Abstract
HIV-1 nucleocapsid protein (NC) is a small basic protein generated by the cleavage of the Gag structural polyprotein precusor by the viral protease during virus assembly in the infected cell. HIV-1 NC possesses two copies of a highly conserved CCHC zinc finger (ZnF), flanked by basic residues. HIV-1 NC and more generally retroviral NC proteins are nucleic acid binding proteins possessing potent nucleic acid condensing and chaperoning activities. As such NC protein drives critical structural rearrangements of the genomic RNA, notably RNA dimerization in the course of virus assembly and viral nucleic acid annealing required for genomic RNA replication by the viral reverse transcriptase (RT).
Here we review the relationships between the 3D structure of HIV-1 NC, notably the central globular domain encompassing the two zinc fingers and the basic linker and NC functions in the early and late phases of virus replication. One of the salient feature of the NC central globular domain is an hydrophobic plateau which appears to orchestrate the NC functions, such as chaperoning the conversion of the genomic RNA into viral DNA by RT during the early phase, and driving the selection and dimerization of the genomic RNA at the initial stage of viral particle assembly. This ensures a bona fide trafficking of early GagNC-genomic RNA complexes to the plasma membrane of the infected cell and ultimately virion formation and budding.
Key words: retroviruses, HIV-1, nucleocapsid protein, zinc fingers, genomic RNA, virus assembly, reverse transcription
Foreword
HIV is a member of the Retrovirus family comprising a large and diverse group of enveloped plus strand RNA viruses defined by common taxonomic features, notably the structure of the viral particle with a dimeric RNA genome, and the general replicative properties, which include the reverse transcription and integration of the genome into the host cell DNA.1–8
Replicating retroviruses are divided into two large subgroups, simple and complex ones. This is based on their genetic composition coding for the Gag structural proteins, the Pol enzymes and the Envelope glycoproteins for the simple ones such as the gammaretrovirus MuLV, while complex ones code for additional proteins and essential factors such as Tat, Rev, Nef, Vif, Vpr and Vpu for HIV-1.9
The HIV-1 viral particle has a globular structure with a mean diameter of about 115–120 nm with an outer phospholipid envelope rich in cholesterol of cellular origin, in which a small number of the viral glycoproteins in a trimeric form are anchored, namely the surface (SU) gp120 and the transmembrane (TM) gp41 encoded by the Env gene.10
The internal part of the viral particle is composed of abundant proteins all issued from Gag known as the structural poly- protein precursor. The mature virion core has a global conical shape thought to be surrounded by molecules of matrix proteins (MA) p17 corresponding to the N-terminal domain of the Gag precursor. The core structure or capsid is composed of about 1,500 molecules of capsid (CA) protein p24 in the form of 250 hexamers surrounding the nucleocapsid structure or virus replicative machinery.11–13
The virus nucleocapsid can be viewed as a ribonucleoparticle or RNP, since the major structural components correspond to the unique genomic RNA coated by molecules of nucleocapsid protein NCp7. A unique feature of the genomic RNA is its dimeric 60S form where the two RNA molecules are in close contact via non-covalent interactions involving the 5′ DLS-DIS (Dimer-Linkage-Structure and Dimerization Initiation Sequence) sequences as well as many other contact sequences. The nucleocapsid protein interacts with the RNA genome via hydrophilic and hydrophobic interactions. Given that about 1,500 NC molecules are present per viral core, and thought to extensively coat the genome, each molecule, if evenly distributed, should bind 11–12 nucleotide residues on the average.14,15 As a molecular replicative machine, the nucleocapsid contains about 50 copies of the viral enzymes RT, protease (PR) and integrase (IN) in dimeric forms. In addition, about 100 to 200 molecules of the viral protein R (VPR) are being incorporated into the nucleocapsid (reviewed in ref. 16 and 17).
Here we briefly review the structure-function relationships of the viral nucleocapsid protein with a special emphasis on its implications in virus assembly in the late phase of the virus replication cycle (reviewed in ref. 16–20).
Structure-Function Relationships of the Retroviral Nucleocapsid Protein
Overview of NC protein 3D structure.
