of myocardial cells. Nat Rev Genet. 2005;6:826–835. doi: 10.1038/nrg1710. [DOI] [PubMed] [Google Scholar]
- 23.Kelly RG, Brown NA, Buckingham ME. The arterial pole of the mouse heart forms from Fgf10-expressing cells in pharyngeal mesoderm. Dev.Cell. 2001;1:435–440. doi: 10.1016/s1534-5807(01)00040-5. [DOI] [PubMed] [Google Scholar]
- 24.Waldo KL, Hutson MR, Ward CC, Zdanowicz M, Stadt HA, Kumiski D, Abu-Issa R, Kirby ML. Secondary heart field contributes myocardium and smooth muscle to the arterial pole of the developing heart. Dev. Biol. 2005;281:78–90. doi: 10.1016/j.ydbio.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 25.Waldo KL, Kumiski DH, Wallis KT, Stadt HA, Hutson MR, Platt DH, Kirby ML. Conotruncal myocardium arises from a secondary heart field. Development. 2001;128:3179–3188. doi: 10.1242/dev.128.16.3179. [DOI] [PubMed] [Google Scholar]
- 26.Mjaatvedt CH, Nakaoka T, Moreno-Rodriguez R, Norris RA, Kern MJ, Eisenberg CA, Turner D, Markwald RR. The outflow tract of the heart is recruited from a novel heart-forming field. Dev. Biol. 2001;238:97–109. doi: 10.1006/dbio.2001.0409. [DOI] [PubMed] [Google Scholar]
- 27.Ward C, Stadt HA, Hutson MR, Kirby ML. Ablation of the secondary heart field leads to tetralogy of Fallot and pulmonary atresia. Dev. Biol. 2005;284:72–83. doi: 10.1016/j.ydbio.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 28.Bajolle F, Zaffran S, Kelly RG, Hadchouel J, Bonnet D, Brown NA, Buckingham ME. Rotation of the myocardial wall of the outflow tract is implicated in the normal positioning of the great arteries. Circ Res. 2006;98:421–428. doi: 10.1161/01.RES.0000202800.85341.6e. [DOI] [PubMed] [Google Scholar]
- 29.Dyer LA, Kirby ML. Sonic hedgehog maintains proliferation in secondary heart field progenitors and is required for normal arterial pole formation. Dev Biol. 2009;330:305–317. doi: 10.1016/j.ydbio.2009.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hutson MR, Zhang P, Stadt HA, Sato A, Li Y-X, Burch J, Creazzo TL, Kirby ML. Cardiac arterial pole alignment is sensitive to FGF8 signaling in the pharynx. Dev. Biol. 2006;295:486–497. doi: 10.1016/j.ydbio.2006.02.052. [DOI] [PubMed] [Google Scholar]
- 31.Christoffels VM, Keijser AGM, Houweling AC, Clout DEW, Moorman AFM. Patterning the embryonic heart: Identification of five mouse Iroquois homeobox genes in the developing heart. Dev. Biol. 2000;224:263–274. doi: 10.1006/dbio.2000.9801. [DOI] [PubMed] [Google Scholar]
- 32.Moorman AF, Christoffels VM. Cardiac chamber formation: development, genes, and evolution. Physiol. Rev. 2003;83:1223–1267. doi: 10.1152/physrev.00006.2003. [DOI] [PubMed] [Google Scholar]
- 33.Snarr BS, O'Neal JL, Chintalapudi MR, Wirrig EE, Phelps AL, Kubalak SW, Wessels A. Isl1 expression at the venous pole identifies a novel role for the second heart field in cardiac development. Circ Res. 2007;101:971–974. doi: 10.1161/CIRCRESAHA.107.162206. [DOI] [PubMed] [Google Scholar]
- 34.Jiang X, Rowitch DH, Soriano P, McMahon AP, Sucov HM. Fate of the mammalian cardiac neural crest. Development. 2000;127:1607–1616. doi: 10.1242/dev.127.8.1607. [DOI] [PubMed] [Google Scholar]
- 35.Waldo K, Miyagawa-Tomita S, Kumiski D, Kirby ML. Cardiac neural crest cells provide new insight into septation of the cardiac outflow tract: Aortic sac to ventricular septal closure. Dev. Biol. 1998;196:129–144. doi: 10.1006/dbio.1998.8860. [DOI] [PubMed] [Google Scholar]
- 36.Lincoln J, Alfieri CM, Yutzey KE. Development of heart valve leaflets and supporting apparatus in chicken and mouse embryos. Dev. Dyn. 2004;230:239–250. doi: 10.1002/dvdy.20051. [DOI] [PubMed] [Google Scholar]
- 37.Manner J, Perez-Pomares JM, Macias D, Munoz-Chapuli R. The origin, formation and developmental significance of the epicardium: a review. Cells Tissues Organs. 2001;169:89–103. doi: 10.1159/000047867. [DOI] [PubMed] [Google Scholar]
- 38.Kalman F, Viragh S, Modis L. Cell surface glycoconjugates and the extracellular matrix of the developing mouse embryo epicardium. Anat Embryol (Berl) 1995;191:451–464. doi: 10.1007/BF00304430. [DOI] [PubMed] [Google Scholar]
- 39.Zhou B, Ma Q, Rajagopal S, Wu SM, Domian I, Rivera-Feliciano J, Jiang D, von Gise A, Ikeda S, Chien KR, Pu WT. Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature. 2008;454:109–113. doi: 10.1038/nature07060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cai CL, Martin JC, Sun Y, Cui L, Wang L, Ouyang K, Yang L, Bu L, Liang X, Zhang X, Stallcup WB, Denton CP, McCulloch A, Chen J, Evans SM. A myocardial lineage derives from Tbx18 epicardial cells. Nature. 2008;454:104–108. doi: 10.1038/nature06969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Christoffels VM, Grieskamp T, Norden J, Mommersteeg MT, Rudat C, Kispert A. Tbx18 and the fate of epicardial progenitors. Nature. 2009;458:E8–9. doi: 10.1038/nature07916. discussion E9-10. [DOI] [PubMed] [Google Scholar]
- 42.Moody SA. Fates of the blastomeres of the 32-cell-stage Xenopus embryo. Dev Biol. 1987;122:300–319. doi: 10.1016/0012-1606(87)90296-x. [DOI] [PubMed] [Google Scholar]
- 43.Dale L, Slack JM. Fate map for the 32-cell stage of Xenopus laevis. Development. 1987;99:527–551. doi: 10.1242/dev.99.4.527. [DOI] [PubMed] [Google Scholar]
- 44.Lee RK, Stainier DY, Weinstein BM, Fishman MC. Cardiovascular development in the zebrafish. II. Endocardial progenitors are sequestered within the heart field. Development. 1994;120:3361–3366. doi: 10.1242/dev.120.12.3361. [DOI] [PubMed] [Google Scholar]
- 45.Stainier DYR, Lee RK, Fishman MC. Cardiovascular development in the zebrafish. I. Myocardial fate map and heart tube formation. Development. 1993;119:31–40. doi: 10.1242/dev.119.1.31. [DOI] [PubMed] [Google Scholar]
- 46.Keegan BR, Meyer D, Yelon D. Organization of cardiac chamber progenitors in the zebrafish blastula. Development. 2004;131:3081–3091. doi: 10.1242/dev.01185. [DOI] [PubMed] [Google Scholar]
- 47.Schoenebeck JJ, Keegan BR, Yelon D. Vessel and blood specification override cardiac potential in anterior mesoderm. Developmental Cell. 2007;13:254–267. doi: 10.1016/j.devcel.2007.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kolker SJ, Tajchman U, Weeks DL. Confocal imaging of early heart development in Xenopus laevis. Dev Biol. 2000;218:64–73. doi: 10.1006/dbio.1999.9558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mohun TJ, Leong LM, Weninger WJ, Sparrow DB. The morphology of heart development in Xenopus laevis. Dev Biol. 2000;218:74–88. doi: 10.1006/dbio.1999.9559. [DOI] [PubMed] [Google Scholar]
