Abstract
Long interspersed element-1 (LINE-1 or L1) is a non-long terminal repeat (LTR) retrotransposon that has amplified to hundreds of thousands of copies in mammalian evolution. A small number of the individual copies of L1 are active retrotransposons which are presently replicating in most species, including humans and mice. L1 retrotransposition begins with transcription of an active element and ends with a newly inserted cDNA copy, a process which requires the two element-encoded proteins to act in cis on the L1 RNA. The ORF1 protein (ORF1p) is a high-affinity, non-sequence-specific RNA binding protein with nucleic acid chaperone activity, whereas the ORF2 protein (ORF2p) supplies the enzymatic activities for cDNA synthesis. This article reviews the nucleic acid chaperone properties of ORF1p in the context of L1 retrotransposition.
Key words: annealing, L1, LINE-1, non-LTR retrotransposon, retroelement, retrotransposition, strand exchange, TPRT
L1 has been and continues to be the dominant dynamic force operating in the genomes of many, if not most, mammals, including humans and mice. It is an autonomous retrotransposon that has amplified to more than 500,000 copies, now comprising 17% of the human genome1 and 19% of the mouse genome.2 In addition to mobilizing and amplifying itself, L1 provides the enzymatic machinery necessary for amplification of short interspersed elements (SINEs,3,4 including primate Alu and rodent B1 and B2), other non-autonomous interspersed repeats such as the primate composite repeat SVA5 (an acronym of SINE-R, VNTR and Alu, where SINE-R is a specific sub-class of SINE, VNTR is an acronym for variable-number-of-tandem-repeats and Alu is the interspersed repeat family released by digestion with the restriction endonuclease AluI, another subclass of SINE) and processed pseudogenes, and is thereby responsible for upwards of 35% of the present day human and mouse genomes.6 Ongoing germline insertions lead to a variety of genetic diseases and somatic activity links to cancer and brain variation.7,8 Recent advances in methods to analyze the genomic locations of L1 and Alu provide robust evidence for substantial variation in the location of individual elements among normal and diseased humans (reviewed in ref. 9).
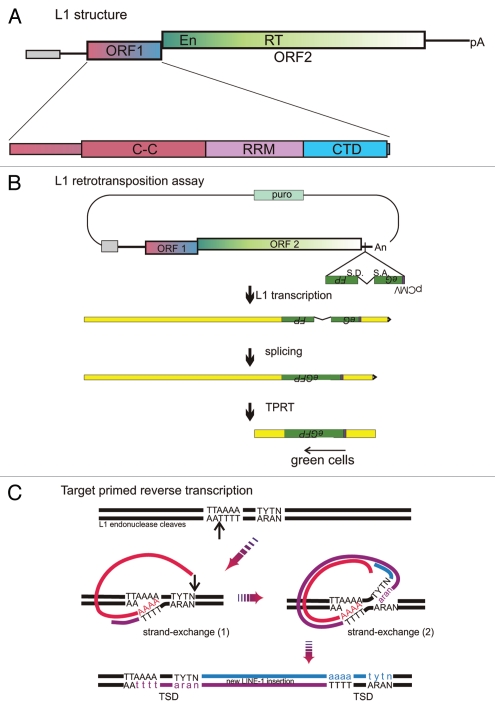
L1, like all retrotransposons, replicates via reverse transcription of an RNA intermediate, albeit with a mechanism distinct from the better-studied retroviruses and LTR-containing retrotransposons. Each replication cycle begins with transcription of one of the full-length, active copies of L1 (Fig. 1A). After moving to the cytoplasm, the L1 dicistronic transcript initially serves as a template for translation of the two L1-encoded essential retrotransposition proteins, ORF1p and ORF2p, which associate in cis with L1 RNA10 to form an L1 ribonucleoprotein particle (L1RNP). From this point on, however, the details of the L1 replication cycle differ dramatically from those of the LTR-containing retroviral-like elements (Fig. 1B). Specifically, the L1RNP juxtaposes the L1 RNA with genomic DNA for target-site primed reverse transcription (TPRT),11 Figure 1C, which converts the RNA to cDNA coincident with genomic insertion. TPRT is best studied for the element R2,12–14 but is likely the universal mechanism for non-LTR retrotransposons, with the main variation being the site of endonuclease cleavage.15 In the specific example of mammalian L1, a primer is formed when the endonuclease domain of ORF2p recognizes and nicks a genomic target site, typically TTTT/AA,16 or similiar.17 The newly released 3′OH at the end of the polyT is annealed to the polyA tail of L1 RNA and then extended by the ORF2p reverse transcriptase, using L1 RNA as the template.18,19 Second strand cDNA is likely made by an analogous TPRT reaction using the newly-synthesized, L1 first-strand cDNA as the template.20,21
Figure 1.
