Abstract
The limited availability of hepatic tissue suitable for the treatment of liver disease and drug discovery research advances the generation of hepatic-like cells from alternative sources as a valuable approach. In this investigation we exploited a unique hepatic differentiation approach to generate hepatocyte-like cells from human embryonic stem cells (hESCs). hESCs were cultured for only 10 days on collagen substrate in highly defined and serum free hepatocyte media. The resulting cell populations exhibited hepatic cell-like morphology and were characterized with a variety of biological endpoint analyses. Real-time PCR analysis demonstrated that mRNA expression of the ‘stemness’ marker genes NANOG and alkaline phosphatase in the differentiated cells was significantly reduced, findings that were functionally validated using alkaline phosphatase activity detection measures. Immunofluorescence studies revealed attenuated levels of the ‘stemness’ markers OCT4, SOX2, SSEA-3, TRA-1-60, and TRA-1-81 in the hepatic-like cell population. The hepatic character of the cells was evaluated additionally by real-time PCR analyses that demonstrated increased mRNA expression of the hepatic transcription factors FOXA1, C/EBPα, and HNF1α, the nuclear receptors CAR, RXRα, PPARα, and HNF4α, the liver-generated plasma proteins α-fetoprotein, transthyretin, transferrin, and albumin, the protease inhibitor α-1-antitrypsin, metabolic enzymes HMGCS2, PEPCK, and biotransformation enzymes CYP3A7, CYP3A4, CYP3A5, and CYP2E1. Indocyanine green uptake and glycogen storage capacity further confirmed acquisition of hepatic function. These studies define an expeditious methodology that facilitates the differentiation of hESCs along a hepatic lineage and provide a framework for their subsequent use in pharmacological and toxicological research applications requiring a renewable supply of human hepatocytes.
Keywords: embryonic stem cells, hepatic, phenotype, transcription
1.0 Introduction
As the largest internal organ in mammals the liver performs myriad functions that are essential to the body including detoxification, production and secretion of plasma proteins, maintenance of cholesterol homeostasis, production and elimination of bile constituents, synthesis and inter-conversion of amino acids, fatty acid processing, and the synthesis and breakdown of glucose [1]. Unfortunately, the liver is the target of a number of diseases processes; in the United States alone more than 40,000 deaths annually may be attributed to liver failure resulting from disease [2]. Both whole liver transplantation, currently the most successful treatment option [3], and hepatocyte transplantation are limited by the paucity of donor tissue, marked donor-to-donor variability in tissue quality, and the risk of infection, tissue rejection, or adverse immune response in the recipient [4]. Primary human hepatocyte cultures also serve as models for drug metabolism research and as predictors of toxicological processes that may be associated with exposures to xenobiotic compounds. However, the scarcity of hepatic tissue, as well as the difficulties inherent in maintaining differentiated hepatocytes in culture, currently limits this vital research. Thus, the promise of procuring a renewable supply of functional hepatocytes from alternative sources, such as from human embryonic stem cells (hESCs), represents an important goal in liver research.
Liver development is governed by a complex series of induction events mediated by extracellular signals. Hepatic induction by these factors initiates intracellular signaling cascades that activate hepatic-enriched transcription factors resulting in global changes in gene expression patterns and manifestation of the hepatic phenotype. In brief, during mouse development the liver bud forms from the ventral foregut endoderm on embryonic day (E) 9 [5;6] in response to fibroblast growth factors (FGFs) secreted from the cardiac mesoderm [7] and bone morphogenetic proteins (BMPs) secreted from the septum transversum mesenchyme [8]. As the liver bud grows the hepatic precursors, or hepatoblasts, delaminate and migrate as cords through the basement membrane which separates the foregut from the adjacent septum transversum [6]. Hepatoblasts in the newly emerged liver bud are bipotential, capable of giving rise to definitive hepatocytes or cholangiocytes [9]. At E10 the migrating hepatoblast cords associate with rudimentary sinusoidal endothelial cells which form basic capillary-like sinusoids that run between the cords [10]. Prior to E12 in mouse development, hepatoblasts are morphologically undifferentiated [11]; compared to the mature hepatocyte, hepatoblasts exhibit irregular shape, have few cellular organelles, and have a large nuclear to cytoplasmic ratio [12]. Although still lacking the spaces and fenestrations that characterize sinusoids in the mature liver, by E14 sinusoidal endothelium has been definitively established [10]. Continuing hepatic differentiation is marked by changes in hepatocyte ultrastructure including increases in the number of Golgi apparati and endoplasmic reticuli which facilitate the necessary enhancement in protein synthesis as well as the emergence of peroxysomes and glycogen rosettes [12]. Hepatocyte polarity is established with the development of bile canaliculi at the apical hepatocyte surface [12].
HESCs, derived from the inner cell mass of the blastocysts of developing embryos, have the potential for indefinite self-renewal yet retain a plasticity which, under appropriate conditions, permits their differentiation into derivatives of all three germ layers [13]. To date a number of studies have undertaken to induce differentiation of hESCs along a hepatic lineage. The majority of these studies have employed defined culture conditions to promote differentiation, supplementing culture media with FGFs [4;14-17], BMPs [15-18], hepatocyte growth factor (HGF) [4;16;17;19], dexamethasone [16;20-22], insulin [15;21;23], oncostatin M (OSM) [16;19;23], activin A [15-18;22;24], wnt3a [25;26], transferrin [15], and sodium butyrate [16;19;27]. Research has also shown the importance of a collagen type I extracellular matrix (ECM) in hepatic specification [4;20;21]. Although promising, these approaches have thus far been insufficient to direct differentiation of hESCs into fully functional, transplantable hepatocytes and subsequently require additional model development.
Primary human hepatocytes rapidly dedifferentiate in culture, losing in vivo hepatic hallmarks such as biotransformation enzyme induction capacity, and studies by our laboratory have been instrumental in defining culture conditions that help to preserve the differentiated hepatic phenotype in culture. By maintaining hepatocytes in the presence of extracellular matrix components [28] and in serum-free media supplemented with physiological concentrations of insulin [29] and the synthetic glucocorticoid dexamethasone [30], we have been able to facilitate retention of markers of the differentiated hepatic phenotype. Building upon a report by Shirahashi et al. [21], where mouse and human ESCs were induced to differentiate along a hepatic lineage in the presence of collagen type I, dexamethasone, and insulin, although cultured in Iscove's Modified Dulbecco's Media supplemented with 20% fetal bovine serum (FBS) and differentiated through an embryoid body intermediate [21], we hypothesized that our previously developed and highly defined culture media - optimized for the maintenance of mature primary human hepatocytes, when coupled with a collagen substrata, would provide an enhanced means of generating hepatic-like cells from hESCs.
Indeed, the hepatic-like cell population derived by culturing hESCs for 10 days under these conditions displayed hepatic cell-like morphology, attenuated expression of pluripotency markers and ‘stemness’ function, increased expression of genes representing a wide range of hepatic functions including transcription factors, nuclear receptors, plasma proteins, and metabolic and biotransformation enzymes, as well as augmented hepatic function. Thus, our results define a unique, expeditious model approach to the differentiation of hESCs along a hepatic lineage.