HIV-1 NCp7 is released from the Gag polyprotein precursor by the protease-mediated cleavage during virus formation at the plasma membrane of the infected cell. In fully mature viral particles, NCp7 is found as 55 and 71 amino acids forms in freshly collected highly infectious viral particles (reviewed in ref. 15 and 16). NCp7 is composed of two highly conserved zinc fingers (ZnF) of the CCHC form, flanked by small domains rich in basic residues. In its apo form NCp7 has a completely disordered conformation, while Zn2+chelation by the two ZnF causes the folding of the central domain while the N- and C-terminal domains remain essentially unfolded. In fact, the NMR solution structure of NCp7 reveals that the two ZnF adopt a similar folding upon Zn2+ binding by the CCHC residues, and that the linker rich in basic residues is responsible for their spatial proximity.21–26 Interestingly, the upper part of the NC central globular domain constitutes an hydrophobic plateau with the hydrophobic residues Val13, Phe16, Ile24 and Ala25 of the first ZnF and the hydrophobic residues Trp37, Gln45 and Met46 of the second ZnF (Figs. 1 and 3). This hydrophobic plateau appears to be essential in the interactions between NC and small oligonucleotides (ODN) since the structures of all ODN/NCp7 complexes resolved today show that this hydrophobic plateau pilots the binding of NCp7 to nucleic acids (NA) via multiple contacts with the nucleotide bases and the phosphate backbone.27–32 In addition, amino acid Trp37 of the second ZnF always stacks with a guanine base, which represents a major contribution to the NCp7/NA binding energy. The nucleoprotein complex is further stabilized by means of numerous electrostatic interactions with the NC basic amino acids present in the N-terminal disordered sequence and the ZnF linker (Fig. 1). Importantly, mutating either one or more of the CCHC residues, that modify the globular folding and prevent formation of the NC hydrophobic plateau, modifies Gag trafficking in the producer cell, alters the virion core structure and causes a complete loss of virus infectivity.15,16,18,33–40
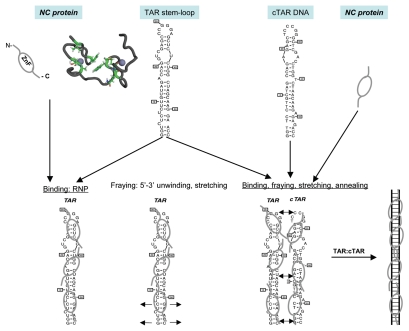
Figure 1.
Schematic illustration of the nucleic acids binding, fraying and annealing properties of the HIV nucleocapsid protein. Top, from left to right: NC protein is represented by an oval corresponding to the hydrophobic plateau (ZnF), flanked by the disordered N- and C-terminal domains shown here as small fragments. The 3D structure of the hydrophobic plateau at the top of the NC zinc-fingers is represented in the CPK mode with Val13, F16, Ile24, Ala25, Trp37, Gln45 and Met46, and the zinc atoms appear as grey spheres. The TAR with its loop and bulges is shown here as a stem-loop. The complementary DNA sequence is synthesized by RT chaperoned by NC at the very beginning of reverse transcription (reviewed in ref. 13 and 14) and shown here as cTAR stem-loop. Bottom, from left to right: Binding of NC to TAR results in the formation of a ribonucleoparticle where the RNA is entirely coated by NC molecules. This causes the transient opening of the 5′ and 3′ end sequences (short arrows), in a process called fraying and strand stretching (Single DNA molecule stretching measures the activity of chemicals that target the HIV-1 nucleocapsid protein).53 Binding of NC to TAR and cTAR causes the formation of nucleoprotein complexes where the nucleic acids are entirely coated by NC molecules. This causes the rapid hybridization of the complementary sequences resulting in the formation of a RNA:DNA hybrid within the nucleoprotein complex. This annealing reaction recapitulates the obligatory 5′-3′ strand transfer that occur at the beginning of reverse transcription.4
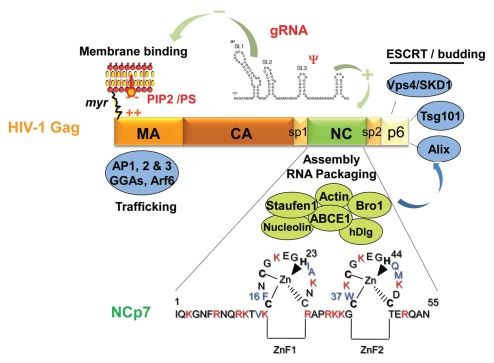
Figure 3.
The Gag polyprotein precursor, its NC domain and the associated cellular factors. HIV-1 Gag is composed of 3 main domains which are MA, CA and NC, one C-terminal peptide p6 and two spacer peptides, sp1 and sp2. The matrix MA domain is responsible for Gag membrane binding via the myristate and the basic (++) region with the PIP2/PS phospholipids. MA domain contains motifs that bind different cellular factors, such as AP1, AP2, AP3, GGA and Arf6 involved in Gag trafficking. The NC domain of Gag is necessary for gRNA recruitment via its interaction with the Psi packaging signal and the NC basic residues (in red) and the hydrophobic plateau encompassing the two conserved zinc fingers. The gRNA is also able to interact with the MA domain by competing with PI(4,5)P2. The NC domain is a key factor for the early assembly complex formation by Gag multimerization on the gRNA. NC and p6 interact with cellular factors; NC with actin, Staufen1, the Bro1 domain of ALIX and ABCE1 as described in this review; the C-terminus of Gag, via the p6 peptide containing the late budding domain is able to recruit the ESCRT cellular proteins, such as Tsg101, Alix and Vps4/SDK1, which are essential for particle budding. Sp1 and Sp2 are linker involved in HIV-1 morphogenesis. The 55 amino acids of mature NC are given; NCp7 is highly basic (in red) and contains two highly conserved zinc CCHC binding motifs and a plateau of hydrophobic amino-acids (in blue) involved in RNA-NC interactions, RNA chaperone activities and particle assembly.