LINE-1 structure and life-cycle. (A) Schematic of L1, depicting from 5′ to 3′: internal promoter (gray box), 5′ untranslated region (UTR, line), ORF1 and ORF2 with endonuclease (En) and reverse transcriptase (RT) domains, 3′UTR (line), polyA (pA) signal which is followed by an A-rich tail. This entire element is typically embedded in a recognizable target site duplication in genomic DNA. The enlargement of ORF1 depicts its three domains: C-C, coiled-coil; RR M, RNA recognition motif; and CTD, C-terminal domain. (B) Assay for retrotransposition of L1. The transfected plasmid contains a selectable gene (puro), origin of replication (not shown) and an intact L1 with an anti-sense intron marked reporter gene embedded in the 3′UTR, in this case eGFP (S.D. and S.A. indicate the splice donor and acceptor sites, respectively). After insertion of the spliced cDNA product of retrotransposition, cells can express eGFP, but not before. The two L1-proteins are required in cis, thus associate with the L1 RNA that encoded them, converting it into the L1RNP that interacts with genomic DNA to insert a new copy of L1 by TPRT. ORF1p is likely present in the RNP at much higher stoichiometry than ORF2p, based upon efforts to detect both proteins in cells.28,58,62 If ORF1p coats the RNA as a trimer binding 50 nt, 140 trimers would be needed for the 7 kb RNA whereas likely just one dimer of ORF2 is needed, predicting a 200-fold excess of ORF1p compared to ORF2p.30 (C) Details of L1 reverse transcription with insertion at the target site by TPRT. The depicted reaction is a sequential TPRT, initially using L1 RNA as the template, followed by L1 first-strand cDNA; but both reactions are primed by the L1 endonuclease and extended by L1 RT. ORF1p may help with the strand transfer required to anneal the genomic primer to the L1 RNA (1) or cDNA (2) templates or facilitate primer extension during reverse transcription or both.20 TSD, target-site duplication.
The precise requirements for ORF1p in the life-cycle of L1 have been challenging to elucidate, but recent structural and functional information is providing insight into the role of ORF1p in L1 retrotransposition. Unlike ORF2 with its recognizable sequence similarity to other endonucleases18 and reverse transcriptases,22 ORF1p is not apparently homologous to any proteins with known function.23 Three distinct structural domains have nevertheless been characterized in the protein, from N- to C-terminus they are: a coiled-coil domain (C-C) which mediates homotrimerization;24,25 an non-canonical RNA recognition motif (RRM) domain;26 and a C-terminal domain, CTD.27 The latter two domains together are necessary for strong binding of single-strand nucleic acids.25–27 ORF1p forms RNPs in vivo28,29 and in vitro,30 and purified ORF1p has potent nucleic acid chaperone activity.20,31,32
The binding and chaperone properties of ORF1p are reminiscent of retroviral nucleocapsid proteins;33 i.e., the protein is an ATP-independent, stoichiometrically-binding nucleic acid chaperone. L1 ORF1p, however, is not related to the better-studied nucleocapsid proteins34 by sequence or by structure. The 43 kDa mouse ORF1p forms a stable trimer of 129 kDa in solution. Consistent with its elution behavior in size-exclusion chromatography, the trimer has an asymmetric dumbbell-shaped structure when imaged by atomic force microscopy.24 Purified ORF1p is a high-affinity, non-sequence-specific RNA binding protein.35 Each trimer interacts with RNA via the large end of the asymmetric dumbbell and occupies 50 nt of RNA.30 Thus, the mass of the protein itself and its apparent footprint on RNA are enormous for ORF1p compared, for example, to the 7 kDa HIV-1 nucleocapsid protein with its binding site for 5–8 nt.33
Purified ORF1p from mouse L1 also functions as a nucleic acid chaperone protein in vitro—it facilitates nucleic acid rearrangements by accelerating both melting and annealing of base-pairs and does so in the absence of ATP and without requiring continuous association between the protein and nucleic acid. Because the life-cycle of L1 is RNA based, ORF1p is very likely an RNA chaperone, although the assays that define its chaperone activity have used DNA substrates.20,31,32 Evidence that the nucleic acid chaperone activity is important for L1 retrotransposition comes mainly from several mutations in ORF1p that abolish L1 retrotransposition without significantly affecting RNA binding;31,32,36 where tested, alteration of in vitro nucleic acid chaperone activity in these mutants correlates with loss of L1 retrotransposition.31,32 Specifically, the mutant proteins alter a delicate balance between the interaction of ORF1p with single- and double-stranded nucleic acids,31,32 and in one case, the mutation clearly slows the kinetics of interaction with mispaired dsDNA oligonucleotides.31 Similar effects have been demonstrated for mutation or truncation variants in other viral nucleic acid chaperone proteins.37–40
