Abstract
Mass spectrometry is an important technique for analyzing proteins and other biomolecular compounds in biological samples. Each of the vendors of these mass spectrometers uses a different proprietary binary output file format, which has hindered data sharing and the development of open source software for downstream analysis. The solution has been to develop, with the full participation of academic researchers as well as software and hardware vendors, an open XML-based format for encoding mass spectrometer output files, and then to write software to use this format for archiving, sharing, and processing. This chapter presents the various components and information available for this format, mzML. In addition to the XML schema that defines the file structure, a controlled vocabulary provides clear terms and definitions for the spectral metadata, and a semantic validation rules mapping file allows the mzML semantic validator to insure that an mzML document complies with one of several levels of requirements. Complete documentation and example files insure that the format may be uniformly implemented. At the time of release there already existed several implementations of the format and vendors have committed to supporting the format in their products.
Keywords: file format, mzML, standards, XML, controlled vocabulary
1. Introduction
Mass spectrometry is an important method to analyse biomolecules by measuring the intact mass-to-charge ratios of their in-situ generated ionised forms or the mass-to-charge ratios of in-situ-generated fragments of these ions. The resulting mass spectra are used for a variety of purposes, among which is the identification, characterization, and absolute or relative quantification of the analysed molecules. The processing steps to achieve these goals typically involve semi-automatic computational analysis of the recorded mass spectra and sometimes also of the associated metadata (e.g., elution characteristics if the instrument is coupled to a chromatography system). The result of the processing can be assigned a score, rank or confidence measure.
Differences inherent in the use of a variety of instruments, different experimental conditions under which analyses are performed, and potential automatic data preprocessing steps by the instrument software can influence the actual measurements and therefore the results after processing. Additionally, most instruments output their acquired data in a very specific and often proprietary format. These proprietary formats are then typically transformed into so-called peak lists to be analysed by identification and characterisation software. Data reduction such as peak centroiding and deisotoping is often performed during this transformation from proprietary formats to peak lists. The peak lists are then used as inputs for subsequent analysis. However, these peak list file formats lack information about the precursor MS signals and about the associated metadata (i.e., instrument settings and description, acquisition mode, etc) compared to the files they were derived from. The many different and often proprietary formats make integration or comparison of mass spectrometer output data difficult or impossible, and the use of the heavily processed and data-poor peak lists is often suboptimal.
The solution has been to create open file formats that can encode the information in these output files in XML (Extensible Markup Language) and then write software to read and write these formats. Several formats were developed and a unified format called mzML emerged. This chapter will first describe the history leading to mzML and then provide an overview of the mzML specification and components, followed by information about the implementations of the format.
2. History
During 2003 – 2005, two data formats to store mass spectrometer output in an open, vendor-neutral, XML format were developed. The mzData format [1] was developed by the Human Proteome Organization (HUPO) Proteomics Standards Initiative (PSI), primarily as a data exchange and archive format. The mzXML format [2] was developed at the Institute for Systems Biology (ISB), primarily in order to streamline data processing software. Both formats are used extensively but having two formats for essentially the same information causes unnecessary confusion in the community and adds complexity to software development as often both formats must be supported. Therefore the designers of mzData and mzXML, including representatives of instrument vendors, analysis software developers and end users, have joined under the auspices of the PSI and jointly developed a single format intended to replace the previous two. This new format is named mzML and is the subject of this chapter.
The main difference between the two original formats, aside from the primary intent described above, is the design philosophy of flexibility. The mzData format was designed to be quite flexible via the extensive use of a controlled vocabulary. It was hoped that the actual XML schema could remain stable for many years while the accompanying controlled vocabulary could be frequently updated to support new technologies, instruments, and methods of acquiring data. However, a significant complication with this philosophy was that flexibility in the format led to a variety of styles of the format, with each different tool representing data in a different dialect of the format, causing significant trouble for reader software.
On the other hand, mzXML was designed with a very strict schema with most auxiliary information described in enumerated attributes. This simplified software implementations as there was only one way to present various attributes and the validity of the documents could be easily checked with industry-standard XML validators. However, virtually any desired change to the format, even adding one attribute, would require a new version number. This led to several closely related version numbers, and different software programs supporting different versions.