- 50.Goetz SC, Brown DD, Conlon FL. TBX5 is required for embryonic cardiac cell cycle progression. Development. 2006;133:2575–2584. doi: 10.1242/dev.02420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Movassagh M, Philpott A. Cardiac differentiation in Xenopus requires the cyclin-dependent kinase inhibitor, p27Xic1. Cardiovasc Res. 2008;79:436–447. doi: 10.1093/cvr/cvn105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Langdon YG, Goetz SC, Berg AE, Swanik JT, Conlon FL. SHP-2 is required for the maintenance of cardiac progenitors. Development. 2007;134:4119–4130. doi: 10.1242/dev.009290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mohun TJ, Leong LM, Weninger WJ, Sparrow DB. The morphology of heart development in Xenopus laevis. 2000;218:74–88. doi: 10.1006/dbio.1999.9559. [DOI] [PubMed] [Google Scholar]
- 54.Kolker SJ, Tajchman U, Weeks DL. Confocal imaging of early heart development in Xenopus laevis. 2000;218:64–73. doi: 10.1006/dbio.1999.9558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yelon D, Horne SA, Stainier DY. Restricted expression of cardiac myosin genes reveals regulated aspects of heart tube assembly in zebrafish. Dev Biol. 1999;214:23–37. doi: 10.1006/dbio.1999.9406. [DOI] [PubMed] [Google Scholar]
- 56.Glickman Holtzman NS, Schoenebeck JJ, Tsai HJ, Yelon D. Endocardium is necessary for cardiomyocyte movement during heart tube assembly. Development. 2007;134:2379–2386. doi: 10.1242/dev.02857. [DOI] [PubMed] [Google Scholar]
- 57.Bakkers J, Verhoeven MC, Abdelilah-Seyfried S. Shaping the zebrafish heart: from left-right axis specification to epithelial tissue morphogenesis. Dev Biol. 2009;330:213–220. doi: 10.1016/j.ydbio.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 58.Bussmann J, Bakkers J, Schulte-Merker S. Early endocardial morphogenesis requires Scl/Tal1. PLoS Genet. 2007;3:e140. doi: 10.1371/journal.pgen.0030140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gessert S, Kuhl M. Comparative gene expression analysis and fate mapping studies suggest an early segregation of cardiogenic lineages in Xenopus laevis. Dev Biol. 2009;334:395–408. doi: 10.1016/j.ydbio.2009.07.037. [DOI] [PubMed] [Google Scholar]
- 60.Brade T, Gessert S, Kuhl M, Pandur P. The amphibian second heart field: Xenopus islet-1 is required for cardiovascular development. Dev Biol. 2007;311:297–310. doi: 10.1016/j.ydbio.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 61.Auman HJ, Coleman H, Riley HE, Olale F, Tsai HJ, Yelon D. Functional Modulation of Cardiac Form through Regionally Confined Cell Shape Changes. PLoS Biol. 2007;5:e53. doi: 10.1371/journal.pbio.0050053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.de Pater E, Clijsters L, Marques SR, Lin YF, Garavito-Aguilar ZV, Yelon D, Bakkers J. Distinct phases of cardiomyocyte differentiation regulate growth of the zebrafish heart. Development. 2009;136:1633–1641. doi: 10.1242/dev.030924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yatskievych TA, Ladd AN, Antin PB. Induction of cardiac myogenesis in avian pregastrula epiblast: the role of the hypoblast and activin. Development. 1997;124:2561–2570. doi: 10.1242/dev.124.13.2561. [DOI] [PubMed] [Google Scholar]
- 64.Rosenquist GC, DeHaan RL. Migration of precardiac cells in the chick embryo: A radioautographic study. Carnegie Inst.Wash.Publ.625, Contrib.Embryol. 1966;38:111–121. [Google Scholar]
- 65.Schoenwolf GC, Garcia-Martinez V, Dias MS. Mesoderm movement and fate during avian gastrulation and neurulation. Dev. Dyn. 1992;193:235–248. doi: 10.1002/aja.1001930304. [DOI] [PubMed] [Google Scholar]
- 66.Auda-Boucher G, Bernard B, Fontaine-Perus J, Rouaud T, Mericksay M, Gardahaut MF. Staging of the commitment of murine cardiac cell progenitors. Dev Biol. 2000;225:214–225. doi: 10.1006/dbio.2000.9817. [DOI] [PubMed] [Google Scholar]
- 67.Mikawa T, Gourdie RG. Pericardial mesoderm generates a population of coronary smooth muscle cells migrating into the heart along with ingrowth of the epicardial organ. Dev Biol. 1996;174:221–232. doi: 10.1006/dbio.1996.0068. [DOI] [PubMed] [Google Scholar]
- 68.Wei Y, Mikawa T. Fate diversity of primitive streak cells during heart field formation in ovo. Dev. Dyn. 2000;219:505–513. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1076>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 69.Sater AK, Jacobson AG. The specification of heart mesoderm occurs during gastrulation in Xenopus laevis. Development. 1989;105:821–830. doi: 10.1242/dev.105.4.821. [DOI] [PubMed] [Google Scholar]
- 70.Sater AK, Jacobson AG. The restriction of the heart morphogenetic field in Xenopus laevis. Dev Biol. 1990;140:328–336. doi: 10.1016/0012-1606(90)90083-u. [DOI] [PubMed] [Google Scholar]
- 71.Nascone N, Mercola M. An inductive role for the endoderm in Xenopus cardiogenesis. Development. 1995;121:515–523. doi: 10.1242/dev.121.2.515. [DOI] [PubMed] [Google Scholar]
- 72.Marques SR, Lee Y, Poss KD, Yelon D. Reiterative roles for FGF signaling in the establishment of size and proportion of the zebrafish heart. Dev Biol. 2008;321:397–406. doi: 10.1016/j.ydbio.2008.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Marques SR, Yelon D. Differential requirement for BMP signaling in atrial and ventricular lineages establishes cardiac chamber proportionality. Dev Biol. 2009;328:472–482. doi: 10.1016/j.ydbio.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Thomas NA, Koudijs M, van Eeden FJ, Joyner AL, Yelon D. Hedgehog signaling plays a cell-autonomous role in maximizing cardiac developmental potential. Development. 2008;135:3789–3799. doi: 10.1242/dev.024083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Twal W, Roze L, Zile MH. Anti-retinoic acid monoclonal antibody localizes all-trans-retinoic acid in target cells and blocks normal development in early quail embryo. Dev. Biol. 1995;168:225–234. doi: 10.1006/dbio.1995.1075. [DOI] [PubMed] [Google Scholar]
- 76.Niederreither K, McCaffery P, Drager UC, Chambon P, Dolle P. Restricted expression and retinoic acid-induced downregulation of the retinaldehyde dehydrogenase type 2 (RALDH-2) gene during mouse development. Mech Dev. 1997;62:67–78. doi: 10.1016/s0925-4773(96)00653-3. [DOI] [PubMed] [Google Scholar]