The first report of nucleic acid chaperone activity associated with L1 ORF1p examined the effect of mouse protein purified from baculovirus-infected insect cells on short DNA oligonucleotides.20 The protein was found to greatly enhance annealing, strand exchange and duplex melting in a time, temperature and concentration-dependent manner. These nucleic acid chaperone activities are sequence-independent and occur at equimolar concentrations of ORF1p and nucleic acid. A model for the role of nucleic acid chaperone activity during TPRT was proposed, but evidence that the chaperone activity is actually required for L1 retrotransposition was lacking.20
A subsequent study examined these biochemical properties in a series of mutants of ORF1p from mouse L1 that either modestly reduce or completely inactivate L1 retrotransposition. Replacement of two highly conserved arginine residues in the CTD (Fig. 1A) of ORF1p with alanine had been shown to abolish retrotransposition of human L1, providing the first evidence that this rapidly evolving protein23 was required for L1 retrotransposition.41 The analogous mouse ORF1p mutant, RR297:298AA, was likewise inactive.32 Substitution of both arginines with lysine, i.e., RR297:298KK and one of the two single-site mutations, R297K, also effectively abolish retrotransposition (≥98 fold reduced), but the other single substitution in this region of ORF1p, R298K, retains 40% of wt activity. The in vitro properties of RNA binding and nucleic acid chaperone activity were tested for the wt and variant ORF1 proteins after purification to apparent homogeneity from baculovirus-infected insect cells. High-affinity RNA binding is indistinguishable among wt, R297K, R298K and RR297:298KK, but significantly (∼25-fold) lower in RR297:298AA. This greatly reduced RNA affinity likely explains the loss of biological function in RR297:298AA, but some other activity of ORF1p must be damaged by R297K and RR297:298KK. Furthermore, that activity remains intact, albeit apparently slightly compromised, in R298K. A similar conclusion was reached based upon co-enrichment of the homologous human ORF1 mutant proteins with RNA in a RNP fraction derived from cell extracts.36
The annealing and melting components of nucleic acid chaperone activity were examined for each of the ORF1p mutations at R297 and R298 using the purified proteins. Annealing assays with complementary short DNA oligonucleotides failed to distinguish among these ORF1p variants, but several other assays of nucleic acid chaperone activity revealed significant differences that mirrored retrotransposition activity. Specifically, wt and R298K lower the melting temperature of a mispaired 29-mer DNA oligonucleotide by 20°C, but the same DNA duplex is actually stabilized in the presence of all of the inactive ORF1 proteins. Wild-type and R298K proteins also increase strand exchange compared to reactions without protein, but at a substantially slower rate than inactive variants. Finally, in single-molecule stretching experiments (for a detailed description of single-molecule DNA stretching results with ORF1p, please see review by Wu, Rouzina and Williams in this issue), all of the variants except for RR297:298AA increase the slope of the helix-coil transition which is a hallmark of nucleic acid chaperone activity.42–45 However, the inactive mutants, RR297:298KK and R297K, aggregate dsDNA too strongly, which undermines their effectiveness as chaperones. Taken together, these data indicate that inactivating mouse L1 by mutation of R297 in ORF1p disrupts the required nucleic acid chaperone activity of this protein during retrotransposition by overly stabilizing DNA duplexes. The results of the single-molecule stretching experiments also provide a direct means to compare the chaperone activity of L1 ORF1p to HIV nucleocapsid, a protein that has been studied extensively by this technique.37,42,43,46 The comparison reveals ORF1p is the more effective nucleic acid chaperone at low protein concentrations.32
Initial studies of mouse ORF1p employed a specific genomic element known as L1spa,47 which is a full-length copy that recently retrotransposed into the glycine β-receptor gene to cause a phenotypic mutation known as spastic.48,49 Given this unmistakable evidence of recent activity, L1spa is surprisingly inefficient, yielding 15-fold fewer events than an element from the X chromosome, TFC, in the autonomous retrotransposition assay.32 The retrotransposition deficit maps to a single nucleotide in the aspartic acid codon at residue 159 of ORF1p, leading to its replacement with histidine.31 Detailed analysis of this isolated mutation, using transfected tissue culture cells and purified ORF1p in vitro, failed to uncover any significant differences between H159 and D159 in the steady-state levels of L1 RNA or ORF1p in cell extracts, or in RNA binding affinity. However, the two proteins differ substantially in several measures of nucleic acid chaperone activity. The less active variant, H159, is able to fully melt a mispaired DNA duplex, whereas the