The main challenge in uniting these two formats was therefore resolving the opposing philosophies rather than fundamental technical issues. The result is a format that contains the best aspects of the two original formats so that it may be widely adopted and resolve the previous problem of two formats.
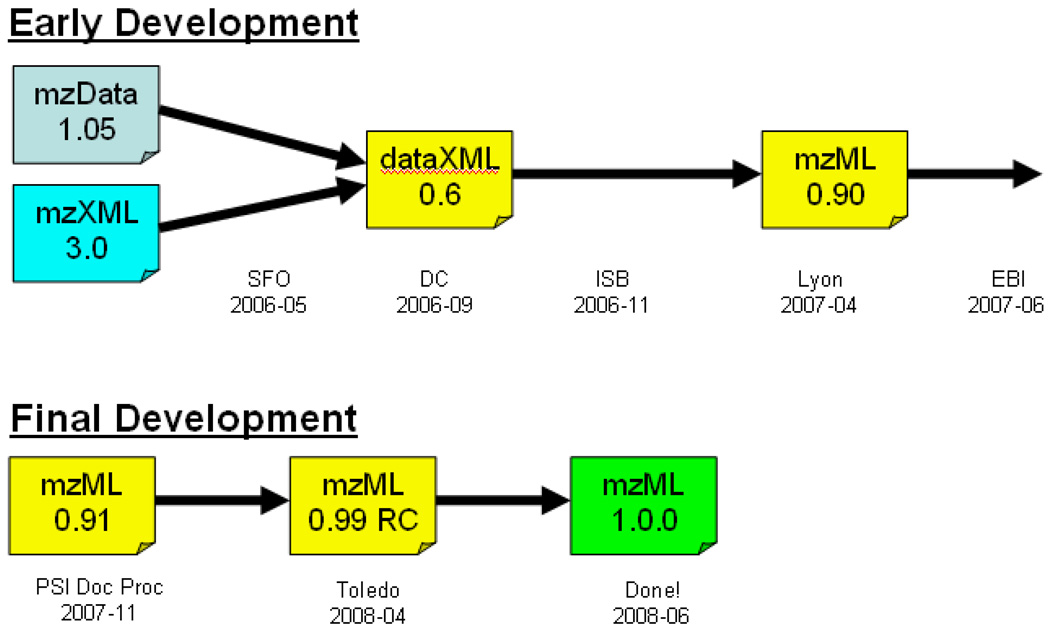
The will to start the unification process was gathered at the spring 2006 PSI workshop in San Francisco, CA. At this meeting the technical differences between the two formats were examined and an agreement to move toward unification was reached. The various vendors voiced their displeasure with the state of having two formats, being almost uniformly unwilling to support both. They all voiced that they would implement a unified format developed by the PSI. For two years a relatively small band of PSI participant volunteers met at workshops and tried to push forward progress between workshops (Figure 1). Finally, two years after the initial agreement, mzML 1.0.0 was released on June 1, 2008, coinciding with the American Society for Mass Spectrometry (ASMS) conference. The PSI MS working group continues to support the format by maintaining the controlled vocabulary, semantic validators, and documentation as described below.
Figure 1.
History of the development of mzML, beginning with the initial unification agreement in May 2006 in San Francisco (SFO), continuing work at PSI workshops in Washington, DC, USA (DC), the Institute for Systems Biology (ISB) in Seattle, USA, Lyon, France, the European Bioinformatics Institute (EBI) in Hinxton, UK, Toledo, Spain, and the 1.0.0 release in June 2008 at the American Society for Mass Spectrometry (ASMS) conference in Denver, USA. Maintenance of mzML will continue with the PSI Mass Spectrometry Standards Working Group. It is expected that the schema will remain stable, but minor updates to the controlled vocabulary and semantic validation rules may be necessary.
3. Design of the format
The design of mzML benefited tremendously from the precursor formats mzXML and mzData. It was not necessary to start from scratch. Rather, the designers could take the best aspects from each of the precursor formats, consider known deficiencies of the previous formats, and apply the lessons learned from years of real-world implementations and use of these formats. In this section, the primary design aspects are presented after a brief discussion of the design philosophy of mzML.