Nucleic acid binding, condensing and chaperoning properties of NC protein.
NC protein binds with high affinity to a very large panel of NA sequences, including in the form of small synthetic ODN (reviewed in ref. 16–18 and 40). This causes the formation of stable nucleoparticles of high molecular weight as seen by electron microscopy41 where both the NA and protein molecules are highly condensed causing a molecular crowding phenomenon (Fig. 1).42 Such a condensation effect results in the protection of the NA, especially the viral DNA, from nuclease degradation and from UV light irradiation.42,43
Regarding binding specificity, NC in the form of the Gag polyprotein precursor appears to specifically recognize the Psi encapsidation sequences, which is thought to account for the selection of the genomic RNA among a vast excess of cellular RNAs, during the initial stage of particle assembly (reviewed in ref. 16, 44–46).
Binding of NC to NA molecules can have important consequences. Firstly NC can drive the structural rearrangement of the NAs into their most stable conformation in an entropy landscape;47,48 secondly when two NA molecules having complementary sequences are present in the nucleoprotein complex, NC provides assistance to their annealing under physiological conditions.49,50 These two main components of the interplay of NC with NA molecules are thought to account for the NC chaperoning activity. The first one is the transient destabilization or fraying, and stretching of the NA structure upon NC binding (Fig. 1).51–53 However, NC-mediated fraying of the NA structure depends on the overall stability of the NA and requires natural mismatches as for wild type TAR in order to take place.46,54,57 This NC-mediated fraying of the NA structure largely relies on the hydrophobic plateau of the ZnF domain.58 The second NC chaperoning component consists in NC-directed annealing of two NAs with complementary sequences when present in the same complex, namely TAR and cTAR sequences.16,18,40 This is illustrated by the rapid annealing of the complementary TAR sequences which appears to require both the fraying and annealing elements of NC, the latter relying on the numerous NC basic residues which neutralize the phosphates of the NA backbone and in turn contribute to the molecular crowding effect
These two main components of NC chaperoning activity are also found in other retroviral NC proteins from gamma and alpharetroviruses,16,17,47,49,50 in the flaviviridae core proteins58 and in cellular proteins such as p50, FMRP and PrP.59
In conclusion, the dynamic interplay of the NC hydrophobic platform and basic domains with the viral nucleic acids via the binding, fraying, condensing and annealing properties of NC may well account for its essential functions in virus replication (Fig. 1). Notably, NC chaperones the complex process of viral DNA synthesis by RT at the levels of the two obligatory strand transfers involving the TAR and PBS sequences, respectively (reviewed in ref. 60), and in virus assembly (reviewed in ref. 19 and 20). In the next paragraph we will review the roles of the NC hydrophobic plateau in the Gag context during particle assembly, from the initial stage of GagNC-vRNA trafficking to the final site of virus formation at the plasma membrane.
Implications of the Nucleocapsid Domain of Gag in Virus Assembly
The late steps of the virus life cycle integrates the expression of the genomic and spliced viral RNA, their nuclear export, the synthesis of the viral proteins and viral particle morphogenesis, including viral assembly. The Env glycoproteins are synthesized on the endoplasmic reticulum polysomes and traffic towards the plasma membrane via the secretory pathway during which they are matured by the cellular furin protein. In contrast the Gag and GagPol protein precursors are synthesized in the cytosol on “free” polysomes.9 Their intracellular trafficking prior to multimerization at the assembly site is currently not completely understood.61–65 Upon assembly with the gRNA and the Env glycoproteins, Gag polyproteins bud as a new virion that undergoes maturation by the proteolytic cleavage of Gag (and GagPol) by the viral protease (reviewed in ref. 9 and 19).