- 77.Lopez-Sanchez C, Climent V, Schoenwolf GC, Alvarez I, Garcia-Martinez V. Induction of cardigenesis by Hensen's node and fibroblast growth factors. Cell Tiss. Res. 2002;309:237–249. doi: 10.1007/s00441-002-0567-2. [DOI] [PubMed] [Google Scholar]
- 78.Alsan BH, Schultheiss TM. Regulation of avian cardiogenesis by Fgf8 signaling. Development. 2002;129:1935–1943. doi: 10.1242/dev.129.8.1935. [DOI] [PubMed] [Google Scholar]
- 79.Saga Y, Kitajima S, Miyagawa-Tomita S. Mesp1 expression is the earliest sign of cardiovascular development. Trends Cardiovasc Med. 2000;10:345–352. doi: 10.1016/s1050-1738(01)00069-x. [DOI] [PubMed] [Google Scholar]
- 80.Raffin M, Leong LM, Rones MS, Sparrow D, Mohun T, Mercola M. Subdivision of the cardiac Nkx2.5 expression domain into myogenic and nonmyogenic compartments. Dev Biol. 2000;218:326–340. doi: 10.1006/dbio.1999.9579. [DOI] [PubMed] [Google Scholar]
- 81.Lough J, Sugi Y. Endoderm and heart development. Dev Dyn. 2000;217:327–342. doi: 10.1002/(SICI)1097-0177(200004)217:4<327::AID-DVDY1>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 82.Foley AC, Gupta RW, Guzzo RM, Korol O, Mercola M. Embryonic heart induction. Ann N Y Acad Sci. 2006;1080:85–96. doi: 10.1196/annals.1380.008. [DOI] [PubMed] [Google Scholar]
- 83.Liberatore CM, Searcy-Schrick RD, Vincent EB, Yutzey KE. Nkx-2.5 gene induction in mice is mediated by a Smad consensus regulatory region. Dev Biol. 2002;244:243–256. doi: 10.1006/dbio.2002.0604. [DOI] [PubMed] [Google Scholar]
- 84.Prall OW, Menon MK, Solloway MJ, Watanabe Y, Zaffran S, Bajolle F, Biben C, McBride JJ, Robertson BR, Chaulet H, Stennard FA, Wise N, Schaft D, Wolstein O, Furtado MB, Shiratori H, Chien KR, Hamada H, Black BL, Saga Y, Robertson EJ, Buckingham ME, Harvey RP. An Nkx2-5/Bmp2/Smad1 negative feedback loop controls heart progenitor specification and proliferation. Cell. 2007;128:947–959. doi: 10.1016/j.cell.2007.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang XM, Ramalho-Santos M, McMahon AP. Smoothened mutants reveal redundant roles for Shh and Ihh signaling including regulation of L/R asymmetry by the mouse node. Cell. 2001;105:781–792. [PubMed] [Google Scholar]
- 86.Reifers F, Walsh EC, Leger S, Stainier DY, Brand M. Induction and differentiation of the zebrafish heart requires fibroblast growth factor 8 (fgf8/acerebellar) Development. 2000;127:225–235. doi: 10.1242/dev.127.2.225. [DOI] [PubMed] [Google Scholar]
- 87.Reiter JF, Verkade H, Stainier DY. Bmp2b and Oep promote early myocardial differentiation through their regulation of gata5. Dev Biol. 2001;234:330–338. doi: 10.1006/dbio.2001.0259. [DOI] [PubMed] [Google Scholar]
- 88.Rones MS, McLaughlin KA, Raffin M, Mercola M. Serrate and Notch specify cell fates in the heart field by suppressing cardiomyogenesis. Development. 2000;127:3865–3876. doi: 10.1242/dev.127.17.3865. [DOI] [PubMed] [Google Scholar]
- 89.Cai CL, Liang X, Shi Y, Chu PH, Pfaff SL, Chen J, Evans S. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell. 2003;5:877–889. doi: 10.1016/s1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lin L, Cui L, Zhou W, Dufort D, Zhang X, Cai CL, Bu L, Yang L, Martin J, Kemler R, Rosenfeld MG, Chen J, Evans SM. Beta-catenin directly regulates Islet1 expression in cardiovascular progenitors and is required for multiple aspects of cardiogenesis. Proc Natl Acad Sci U S A. 2007;104:9313–9318. doi: 10.1073/pnas.0700923104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cohen ED, Wang Z, Lepore JJ, Lu MM, Taketo MM, Epstein DJ, Morrisey EE. Wnt/beta-catenin signaling promotes expansion of Isl-1-positive cardiac progenitor cells through regulation of FGF signaling. J Clin Invest. 2007;117:1794–1804. doi: 10.1172/JCI31731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ai D, Fu X, Wang J, Lu MF, Chen L, Baldini A, Klein WH, Martin JF. Canonical Wnt signaling functions in second heart field to promote right ventricular growth. Proc Natl Acad Sci U S A. 2007;104:9319–9324. doi: 10.1073/pnas.0701212104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kwon C, Arnold J, Hsiao EC, Taketo MM, Conklin BR, Srivastava D. Canonical Wnt signaling is a positive regulator of mammalian cardiac progenitors. Proc Natl Acad Sci U S A. 2007;104:10894–10899. doi: 10.1073/pnas.0704044104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cohen ED, Tian Y, Morrisey EE. Wnt signaling: an essential regulator of cardiovascular differentiation, morphogenesis and progenitor self-renewal. Development. 2008;135:789–798. doi: 10.1242/dev.016865. [DOI] [PubMed] [Google Scholar]
- 95.Eisenberg LM, Eisenberg CA. Wnt signal transduction and the formation of the myocardium. Dev Biol. 2006;293:305–315. doi: 10.1016/j.ydbio.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 96.Kang J, Nathan E, Xu SM, Tzahor E, Black BL. Isl1 is a direct transcriptional target of Forkhead transcription factors in second heart field-derived mesoderm. Dev Biol. 2009 doi: 10.1016/j.ydbio.2009.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Park EJ, Ogden LA, Talbot A, Evans S, Cai CL, Black BL, Frank DU, Moon AM. Required, tissue-specific roles for Fgf8 in outflow tract formation and remodeling. Development. 2006;133:2419–2433. doi: 10.1242/dev.02367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Abu-Issa R, Smyth G, Smoak I, Yamamura K-I, Meyers E. Fgf8 is required for pharyngeal arch and cardiovascular development in the mouse. Development. 2002;129:4613–4625. doi: 10.1242/dev.129.19.4613. [DOI] [PubMed] [Google Scholar]
- 99.Hutson MR, Zeng L, Kim AJ, Antoon E, Harward S, Kirby ML. Cardiac stem cells in the secondary heart field are governed by opposing FGF/BMP signals. Development. 2010 in press. [Google Scholar]
- 100.Reiter JF, Alexander J, Rodaway A, Yelon D, Patient R, Holder N, Stainier DY. Gata5 is required for the development of the heart and endoderm in zebrafish. Genes Dev. 1999;13:2983–2995. doi: 10.1101/gad.13.22.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Latinkic BV, Kotecha S, Mohun TJ. Induction of cardiomyocytes by GATA4 in Xenopus ectodermal explants. Development. 2003;130:3865–3876. doi: 10.1242/dev.00599. [DOI] [PubMed] [Google Scholar]
- 102.Takeuchi JK, Bruneau BG. Directed transdifferentiation of mouse mesoderm to heart tissue by defined factors. Nature. 2009;459:708–711. doi: 10.1038/nature08039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yutzey KE, Rhee JT, Bader D. Expression of the atrial-specific myosin heavy chain AMHC1 and the establishment of anteroposterior polarity in the developing chicken heart. Development. 1994;120:871–883. doi: 10.1242/dev.120.4.871. [DOI] [PubMed] [Google Scholar]