more active variant, D159, can only partially melt the duplex in the absence of strand exchange. In single molecule studies of force induced melting, H159 interacts more strongly with ssDNA than D159. This strong interaction reduces the width of the helix-coil transition, indicative of reduced nucleic acid chaperone activity. Finally, the D159 and H159 ORF1p variants interact differently with mispaired DNA duplex in surface plasmon resonance experiments. Significantly, the rates of association and dissociation of the two proteins with ssDNA and perfect duplex DNA are effectively identical, but H159 dissociates from the mispaired duplex 10-times more slowly than D159. Thus, the reduced retrotransposition capability of L1spa appears to result from decreased efficacy of the nucleic acid chaperone function of H159 ORF1p caused by the slowed kinetics of interaction with a breathing DNA duplex and ssDNA; this reduced mobility of the protein-nucleic acid complex leads to reduced annealing activity.31 Reduced kinetics of nucleic acid binding and dissociation is also correlated with reduced strand annealing and nucleic acid chaperone activity in variants of HIV nucleocapsid protein.37
Effective nucleic acid chaperones must balance opposing activities of strand annealing and duplex destabilization, i.e., annealing and melting of nucleic acid basepairs. Single residue substitutions in ORF1p, distantly located in the primary sequence of the protein, can significantly alter its nucleic acid chaperone activity and diminish or abolish L1 retrotransposition. Recently, Evans et al.50 devised a method to quantify this balance between annealing and destabilization and tested the approach on new mutations of other highly-conserved residues in ORF1p that, when replaced with alanine, abolish retrotransposition. Specifically, R284A and Y318A inactivate L1 retrotransposition in the cultured cell assay. The retrotransposition defect of these two alanine substitutions can be partially or fully restored by further substitution to biochemically more conservative residues, R284K or Y318F, respectively, providing matched pairs of active and inactive variants in ORF1p. Changes in the steady-state levels of L1 RNA or ORF1p in transfected cells at any time during the timecourse of the retrotransposition assay were eliminated as the cause of retrotransposition defects in these mutants, as were changes in the quaternary structure (e.g., trimer formation) or RNA affinity of the purified proteins. Single-molecule stretching results confirm that the active variants of each mutant pair have the greatest effect on the width of the helix-coil transition, consistent with their being functional nucleic acid chaperones. However, the change in transition width fails to distinguish the inactive Y318A from the active R284K because of excessive aggregation of ssDNA by R284K. To define a quantitative measure that distinguishes between all of the active and inactive ORF1p nucleic acid chaperone variants, kinetics measurements of both annealing and strand exchange were obtained for each protein. These data were used to derive the energetic barriers to annealing and melting using transition state theory. When the reduction of the energetic barrier to annealing (compared to reactions without added protein) is plotted against the reduction of the energetic barrier to melting, the active variants, wt, R284K and Y318F, cluster together and define a line. The inactive variants, R284A and Y318A, fall far off of this line.50 This analysis provides a quantitative assessment of the balance between the competing activities of strand annealing and melting that characterizes functional nucleic acid chaperones. Application of this approach in future studies should prove useful for analysis of additional mutations in ORF1p to aid functional assignment of the two chaperone activities to specific regions of the proteins. In addition, the method should be generally applicable to studies of nucleic acid chaperone structure and function.
Like many other retroelements,34,51–56 L1 encodes a protein with nucleic acid chaperone activity and all available data indicates that the nucleic acid chaperone function of ORF1p is essential for L1 retrotransposition.31,32,50 The data are not extensive, however, and the precise role of the chaperone activity in the life-cycle of L1 is presently speculative. ORF1p likely serves two functions during retrotransposition: first, its high affinity RNA binding activity facilitates formation of a stable RNP intermediate containing L1 RNA and both L1-encoded proteins,57 and second, as a nucleic acid chaperone.31,32 When RNA binding capability is lost, RNP complex formation is compromised and retrotransposition does not occur.36 L1 RNP complexes are associated with stress granules in cultured cells; mutations in ORF1p can alter the subcellular distribution of L1 RNA,58 but the relationship between the association of L1 RNPs with stress granules and active retrotransposition is not understood.