The retroviral Gag structural polyprotein orchestrates viral particle assembly (Fig. 2) (reviewed in ref. 19, 20, 61, 62 and 65). The Gag polyprotein is usually composed of 3 main domains, namely Matrix (MA), Capsid (CA) and Nucleocapsid (NC) but other domains can be found such as in HIV-1 Gag which is composed of Matrix, Capsid, SP1, Nucleocapsid, SP2 and the C-terminal p6 domain (Fig. 3).66,67 MA is anchored into the cellular membrane via its N-terminus and is responsible for Gag membrane targeting and attachment; the CA domain promotes Gag-Gag dimerization and interaction to form the viral capsid;66 the NC protein in the C-terminus of all the retroviral Gag polyproteins is critical for the selection of the viral genomic RNA and the assembly of the virion core. In the case of HIV-1 Gag, the p6 domain interacts with cellular factors necessary for Gag budding such as the ESCRT machinery (reviewed in ref. 68 and 69). In the case of other retroviruses, such as MuLV with the p12 domain and RSV with the p10 domain located in between MA and CA, the function of the intervening peptides is associated with late assembly events and core morphogenesis.70–73 At the end of RSV Gag is located the viral protease (PR) necessary for viral particle maturation (reviewed in ref. 9). These intervening peptides between the main domains of Gag can have different role in particle assembly and morphogenesis, not always well characterized, but often containing the “late budding domain” which can vary position within Gag and which is the docking site for cellular factors involved in particle budding.19,61,68,69
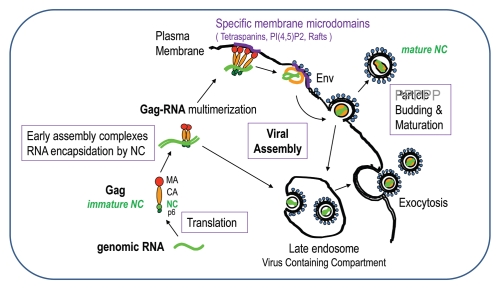
Figure 2.
The late steps of retroviral particle formation: traffic, assembly and budding. The different stages of NC: immature NC in Gag and mature NC in the viral particle. Scheme of Gag-gRNA complexes trafficking within the cell and viral particle assembly and release. Upon export of the genomic RNA from the nucleus, the gRNA is translated into Gag and GagPol proteins that recruit the genome and traffic through the cytoplasm onto the assembly site et the plasma membrane; few Gag complexes with the viral genome anchor into the plasma membrane; Gag recognizes the PM phospholipids PI(4,5)P2 which triggers the anchoring of the myristate into the inner leaflet of the membrane, multimers of Gag thus accumulate at the assembly site which can be rafts or tetraspanins enriched microdomains and will serve as a plateform for particle budding; other cellular determinant can intervene to promote budding and particle release, such as the ESCRT machinery. Some preformed or formed virions can undergo endocytosis from the plasma membrane and create new virus-containing compartment (VVC) in the cell that could be released by exocytosis upon cellular signal. During release, the particle undergoes a maturation process due to the viral protease that cuts Gag and GagPol to specific sites which releases the structural proteins and enzymes in order to form the mature infectious particle, in which free NC proteins condensed on the genomic RNA inside the viral mature core. This phenomenon is a key determinant for particle morphogenesis and infectivity.
The NC domain is involved in Gag-Gag multimerisation through its association with the genomic RNA (reviewed in ref. 19, 44, 74–86). Mutations in the polybasic region of NC or the zinc fingers impair and even abrogate genomic RNA packaging with the notion that the ZnF and hydrophobic plateau are driving the specificity of gRNA encapsidation, while the basic residues are more involved in Gag-Gag multimerization and assembly.19,75–78 These NC motifs are involved in specific Gag-gRNA interactions via the Psi signal located in the 5′ leader region of the gRNA (Fig. 3). Indeed, NC mutants of these motifs are viruses defective in the efficacy of Gag assembly, budding and particle morphogenesis for many retroviruses (HIV, SIV, MLV, RSV, BLV, MPMV).19,74–78,81–83,85 However, in most cases, some mutant Gag are still targeted to the assembly sites, i.e., at the plasma membrane, but Gag oligomer formation or viral particle budding is, at least in part, prevented and aberrant budding particles are observed in many instances.76,82–84 In the case of HIV-1, the situation is rather complex due to the fact that NC is located between sp1 and sp2 with the p6 domain at the extreme C-terminus (Fig. 3). It appears as if these domains are mutually interdependent and involved in more than one step of the late process of particle formation; thus mutating NC might affect several steps of the assembly and budding processes. For example, it was recently reported that NC cooperates with the budding pathway also dependent on the sp2 and p6 domain and its association with the ESCRT proteins Tsg101 and Alix are required for retrovirus budding.68,69 Indeed the N-terminal Bro1 domain of Alix interacts with the N-terminal and first zinc finger of HIV-1 NC, which helps to recruit the budding machinery components, ESCRT-III and Vps4, to promote virus release.87–89
The role of RNA as a favorite NC partner in Gag assembly.