3.1. Design Principles
Since the development of mzML brought together different philosophies, the primary mzML designers agreed on the following design principles that would guide its development:
Keep the format simple. Many elaborate extensions were proposed but most were rejected in favour of a simple implementation.
Eliminate alternate ways of encoding the same information. Such flexibility, while sometimes touted as a benefit for some products, is bad for data formats.
Build in some flexibility for encoding new important information but keep the format stable. There is a strong desire from companies that develop software for their customers to keep the data format stable over long periods of time with updates to an auxiliary file.
Support the features of mzData and mzXML but not much more in version 1.0. The only major new feature deemed crucial was support for selected reaction monitoring (SRM) data (which is also supported in the latest mzXML 3.1 and thus not really new).
Finish version 1.0 of the format soon with the resources available. It was felt that the greatest community benefit would be to resolve the mzData/mzXML duality rather than expend limited resources on new features.
Validate the new format by implementing software to read and write the format before its release.
There was great temptation to add support for many new kinds of data and representation possibilities. There are many enhancements that have been suggested, but the small group of volunteers that have actively developed this format have opted to focus on the primary goal set before them: develop a single format that the vendors and current software can easily support and thereby obsolete mzData and mzXML. The enhancements not considered compatible with this goal will be entertained for mzML 2.0
One of the aspects of mzXML that enabled its very swift adoption as a de facto standard was an immediately available set of open source tools that implemented the format. With these tools many users were able to begin using the format immediately without coding their own software. Therefore, to insure that mzML is a format that will be adopted quickly and implemented uniformly, the format was to be released along with several tools that write, read, and validate the format. It was deemed crucial that at submission to the PSI document process, the following minimum software would implement mzML:
Two or more converters that convert from vendor formats to mzML.
The popular RAMP parser library that currently supports mzData and mzXML.
An mzML semantic validator that checks for correct implementation of files.
These implementations are discussed in section 4.
3.2. XML Schema
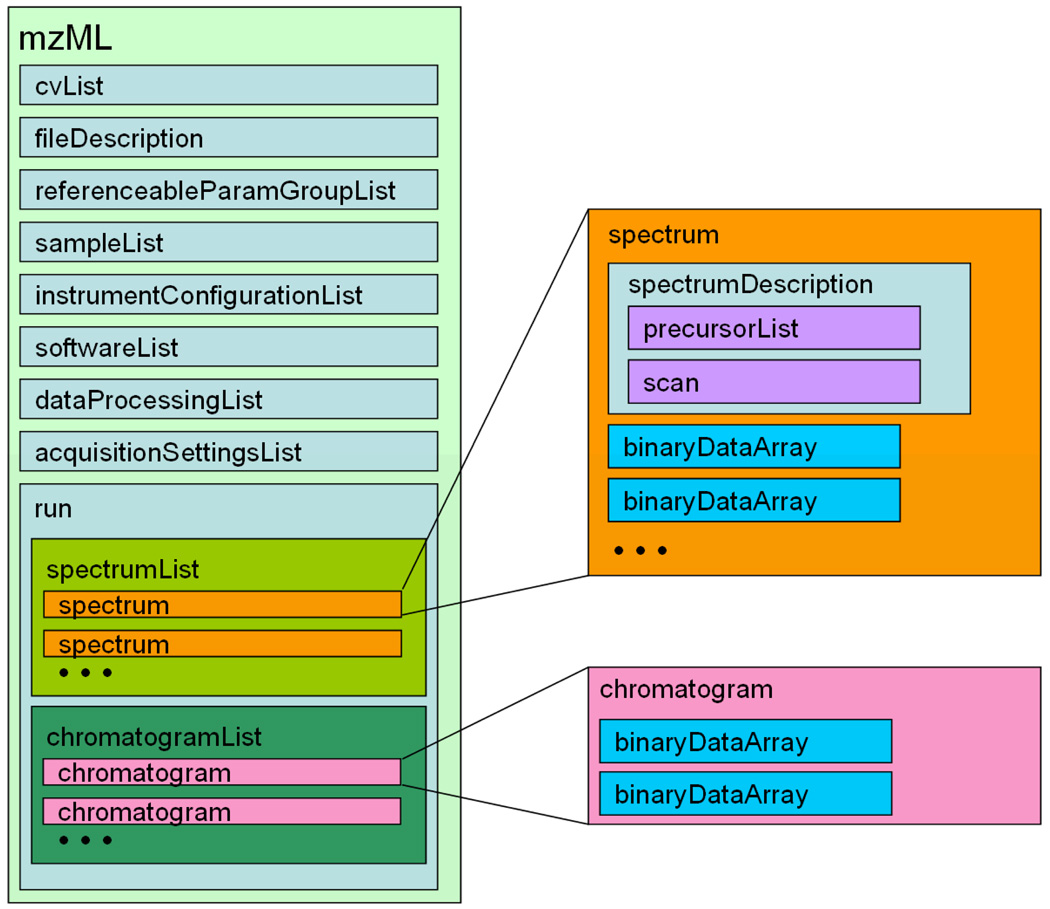