There are several known mutations in ORF1p that do not prevent RNP formation but nevertheless abolish retrotransposition, indicating that a later step in the L1 life-cycle is affected, most likely TPRT.31,32,36,50 Reverse transcription by ORF2p in vitro does not depend upon ORF1p.57,59,60 However, the absence of ORF1p in L1 RNA-ORF2p complexes enriched from cells transfected with L1 containing mutant (non-RNA binding) ORF1p reduces the quantity and length of reverse transcripts in an in vitro L1 amplification protocol (LEAP) and alters their start sites, which is consistent with a role for ORF1p in TPRT.57 Given that L1 amplifies SINEs, ORF1p is paradoxically not required for transmobilization of the primate SINE, Alu, in cultured cells, even though the Alu element clearly uses L1 ORF2 for insertion by TPRT.3 It is possible that a nucleic acid chaperone requirement during TPRT of Alu is fulfilled instead by a cellular protein. Interestingly, when Alu is transcribed by RNA polymerase II rather than polymerase III in this retrotransposition assay, ORF1p is required.61
In summary, the nucleic acid chaperone activity of ORF1p is required for L1 retrotransposition. ORF1p is sensitive to mutation that disturbs a finely tuned balance between the two competing components of nucleic acid chaperone activity, DNA melting and annealing. Additional studies of wt and mutant proteins, including a structure of the protein at atomic level resolution with and without nucleic acids, will be required to elucidate the binding interactions between DNA, RNA and ORF1p and the precise mechanism of chaperone activity during L1 replication. It is likely that a precise balance between annealing and melting activities will likewise be critical for the biological function of all nucleic acid chaperone proteins, but that this relationship will vary quantitatively depending upon specific details of the nucleic acid rearrangements catalyzed.
Acknowledgements
I thank Dr. Suresh Peddigari and James Evans for helpful discussion and comments on the manuscript and NIH GM40367 for support.
Abbreviations
- C-C
coiled coil
- CTD
C-terminal domain
- L1 or LINE-1
long interspersed element one
- LEAP
L1 element amplification protocol
- LTR
long terminal repeat
- ORF
open reading frame
- ORF1p
the ORF1 protein from LINE-1
- ORF2p
the ORF2 protein from LINE-1
- RNP
ribonucleoprotein particle
- RRM
RNA recognition motif
- SINE
short interspersed element
- ssDNA
single-strand DNA
- SVA
a composite interspersed repeat (comprised of SINE-R, VNTR and Alu)
- TPRT
target-site primed reverse transcription
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/13766
References
- 1.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Consortium HG, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:7–8. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Consortium MGS. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 3.Dewannieux M, Esnault C, Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nat Genet. 2003;35:41–48. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- 4.Dewannieux M, Heidmann T. L1-mediated Retrotransposition of Murine B1 and B2 SINEs Recapitulated in Cultured Cells. J Mol Biol. 2005;349:241–247. doi: 10.1016/j.jmb.2005.03.068. [DOI] [PubMed] [Google Scholar]
- 5.Ostertag EM, Goodier JL, Zhang Y, Kazazian HH., Jr SVA Elements Are Nonautonomous Retrotransposons that Cause Disease in Humans. The American Journal of Human Genetics. 2003;73:1444–1451. doi: 10.1086/380207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat Rev Genet. 2009;10:691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goodier JL, Kazazian HH., Jr Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell. 2008;135:23–35. doi: 10.1016/j.cell.2008.09.022. [DOI] [PubMed] [Google Scholar]
- 8.Belancio VP, Deininger PL, Roy-Engel AM. LINE dancing in the human genome: transposable elements and disease. Genome Med. 2009;1:97. doi: 10.1186/gm97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lupski JR. Retrotransposition and structural variation in the human genome. Cell. 2010;141:1110–1112. doi: 10.1016/j.cell.2010.06.014. [DOI] [PubMed] [Google Scholar]