- 104.Ryckebusch L, Wang Z, Bertrand N, Lin SC, Chi X, Schwartz R, Zaffran S, Niederreither K. Retinoic acid deficiency alters second heart field formation. Proc Natl Acad Sci U S A. 2008;105:2913–2918. doi: 10.1073/pnas.0712344105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sirbu IO, Zhao X, Duester G. Retinoic acid controls heart anteroposterior patterning by down-regulating Isl1 through the Fgf8 pathway. Dev Dyn. 2008;237:1627–1635. doi: 10.1002/dvdy.21570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Keegan BR, Feldman JL, Begemann G, Ingham PW, Yelon D. Retinoic acid signaling restricts the cardiac progenitor pool. Science. 2005;307:247–249. doi: 10.1126/science.1101573. [DOI] [PubMed] [Google Scholar]
- 107.Waxman JS, Keegan BR, Roberts RW, Poss KD, Yelon D. Hoxb5b acts downstream of retinoic acid signaling in the forelimb field to restrict heart field potential in zebrafish. Developmental Cell. 2008;15:923–934. doi: 10.1016/j.devcel.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ueno S, Weidinger G, Osugi T, Kohn AD, Golob JL, Pabon L, Reinecke H, Moon RT, Murry CE. Biphasic role for Wnt/beta-catenin signaling in cardiac specification in zebrafish and embryonic stem cells. Proc Natl Acad Sci U S A. 2007;104:9685–9690. doi: 10.1073/pnas.0702859104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.King T, Brown NA. Left-right asymmetry: The embryo's one-sided genes. Curr Biol. 1995;5:1364–1366. [PubMed] [Google Scholar]
- 110.Levin M, Johnson RL, Stern CD, Kuehn M, Tabin C. A molecular pathway determining left-right asymmetry in chick embryogenesis. Cell. 1995;82:803–814. doi: 10.1016/0092-8674(95)90477-8. [DOI] [PubMed] [Google Scholar]
- 111.Collignon J, Varlet I, Robertson EJ. Relationship between asymmetric nodal expression and the direction of embryonic turning. Nature. 1996;381:155–158. doi: 10.1038/381155a0. [DOI] [PubMed] [Google Scholar]
- 112.Meno C, Saijoh Y, Fujii H, Ikeda M, Yokoyama T, Yokoyama M, Toyoda Y, Hamada H. Left-right asymmetric expression of the TGF beta-family member lefty in mouse embryos. Nature. 1996;381:151–155. doi: 10.1038/381151a0. [DOI] [PubMed] [Google Scholar]
- 113.Christine KS, Conlon FL. Vertebrate CASTOR is required for differentiation of cardiac precursor cells at the ventral midline. Dev Cell. 2008;14:616–623. doi: 10.1016/j.devcel.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Campione M, Ros MA, Icardo JM, Piedra E, Christoffels VM, Schweickert A, Blum M, Franco D, Moorman AF. Pitx2 expression defines a left cardiac lineage of cells: evidence for atrial and ventricular molecular isomerism in the iv/iv mice. Dev Biol. 2001;231:252–264. doi: 10.1006/dbio.2000.0133. [DOI] [PubMed] [Google Scholar]
- 115.Franco D, Campione M, Kelly R, Zammit PS, Buckingham M, Lamers WH, Moorman AF. Multiple transcriptional domains, with distinct left and right components, in the atrial chambers of the developing heart. Circ Res. 2000;87:984–991. doi: 10.1161/01.res.87.11.984. [DOI] [PubMed] [Google Scholar]
- 116.Liu C, Liu W, Palie J, Lu MF, Brown NA, Martin JF. Pitx2c patterns anterior myocardium and aortic arch vessels and is required for local cell movement into atrioventricular cushions. Development. 2002;129:5081–5091. doi: 10.1242/dev.129.21.5081. [DOI] [PubMed] [Google Scholar]
- 117.Galli D, Dominguez JN, Zaffran S, Munk A, Brown NA, Buckingham ME. Atrial myocardium derives from the posterior region of the second heart field, which acquires left-right identity as Pitx2c is expressed. Development. 2008;135:1157–1167. doi: 10.1242/dev.014563. [DOI] [PubMed] [Google Scholar]
- 118.Cai CL, Liang X, Shi Y, Chu PH, Pfaff SL, Chen J, Evans S. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell. 2003;5:877–889. doi: 10.1016/s1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hoffmann AD, Peterson MA, Friedland-Little JM, Anderson SA, Moskowitz IP. sonic hedgehog is required in pulmonary endoderm for atrial septation. Development. 2009;136:1761–1770. doi: 10.1242/dev.034157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sizarov A, Anderson RH, Christoffels VM, Moorman AF. Three-Dimensional and Molecular Analysis of the Venous Pole of the Developing Human Heart. Circulation. 2010 doi: 10.1161/CIRCULATIONAHA.110.953844. [DOI] [PubMed] [Google Scholar]
- 121.Ai D, Liu W, Ma L, Dong F, Lu MF, Wang D, Verzi MP, Cai C, Gage PJ, Evans S, Black BL, Brown NA, Martin JF. Pitx2 regulates cardiac left-right asymmetry by patterning second cardiac lineage-derived myocardium. Dev Biol. 2006;296:437–449. doi: 10.1016/j.ydbio.2006.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tessari A, Pietrobon M, Notte A, Cifelli G, Gage PJ, Schneider MD, Lembo G, Campione M. Myocardial Pitx2 differentially regulates the left atrial identity and ventricular asymmetric remodeling programs. Circ Res. 2008;102:813–822. doi: 10.1161/CIRCRESAHA.107.163188. [DOI] [PubMed] [Google Scholar]
- 123.Hildreth V, Webb S, Chaudhry B, Peat JD, Phillips HM, Brown N, Anderson RH, Henderson DJ. Left cardiac isomerism in the Sonic hedgehog null mouse. J Anat. 2009;214:894–904. doi: 10.1111/j.1469-7580.2009.01087.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhou W, Lin L, Majumdar A, Li X, Zhang X, Liu W, Etheridge L, Shi Y, Martin J, Van de Ven W, Kaartinen V, Wynshaw-Boris A, McMahon AP, Rosenfeld MG, Evans SM. Modulation of morphogenesis by noncanonical Wnt signaling requires ATF/CREB family-mediated transcriptional activation of TGFbeta2. Nat Genet. 2007;39:1225–1234. doi: 10.1038/ng2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Phillips HM, Murdoch JN, Chaudhry B, Copp AJ, Henderson DJ. Vangl2 acts via RhoA signaling to regulate polarized cell movements during development of the proximal outflow tract. Circ Res. 2005;96:292–299. doi: 10.1161/01.RES.0000154912.08695.88. [DOI] [PubMed] [Google Scholar]
- 126.Nowotschin S, Liao J, Gage PJ, Epstein JA, Campione M, Morrow BE. Tbx1 affects asymmetric cardiac morphogenesis by regulating Pitx2 in the secondary heart field. Development. 2006;133:1565–1573. doi: 10.1242/dev.02309. [DOI] [PubMed] [Google Scholar]