An mzML document is designed to contain all the information for a single MS run, including metadata about the spectra plus all the spectra themselves, either in centroided (peak list) or profile mode. At the top of the file, the <cvList> element contains information about the controlled vocabularies referenced in the rest of the file. The <fileDescription> element contains some basic information about the type of spectra to be found in the file. The optional <referenceableParamGroupList> element contains a list of groups of controlled vocabulary terms that are used frequently in the file and may simply be defined once and referenced thereafter. Following this basic housekeeping information, the <sampleList> element may optionally contain information about samples that are referenced throughout the file. The <instrumentConfiguration> element contains information about the instrument used for the run (possibly in more than one configuration for hybrid instruments). The <softwareList> and <dataProcessingList> elements provide a history of data processing that may have occurred since raw acquisition. An optional <acquisitionSettingsList> element can hold special input parameters to mass spectrometers such as inclusion lists. This is followed by the actual spectra and optionally some chromatograms. The high level outline of the schema structure is shown in Figure 1.
As with its predecessors, mzML is encoded in an XML format. The structure of the format is defined by an XSD (XML schema definition), which is used to insure that documents are properly formed. XML is both easily parsed by computer programs, using well established libraries, and is also relatively easily readable by humans since it is a text-based format. This is a benefit during the design process, and aids in troubleshooting file problems, although it does come at the cost of larger file sizes than with binary formats. However, XML documents do compress well.
One of the requirements for mzML was that it provide a standardized mechanism for a random access index in the same way as mzXML. The use case is that when writing software to process mzML or view individual spectra, it is often necessary to quickly pull out an arbitrary spectrum. If a program needs to display spectrum number 18,345 to display to a user, it must be able to seek to that spectrum in the file rather than read it sequentially if a fast user experience can be expected.
Some have argued that providing a random access index into XML is an anathema to the intent of XML and list the many possible ways in which an index could become broken. However, several years of use of such an index in the mzXML format has shown that the indexing problems are few and the benefits are enormous. Reader software can (and has been) easily written to make use of the index, but automatically rebuild the index if it is noticed to be incorrect.
Since many are not interested in an index, mzML has been designed such that the main part of an mzML document does not contain an index, but that the document may be enclosed in a wrapper schema that includes an index. Therefore an .mzML file may contain a plain mzML document or an indexed mzML document. Reader software is designed to handle either. A sample snippet of XML showing the wrapper indexing schema is show in Figure 3.
Figure 3.
Example top and bottom of an mzML document with the middle segment removed for display purposes. The main part of the mzML document is contained within the <mzML></mzML> tags. It is wrapped within an <indexedmzML></indexedmzML> construct, which contains the random access index at the bottom.