- 10.Wei W, Gilbert N, Ooi SL, Lawler JF, Ostertag EM, Kazazian HH, et al. Human L1 retrotransposition: cis preference versus trans complementation. Mol Cell Biol. 2001;21:1429–1439. doi: 10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luan DD, Korman MH, Jakubczak JL, Eickbush TH. Reverse transcription of R2Bm RNA is primed by a nick at the chomosomal target site: a mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- 12.Christensen SM, Eickbush TH. R2 target-primed reverse transcription: ordered cleavage and polymerization steps by protein subunits asymmetrically bound to the target DNA. Mol Cell Biol. 2005;25:6617–6628. doi: 10.1128/MCB.25.15.6617-6628.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christensen SM, Ye J, Eickbush TH. RNA from the 5′ end of the R2 retrotransposon controls R2 protein binding to and cleavage of its DNA target site. Proc Natl Acad Sci USA. 2006;103:17602–17607. doi: 10.1073/pnas.0605476103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kurzynska-Kokorniak A, Jamburuthugoda VK, Bibillo A, Eickbush TH. DNA-directed DNA polymerase and strand displacement activity of the reverse transcriptase encoded by the R2 retrotransposon. J Mol Biol. 2007;374:322–333. doi: 10.1016/j.jmb.2007.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eickbush TH, Jamburuthugoda VK. The diversity of retrotransposons and the properties of their reverse transcriptases. Virus Res. 2008;134:221–234. doi: 10.1016/j.virusres.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jurka J. Sequence patterns indicate an enzymatic involvement in integration of mammalian retroposons. Proc Natl Acad Sci USA. 1997;94:1872–1877. doi: 10.1073/pnas.94.5.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cost GJ, Boeke JD. Targeting of human retrotransposon integration is directed by the specificity of the L1 endonuclease for regions of unusual DNA structure. Biochemistry. 1998;37:18081–18093. doi: 10.1021/bi981858s. [DOI] [PubMed] [Google Scholar]
- 18.Feng Q, Moran JV, Kazazian HHJ, Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- 19.Cost GJ, Feng Q, Jacquier A, Boeke JD. Human L1 element target-primed reverse transcription in vitro. EMBO J. 2002;21:5899–5910. doi: 10.1093/emboj/cdf592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin SL, Bushman FD. Nucleic acid chaperone activity of the ORF1 protein from the mouse LINE-1 retrotransposon. Mol Cell Biol. 2001;21:467–475. doi: 10.1128/MCB.21.2.467-475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin SL, Li PW-l, Furano AV, Boissinot S. The structures of mouse and human L1 elements reflect their insertion mechanism. Cytogenet Genome Res. 2005;110:223–228. doi: 10.1159/000084956. [DOI] [PubMed] [Google Scholar]
- 22.Loeb DD, Padgett RW, Hardies SC, Shehee WR, Comer MB, Edgell MH, et al. The sequence of a large L1Md element reveals a tandemly repeated 5′ end and several features found in retrotransposons. Mol Cell Biol. 1986;6:168–182. doi: 10.1128/mcb.6.1.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin SL. The ORF1 protein encoded by LINE-1: structure and function during L1 retrotransposition. J Biomed Biotechnol. 2006;2006:45621–45626. doi: 10.1155/JBB/2006/45621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin SL, Branciforte D, Keller D, Bain DL. Trimeric structure for an essential protein in L1 retrotransposition. Proc Natl Acad Sci USA. 2003;100:13815–13820. doi: 10.1073/pnas.2336221100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin SL, Li J, Weisz JA. Deletion analysis defines distinct functional domains for protein-protein and nucleic acid interactions in the ORF1 protein of mouse LINE-1. J Mol Biol. 2000;304:11–20. doi: 10.1006/jmbi.2000.4182. [DOI] [PubMed] [Google Scholar]
- 26.Khazina E, Weichenrieder O. Non-LTR retrotransposons encode noncanonical RRM domains in their first open reading frame. Proc Natl Acad Sci USA. 2009;106:731–736. doi: 10.1073/pnas.0809964106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Januszyk K, Li PW, Villareal V, Branciforte D, Wu H, Xie Y, et al. Identification and Solution Structure of a Highly Conserved C-terminal Domain within ORF1p Required for Retrotransposition of Long Interspersed Nuclear Element-1. J Biol Chem. 2007;282:24893–24904. doi: 10.1074/jbc.M702023200. [DOI] [PubMed] [Google Scholar]
- 28.Martin SL. Ribonucleoprotein particles with LINE-1 RNA in mouse embryonal carcinoma cells. Mol Cell Biol. 1991;11:4804–4807. doi: 10.1128/mcb.11.9.4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hohjoh H, Singer MF. Cytoplasmic ribonucleoprotein complexes containing human LINE-1 protein and RNA. EMBO J. 1996;15:630–639. [PMC free article] [PubMed] [Google Scholar]
- 30.Basame S, Li PW-l, Howard G, Branciforte D, Keller D, Martin SL. Spatial assembly and RNA binding stoichiometry of a protein essential for LINE-1 retrotransposition. J Mol Biol. 2006;357:351–357. doi: 10.1016/j.jmb.2005.12.063. [DOI] [PubMed] [Google Scholar]