- 127.Theveniau-Ruissy M, Dandonneau M, Mesbah K, Ghez O, Mattei MG, Miquerol L, Kelly RG. The del22q11.2 candidate gene Tbx1 controls regional outflow tract identity and coronary artery patterning. Circ Res. 2008;103:142–148. doi: 10.1161/CIRCRESAHA.108.172189. [DOI] [PubMed] [Google Scholar]
- 128.Bruneau BG, Logan M, Davis N, Levi T, Tabin CJ, Seidman JG, Seidman CE. Chamber-specific cardiac expression of Tbx5 and heart defects in Holt-Oram syndrome. Dev. Biol. 1999;211:100–108. doi: 10.1006/dbio.1999.9298. [DOI] [PubMed] [Google Scholar]
- 129.Liberatore CM, Searcy-Schrick RD, Yutzey KE. Ventricular expression of tbx5 inhibits normal heart chamber development. Dev. Biol. 2000;223:169–180. doi: 10.1006/dbio.2000.9748. [DOI] [PubMed] [Google Scholar]
- 130.Takeuchi JK, Ohgi M, Koshiba-Takeuchi K, Shiratori H, Sakaki I, Ogura K, Saijoh Y, Ogura T. Tbx5 specifies the left/right ventricles and ventricular septum position during cardiogenesis. Development. 2003;130:5953–5964. doi: 10.1242/dev.00797. [DOI] [PubMed] [Google Scholar]
- 131.Koshiba-Takeuchi K, Mori AD, Kaynak BL, Cebra-Thomas J, Sukonnik T, Georges RO, Latham S, Beck L, Henkelman RM, Black BL, Olson EN, Wade J, Takeuchi JK, Nemer M, Gilbert SF, Bruneau BG. Reptilian heart development and the molecular basis of cardiac chamber evolution. Nature. 2009;461:95–98. doi: 10.1038/nature08324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Guris DL, Duester G, Papaioannou VE, Imamoto A. Dose-dependent interaction of Tbx1 and Crkl and locally aberrant RA signaling in a model of del22q11 syndrome. Dev Cell. 2006;10:81–92. doi: 10.1016/j.devcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 133.Roberts C, Ivins S, Cook AC, Baldini A, Scambler PJ. Cyp26 genes a1, b1 and c1 are down-regulated in Tbx1 null mice and inhibition of Cyp26 enzyme function produces a phenocopy of DiGeorge Syndrome in the chick. Hum Mol Genet. 2006;15:3394–3410. doi: 10.1093/hmg/ddl416. [DOI] [PubMed] [Google Scholar]
- 134.Yang L, Cai CL, Lin L, Qyang Y, Chung C, Monteiro RM, Mummery CL, Fishman GI, Cogen A, Evans S. Isl1Cre reveals a common Bmp pathway in heart and limb development. Development. 2006;133:1575–1585. doi: 10.1242/dev.02322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Mandel EM, Kaltenbrun E, Callis TE, Zeng XX, Marques SR, Yelon D, Wang DZ, Conlon FL. The BMP pathway acts to directly regulate Tbx20 in the developing heart. Development. 2010;137:1919–1929. doi: 10.1242/dev.043588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Walters MJ, Wayman GA, Christian JL. Bone morphogenetic protein function is required for terminal differentiation of the heart but not for early expression of cardiac marker genes. Mech Dev. 2001;100:263–273. doi: 10.1016/s0925-4773(00)00535-9. [DOI] [PubMed] [Google Scholar]
- 137.Shi Y, Katsev S, Cai C, Evans S. BMP signaling is required for heart formation in vertebrates. Dev Biol. 2000;224:226–237. doi: 10.1006/dbio.2000.9802. [DOI] [PubMed] [Google Scholar]
- 138.Liao J, Aggarwal VS, Nowotschin S, Bondarev A, Lipner S, Morrow BE. Identification of downstream genetic pathways of Tbx1 in the second heart field. Dev Biol. 2008;316:524–537. doi: 10.1016/j.ydbio.2008.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Song MR, Shirasaki R, Cai CL, Ruiz EC, Evans SM, Lee SK, Pfaff SL. T-Box transcription factor Tbx20 regulates a genetic program for cranial motor neuron cell body migration. Development. 2006;133:4945–4955. doi: 10.1242/dev.02694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Singh MK, Christoffels VM, Dias JM, Trowe MO, Petry M, Schuster-Gossler K, Burger A, Ericson J, Kispert A. Tbx20 is essential for cardiac chamber differentiation and repression of Tbx2. Development. 2005;132:2697–2707. doi: 10.1242/dev.01854. [DOI] [PubMed] [Google Scholar]
- 141.Stennard FA, Costa MW, Lai D, Biben C, Furtado MB, Solloway MJ, McCulley DJ, Leimana C, Preis JI, Dunwoodie SL, Elliot DE, Prall OW, Black BL, Fatkin D, Harvey RP. Murine T-box transcription factor Tbx20 acts as a repressor during heart development, and is essential for adult heart integrity, function and adaptation. Development. 2005;132:2451–2462. doi: 10.1242/dev.01799. [DOI] [PubMed] [Google Scholar]
- 142.Cai CL, Zhou W, Yang L, Bu L, Qyang Y, Zhang X, Li X, Rosenfeld MG, Chen J, Evans S. T-box genes coordinate regional rates of proliferation and regional specification during cardiogenesis. Development. 2005;132:2475–2487. doi: 10.1242/dev.01832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Brown DD, Martz SN, Binder O, Goetz SC, Price BMJ, Smith JC, Conlon FL. Tbx5 and Tbx20 act synergistically to control vertebrate heart morphogenesis. Development. 2005:132. doi: 10.1242/dev.01596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Goddeeris MM, Rho S, Petiet A, Davenport CL, Johnson GA, Meyers EN, Klingensmith J. Intracardiac septation requires hedgehog-dependent cellular contributions from outside the heart. Development. 2008;135:1887–1895. doi: 10.1242/dev.016147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Qyang Y, Martin-Puig S, Chiravuri M, Chen S, Xu H, Bu L, Jiang X, Lin L, Granger A, Moretti A, Caron L, Wu X, Clarke J, Taketo MM, Laugwitz KL, Moon RT, Gruber P, Evans SM, Ding S, Chien KR. The renewal and differentiation of Isl1+ cardiovascular progenitors are controlled by a Wnt/beta-catenin pathway. Cell Stem Cell. 2007;1:165–179. doi: 10.1016/j.stem.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 146.Niu Z, Li A, Zhang SX, Schwartz RJ. Serum response factor micromanaging cardiogenesis. Curr Opin Cell Biol. 2007;19:618–627. doi: 10.1016/j.ceb.2007.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- 148.Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.van Rooij E, Marshall WS, Olson EN. Toward microRNA-based therapeutics for heart disease: the sense in antisense. Circ Res. 2008;103:919–928. doi: 10.1161/CIRCRESAHA.108.183426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Callis TE, Wang DZ. Taking microRNAs to heart. Trends Mol Med. 2008;14:254–260. doi: 10.1016/j.molmed.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 151.Srivastava D, Thomas T, Lin Q, Kirby ML, Brown D, Olson EN. Regulation of cardiac mesodermal and neural crest development by the bHLH transcription factor, dHAND. Nat Genet. 1997;16:154–160. doi: 10.1038/ng0697-154. [DOI] [PubMed] [Google Scholar]