2.0 Materials and Methods
2.1 Transformed cell line culture
All cell culture reagents were obtained from Gibco (Grand Island, NY) unless otherwise indicated. Human foreskin fibroblasts (hFFs), acquired from ATCC (Manassas, VA), were cultured in Dulbecco's Modified Eagle Media + GlutaMAX supplemented with 0.75 g/l sodium bicarbonate and 15% fetal bovine serum (FBS). Hek 293T/17 transformed human embryonic kidney cells (ATCC) were maintained in Dulbecco's Modified Eagle Media + GlutaMAX supplemented with 0.1 mM non-essential amino acids, 0.75 g/l sodium bicarbonate, 1 mM sodium pyruvate, 10 mM HEPES, 100 units/ml penicillin, 100 μg/ml streptomycin, and 10% FBS. HepG2 human hepatoma-derived cells (ATCC) were cultured in Minimum Essential Media + Earle's Salts + ι-glutamine supplemented with 0.1 mM non-essential amino acids, 0.75 g/l sodium bicarbonate, 1 mM sodium pyruvate, 10 mM HEPES, 100 units/ml penicillin, 100 μg/ml streptomycin, and 10% FBS.
2.2 Primary human hepatocyte culture
Primary human hepatocyte cultures, secured through the Liver Tissue Procurement and Distribution System (NIH Contract #N01-DK-9-2310), were isolated by collagenase perfusion and plated on rat-tail collagen as described previously [31]. Hepatocytes were cultured in our hepatocyte media: William's E Media supplemented with 10 mM HEPES, 2 mM GlutaMAX, 100 units/ml penicillin, 100 μg/ml streptomycin, 25 nM dexamethasone (Sigma; St. Louis, MO), 10 nM insulin (Sigma), 5 ng/ml selenium (Sigma), 5 μg/ml transferrin (Sigma), and 1% linoleic acid/albumin (Sigma). Hepatocyte media was replenished every other day.
2.3 Human embryonic stem cell culture
WA09 (H9) human embryonic stem cells, acquired through the National Stem Cell Bank at the WiCell Research Institute (Madison, WI), were maintained on irradiated hFF feeder layer cells in hESC media: Dulbecco's Modified Eagle Media F-12 supplemented with 20% knock-out serum replacement, 0.1 mM non-essential amino acids, 1 mM GlutaMAX, 100 ng/ml basic fibroblast growth factor (National Cancer Institute; Bethesda, MD), and 0.1 mM β-mercaptoethanol (Sigma). Media was replenished daily, differentiated colonies were removed 2-3 times per week by manual dissociation, and cells were passaged weekly and plated on fresh hFF feeder layers.
2.4 Hepatic differentiation of human embryonic stem cells and treatments
To induce hepatic differentiation, human embryonic stem cells were plated in wells coated with ~3 μg/cm2 of rat tail type I collagen (Sigma) in hESC media. After 2 days media was switched to hepatocyte media and cells were maintained for either 8 or 18 additional days with daily replenishment of hepatocyte media for a total of 10, or 20 days in culture. Treatments were carried out with phenobarbital (Sigma) and 6-(4-chlorophenyl)imidazo[2,1–b] [1,3] thiazole-5-carbaldehyde O-(3,4-dichlorobenzyl)oxime (CITCO) (BioMol; Plymouth Meeting, PA).
2.5 RNA isolation, cDNA archiving, and real-time PCR
RNA was isolated using TRIzol Reagent (Invitrogen; Carlsbad, CA) and converted to cDNA using High Capacity cDNA Archive Kit (Applied Biosystems; Foster City, CA), both according to manufacturers' instructions. Real-time PCR was performed using Assays-on-Demand Gene Expression Products (Applied Biosystems), according to manufacturer's protocol. Briefly, 100 ng of cDNA template, 25 μl 2X Taqman Universal Master Mix, and 2.5 μl 20X Target Assay Mix were combined into 50 μl reactions. Reactions were divided in half and run as technical replicates on an ABI 7300 Real-time PCR System. Data were analyzed using the ΔΔCT method as previously described [32], with all target gene values normalized to 18S. Due to variability in the gene expression between hESCs of different passages, all data were normalized to a low-passage number (passage 33) hESC control population.
2.6 Immunofluorescence
Immunofluorescence was carried out using Human Embryonic Stem/Induced Pluripotent Stem Cell Characterization Kit (Applied StemCell Inc; Sunnyvale, CA) and all reagents were obtained from Applied StemCell Inc. unless otherwise indicated. A modified manufacturer's protocol was followed for immunofluorescence analysis. Briefly, cells were grown in 12-well plates and immersed in cell fixation solution for 1 h, washed 3 times with PBS for 5 min each, incubated in permeabilization solution for 30 minutes, and immersed in blocking solution for 1 h. 150 μl of pre-diluted primary antibody solution ( OCT4 anti-rabbit, SOX2 anti-rabbit, SSEA-3 anti-rat, TRA-1-60 anti-mouse, or TRA-1-81 anti-mouse) was added and cells were incubated overnight in a humidified 4° chamber. Cells were washed twice with PBS for 1 0 min each, incubated in 200 μl of either Alexa Fluor 546 goat anti-rabbit, Alexa Fluor 546 goat anti-rat, or Alexa Fluor 488 goat anti-mouse secondary antibody (Invitrogen) diluted 1:200 in PBS for 1 h in the dark, and washed in PBS for 10 min in the dark. Cells were examined microscopically on a Nikon inverted fluorescent microscope (Melville, NY) using a Nikon B-2E/C or G-2E/C filter and images were taken using a digital camera and SpotRT software (Diagnostic Instruments; Sterling Heights, MI).
2.7 Alkaline phosphatase staining
Alkaline phosphatase staining was performed using Alkaline Phosphatase Detection Kit (Millipore; Billerica, MA) according to manufacturer's protocol. Briefly, cells were fixed with 4% formaldehyde in PBS for 1-2 min, washed with TBST (20mM Tris-HCl, 0.15M NaCl, 0.05% Tween-20), stained in the dark for 15 min with a 2:1:1 ratio of Fast Red Violet:Naphthol:water, and washed with TBST prior to microscopic examination and imaging.
2.8 Indocyanine green uptake
Indocyanine green (ICG) uptake analysis was carried out essentially as described by Yamada et al. except that a final ICG concentration of 100 μg/ml was use d due to toxicity observed in hESC cultures at the recommended 1 mg/ml concentration [33]. Briefly, Cardiogreen (Sigma) was dissolved in water to make a fresh 5 mg/ml stock solution and then diluted in culture medium to a final concentration of 100 μg/ml. Cells were immersed in the diluted ICG solution, incubated for 15 min at 37°C, washed with PBS, and observed and imaged microscopically.
2.9 Periodic acid-Schiff staining
Periodic acid-Schiff (PAS) staining was performed using Periodic Acid-Schiff Kit (Sigma) according to manufacturer's instructions with minor adaptations. Briefly, cells were fixed in 4% formaldehyde in PBS for 1-2 min, washed with TBST, stained for 5 min with periodic acid, and washed 3x with PBS. Cells were then stained with Schiff's reagent for 15 min, washed 3 times with PBS, counterstained for 90 sec with hematoxylin solution, and washed 3 times with PBS prior to microscopic examination and imaging.