The RNA is thought to play a role in scaffolding particle formation via either the gRNA or cellular RNAs by creating a platform onto which the NC can find support for Gag multimerization.19,44,46 It probably induces a conformation change in Gag that reinforces the anchoring of the Gag MA domain into the membrane.90,91 Thus the initial step of NC-RNA recognition, early after Gag translation, seems to be essential for initiating the assembly process,85,92 and the specific routing of Gag towards its assembly site (Fig. 2). The efficacy of HIV-1 Gag assembly depends also on MA65,93 and on the precise pathway of Gag mRNA export from the nucleus,94,95 where HIV MA function is regulated by the trafficking pathway of the encoding mRNA.95 These results support an RNA-binding requirement for Gag assembly, which relies on binding of RNA by MA or NC sequences to condense, organize, and stabilize the HIV-1 Gag-Gag interactions that form the virion. Indeed the initiation of particle production was delayed by NC-deleted Gag in comparison to wild-type Gag,85 indicating that the initial recognition of NC with the gRNA, most probably via the 5′RNA psi signal, is a key determinant in Gag assembly. Thus NC is critical in nucleating the assembly process of Gag. Actually, this could be concomitant with the end of Gag translation, and once a certain level of Gag is reached, there could be a competition between Gag and the ribosomes for Gag translation since the same gRNA serves as a genome and a template for Gag translation.96–98
Recently, several lines of evidence indicate that the beginning of assembly occurs once few Gag molecules are bound to the genomic RNA, which then travel to the plasma membrane to dock onto specific sites (Fig. 2). These Gag assembly sites are composed of PIP2, rafts or tetraspanin enriched microdomains associated with the plasma membrane (Fig. 2) (reviewed in ref. 65). Such small and early Gag-gRNA complexes then anchor in these specific plasma membrane microdomains in order to recruit and accumulate Gag and/or other Gag-RNA complexes to form new viral buds.46,85 In the absence of NC, MLV Gag is still targeted and associated to the plasma membrane but no Gag-RNA complexes accumulate and no viral buds are visible by microscopy.76 As for HIV-1, a multimerization-defective NC mutant still binds to the plasma membrane raft microdomains,86 suggesting that NC is dispensable for Gag membrane association. In contrast, basic residues in MA are required for Gag membrane binding via their interactions with the phosphinositide PI(4,5)P2.99–101 In addition for HIV-1, these MA basic residues are also a site for gRNA binding. Thus, it was proposed that RNA would serve as a regulator of Gag assembly. The MA domain of Gag interacting with the gRNA, represents a competitor for MA binding to PI(4,5)P2 at the plasma membrane.93,102 Consequently, the gRNA interacting with MA and NC domains would regulate viral particle assembly via its ability to mask PI(4,5)P2 binding site and to be a scaffold for Gag multimerization102 (Fig. 3).
In conclusion, very close interactions between Gag-NC, and most probably the zinc fingers with the hydrophobic plateau of NC, and the gRNA appear necessary for promoting efficient virus assembly.
Role of NC in intracellular Gag trafficking.
Gag trafficking is dependent upon host cell factors interacting mainly with the MA domain. Briefly, Gag can interact with the adaptor proteins AP1, AP2, AP3 or GGA and Arf proteins103–106 and the inhibition or overexpression of these factors impair viral assembly by disrupting the endosomal sorting pathway. NC does not appear to have a definite role in Gag trafficking to the plasma membrane as total deletion of the NC domain of Gag does not prevent some Gag molecules from reaching the plasma membrane but they are deficient in Gag-Gag multimerization and viral assembly.74,76,83 However, several groups report that some NC mutants present defect in early assembly complexes,85 in reaching endosomes83 and even in Gag cellular membrane binding.84 Gag and the genomic RNA undergo trafficking through endosomes that appear to be implicated in viral egress.107–109 Indeed, deleting both NC zinc fingers alters the location of HIV-1 Gag in late endosomes, potential sites for viral assembly.83 The mechanism by which these Gag(NC)-RNA ribonucleic protein complexes traffic and lead to assembly will need future characterizations. Along this line, it has been recently suggested that endosomal trafficking of Gag/RNA complexes could regulate virion formation at specific sites, such as the virological synapse involved in virus cell-to-cell transmission.108
Host cell factors promoting assembly by interacting with NC.