- 152.Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, Tsuchihashi T, McManus MT, Schwartz RJ, Srivastava D. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 153.Liu N, Bezprozvannaya S, Williams AH, Qi X, Richardson JA, Bassel-Duby R, Olson EN. microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev. 2008;22:3242–3254. doi: 10.1101/gad.1738708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Krishnan J, Ahuja P, Bodenmann S, Knapik D, Perriard E, Krek W, Perriard JC. Essential role of developmentally activated hypoxia-inducible factor 1alpha for cardiac morphogenesis and function. Circ Res. 2008;103:1139–1146. doi: 10.1161/01.RES.0000338613.89841.c1. [DOI] [PubMed] [Google Scholar]
- 155.Christoffels VM, Hoogaars WM, Tessari A, Clout DE, Moorman AF, Campione M. T-box transcription factor Tbx2 represses differentiation and formation of the cardiac chambers. Dev Dyn. 2004;229:763–770. doi: 10.1002/dvdy.10487. [DOI] [PubMed] [Google Scholar]
- 156.Hoogaars WM, Barnett P, Moorman AF, Christoffels VM. T-box factors determine cardiac design. Cell Mol Life Sci. 2007;64:646–660. doi: 10.1007/s00018-007-6518-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Habets PE, Moorman AF, Clout DE, van Roon MA, Lingbeek M, van Lohuizen M, Campione M, Christoffels VM. Cooperative action of Tbx2 and Nkx2.5 inhibits ANF expression in the atrioventricular canal: implications for cardiac chamber formation. Genes Dev. 2002;16:1234–1246. doi: 10.1101/gad.222902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Dupays L, Kotecha S, Angst B, Mohun TJ. Tbx2 misexpression impairs deployment of second heart field derived progenitor cells to the arterial pole of the embryonic heart. Dev Biol. 2009;333:121–131. doi: 10.1016/j.ydbio.2009.06.025. [DOI] [PubMed] [Google Scholar]
- 159.Aanhaanen WT, Brons JF, Dominguez JN, Rana MS, Norden J, Airik R, Wakker V, de Gier-de Vries C, Brown NA, Kispert A, Moorman AF, Christoffels VM. The Tbx2+ primary myocardium of the atrioventricular canal forms the atrioventricular node and the base of the left ventricle. Circ Res. 2009;104:1267–1274. doi: 10.1161/CIRCRESAHA.108.192450. [DOI] [PubMed] [Google Scholar]
- 160.Mesbah K, Harrelson Z, Theveniau-Ruissy M, Papaioannou VE, Kelly RG. Tbx3 is required for outflow tract development. Circ Res. 2008;103:743–750. doi: 10.1161/CIRCRESAHA.108.172858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Bakker ML, Boukens BJ, Mommersteeg MT, Brons JF, Wakker V, Moorman AF, Christoffels VM. Transcription factor Tbx3 is required for the specification of the atrioventricular conduction system. Circ Res. 2008;102:1340–1349. doi: 10.1161/CIRCRESAHA.107.169565. [DOI] [PubMed] [Google Scholar]
- 162.Stennard FA, Costa MW, Lai D, Biben C, Furtado MB, Solloway MJ, McCulley DJ, Leimena C, Preis JI, Dunwoodie SL, Elliott DE, Prall OW, Black BL, Fatkin D, Harvey RP. Murine T-box transcription factor Tbx20 acts as a repressor during heart development, and is essential for adult heart integrity, function and adaptation. Development. 2005;132:2451–2462. doi: 10.1242/dev.01799. [DOI] [PubMed] [Google Scholar]
- 163.Bruneau BG, Nemer G, Schmitt JP, Charron F, Robitaille L, Caron S, Conner DA, Gessler M, Nemer M, Seidman CE, Seidman JG. A murine model of Holt-Oram syndrome defines roles of the T-box transcription factor Tbx5 in cardiogenesis and disease. Cell. 2001;106:709–721. doi: 10.1016/s0092-8674(01)00493-7. [DOI] [PubMed] [Google Scholar]
- 164.Wiese C, Grieskamp T, Airik R, Mommersteeg MT, Gardiwal A, de Gier-de Vries C, Schuster-Gossler K, Moorman AF, Kispert A, Christoffels VM. Formation of the sinus node head and differentiation of sinus node myocardium are independently regulated by Tbx18 and Tbx3. Circ Res. 2009;104:388–397. doi: 10.1161/CIRCRESAHA.108.187062. [DOI] [PubMed] [Google Scholar]
- 165.Jiao K, Kulessa H, Tompkins K, Zhou Y, Batts L, Baldwin HS, Hogan BL. An essential role of Bmp4 in the atrioventricular septation of the mouse heart. Genes Dev. 2003;17:2362–2367. doi: 10.1101/gad.1124803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Ma L, Lu MF, Schwartz RJ, Martin JF. Bmp2 is essential for cardiac cushion epithelial-mesenchymal transition and myocardial patterning. Development. 2005;132:5601–5611. doi: 10.1242/dev.02156. [DOI] [PubMed] [Google Scholar]
- 167.Chi NC, Shaw RM, De Val S, Kang G, Jan LY, Black BL, Stainier DY. Foxn4 directly regulates tbx2b expression and atrioventricular canal formation. Genes Dev. 2008;22:734–739. doi: 10.1101/gad.1629408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Park C, Lavine K, Mishina Y, Deng CX, Ornitz DM, Choi K. Bone morphogenetic protein receptor 1A signaling is dispensable for hematopoietic development but essential for vessel and atrioventricular endocardial cushion formation. Development. 2006;133:3473–3484. doi: 10.1242/dev.02499. [DOI] [PubMed] [Google Scholar]
- 169.Goldman DC, Donley N, Christian JL. Genetic interaction between Bmp2 and Bmp4 reveals shared functions during multiple aspects of mouse organogenesis. Mech Dev. 2009;126:117–127. doi: 10.1016/j.mod.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Chen YH, Ishii M, Sucov HM, Maxson RE., Jr Msx1 and Msx2 are required for endothelial-mesenchymal transformation of the atrioventricular cushions and patterning of the atrioventricular myocardium. BMC Dev Biol. 2008;8:75. doi: 10.1186/1471-213X-8-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Boogerd KJ, Wong LY, Christoffels VM, Klarenbeek M, Ruijter JM, Moorman AF, Barnett P. Msx1 and Msx2 are functional interacting partners of T-box factors in the regulation of Connexin43. Cardiovasc Res. 2008;78:485–493. doi: 10.1093/cvr/cvn049. [DOI] [PubMed] [Google Scholar]
- 172.Rutenberg JB, Fischer A, Jia H, Gessler M, Zhong TP, Mercola M. Developmental patterning of the cardiac atrioventricular canal by Notch and Hairy-related transcription factors. Development. 2006;133:4381–4390. doi: 10.1242/dev.02607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Kokubo H, Tomita-Miyagawa S, Hamada Y, Saga Y. Hesr1 and Hesr2 regulate atrioventricular boundary formation in the developing heart through the repression of Tbx2. Development. 2007;134:747–755. doi: 10.1242/dev.02777. [DOI] [PubMed] [Google Scholar]