2.10 Albumin secretion ELISA assay
Conditioned media from the differentiated hESCs was collected at day 0, 10, and 20 and stored at −20°C until assayed. The concentration of human albumin secreted into the cell culture medium was determined using the human albumin ELISA quantitation kit (Bethyl Laboratory, Montgomery, TX, USA), according to the manufacturer's instructions. Briefly, the plate was prepared by incubating with the human albumin coating antibody for 1 h, washed 5 times, blocked by blocking solution containing 1% BSA for 30 min, and then washed 5 times. 100 μl of each standard, contr ol, and samples were loaded to each well and incubated for 1 h, followed by 5 washes. The plate was incubated with HRP-conjugated human albumin detection antibody for 1 h, washed 5 times, and immersed in tetramethylbenzidine (TMB) substrate solution for 15 min in the dark. The color development was stopped by addition of 0.18 M H2SO4. The plate was read at 450nm using a Packard Spectra Count (Meriden, CT) reader. The concentration of human albumin was normalized to the number of total cells determined from each well.
2.11 Statistical analyses
A Student's t-test (one-tailed; two-sample, unequal variance) was used for all statistical analyses.
3.0 Results
3.1 HESCs subjected to hepatic differentiation exhibit hepatic cell-like morphology
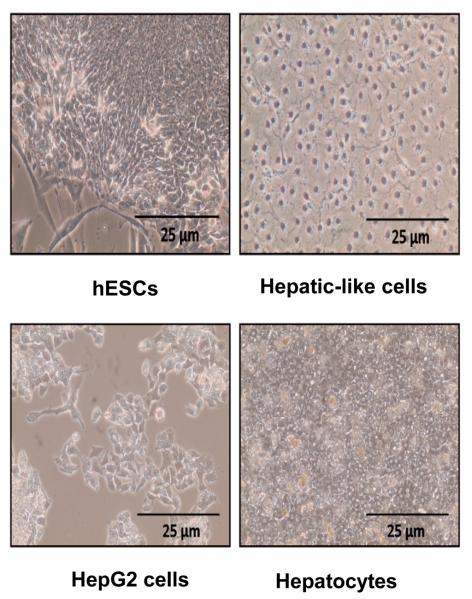
We subjected the H9 hESC line – selected because it has been extensively characterized and employed for numerous studies of hepatic differentiation – to a unique hepatic differentiation approach in which the cells were maintained on rat-tail collagen type I substrate in our hepatocyte media (William's E Media supplemented with HEPES, glutamine, antibiotics, dexamethasone, insulin, transferrin, selenium, and linoleic acid/albumin) for 10 days. Microscopic evaluation revealed that a large subset (~60%) of the resulting cell population displayed a uniform morphology similar to that of hepatic-like cells including the hepatoma-derived HepG2 cell line, a model widely employed in hepatic studies, and cultures of primary human hepatocytes (Fig. 1). In contrast to hESCs which exhibit a small, flat morphology and grow in culture in distinct colonies, the hepatic-like cells exhibit a larger, more cuboidal shape and a homogenous distribution more similar to that of cultures of established hepatoma lines or primary hepatocytes (Fig. 1).
Figure 1. HESCs subjected to hepatic differentiation exhibit hepatic cell-like morphology.
Images depict phase-contrast micrographs of an undifferentiated hESC colony (hESCs) and hESC-derived hepatic-like cells generated by culturing passage-matched hESCs for 10 days on type I rat-tail collagen in hepatocyte media (Hepatic-like cells), in parallel to the undifferentiated hESC negative controls. HepG2 hepatoma cells (HepG2 cells) and cultures of primary human hepatocytes (Hepatocytes) are included as positive controls.
3.2 ‘Stemness’ marker mRNA expression is significantly decreased in hepatic-like cells
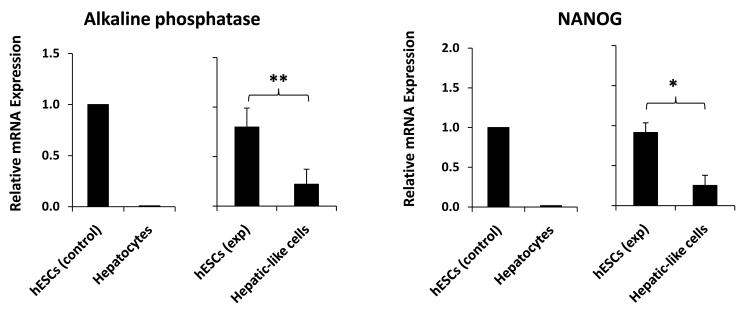
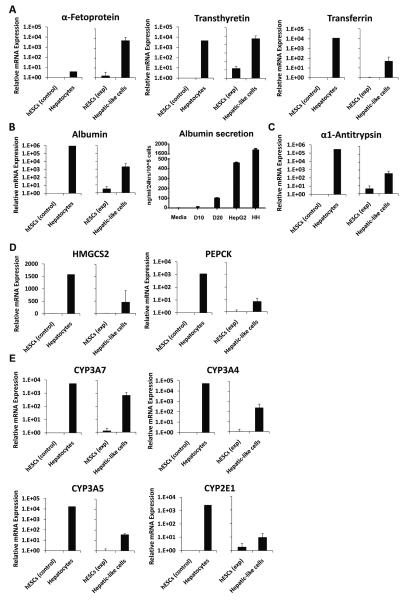
Differentiation of hESCs is accompanied by the loss of expression of markers of pluripotency or ‘stemness,’ such as the transcription factor nanog homeobox (NANOG) and the alkaline phosphatase enzyme. Real-time PCR analyses on the hepatic-like cell population resulting from our differentiation protocol revealed a significant reduction in mRNA expression of both NANOG – which together with octamer-binding transcription factor 4 (OCT4) is widely considered to be the key factor in the conferral of stem cell pluripotency [34] – and alkaline phosphatase, to levels approximately one-third of those of hESC controls (Fig. 2).
Figure 2. ‘Stemness’ marker mRNA expression is significantly decreased in hepatic-like cells.
Passage-matched HESCs were cultured in parallel on either hFF feeder layers in hESC media (hESCs exp) or on collagen in hepatocyte media (Hepatic-like cells). After 10 days in culture cells were harvested and RNA was isolated, converted to cDNA, subjected to real-time PCR, and the data analyzed using the ΔΔCT method to determine fold expression levels of NANOG and alkaline phosphatase relative to low-passage hESCs. Control data (left graph) depict means of expression levels of replicate determinations of a typical low-passage culture of hESCs (hESCs control), in contrast to mean expression levels of replicate determinations performed using a pooled sample of six adult primary human hepatocyte donors (Hepatocytes). In these control experiments, the replicate values were reproducible within a ≤ 5% error range. Experimental data (right graph) depict expression levels of hESCs (hESCs exp) and passage-matched hESCs subject to our hepatic differentiation protocol (Hepatic-like cells). Experimental data are expressed as mean +/− standard deviations of at least two independent trials using hESCs from different passages. * p < 0.05; ** p < 0.01.