The NC domain of Gag, in association with the genomic RNA, is a key determinant in the recruitment of host cellular factors that are involved at different stages of viral assembly (e.g., Staufen1/ABCE1/Bro1 etc.,), however, the timing of these interactions and cellular location are not well defined. Several cellular proteins were described to interact with the Gag NC domain, which in turn play a role in particle assembly or budding, suggesting that the presence of sequential Gag assembly intermediates.110,111 Actually, these cellular factors can be eventually incorporated into the nascent budding virions suggesting a dynamic interaction with the assembly complex.112
The human Staufen 1protein is found in early ribonucleoprotein assembly complexes of HIV-1 as cytoplasmic granules. It binds the HIV-1 genomic RNA and Gag via its NC domain allowing its incorporation into the virion.112–115 Staufen 1 was found to be involved in RNA trafficking and metabolism, and as such it appears to have a central role in favoring encapsidation of HIV-1 gRNA and its interaction with Gag.113 Recent live imaging data reinforce the notion that Gag interacts with Staufen 1 in the cytoplasm and also on lipid raft microdomains at the plasma membrane where HIV-1 assembly occurs. Gag and Staufen1 are thought to actively recruit protein partners of the assembly complex in association with the gRNA.115
Nucleolin is an RNA binding chaperoning protein that is usually nuclear but has been recently found at the plasma membrane of HIV-1 infected cells.116 It could have a role in retroviral assembly, such as nucleoplasmic transport of the gRNA to the translation site because Nucleolin directs ribosome assembly and nucleocytoplasmic transport.112 Nucleolin binds the genomic RNA of HIV and MLV and is recruited in virions suggesting the presence of this protein at the assembly site. The C-terminus of nucleolin interacts with the NC domain of Gag117 and as such is able to inhibit virion assembly. On the other hand, nucleolin is able to enhance HIV-1 release by favouring the budding of virions containing both the gRNA and the cellular protein.117
The cellular protein ABCE1 binds early HIV-1 Gag-RNA complexes.118 This protein is a member of the superfamily of ATP-binding cassette (ABC) transporters, which transport various molecules across cellular membranes. Also referred to as the RNase L inhibitor or HP68 this protein functions to block the activity of ribonuclease L. Activation of ribonuclease L leads to inhibition of protein synthesis in the central pathway for viral interferon action. ABCE1 is found at HIV-1 assembly sites at the plasma membrane and is also found associated with Gag early after Gag translation. A critical role for ABCE1 and its ATP dependent function is to assist the early stage of virus assembly in human T cells infected by HIV-1.118 More precisely, the basic residues of NC are required for ABCE1/Gag interaction.119 However, ABCE1 is not incorporated into virions and dissociates from Gag during virus production.120
Bro1 is the N-terminal domain of ALIX. ALIX was found to interact with the LYPXnL motif of the L domain of the Gag p6 that recruits the ESCRT machinery to assist virus budding.61,68 ALIX and Tsg101 are two components necessary for the MVB biogenesis and for retroviral budding that associate with p6 for Tsg101 and for p6 in cooperation with NC for ALIX.87,88 As mentioned before, the Bro1 domain of Alix interacts with the NC domain of HIV-1 Gag and in that manner cooperates to recruit the ESCRT via the p6 domain for retroviral budding.87–89
Actin and actin-binding proteins are found inside HIV-1 virions15,121 raising the question of a potential role of actin cytoskeleton dynamic in either Gag trafficking or in virion budding.20,122–125 The Gag NC domain of several retroviruses (HIV, MLV, EIAV) is known to interact with actin. Indeed, Gag multimerization is required for filamentous actin-Gag association during EIAV assembly.124 Is the actin cytoskeleton a cellular structural component on which Gag multimers are located for assembly? Is actin dynamics regulated by HIV for promoting budding and virion release?125 These questions remain to be addressed.
Host restriction factors associated with NC?
The cellular prion protein. Although the disease related prion protein has a central role in the transmissible spongiform encephalopathies (TSEs), the biological function of the normal cellular prion protein, PrP, remains enigmatic. Recent evidence suggests that PrP is likely to play a role as a cellular RNA chaperone similar to p50 or FMRP that are ubiquitous chaperones.41,126 In fact, PrP is as active as HIV NC in binding the genomic RNA and providing assistance to the RT enzyme during viral DNA synthesis.41,126 Interestingly, co-expression of PrP and HIV-1 in human cells has a large negative impact on virion production,126 probably through interactions of PrP with the genomic RNA and the NC domain of Gag.59 More generally, in vivo expression of an endogenous murine leukaemia virus causes overexpression of PrP, which in turn has a negative impact on MLV production.127
The human Discs Large (hDlg1) is a cytosolic protein, recruited beneath the plasma membrane allowing the assembly of multiprotein complexes, capable of negatively modulating HIV-1 infectivity.128 Although it does not have an effect on viral production, the human hDlg1 protein was found to interact with the NC domain of Gag and to have a role in Gag subcellular distribution since both proteins accumulate at the plasma membrane where they partially colocalize with Gag in HIV-1 infected T cells.128
These findings favor the notion that the cellular prion protein and the human Dlg1 could interact with the NC domain of Gag during assembly and thus would act as anti-viral restriction factors targeting NC.