- 174.Morton SU, Scherz PJ, Cordes KR, Ivey KN, Stainier DY, Srivastava D. microRNA-138 modulates cardiac patterning during embryonic development. Proc Natl Acad Sci U S A. 2008;105:17830–17835. doi: 10.1073/pnas.0804673105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Mikawa T, Hurtado R. Development of the cardiac conduction system. Semin Cell Dev Biol. 2007;18:90–100. doi: 10.1016/j.semcdb.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 176.Jongbloed MR, Mahtab EA, Blom NA, Schalij MJ, Gittenberger-de Groot AC. Development of the cardiac conduction system and the possible relation to predilection sites of arrhythmogenesis. ScientificWorldJournal. 2008;8:239–269. doi: 10.1100/tsw.2008.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.McNally EM, Svensson EC. Setting the pace: Tbx3 and Tbx18 in cardiac conduction system development. Circ Res. 2009;104:285–287. doi: 10.1161/CIRCRESAHA.109.193680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Kreuzberg MM, Willecke K, Bukauskas FF. Connexin-mediated cardiac impulse propagation: connexin 30.2 slows atrioventricular conduction in mouse heart. Trends Cardiovasc Med. 2006;16:266–272. doi: 10.1016/j.tcm.2006.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Bernstein SA, Morley GE. Gap junctions and propagation of the cardiac action potential. Adv Cardiol. 2006;42:71–85. doi: 10.1159/000092563. [DOI] [PubMed] [Google Scholar]
- 180.Gourdie RG. A map of the heart: gap junctions, connexin diversity and retroviral studies of conduction myocyte lineage. Clin Sci (Lond) 1995;88:257–262. doi: 10.1042/cs0880257. [DOI] [PubMed] [Google Scholar]
- 181.Hatcher CJ, Basson CT. Specification of the cardiac conduction system by transcription factors. Circ Res. 2009;105:620–630. doi: 10.1161/CIRCRESAHA.109.204123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Christoffels VM, Smits GJ, Kispert A, Moorman AF. Development of the pacemaker tissues of the heart. Circ Res. 2010;106:240–254. doi: 10.1161/CIRCRESAHA.109.205419. [DOI] [PubMed] [Google Scholar]
- 183.Gourdie RG, Mima T, Thompson RP, Mikawa T. Terminal diversification of the myocyte lineage generates Purkinje fibers of the cardiac conduction system. Development. 1995;121:1423–1431. doi: 10.1242/dev.121.5.1423. [DOI] [PubMed] [Google Scholar]
- 184.Cheng G, Litchenberg WH, Cole GJ, Mikawa T, Thompson RP, Gourdie RG. Development of the cardiac conduction system involves recruitment within a multipotent cardiomyogenic lineage. Development. 1999;126:5041–5049. doi: 10.1242/dev.126.22.5041. [DOI] [PubMed] [Google Scholar]
- 185.Gourdie RG, Harris BS, Bond J, Edmondson AM, Cheng G, Sedmera D, O'Brien TX, Mikawa T, Thompson RP. His-Purkinje lineages and development. Novartis Found Symp. 2003;250:110–122. discussion 122-114, 276-119. [PubMed] [Google Scholar]
- 186.Miquerol L, Kelly RG. Monitoring clonal growth in the developing ventricle. Pediatr Cardiol. 2009;30:603–608. doi: 10.1007/s00246-008-9371-4. [DOI] [PubMed] [Google Scholar]
- 187.Takebayashi-Suzuki K, Yanagisawa M, Gourdie RG, Kanzawa N, Mikawa T. In vivo induction of cardiac Purkinje fiber differentiation by coexpression of preproendothelin-1 and endothelin converting enzyme-1. Development. 2000;127:3523–3532. doi: 10.1242/dev.127.16.3523. [DOI] [PubMed] [Google Scholar]
- 188.Hall CE, Hurtado R, Hewett KW, Shulimovich M, Poma CP, Reckova M, Justus C, Pennisi DJ, Tobita K, Sedmera D, Gourdie RG, Mikawa T. Hemodynamic-dependent patterning of endothelin converting enzyme 1 expression and differentiation of impulse-conducting Purkinje fibers in the embryonic heart. Development. 2004;131:581–592. doi: 10.1242/dev.00947. [DOI] [PubMed] [Google Scholar]
- 189.Rentschler S, Zander J, Meyers K, France D, Levine R, Porter G, Rivkees SA, Morley GE, Fishman GI. Neuregulin-1 promotes formation of the murine cardiac conduction system. Proc Natl Acad Sci U S A. 2002;99:10464–10469. doi: 10.1073/pnas.162301699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Milan DJ, Giokas AC, Serluca FC, Peterson RT, MacRae CA. Notch1b and neuregulin are required for specification of central cardiac conduction tissue. Development. 2006;133:1125–1132. doi: 10.1242/dev.02279. [DOI] [PubMed] [Google Scholar]
- 191.Lyons GE, Ontell M, Cox R, Sassoon D, Buckingham M. The expression of myosin genes in developing skeletal muscle in the mouse embryo. J Cell Biol. 1990;111:1465–1476. doi: 10.1083/jcb.111.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Kubalak SW, Miller-Hance WC, O'Brien TX, Dyson E, Chien KR. Chamber specification of atrial myosin light chain-2 expression precedes septation during murine cardiogenesis. J Biol Chem. 1994;269:16961–16970. [PubMed] [Google Scholar]
- 193.Kelly RG, Zammit PS, Buckingham ME. Cardiosensor mice and transcriptional subdomains of the vertebrate heart. Trends Cardiovasc Med. 1999;9:3–10. doi: 10.1016/s1050-1738(98)00034-6. [DOI] [PubMed] [Google Scholar]
- 194.Franco D, Kelly R, Lamers WH, Buckingham M, Moorman AF. Regionalized transcriptional domains of myosin light chain 3f transgenes in the embryonic mouse heart: morphogenetic implications. Dev Biol. 1997;188:17–33. doi: 10.1006/dbio.1997.8622. [DOI] [PubMed] [Google Scholar]
- 195.Bao ZZ, Bruneau BG, Seidman JG, Seidman CE, Cepko CL. Regulation of chamber-specific gene expression in the developing heart by Irx4. Science. 1999;283:1161–1164. doi: 10.1126/science.283.5405.1161. [DOI] [PubMed] [Google Scholar]
- 196.Wang GF, Nikovits W, Jr., Bao ZZ, Stockdale FE. Irx4 forms an inhibitory complex with the vitamin D and retinoic X receptors to regulate cardiac chamber-specific slow MyHC3 expression. J Biol Chem. 2001;276:28835–28841. doi: 10.1074/jbc.M103716200. [DOI] [PubMed] [Google Scholar]
- 197.Xin M, Small EM, van Rooij E, Qi X, Richardson JA, Srivastava D, Nakagawa O, Olson EN. Essential roles of the bHLH transcription factor Hrt2 in repression of atrial gene expression and maintenance of postnatal cardiac function. Proc Natl Acad Sci U S A. 2007;104:7975–7980. doi: 10.1073/pnas.0702447104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.McFadden DG, Charite J, Richardson JA, Srivastava D, Firulli AB, Olson EN. A GATA-dependent right ventricular enhancer controls dHAND transcription in the developing heart. Development. 2000;127:5331–5341. doi: 10.1242/dev.127.24.5331. [DOI] [PubMed] [Google Scholar]
- 199.Togi K, Kawamoto T, Yamauchi R, Yoshida Y, Kita T, Tanaka M. Role of Hand1/eHAND in the dorso-ventral patterning and interventricular septum formation in the embryonic heart. Mol Cell Biol. 2004;24:4627–4635. doi: 10.1128/MCB.24.11.4627-4635.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.McFadden DG, Barbosa AC, Richardson JA, Schneider MD, Srivastava D, Olson EN. The Hand1 and Hand2 transcription factors regulate expansion of the embryonic cardiac ventricles in a gene dosage-dependent manner. Development. 2005;132:189–201. doi: 10.1242/dev.01562. [DOI] [PubMed] [Google Scholar]