3.3. Controlled Vocabulary
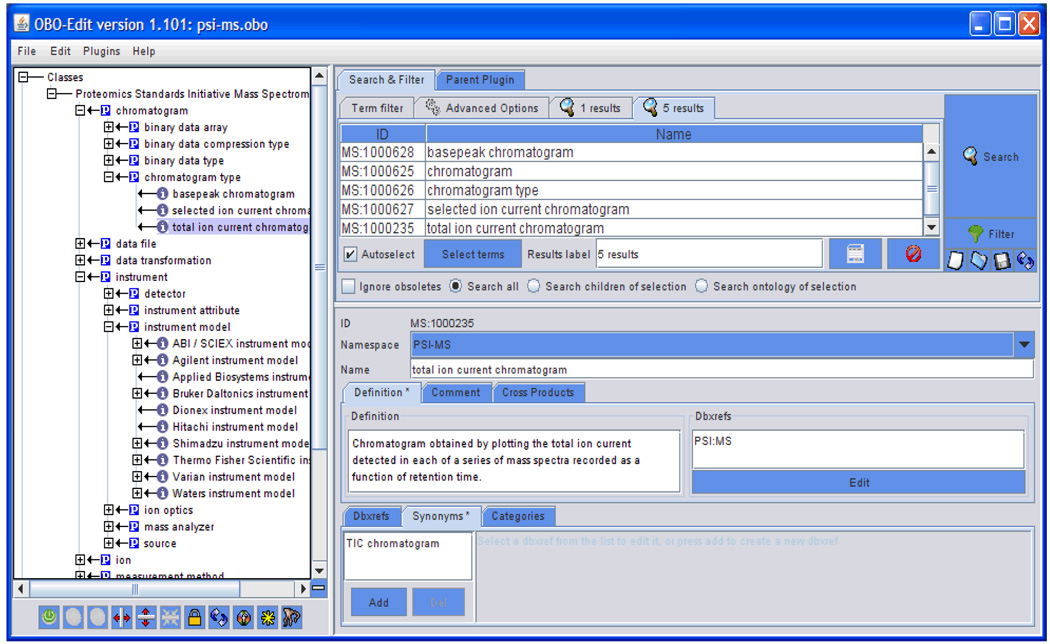
Much of the metadata encoded in the mzML is in the form of a cvParam, an XML element that provides a reference to a specific concept within the PSI MS controlled vocabulary. Each term has an explicit and detailed definition, and may have information about its data type and what kind of units it requires, if any. The controlled vocabulary is edited in OBO format with the OBO-Edit software [3] (Figure 4) and is used by most readers and writers of mzML. The controlled vocabulary can be easily adjusted and extended without modifying the mzML schema.
Figure 4.
A view of the PSI MS controlled vocabulary as seen in the OBO-Edit program, currently highlighting the term “total ion current chromatogram”.
3.4. Semantic Validator
The mzData format was a far more flexible format than mzXML. The support of new technologies could be added to mzData files by adding new controlled vocabulary terms, while mzXML often required a full schema revision. However, mzData did suffer from a problem of inconsistently used vocabulary terms and there appeared several different dialects of mzData, encoding the same information in subtly different ways. This was not usually a problem for human inspection of the file, but caused difficulty writing and maintaining reader software.
This problem has been solved (it is hoped) for mzML by releasing a semantic validator with the data format. This semantic validator enforces many rules as to how controlled vocabulary terms are used, not only making sure that the terms are in the CV, but also that the correct terms are used in the correct location in the document and the required terms are present the correct number of times. This allows greater flexibility in the schema, but enforces order in how the CV terms are used. This will require the discipline to use the semantic validator, not just an XML validator, to validate new or updated mzML writer code. The result is that new technologies or information can be accommodated with adjustments to the controlled vocabulary and validator, not to the schema. Opinions differ on whether this is a benefit or not.
Another benefit of using the semantic validator is that different levels of compliance can be defined. For example, if configured with a rules file for basic mzML, the validator will point out any problems that prevent the mzML from being correct at a basic level that should be expected by all parsers. However, for submission to a journal that requires the MIAPE-MS guidelines[4], the validator can be configured to use the MIAPE-MS rules file to check if an mzML file is fully compliant at the MIAPE-MS level. This level of compliance insures significant additional metadata that should be produced for new data and publications but cannot be expected from older data.
Another benefit is that metadata requirements can be adjusted for different types of data. For example, photodiode array (PDA) spectra generated from a mass spectrometer instrument can be encoded in mzML, but the requirements will be different than for mass spectra. These different types of spectra can be encoded using the same tags, just with different metadata, and this can be enforced through the semantic validator.