3.3 ‘Stemness’ marker protein expression is reduced in hepatic-like cells
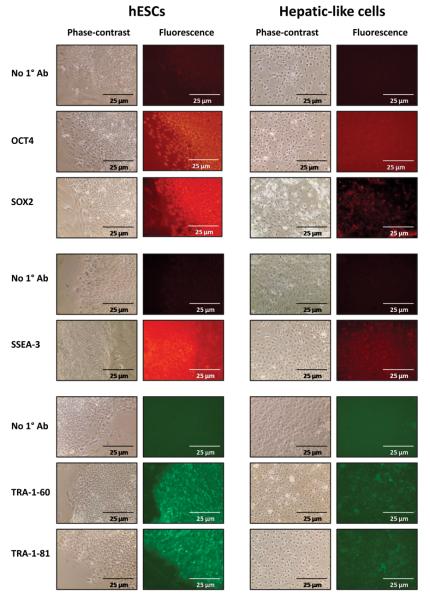
Changes in gene expression at the protein level were assessed by immunofluorescence analysis which showed that while hESCs exhibited robust, uniform expression of the ‘stemness’ transcription factors OCT4 and SRY-box containing gene 2 (SOX2) and the surface antigens stage-specific embryonic antigen 3 (SSEA-3), tumor rejection antigen 1-60 (TRA-1-60), and tumor rejection antigen 1-81 (TRA-1-81), expression of these markers in the hepatic-like cells was noticeably attenuated (Fig. 3). Of the markers assessed, expression of the key pluripotency factor OCT4 exhibited a dramatic decrease upon hepatic differentiation (Fig. 3, second row), as did SOX2 (Fig. 3, third row) and SSEA-2 (Fig. 3, fifth row).
Figure 3. ‘Stemness’ marker protein expression is reduced in hepatic-like cells.
Passage-matched hESCs were cultured in parallel on either hFF feeder layers in hESC media (hESCs) or on collagen in hepatocyte media (Hepatic-like cells). At 10 days in culture cells were probed for OCT4, SOX2, SSEA-3, TRA-1-60, and TRA-1-81 protein expression by immunofluorescence using antibodies specified in the Methods. Cells were examined microscopically and phase-contrast and fluorescence images were captured.
3.4 Hepatic-like cells exhibit decreased ‘stemness’ function
To assess functional changes in the hESC-derived hepatic-like cells we utilized alkaline phosphatase activity staining. Staining revealed robust enzymatic activity in the hESCs (pink), activity which was undetectable in both the hepatic-like cell population and primary human hepatocytes (Fig. 4). These data confirm the functional relevance of the reduced alkaline phosphatase mRNA expression observed previously in the hepatic-like cells and, together with the data indicating reduction in mRNA and protein expression of an array of select other markers of ‘stemness,’ demonstrating that the hepatic-like cell population has indeed transitioned from a pluripotent state.
Figure 4. Hepatic-like cells exhibit decreased ‘stemness’ function.
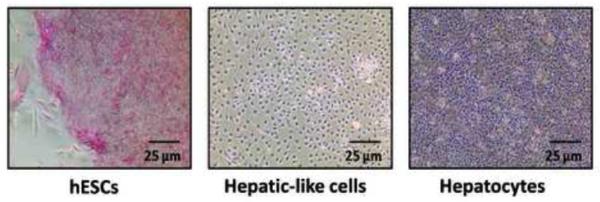
Passage-matched hESCs were cultured in parallel on either hFF feeder layers in hESC media (hESCs) or on collagen in hepatocyte media (Hepatic-like cells). At 10 days in culture cells were stained for alkaline phosphatase activity (pink) and phase-contrast images were captured. Primary adult human hepatocytes (Hepatocytes) stained in parallel serve as a negative control.
3.5 Hepatic transcription factor mRNA expression is increased in hepatic-like cells
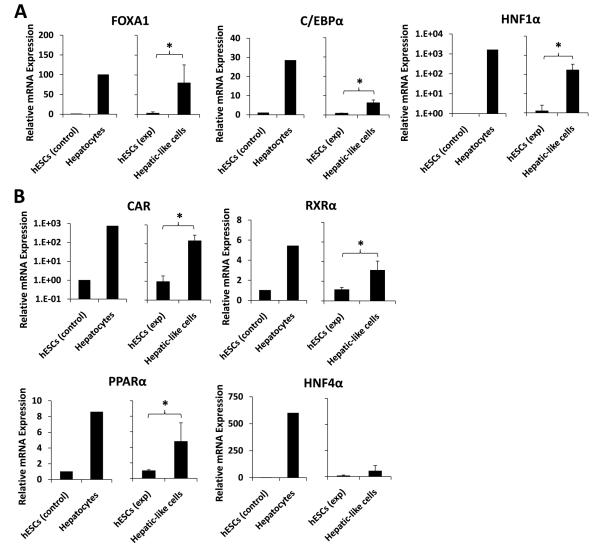
To determine the extent of acquisition of hepatic character in the hepatic-like cell population we assessed mRNA transcript levels of select hepatic-enriched transcription factors by quantitative real-time PCR analyses. Compared to hESCs, the hepatic-like cells exhibited significantly increased expression of forkhead box A1 (FOXA1), CCAAT/enhancer binding protein alpha (C/EBPα), and hepatic nuclear factor 1 alpha (HNF1α) (Fig. 5A), integral members of the combinatorial regulatory network that controls transcription of most genes in the hepatic program [35], as well as augmented expression of several hepatic nuclear receptors, including the constitutive androstane receptor (CAR), retinoid X receptor alpha (RXRα), peroxisome proliferator-activated receptor alpha (PPARα), and hepatic nuclear factor 4 alpha (HNF4α) (Fig. 5B). Levels of the pregnane X-receptor were also evaluated but were low and near the detection limits of these assays (data not shown). Notably, expression of FOXA1, a transcription factor vital for the onset of hepatogenesis [36], was enhanced an average of 80-fold in hepatic-like cells compared to hESCs, to levels comparable to those of primary human hepatocytes (Fig. 5A, left).
Figure 5. Hepatic transcription factor mRNA expression is increased in hepatic-like cells.
Passage-matched hESCs were cultured in parallel on either hFF feeder layers in hESC media (hESCs exp) or on collagen in hepatocyte media (Hepatic-like cells). After 10 days in culture, cells were harvested and RNA was isolated, converted to cDNA, subjected to real-time PCR, and the data analyzed using the ΔΔCT method [32] to determine fold expression levels relative to low-passage hESCs (hESCs control) of: (A), the hepatic transcription factors FOXA1, C/EBPα, and HNF1α; and (B), the hepatic nuclear receptors CAR, RXRα, PPARα, and HNF4α. Control data (left graph for each endpoint) depict means of expression levels of replicate determinations of a typical low-passage culture of hESCs (hESCs control), in contrast to mean expression levels of replicate determinations performed using a pooled sample of six adult primary human hepatocyte donors (Hepatocytes). In these control experiments, the replicate values were reproducible within a ≤ 5% error range. Experimental data (right graph) depict expression levels of hESCs (hESCs exp) and passage-matched hESCs subject to our hepatic differentiation protocol (Hepatic-like cells). Experimental data are expressed as mean +/− standard deviations of at least three independent trials using hESCs from different passages. * p < 0.05.