- 31.Martin SL, Bushman D, Wang F, Li PWL, Walker A, Cummiskey J, et al. A single amino acid substitution in ORF1 dramatically decreases L1 retrotransposition and provides insight into nucleic acid chaperone activity. Nucl Acids Res. 2008;18:5845–5854. doi: 10.1093/nar/gkn554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin SL, Cruceanu M, Branciforte D, Wai-Lun Li P, Kwok SC, Hodges RS, et al. LINE-1 retrotransposition requires the nucleic acid chaperone activity of the ORF1 protein. J Mol Biol. 2005;348:549–561. doi: 10.1016/j.jmb.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 33.Levin JG, Guo J, Rouzina I, Musier-Forsyth K. Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: critical role in reverse transcription and molecular mechanism. Prog Nucleic Acid Res Mol Biol. 2005;80:217–286. doi: 10.1016/S0079-6603(05)80006-6. [DOI] [PubMed] [Google Scholar]
- 34.Stewart-Maynard KM, Cruceanu M, Wang F, Vo MN, Gorelick RJ, Williams MC, et al. Retroviral nucleocapsid proteins display nonequivalent levels of nucleic acid chaperone activity. J Virol. 2008;82:10129–10142. doi: 10.1128/JVI.01169-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kolosha VO, Martin SL. High affinity, non-sequence-specific RNA binding by the open reading frame 1 (ORF1) protein from long interspersed nuclear element 1 (LINE-1) J Biol Chem. 2003;278:8112–8117. doi: 10.1074/jbc.M210487200. [DOI] [PubMed] [Google Scholar]
- 36.Kulpa DA, Moran JV. Ribonucleoprotein particle formation is necessary but not sufficient for LINE-1 retrotransposition. Hum Mol Gen. 2005;14:3237–3248. doi: 10.1093/hmg/ddi354. [DOI] [PubMed] [Google Scholar]
- 37.Cruceanu M, Gorelick RJ, Musier-Forsyth K, Rouzina I, Williams MC. Rapid Kinetics of Protein-Nucleic Acid Interaction is a Major Component of HIV-1 Nucleocapsid Protein's Nucleic Acid Chaperone Function. J Mol Biol. 2006;363:867–877. doi: 10.1016/j.jmb.2006.08.070. [DOI] [PubMed] [Google Scholar]
- 38.Hargittai MRS, Gorelick RJ, Rouzina I, Musier-Forsyth K. Mechanistic insights into the kinetics of HIV-1 Nucleocapsid Protein-facilitated tRNA annealing to the Primer Binding Site. J Mol Biol. 2004;337:951–968. doi: 10.1016/j.jmb.2004.01.054. [DOI] [PubMed] [Google Scholar]
- 39.Qualley DF, Stewart-Maynard KM, Wang F, Mitra M, Gorelick RJ, Rouzina I, et al. C-terminal domain modulates the nucleic acid chaperone activity of human T-cell leukemia virus type 1 nucleocapsid protein via an electrostatic mechanism. J Biol Chem. 2010;285:295–307. doi: 10.1074/jbc.M109.051334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharma K, Didier P, Darlix JL, de Rocquigny H, Bensikaddour H, Lavergne JP, et al. Kinetic analysis of the nucleic acid chaperone activity of the hepatitis C virus core protein. Nucleic Acids Res. 2010;38:3632–3642. doi: 10.1093/nar/gkq094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH., Jr High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 42.Williams MC, Gorelick RJ, Musier-Forsyth K. Specific zinc-finger architecture required for HIV-1 nucleocapsid protein's nucleic acid chaperone function. Proc Natl Acad Sci USA. 2002;99:8614–8619. doi: 10.1073/pnas.132128999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Williams MC, Rouzina I, Wenner JR, Gorelick RJ, Musier-Forsyth K, Bloomfield VA. Mechanism for nucleic acid chaperone activity of HIV-1 nucleocapsid protein revealed by single molecule stretching. Proc Natl Acad Sci USA. 2001;98:6121–6126. doi: 10.1073/pnas.101033198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCauley MJ, Williams MC. Mechanisms of DNA binding determined in optical tweezers experiments. Biopolymers. 2007;85:154–168. doi: 10.1002/bip.20622. [DOI] [PubMed] [Google Scholar]
- 45.Williams MC, Rouzina I. Force spectroscopy of single DNA and RNA molecules. Curr Opin Struct Biol. 2002;12:330–336. doi: 10.1016/s0959-440x(02)00340-8. [DOI] [PubMed] [Google Scholar]
- 46.Cruceanu M, Urbaneja MA, Hixson CV, Johnson DG, Datta SA, Fivash MJ, et al. Nucleic acid binding and chaperone properties of HIV-1 Gag and nucleocapsid proteins. Nucleic Acids Res. 2006;34:593–605. doi: 10.1093/nar/gkj458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Naas TP, DeBerardinis RJ, Moran JV, Ostertag EM, Kingsmore SF, Seldin MF, et al. An actively-retrotransposing, novel subfamily of mouse L1 elements. EMBO J. 1998;17:590–597. doi: 10.1093/emboj/17.2.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kingsmore SF, Giros B, Suh D, Bieniarz M, Caron MG, Seldin MF. Glycine receptor beta-subunit gene mutation in spastic mouse associated with LINE-1 element insertion. Nat Genet. 1994;7:136–142. doi: 10.1038/ng0694-136. [DOI] [PubMed] [Google Scholar]
- 49.Mullhard C, Fischer M, Gass P, Simon-Chazottes D, Guenet JL, Becker CM. The spastic mouse: aberrant splicing of glycine receptor beta subunit mRNA caused by intronic insertion of L1 element. Neuron. 1994;13:1003–1015. doi: 10.1016/0896-6273(94)90265-8. [DOI] [PubMed] [Google Scholar]
- 50.Evans JD, Peddigari S, Chaurasiya KR, Williams MC, Martin SL. Long Interspersed Element-1 (LINE-1) retrotransposition depends on the balanced catalytic properties of its nucleic acid chaperone. 2010. In review.