Semantic rules are encoded in one or more (for different compliance levels) rules mapping files and these can be updated along with the controlled vocabulary without changing the schema. The validator is available as a web page to which any file can be uploaded or as a standalone tool that can validate local files.
3.5. Documentation
The full mzML specification is available as a PDF document at the mzML web site. It describes many of the details of the design of the format and then describes each of the elements of the format in detail, along with figures depicting the structure graphically. The element documentation is autogenerated by some custom software that reads the XSD file, a sample document, the controlled vocabulary, and the rules mapping file and writes out an HTML representation of the information contained within these files. This representation is also imported into the full specification document.
In order to exercise the schema and demonstrate that the various use cases have been adequately modeled, we have developed several example instance documents. Some of the documents are hand-crafted with an ordinary editor, while others are written out as a software test as part of the ProteoWizard reference implementation. In addition, several instance documents are conversions of real data files using ProteoWizard or other implementations of converters.
4. Implementations of the format
The best way to test a new format is by implementing it in software. Inevitably as a format is implemented, one finds minor inconsistencies or missing features. The initial release of mzML is strengthened by the breadth of implementations that coexisted with the release and have exercised the various use cases.
4.1. Reference Implementation
The ProteoWizard software project[5, 6], initiated by the Spielberg Family Center for Applied Proteomics at the Cedars-Sinai Medical Center, provides a modular and extensible set of open-source, cross-platform tools and libraries. The tools perform proteomics data analyses; the libraries enable rapid tool creation by providing a robust, pluggable development framework that simplifies and unifies data file access, and performs standard chemistry and LCMS dataset computations. During the final stages of mzML development, refinement, and testing, the ProteoWizard library has provided the necessary framework for testing and reference implementation of mzML.
ProteoWizard is modular C++ library with an internal data model that has a one-to-one translation of mzML data elements to C++ data structures. It builds with native compilers on all major platforms (MSVC on Windows, gcc on Linux, XCode on OSX) and is available under the Apache Version 2 license. ProteoWizard has a plug-in Reader interface for reading both open and vendor proprietary data formats: mzML, mzXML, Thermo RAW, MGF; there are additional Readers in development. CLI binding allows use of ProteoWizard libraries from .NET languages (C++/CLI, C#, VB.NET), and SWIG bindings for scripting (from Java, Python, Perl, R) are in development.
ProteoWizard also comes with several tools that make use of the library to perform various processing or display tasks. The msconvert tool provides general file format conversion, including native centroiding and zlib compression. The SeeMS and mspicture tools allow visualization of mass spectral data.
It is worth emphasizing that ProteoWizard has been released under a very permissive license, the Apache Version 2 license, which allows the library to be used in commercial software without influencing the licensing terms of that software. This is in contrast to some other open-source licenses which require that software that uses such a library also be open source.
4.2. Other Implementations
At the time of this writing, there are many software implementations of the mzML format already in place or emerging. A data format is only as usable as the software that implements it. One of the strengths of mzML is this wide variety of software available that uses mzML. Table 1 lists the available software at the time of this writing. An up-to-date table is available at the mzML web site.
Table 1.
A list of software supporting mzML at the time of this writing. An updated list is available at the mzML web site.