3.6 Hepatic marker mRNA expression is augmented in hepatic-like cells
While expression of hepatic transcription factors is indicative of differentiation along a hepatic lineage, hepatic character is ultimately defined by acquisition of hepatic function. Thus, we performed further real-time PCR analyses to assess expression of hepatic functional marker genes. Indeed, compared to hESCs, the hepatic-like cells exhibited increased mRNA expression of the liver-generated plasma proteins α-fetoprotein, transthyretin and transferrin, (Fig. 6A), Serum albumin transcript levels and direct secretion of albumin were also assessed (Fig. 6B). The hepatic cells exhibited clearly elevated levels of albumin mRNA following 10 days in culture, and exhibited further elevation of albumin secretion following 20 days in culture (Fig. 6). Although the secreted levels detected in the day 20 cultures were clearly less than those measured in either HepG2 hepatoma cells or primary human hepatocytes (Fig. 6B), other reports have indicated that serum albumin secretion in HepG2 cells is robust in comparison to primary hepatocytes [37;38], such that this comparison is a relatively high standard to attain. Transcript levels of a the protease inhibitor, α-1-antitrypsin (Fig. 6C), levels of the metabolic enzymes, 3-hydroxy-3-methylglutaryl-coenzyme A synthase 2 (HMGCS2) and phosphoenolpyruvate carboxykinase (PEPCK) (Fig. 6D), as well as members of the cytochrome P450 (CYP) family of biotransformation enzymes, including CYP3A7, CYP3A4, CYP3A5, and CYP2E1 (Fig. 6E) were all substantially upregulated in the hepatic-like cell population following 10 days in culture.
Figure 6. Hepatic marker mRNA expression is augmented in hepatic-like cells.
Passage-matched hESCs were cultured in parallel on either hFF feeder layers in hESC media (hESCs exp) or on collagen in hepatocyte media (Hepatic-like cells). After either 10 or 20 days in culture, cells were harvested and RNA was isolated, converted to cDNA, subjected to real-time PCR, and the data analyzed using the ΔΔCT method [32] to determine fold expression levels relative to low-passage hESCs (hESCs control) of: (A) the plasma proteins α-fetoprotein, transthyretin, and transferrin; (B) albumin transcript expression and Elisa-based protein secretion analysis; (C) the protease inhibitor α-1-antitrypsin; (D), the metabolic enzymes HMGCS2 and PEPCK; and (E), the biotransformation enzymes CYP3A7, CYP3A4, CYP3A5, and CYP2E1. Control data (left graph) depict means of expression levels of replicate determinations of a typical low-passage culture of hESCs (hESCs control), in contrast to mean expression levels of replicate determinations performed using a pooled sample of six adult primary human hepatocyte donors (Hepatocytes). In these control experiments, the replicate values were reproducible within a ≤ 5% error range. Experimental data (right graph) depict expression levels of hESCs (hESCs exp) and passage-matched hESCs subject to our hepatic differentiation protocol (Hepatic-like cells). Experimental data are expressed as mean +/− standard deviations of at least three independent trials using hESCs from different passages.
3.7 Hepatic-like cells exhibit the hepatic-specific functions of ICG uptake and glycogen storage
We assessed cellular uptake of indocyanine green (ICG), an organic anion taken up and eliminated by hepatic transporter systems, used as an indicator of hepatic function [33], to further assess the hepatic character of the hESC-derived hepatic-like cells. Despite the differences in confluence between the cell populations, microscopic examination of hepatic-like cells exposed to ICG solution revealed levels of uptake (cyan-green) approaching those of primary human hepatocyte cultures, while hESC cultures wholly excluded the anionic solution (Fig. 7, top two rows). Levels of ICG uptake appeared further enhanced at 20 days in culture when compared to the 10 day cultures. We also assessed the ability of the hepatic-like cells to store glycogen, a unique function of the specialized hepatocyte, using periodic acid-Schiff (PAS) staining. Again, although levels of confluence varied between the cell populations, microscopic analysis indicated clearly that the hepatic-like cell populations exhibited levels of glycogen accumulation (pink) approaching those of primary human hepatocyte cultures (Fig. 7, bottom two rows). Again, PAS staining was enhanced in the 20 day cultures compared with the 10 day cultures. Although hESCs exhibited limited PAS staining as well, the stain was distributed uniformly throughout the cells, unlike the hepatic-like cells and hepatocytes where glycogen was detected only in the cytoplasm, suggesting that the staining observed in the hESCs may be artifactual in nature. The cytoplasmic glycogen stores in the hepatic-like cells serve to further highlight the hepatic cells' enhanced nuclear-to-cytoplasmic ratio, cuboidal morphology, and prominent nucleoli, characteristics of a mature hepatic phenotype.
Figure 7. Hepatic-like cells exhibit the hepatic-specific functions of ICG uptake and glycogen storage.
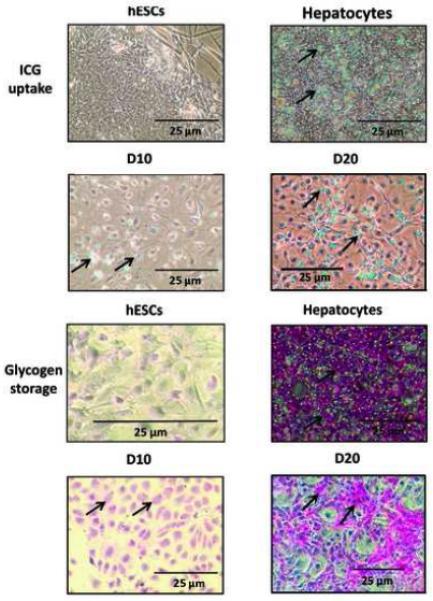
Passage-matched hESCs were cultured in parallel on either hFF feeder layers in hESC media (hESCs) or on collagen in hepatocyte media (Hepatic-like cells) for either 10 or 20 days. Cells were evaluated for ICG uptake (cyan-green) by incubation with 100 μg/ml ICG and for glycogen storage capacity (pink) using periodic acid-Schiff (PAS) staining, both as described in Materials and Methods. Cells were examined microscopically and phase-contrast images were captured. Cultures of adult primary human hepatocytes (Hepatocytes) assessed in parallel were included as positive controls.
4.0 Discussion
The hepatic differentiation approach defined in this study – culture in hepatocyte media (William's E media supplemented with HEPES, glutamine, antibiotics, dexamethasone, insulin, transferrin, selenium, and linoleic acid/albumin) on collagen type I substrate for 10 days – provides a simple, straight-forward methodology by which hESCs may be directed to differentiate along a hepatic lineage. Extensive characterization of the hESC-derived hepatic-like cell population revealed attenuated mRNA expression of markers of pluripotency including the transcription factor NANOG and the alkaline phosphatase enzyme (validated functionally by activity staining), reduced protein expression of the ‘stemness’ transcription factors OCT4 and SOX2, as well as marked reductions in levels of the surface antigens, SSEA-3, TRA-1-60, and TRA-1-81. In contrast, the hepatic-like cells exhibited increased mRNA expression levels of the hepatic-enriched transcription factors, FOXA1, C/EBPα, and HNF1α, nuclear receptors CAR, RXRα, PPARα, and HNF4α; the plasma proteins α-fetoprotein, transthyretin, transferrin, and albumin; the protease inhibitor, α-1-antitrypsin; the metabolic enzymes, HMGCS2, PEPCK; and mRNAs encoding the biotransformation enzymes, CYP3A7, CYP3A4, CYP3A5, and CYP2E1. Indocyanine green uptake and glycogen storage capacity by the differentiated cells confirmed acquisition of hepatic function, properties that were enhanced following extension of the culture time from 10 days to 20 days.