Other components interacting with Gag-RNA complexes and implication for virus assembly and release.
Other viral components are required for the production of infectious viral particles. For their incorporation, the Env glycoproteins need to be at the assembly site of Gag/GagPol with the genomic RNA (reviewed in ref. 19). Actually, the Env glycoproteins segregate to lipid rafts in order to associate with HIV-1 Gag129 while Gag and the gRNA nucleate early after synthesis, thanks to the NC domain, to be recruited to the assembly sites.46
In the case of HIV-1, the regulatory proteins Vpu and Vif are necessary for optimal particle production and infectivity. The viral protein Vpr is also part of the Gag-gRNA complex: its incorporation into the viral particle is mediated by the C-terminus of Gag and the gRNA (reviewed in ref. 19).
The viral infectivity factor Vif contributes to virion morphogenesis and core stability, as well as reverse transcription.130 Recent studies revealed that Vif counteracts the human restriction factors APOBEC-3G (hA3G) and APOBEC-3F (hA3F) that have strong anti-HIV-1 activity. APOBEC3G is incorporated in HIV-1 virions131 in an NC dependent manner.132 Actually, Vif is part of an RNA nucleoprotein complex with Gag, the gRNA and the ABCE1-Gag complexes present at the assembly site.133–135 Thus it was suggested that the unresolved role of Vif in virus assembly could be attributed to hA3G and hA3F partially exerting their antiviral activity by destabilizing the viral core and the reverse transcription complex, possibly by interfering with the assembly and/or maturation of the viral particles.130 Vif could then counteract hA3G and hA3F by excluding them from the viral assembly intermediates through competition for the viral genomic RNA, by regulating the proteolytic processing of Gag, and by enhancing the efficiency of the reverse transcription reaction.
Finally, the viral protein Vpu dramatically enhances virus production by counteracting the host factor tetherin at a post-assembly step of the HIV-1 replication cycle, which prevents fully assembled virion from being released from the plasma membrane.136–138
Other cell factors might be involved in assisting viral assembly, budding or release such as the calcium and proteins associated with calcium metabolism.107,139,140
Concluding Remarks
HIV-1 NC has received much attention over the past 20 years and a large number of studies have clearly shown that this small viral protein is much more than a simple NA binding protein. In fact HIV NC has the ability to coat and protect the NA, at the same time driving the condensation of the nucleoprotein complexes causing a molecular crowding effect. In such highly compact nucleoprotein complexes, the dynamic interplay of NC, most probably as oligomers, with the nucleic acids causes formation of the most stable structures, including hybridization of two distinct NA molecules with complementary sequences such as the HIV TAR and cTAR at the early step of reverse transcription (reviewed in ref. 4 and 60). Such NC properties whereby NC provides assistance to the folding of NA molecules have been named chaperoning that largely rely on the ZnF hydrophobic plateau and the flanking basic residues (Fig. 3). The mechanism of NC-mediated assistance to NA folding has been proposed to be by an entropy exchange,48 but this is still a matter of debate. Other avenues of research on HIV-1 NC should include how NC oligomers are structured along a NA template, since only the structure of a single NC molecule bound to a very small NA molecule is known (reviewed in ref. 16).
The relationships between the NC chaperoning properties and its role in viral DNA synthesis by RT appear rather straightforward, notably in the two obligatory strand transfers to generate the LTRs. However, the extent to which NC chaperoning activities are involved in virus assembly are far less clear. In fact, NC is part of the Gag precursor, which, in the form of GagΔp6, appears to tightly and stably bind the genomic RNA, preferably the Psi packaging sequences, and exhibits chaperoning activity in vitro and in assembling virions.16 NC preference for the viral packaging sequences may account for the genomic RNA selection in the assembly process and the particle assembly itself, but when and where Gag-NC operates is still a matter of debate. Selection could take place soon after Gag translation of the genome length RNA by the ribosome machinery, which would then occur on translating polysomes in a cis-dependent manner and could involve a folding switch of the genomic leader region encompassing the packaging sequences and the internal ribosome entry segment.141,142 Once Gag-NC-genomic RNA complexes start to be formed, when and how are they targeted to the site of virus assembly (at the plasma membrane or other cellular membranes) is poorly understood. The recent advances in the visualization of Gag trafficking and virus assembly in live cells46,143–148 should allow in the future the discovery of the sequential events that guide particle assembly. The challenge will be to define which cellular factors act during the pathway from Gag synthesis to particle assembly, and where and how they act in this pathway. Although there is no doubt that NC's hydrophobic plateau contributes to Gag trafficking and assembly, many open questions remain regarding the mechanism by which NC directs gRNA selection, RNA remodelling and dimerization. In addition, NC folding assistance could be modulated by cellular factors such as Staufen1, ABCE1 and the Bro1 domain of ALIX. All these issues represent novel exciting avenues of research on the multiple roles of HIV-1 NC in virus replication.