- 201.Kelly RG, Lemonnier M, Zaffran S, Munk A, Buckingham ME. Cell history determines the maintenance of transcriptional differences between left and right ventricular cardiomyocytes in the developing mouse heart. J Cell Sci. 2003;116:5005–5013. doi: 10.1242/jcs.00824. [DOI] [PubMed] [Google Scholar]
- 202.Reese DE, Mikawa T, Bader DM. Development of the coronary vessel system. Circ Res. 2002;91:761–768. doi: 10.1161/01.res.0000038961.53759.3c. [DOI] [PubMed] [Google Scholar]
- 203.Lie-Venema H, van den Akker NM, Bax NA, Winter EM, Maas S, Kekarainen T, Hoeben RC, deRuiter MC, Poelmann RE, Gittenberger-de Groot AC. Origin, fate, and function of epicardium-derived cells (EPDCs) in normal and abnormal cardiac development. ScientificWorldJournal. 2007;7:1777–1798. doi: 10.1100/tsw.2007.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Ieda M, Tsuchihashi T, Ivey KN, Ross RS, Hong TT, Shaw RM, Srivastava D. Cardiac fibroblasts regulate myocardial proliferation through beta1 integrin signaling. Dev Cell. 2009;16:233–244. doi: 10.1016/j.devcel.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Olivey HE, Svensson EC. Epicardial-myocardial signaling directing coronary vasculogenesis. Circ Res. 2010;106:818–832. doi: 10.1161/CIRCRESAHA.109.209197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Gittenberger-de Groot AC, Mahtab EA, Hahurij ND, Wisse LJ, Deruiter MC, Wijffels MC, Poelmann RE. Nkx2.5-negative myocardium of the posterior heart field and its correlation with podoplanin expression in cells from the developing cardiac pacemaking and conduction system. Anat Rec (Hoboken) 2007;290:115–122. doi: 10.1002/ar.20406. [DOI] [PubMed] [Google Scholar]
- 207.Mommersteeg MT, Dominguez JN, Wiese C, Norden J, de Gier-de Vries C, Burch JB, Kispert A, Brown NA, Moorman AF, Christoffels VM. The sinus venosus progenitors separate and diversify from the first and second heart fields early in development. Cardiovasc Res. 2010 doi: 10.1093/cvr/cvq033. [DOI] [PubMed] [Google Scholar]
- 208.Schlueter J, Manner J, Brand T. BMP is an important regulator of proepicardial identity in the chick embryo. Dev Biol. 2006;295:546–558. doi: 10.1016/j.ydbio.2006.03.036. [DOI] [PubMed] [Google Scholar]
- 209.Kruithof BP, van Wijk B, Somi S, Kruithof-de Julio M, Perez Pomares JM, Weesie F, Wessels A, Moorman AF, van den Hoff MJ. BMP and FGF regulate the differentiation of multipotential pericardial mesoderm into the myocardial or epicardial lineage. Dev Biol. 2006;295:507–522. doi: 10.1016/j.ydbio.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 210.van Wijk B, van den Berg G, Abu-Issa R, Barnett P, van der Velden S, Schmidt M, Ruijter JM, Kirby ML, Moorman AF, van den Hoff MJ. Epicardium and myocardium separate from a common precursor pool by crosstalk between bone morphogenetic protein- and fibroblast growth factor-signaling pathways. Circ Res. 2009;105:431–441. doi: 10.1161/CIRCRESAHA.109.203083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Grieskamp T, Rudat C, Norden J, Kispert A. Epicardial fate and notch signaling; Paper presented at: Weinstein Cardiovascular Development Conference; Amsterdam. May 20-22, 2010. [Google Scholar]
- 212.Lee KF, Simon H, Chen H, Bates B, Hung MC, Hauser C. Requirement for neuregulin receptor erbB2 in neural and cardiac development. Nature. 1995;378:394–398. doi: 10.1038/378394a0. [DOI] [PubMed] [Google Scholar]
- 213.Meyer D, Birchmeier C. Multiple essential functions of neuregulin in development. Nature. 1995;378:386–390. doi: 10.1038/378386a0. [DOI] [PubMed] [Google Scholar]
- 214.Erickson SL, O'Shea KS, Ghaboosi N, Loverro L, Frantz G, Bauer M, Lu LH, Moore MW. ErbB3 is required for normal cerebellar and cardiac development: a comparison with ErbB2-and heregulin-deficient mice. Development. 1997;124:4999–5011. doi: 10.1242/dev.124.24.4999. [DOI] [PubMed] [Google Scholar]
- 215.Nebigil CG, Maroteaux L. A novel role for serotonin in heart. Trends Cardiovasc Med. 2001;11:329–335. doi: 10.1016/s1050-1738(01)00135-9. [DOI] [PubMed] [Google Scholar]
- 216.Grego-Bessa J, Luna-Zurita L, del Monte G, Bolos V, Melgar P, Arandilla A, Garratt AN, Zang H, Mukouyama YS, Chen H, Shou W, Ballestar E, Esteller M, Rojas A, Perez-Pomares JM, de la Pompa JL. Notch signaling is essential for ventricular chamber development. Dev Cell. 2007;12:415–429. doi: 10.1016/j.devcel.2006.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Chen H, Shi S, Acosta L, Li W, Lu J, Bao S, Chen Z, Yang Z, Schneider MD, Chien KR, Conway SJ, Yoder MC, Haneline LS, Franco D, Shou W. BMP10 is essential for maintaining cardiac growth during murine cardiogenesis. Development. 2004;131:2219–2231. doi: 10.1242/dev.01094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Pashmforoush M, Lu JT, Chen H, Amand TS, Kondo R, Pradervand S, Evans SM, Clark B, Feramisco JR, Giles W, Ho SY, Benson DW, Silberbach M, Shou W, Chien KR. Nkx2-5 pathways and congenital heart disease; loss of ventricular myocyte lineage specification leads to progressive cardiomyopathy and complete heart block. Cell. 2004;117:373–386. doi: 10.1016/s0092-8674(04)00405-2. [DOI] [PubMed] [Google Scholar]
- 219.Lavine KJ, Yu K, White AC, Zhang X, Smith C, Partanen J, Ornitz DM. Endocardial and epicardial derived FGF signals regulate myocardial proliferation and differentiation in vivo. Dev Cell. 2005;8:85–95. doi: 10.1016/j.devcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 220.Lu SY, Sheikh F, Sheppard PC, Fresnoza A, Duckworth ML, Detillieux KA, Cattini PA. FGF-16 is required for embryonic heart development. Biochem Biophys Res Commun. 2008;373:270–274. doi: 10.1016/j.bbrc.2008.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Stankunas K, Hang CT, Tsun ZY, Chen H, Lee NV, Wu JI, Shang C, Bayle JH, Shou W, Iruela-Arispe ML, Chang CP. Endocardial Brg1 represses ADAMTS1 to maintain the microenvironment for myocardial morphogenesis. Dev Cell. 2008;14:298–311. doi: 10.1016/j.devcel.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Abu-Issa R, Kirby ML. Heart field: from mesoderm to heart tube. Annu Rev Cell Dev Biol. 2007;23:45–68. doi: 10.1146/annurev.cellbio.23.090506.123331. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.