In the differentiation protocol described here, hESCs were cultured on collagen type I, a protein component of ECM that along with soluble signaling factors such as those found in our defined hepatocyte media, influences hepatocyte development and function in the liver microenvironment. Liver ECM is comprised primarily of collagen type I and fibronectin, as well as collagen types III, IV, and VI, tenascin, and laminin [39]. While primary human hepatocytes maintained on plastic dishes rapidly de-differentiate in culture, a number of studies have shown that primary hepatocytes cultured on collagen type I gel retain their differentiated functions, including expression of liver-specific genes, albumin secretion, and urea production [39]. While culturing of hESCs on collagen substrate in the absence of hepatocyte media resulted in limited expression of hepatic differentiation markers (data not shown), the combination of collagen substrate and hepatocyte media was necessary to achieve maximal hepatic induction.
The unique hepatocyte media used in these studies is comprised of a number of factors that likely act in concert to facilitate hepatic differentiation of hESCs. In particular, dexamethasone and insulin play integral roles in normal liver development and thus are potentially the primary factors inducing differentiation in our approach. For example, dexamethasone is required for hepatic induction by both OSM and HGF [40;41]. Using cultures of murine fetal hepatocytes, Kamiya et al. demonstrated that treatment with a combination of dexamethasone and OSM results in morphological changes consistent with fully differentiated hepatocytes, augmented expression of hepatic marker genes, and intracellular glycogen accumulation [40]. In contrast to OSM stimulation alone, treatment with dexamethasone alone promoted a number of hepatic specification markers, albeit to a much lesser extent than the dexamethasone/OSM combination [42]. Studies assessing the role of HGF in hepatic differentiation yielded similar results; in the absence of dexamethasone, HGF is unable to induce differentiation, as measured by the up-regulation of hepatic marker genes and intracellular glycogen accumulation [41]. The liver is also subject to continual exposure to insulin secreted by the pancreatic islets through the portal vein, the importance of which is underscored by the liver atrophy which results from the removal of this signal [43].
While the hepatic-like cells derived from the protocol described here expressed impressive mRNA levels for a broad array of hepatic differentiation markers, in general their respective levels were lower than those present in primary human hepatocytes. These latter findings are largely consistent with those of others, in studies assessing a variety of differentiation procedures [4;21;44;45]. However, our results involved direct comparisons to a robust primary hepatocyte model that exhibits a highly differentiated phenotype [46;47], and therefore represents a high standard of comparison. Results from high quality primary hepatocytes are arguably more closely relevant than comparisons relying principally on HepG2 or hepatoma cell-derived cultures, models that report comparative deficiencies in expression of selected differentiation markers within hESC-derived hepatic-like cell populations [20;23]. It is important to note that although an estimated 60% of the cells derived from our differentiation conditions exhibited hepatic-like morphology, the mRNA expression data obtained were derived from extracts of the entire cell population, and therefore would be expected to include cells representing other phases of lineage development. Populations more highly enriched for hepatic-like endpoint morphology likely would exhibit higher relative expression levels of the relevant hepatic markers. Additionally, the derived hepatic-like cell population expressed the fetal liver markers, α-fetoprotein and CYP3A7, and exhibited increased levels of CYP3A4 as compared to hESCs, However, treatment with 500 μM phenobarbital or 100 nM CITCO did not result in further induction of CYP3A4 (data not shown), a feature otherwise noted as a hallmark of a fully differentiated hepatocyte. These findings suggest that the hepatic-like cell population propagated under these experimental conditions is not yet fully mature and consequently might be better characterized as hepatoblasts or as hepatic-progenitor cells. An additional consideration is that although gene expression profile distinctions between visceral endoderm and definitive endoderm (from which the liver is derived), are difficult to truly discriminate on an mRNA expression basis [48], the complementary hepatic functional analyses included in this study demonstrate more definitively that the hepatoblast cell populations generated with these methods are indeed hepatocyte-like in nature. Future studies will be directed toward the elucidation of the full composite of factors and culture conditions required to generate completely differentiated human hepatocytes, clearly a highly complex developmental program that ultimately mediates hepatic differentiation.
5.0 Conclusions
In summary, the results presented in this study underscore the importance of collagen type I together with dexamethasone and insulin as facilitators of the hepatic developmental lineage. These factors had previously been assumed to act at a later developmental stage than FGFs, BMP, OSM, HGF, or activin A in hepatic differentiation [49] and therefore largely unrecognized as directors of hepatogenesis. We posit that the differentiation approach described here, when combined with other hepatic induction methodologies, may provide the supplementary factors needed to generate fully functional hepatocytes from human embryonic stem cells and ultimately provide a valuable source of hepatocytes for therapeutic transplantations and for direct application in pharmacological and toxicological research investigations.
Acknowledgements
The authors wish to thank Ms. Mary Hutchinson for expert technical assistance. This research was supported by a grant from the National Institute of General Medical Sciences, USPHS GM066411 (C.J.O).