- 51.Dawson A, Hartswood E, Paterson T, Finnegan DJ. A LINE-like transposable element in Drosophila, the I factor, encodes a protein with properties similar to those of retroviral nucleocapsids. EMBO J. 1997;16:4448–4455. doi: 10.1093/emboj/16.14.4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Heras SR, López MC, García-Pérez JL, Martin SL, Thomas MC. The L1Tc C-terminal domain from a Trypanosoma cruzi non-LTR retrotransposon codes for a protein that bears two C2H2 zinc-finger motifs and is endowed with nucleic acid chaperone activity. Mol Cell Biol. 2005;25:9209–9220. doi: 10.1128/MCB.25.21.9209-9220.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Heras SR, Thomas MC, Macias F, Patarroyo ME, Alonso C, Lopez MC. Nucleic-acid-binding properties of the C2-L1Tc nucleic acid chaperone encoded by L1Tc retrotransposon. Biochem J. 2009;424:479–490. doi: 10.1042/BJ20090766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gabus C, Ivanyi-Nagy R, Depollier J, Bucheton A, Pelisson A, Darlix JL. Characterization of a nucleocapsid-like region and of two distinct primer tRNALys,2 binding sites in the endogenous retrovirus Gypsy. Nucleic Acids Res. 2006;34:5764–5777. doi: 10.1093/nar/gkl722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cristofari G, Ficheux D, Darlix JL. The gag-like protein of the yeast Ty1 retrotransposon contains a nucleic acid chaperone domain analogous to retroviral nucleocapsid proteins. J Biol Chem. 2000;275:19210–19217. doi: 10.1074/jbc.M001371200. [DOI] [PubMed] [Google Scholar]
- 56.Rajkowitsch L, Chen D, Stampfl S, Semrad K, Waldsich C, Mayer O, et al. RNA chaperones, RNA annealers and RNA helicases. RNA Biol. 2007;4:118–130. doi: 10.4161/rna.4.3.5445. [DOI] [PubMed] [Google Scholar]
- 57.Kulpa DA, Moran JV. Cis-preferential LINE-1 reverse transcriptase activity in ribonucleoprotein particles. Nat Struct Mol Biol. 2006;13:655–660. doi: 10.1038/nsmb1107. [DOI] [PubMed] [Google Scholar]
- 58.Goodier JL, Mandal PK, Zhang L, Kazazian HH., Jr Discrete subcellular partitioning of human retrotransposon RNAs despite a common mechanism of genome insertion. Hum Mol Genet. 2010;19:1712–1725. doi: 10.1093/hmg/ddq048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Piskareva O, Denmukhametova S, Schmatchenko V. Functional reverse transcriptase encoded by the human LINE-1 from baculovirus-infected insect cells. Protein Expression and Purification. 2003;28:125–130. doi: 10.1016/s1046-5928(02)00655-1. [DOI] [PubMed] [Google Scholar]
- 60.Piskareva O, Schmatchenko V. DNA polymerization by the reverse transcriptase of the human L1 retrotransposon on its own template in vitro. FEBS Lett. 2006;580:661–668. doi: 10.1016/j.febslet.2005.12.077. [DOI] [PubMed] [Google Scholar]
- 61.Kroutter EN, Belancio VP, Wagstaff BJ, Roy-Engel AM. The RNA Polymerase Dictates ORF1 Requirement and Timing of LINE and SINE Retrotransposition. PLoS Genet. 2009;5:e1000458. doi: 10.1371/journal.pgen.1000458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Branciforte D, Martin SL. Developmental and cell type specificity of LINE-1 expression in mouse testis: implications for transposition. Mol Cell Biol. 1994;14:2584–2592. doi: 10.1128/mcb.14.4.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]