| Author | Product | mzML support |
|---|---|---|
| CSHS | ProteoWizard | Full mzML support available today http://proteowizard.sourceforge.net/ |
| ISB | TPP | Full mzML support available today http://tools.proteomecenter.org/ |
| ISB | RAMP, JRAP | Full mzML support available today http://tools.proteomecenter.org/ |
| Insilicos | Insilicos Viewer | Full mzML support available today http://www.insilicos.com/viewer_download.html |
| GeneBio | Phenyx | Full mzML support available today http://www.phenyx-ms.com/ |
| Vanderbilt | MyriMatch | Full mzML support available today http://fenchurch.mc.vanderbilt.edu/lab/software.php |
| Thermo Scientific | RAW->mzML conv | beta |
| Applied Biosystems | WIFF->mzML conv | beta |
| NCBI | NCBI C++ toolkit | beta http://www.ncbi.nlm.nih.gov/IEB/ToolBox/CPP_DOC/ |
| Univ. of Lund | Proteios | Full mzML support available today http://www.proteios.org/ |
| SIB | InSilicoSpectro | Full mzML support available today http://insilicospectro.vital-it.ch/ |
5. Conclusion
The mzML format is an open, XML-based format for mass spectrometer output files, developed with the full participation of vendors and researchers in order to create a single open format that would be supported by all software. The format includes the best features from pre-existing open formats and has additional support for chromatograms and some other features deemed highly desirable. It is expected that the schema will remain stable for at least a year, hopefully more. However, the controlled vocabulary and semantic validation rules will continue to be updated and refined as all authors and vendors finish implementing their software for mzML.
Additional feature requests that cannot be accommodated using the existing schema will be collected and considered for an update release in the next few years.
During the early design phase, RDF (resource description framework) was considered as an alternative to XML. In many ways, the type of flexibility that has been worked into mzML, notably the adapting controlled vocabulary and the semantic validation, are concepts that RDF has the potential to solve nicely. However, it was determined that the developers and the implementer community were not yet ready to try to implement a standard in RDF, a significant departure from custom XML schema. Moreover, the primary goal of the designers was to fix the two-format problem, rather than set out a bold new course. The new mzML 1.0 release fulfills the goals set before the designers. It may well be that the next major release, mzML 2.0, not expected for several years, will be designed using RDF, at a time when the designers and implementers are ready to use this newer platform.
Figure 2.
Overview of the mzML schema. The root mzML element contains elements that provide metadata about a file and run followed by the spectral data itself with optional chromatograms.
Acknowledgements
The design of mzML was a long process that involved a great many contributors. The author would like to thank the following for their contributions to mzML: Lennart Martens, Pierre-Alain Binz, Darren Kessner, Matt Chambers, Luisa Montecchi-Palazzi, Jim Shofstahl, Josh Tasman, Randall K Julian, Fredrik Levander, Puneet Souda, Jari Häkkinen, Brian Pratt, Erik Nilsson, Mike Coleman, Luis Mendoza, David Shteynberg, Lars Nilse, Benito Cañas, Lola Gutierrez, Alberto Medina, Trish Wheztel, Eva Duchoslav, Henning Hermjakob, Angel Pizarro, Phil Jones, Jimmy Eng, Kent Laursen, Sandra Orchard, Chris Taylor, Patrick Pedrioli, Sean Seymour, David Creasy, Howard Read, Jim Langridge, Jayson Falkner, David Horn, Ruth McNally, Ron Beavis, Norman Paton, Marc Sturm, Parag Mallick, Rune Philosof, David Sparkman, Wilfred Tang, Marius Kallhardt, and Ruedi Aebersold.
EWD has been funded in part with Federal funds from the National Heart, Lung, and Blood Institute, National Institutes of Health, under contract No. N01-HV-28179, and from PM50 GMO76547/Center for Systems Biology.
References
- 1.mzData. http://psidev.info/index.php?q=node/80#mzdata.
- 2.Pedrioli PG, et al. A common open representation of mass spectrometry data and its application to proteomics research. Nat Biotechnol. 2004;22(11):1459–1466. doi: 10.1038/nbt1031. [DOI] [PubMed] [Google Scholar]
- 3.Day-Richter J, et al. OBO-Edit--an ontology editor for biologists. Bioinformatics. 2007;23(16):2198–2200. doi: 10.1093/bioinformatics/btm112. [DOI] [PubMed] [Google Scholar]
- 4.Taylor CF, et al. Guidelines for reporting the use of mass spectrometry in proteomics. Nat Biotechnol. 2008;26(8):860–861. doi: 10.1038/nbt0808-860. [DOI] [PubMed] [Google Scholar]
- 5.Kessner D, et al. ProteoWizard: Open Source Software for Rapid Proteomics Tools Development. Bioinformatics. 2008 doi: 10.1093/bioinformatics/btn323. accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.ProteoWizard. http://proteowizard.sourceforge.net.