Abbreviations
- BMPs
bone morphogenetic proteins
- CAR
constitutive androstane receptor
- C/EBPα
CCAAT/enhancer binding protein alpha
- CITCO
6-(4-chlorophenyl)imidazo[2,1–b] [1,3] thiazole-5-carbaldehyde O-(3,4-dichlorobenzyl)oxime
- CYP
cytochrome P450
- ECM
extracellular matrix
- FBS
fetal bovine serum
- FGFs
fibroblast growth factors
- FOXA1
forkhead box A1
- HGF
hepatocyte growth factor
- hESCs
human embryonic stem cells
- hFFs
human foreskin fibroblasts
- HMGCS2
3-hydroxy-3-methylglutaryl-coenzyme A synthase 2
- HNF1α
hepatic nuclear factor 1 alpha
- HNF4α
hepatic nuclear factor 4 alpha
- ICG
indocyanine green
- NANOG
nanog homeobox
- OCT4
octamer-binding transcription factor 4
- OSM
oncostatin M
- PAS
periodic acid-Schiff
- PEPCK
phosphoenolpyruvate carboxykinase
- PPARα
peroxisome proliferator-activated receptor alpha
- RXRα
retinoid X receptor alpha
- SOX2
SRY-box containing gene 2
- SSEA-3
stage-specific embryonic antigen 3
- TRA-1-60
tumor rejection antigen 1-60
- TRA-1-81
tumor rejection antigen 1-81
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Spear BT, Jin L, Ramasamy S, Dobierzewska A. Transcriptional control in the mammalian liver: liver development, perinatal repression, and zonal gene regulation. Cell Mol Life Sci. 2006;63:2922–38. doi: 10.1007/s00018-006-6258-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cao S, Esquivel CO, Keeffe EB. New approaches to supporting the failing liver. Annu Rev Med. 1998;49:85–94. doi: 10.1146/annurev.med.49.1.85. [DOI] [PubMed] [Google Scholar]
- 3.Malhi H, Gupta S. Hepatocyte transplantation: new horizons and challenges. J Hepatobiliary Pancreat Surg. 2001;8:40–50. doi: 10.1007/s005340170049. [DOI] [PubMed] [Google Scholar]
- 4.Schwartz RE, Linehan JL, Painschab MS, Hu WS, Verfaillie CM, Kaufman DS. Defined conditions for development of functional hepatic cells from human embryonic stem cells. Stem Cells Dev. 2005;14:643–55. doi: 10.1089/scd.2005.14.643. [DOI] [PubMed] [Google Scholar]
- 5.Lemaigre F, Zaret KS. Liver development update: new embryo models, cell lineage control, and morphogenesis. Curr Opin Genet Dev. 2004;14:582–90. doi: 10.1016/j.gde.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 6.Douarin NM. An experimental analysis of liver development. Med Biol. 1975;53:427–55. [PubMed] [Google Scholar]
- 7.Jung J, Zheng M, Goldfarb M, Zaret KS. Initiation of mammalian liver development from endoderm by fibroblast growth factors. Science. 1999;284:1998–2003. doi: 10.1126/science.284.5422.1998. [DOI] [PubMed] [Google Scholar]
- 8.Rossi JM, Dunn NR, Hogan BL, Zaret KS. Distinct mesodermal signals, including BMPs from the septum transversum mesenchyme, are required in combination for hepatogenesis from the endoderm. Genes Dev. 2001;15:1998–2009. doi: 10.1101/gad.904601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blouin MJ, Lamy I, Loranger A, Noel M, Corlu A, Guguen-Guillouzo C, Marceau N. Specialization switch in differentiating embryonic rat liver progenitor cells in response to sodium butyrate. Exp Cell Res. 1995;217:22–30. doi: 10.1006/excr.1995.1059. [DOI] [PubMed] [Google Scholar]
- 10.Enzan H, Himeno H, Hiroi M, Kiyoku H, Saibara T, Onishi S. Development of hepatic sinusoidal structure with special reference to the Ito cells. Microsc Res Tech. 1997;39:336–49. doi: 10.1002/(SICI)1097-0029(19971115)39:4<336::AID-JEMT4>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 11.Medlock ES, Haar JL. The liver hemopoietic environment: I. Developing hepatocytes and their role in fetal hemopoiesis. Anat Rec. 1983;207:31–41. doi: 10.1002/ar.1092070105. [DOI] [PubMed] [Google Scholar]
- 12.Luzzatto AC. Hepatocyte differentiation during early fetal development in the rat. Cell Tissue Res. 1981;215:133–42. doi: 10.1007/BF00236254. [DOI] [PubMed] [Google Scholar]
- 13.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–7. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 14.Lavon N, Yanuka O, Benvenisty N. Differentiation and isolation of hepatic-like cells from human embryonic stem cells. Differentiation. 2004;72:230–8. doi: 10.1111/j.1432-0436.2004.07205002.x. [DOI] [PubMed] [Google Scholar]
- 15.Brolen G, Sivertsson L, Bjorquist P, Eriksson G, Ek M, Semb H, Johansson I, Andersson TB, Ingelman-Sundberg M, Heins N. Hepatocyte-like cells derived from human embryonic stem cells specifically via definitive endoderm and a progenitor stage. J Biotechnol. 2010;145:284–94. doi: 10.1016/j.jbiotec.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 16.Duan Y, Ma X, Zou W, Wang C, Bahbahan IS, Ahuja TP, Tolstikov V, Zern MA. Differentiation and characterization of metabolically functioning hepatocytes from human embryonic stem cells. Stem Cells. 2010;28:674–86. doi: 10.1002/stem.315. [DOI] [PubMed] [Google Scholar]
- 17.Touboul T, Hannan NR, Corbineau S, Martinez A, Martinet C, Branchereau S, Mainot S, Strick-Marchand H, Pedersen R, Di SJ, Weber A, Vallier L. Generation of functional hepatocytes from human embryonic stem cells under chemically defined conditions that recapitulate liver development. Hepatology. 2010;51:1754–65. doi: 10.1002/hep.23506. [DOI] [PubMed] [Google Scholar]
- 18.Cai J, Zhao Y, Liu Y, Ye F, Song Z, Qin H, Meng S, Chen Y, Zhou R, Song X, Guo Y, Ding M, Deng H. Directed differentiation of human embryonic stem cells into functional hepatic cells. Hepatology. 2007;45:1229–39. doi: 10.1002/hep.21582. [DOI] [PubMed] [Google Scholar]
- 19.Hay DC, Zhao D, Fletcher J, Hewitt ZA, McLean D, Urruticoechea-Uriguen A, Black JR, Elcombe C, Ross JA, Wolf R, Cui W. Efficient differentiation of hepatocytes from human embryonic stem cells exhibiting markers recapitulating liver development in vivo. Stem Cells. 2008;26:894–902. doi: 10.1634/stemcells.2007-0718. [DOI] [PubMed] [Google Scholar]
- 20.Agarwal S, Holton KL, Lanza R. Efficient Differentiation of Functional Hepatocytes from Human Embryonic Stem Cells. Stem Cells. 2008 doi: 10.1634/stemcells.2007-1102. [DOI] [PubMed] [Google Scholar]
- 21.Shirahashi H, Wu J, Yamamoto N, Catana A, Wege H, Wager B, Okita K, Zern MA. Differentiation of human and mouse embryonic stem cells along a hepatocyte lineage. Cell Transplant. 2004;13:197–211. doi: 10.3727/000000004783984016. [DOI] [PubMed] [Google Scholar]
- 22.Kubo A, Kim YH, Irion S, Kasuda S, Takeuchi M, Ohashi K, Iwano M, Dohi Y, Saito Y, Snodgrass R, Keller G. The homeobox gene Hex regulates hepatocyte differentiation from embryonic stem cell-derived endoderm. Hepatology. 2010;51:633–41. doi: 10.1002/hep.23293. [DOI] [PubMed] [Google Scholar]
- 23.Baharvand H, Hashemi SM, Kazemi AS, Farrokhi A. Differentiation of human embryonic stem cells into hepatocytes in 2D and 3D culture systems in vitro. Int J Dev Biol. 2006;50:645–52. doi: 10.1387/ijdb.052072hb. [DOI] [PubMed] [Google Scholar]
- 24.D'Amour KA, Agulnick AD, Eliazer S, Kelly OG, Kroon E, Baetge EE. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat Biotechnol. 2005;23:1534–41. doi: 10.1038/nbt1163. [DOI] [PubMed] [Google Scholar]
- 25.Hay DC, Fletcher J, Payne C, Terrace JD, Gallagher RC, Snoeys J, Black JR, Wojtacha D, Samuel K, Hannoun Z, Pryde A, Filippi C, Currie IS, Forbes SJ, Ross JA, Newsome PN, Iredale JP. Highly efficient differentiation of hESCs to functional hepatic endoderm requires ActivinA and Wnt3a signaling. Proc Natl Acad Sci U S A. 2008;105:12301–6. doi: 10.1073/pnas.0806522105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moore RN, Moghe PV. Expedited growth factor-mediated specification of human embryonic stem cells toward the hepatic lineage. Stem Cell Res. 2009 doi: 10.1016/j.scr.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 27.Rambhatla L, Chiu CP, Kundu P, Peng Y, Carpenter MK. Generation of hepatocyte-like cells from human embryonic stem cells. Cell Transplant. 2003;12:1–11. doi: 10.3727/000000003783985179. [DOI] [PubMed] [Google Scholar]
- 28.Sidhu JS, Farin FM, Omiecinski CJ. Influence of extracellular matrix overlay on phenobarbital-mediated induction of CYP2B1, 2B2, and 3A1 genes in primary adult rat hepatocyte culture. Arch Biochem Biophys. 1993;301:103–13. doi: 10.1006/abbi.1993.1121. [DOI] [PubMed] [Google Scholar]
- 29.Sidhu JS, Omiecinski CJ. Insulin-mediated modulation of cytochrome P450 gene induction profiles in primary rat hepatocyte cultures. J Biochem Mol Toxicol. 1999;13:1–9. doi: 10.1002/(sici)1099-0461(1999)13:1<1::aid-jbt1>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 30.Sidhu JS, Omiecinski CJ. Modulation of xenobiotic-inducible cytochrome P450 gene expression by dexamethasone in primary rat hepatocytes. Pharmacogenetics. 1995;5:24–36. doi: 10.1097/00008571-199502000-00003. [DOI] [PubMed] [Google Scholar]
- 31.Strom SC, Pisarov LA, Dorko K, Thompson MT, Schuetz JD, Schuetz EG. Use of human hepatocytes to study P450 gene induction. Methods Enzymol. 1996;272:388–401. doi: 10.1016/s0076-6879(96)72044-x. [DOI] [PubMed] [Google Scholar]
- 32.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 33.Yamada T, Yoshikawa M, Kanda S, Kato Y, Nakajima Y, Ishizaka S, Tsunoda Y. In vitro differentiation of embryonic stem cells into hepatocyte-like cells identified by cellular uptake of indocyanine green. Stem Cells. 2002;20:146–54. doi: 10.1634/stemcells.20-2-146. [DOI] [PubMed] [Google Scholar]
- 34.Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, Wong KY, Sung KW, Lee CW, Zhao XD, Chiu KP, Lipovich L, Kuznetsov VA, Robson P, Stanton LW, Wei CL, Ruan Y, Lim B, Ng HH. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–40. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 35.Kyrmizi I, Hatzis P, Katrakili N, Tronche F, Gonzalez FJ, Talianidis I. Plasticity and expanding complexity of the hepatic transcription factor network during liver development. Genes Dev. 2006;20:2293–305. doi: 10.1101/gad.390906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee CS, Friedman JR, Fulmer JT, Kaestner KH. The initiation of liver development is dependent on Foxa transcription factors. Nature. 2005;435:944–7. doi: 10.1038/nature03649. [DOI] [PubMed] [Google Scholar]
- 37.Nakamura K, Mizutani R, Sanbe A, Enosawa S, Kasahara M, Nakagawa A, Ejiri Y, Murayama N, Miyamoto Y, Torii T, Kusakawa S, Yamauchi J, Fukuda M, Yamazaki H, Tanoue A. Evaluation of drug toxicity with hepatocytes cultured in a micro-space cell culture system. J Biosci Bioeng. 2010 doi: 10.1016/j.jbiosc.2010.08.008. [DOI] [PubMed] [Google Scholar]
- 38.Peters SJ, Haagsman HP, van NK. Arginase release by primary hepatocytes and liver slices results in rapid conversion of arginine to urea in cell culture media. Toxicol In Vitro. 2008;22:1094–8. doi: 10.1016/j.tiv.2008.01.016. [DOI] [PubMed] [Google Scholar]
- 39.Hansen LK, Wilhelm J, Fassett JT. Regulation of hepatocyte cell cycle progression and differentiation by type I collagen structure. Curr Top Dev Biol. 2006;72:205–36. doi: 10.1016/S0070-2153(05)72004-4. [DOI] [PubMed] [Google Scholar]
- 40.Kamiya A, Kinoshita T, Ito Y, Matsui T, Morikawa Y, Senba E, Nakashima K, Taga T, Yoshida K, Kishimoto T, Miyajima A. Fetal liver development requires a paracrine action of oncostatin M through the gp130 signal transducer. EMBO J. 1999;18:2127–36. doi: 10.1093/emboj/18.8.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kamiya A, Kinoshita T, Miyajima A. Oncostatin M and hepatocyte growth factor induce hepatic maturation via distinct signaling pathways. FEBS Lett. 2001;492:90–4. doi: 10.1016/s0014-5793(01)02140-8. [DOI] [PubMed] [Google Scholar]
- 42.Kinoshita T, Miyajima A. Cytokine regulation of liver development. Biochim Biophys Acta. 2002;1592:303–12. doi: 10.1016/s0167-4889(02)00323-3. [DOI] [PubMed] [Google Scholar]
- 43.Michalopoulos GK, DeFrances MC. Liver regeneration. Science. 1997;276:60–6. doi: 10.1126/science.276.5309.60. [DOI] [PubMed] [Google Scholar]
- 44.Ek M, Soderdahl T, Kuppers-Munther B, Edsbagge J, Andersson TB, Bjorquist P, Cotgreave I, Jernstrom B, Ingelman-Sundberg M, Johansson I. Expression of drug metabolizing enzymes in hepatocyte-like cells derived from human embryonic stem cells. Biochem Pharmacol. 2007;74:496–503. doi: 10.1016/j.bcp.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 45.Shiraki N, Umeda K, Sakashita N, Takeya M, Kume K, Kume S. Differentiation of mouse and human embryonic stem cells into hepatic lineages. Genes Cells. 2008;13:731–46. doi: 10.1111/j.1365-2443.2008.01201.x. [DOI] [PubMed] [Google Scholar]
- 46.Olsavsky KM, Page JL, Johnson MC, Zarbl H, Strom SC, Omiecinski CJ. Gene expression profiling and differentiation assessment in primary human hepatocyte cultures, established hepatoma cell lines, and human liver tissues. Toxicol Appl Pharmacol. 2007;222:42–56. doi: 10.1016/j.taap.2007.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Page JL, Johnson MC, Olsavsky KM, Strom SC, Zarbl H, Omiecinski CJ. Gene expression profiling of extracellular matrix as an effector of human hepatocyte phenotype in primary cell culture. Toxicol Sci. 2007;97:384–97. doi: 10.1093/toxsci/kfm034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sherwood RI, Jitianu C, Cleaver O, Shaywitz DA, Lamenzo JO, Chen AE, Golub TR, Melton DA. Prospective isolation and global gene expression analysis of definitive and visceral endoderm. Dev Biol. 2007;304:541–55. doi: 10.1016/j.ydbio.2007.01.011. [DOI] [PubMed] [Google Scholar]
- 49.Snykers S, De Kock J, Rogiers V, Vanhaecke T. In vitro differentiation of embryonic and adult stem cells into hepatocytes: state of the art. Stem Cells. 2009;27:577–605. doi: 10.1634/stemcells.2008-0963. [DOI] [PMC free article] [PubMed] [Google Scholar]