Acknowledgements
Thanks are due to INSERM, ANRS, CNRS, ENS LYON and Sidaction for their continuous support.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/14065
References
- 1.Baltimore D. RNA-dependent DNA polymerase in virions of RNA tumour viruses. Nature. 1970;226:1209–1211. doi: 10.1038/2261209a0. [DOI] [PubMed] [Google Scholar]
- 2.Gilboa E, Mitra SW, Goff S, Baltimore D. A detailed model of reverse transcription and tests of crucial aspects. Cell. 1979;18:93–100. doi: 10.1016/0092-8674(79)90357-x. [DOI] [PubMed] [Google Scholar]
- 3.Coffin JM. In: Virology. Fields BN, Knipe DM, et al., editors. Vol. 2. New York: Raven Press Ltd.; 1990. pp. 1437–500. [Google Scholar]
- 4.Mougel M, Houzet L, Darlix JL. When is it time for reverse transcription to start and go? Retrovirology. 2009;6:24. doi: 10.1186/1742-4690-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Temin HM, Mizutani S. RNA-dependent DNA polymerase in virions of Rous sarcoma virus. Nature. 1970;226:1211–1213. doi: 10.1038/2261211a0. [DOI] [PubMed] [Google Scholar]
- 6.Mizutani S, Boettiger D, Temin HM. A DNAdependent DNA polymerase and a DNA endonuclease in virions of Rous sarcoma virus. Nature. 1970;228:424–427. doi: 10.1038/228424a0. [DOI] [PubMed] [Google Scholar]
- 7.Delelis O, Carayon K, Saib A, Deprez E, Mouscadet JF. Integrase and integration: biochemical activities of HIV-1 integrase. Retrovirology. 2008;5:114. doi: 10.1186/1742-4690-5-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lewinski MK, Bushman FD. Retroviral DNA integration—mechanism and consequences. Adv Genet. 2005;55:147–181. doi: 10.1016/S0065-2660(05)55005-3. [DOI] [PubMed] [Google Scholar]
- 9.Coffin J, Hughes S, Varmus H. Retroviruses. Cold Spring Harbor, New York: Cold Spring Harbor laboratory Press; 1997. [PubMed] [Google Scholar]
- 10.Ganser-Pornillos BK, Yeager M, Sundquist WI. The structural biology of HIV assembly. Curr Opin Struct Biol. 2008;18:203–217. doi: 10.1016/j.sbi.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mateu MG. The capsid protein of human immunodeficiency virus: intersubunit interactions during virus assembly. FEBS J. 2009;276:6098–6109. doi: 10.1111/j.1742-4658.2009.07313.x. [DOI] [PubMed] [Google Scholar]
- 12.Pornillos O, Ganser-Pornillos BK, Kelly BN, Hua Y, Whitby FG, Stout CD, et al. X-ray structures of the hexameric building block of the HIV capsid. Cell. 2009;137:1282–1292. doi: 10.1016/j.cell.2009.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vogt VM, Simon MN. Mass determination of rous sarcoma virus virions by scanning transmission electron microscopy. J Virol. 1999;73:7050–7055. doi: 10.1128/jvi.73.8.7050-7055.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen J, Nikolaitchik O, Singh J, Wright A, Bencsics CE, Coffin JM, et al. High efficiency of HIV-1 genomic RNA packaging and heterozygote formation revealed by single virion analysis. Proc Natl Acad Sci USA. 2009;106:13535–13540. doi: 10.1073/pnas.0906822106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chertova E, Chertov O, Coren LV, Roser JD, Trubey CM, Bess JWJ, et al. Proteomic and biochemical analysis of purified human immunodeficiency virus type 1 produced from infected monocyte-derived macrophages. J Virol. 2006;80:9039–9052. doi: 10.1128/JVI.01013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Darlix JL, Garrido JL, Morellet N, Mély Y, de Rocquigny H. Properties, functions and drug targeting of the multifunctional nucleocapsid protein of the human immunodeficiency virus. Adv Pharmacol. 2007;55:299–346. doi: 10.1016/S1054-3589(07)55009-X. [DOI] [PubMed] [Google Scholar]
- 17.Darlix JL, Lapadat-Tapolsky M, de Rocquigny H, Roques BP. First glimpses at structure-function relationships of the nucleocapsid protein of retroviruses. J Mol Biol. 1995;254:523–537. doi: 10.1006/jmbi.1995.0635. [DOI] [PubMed] [Google Scholar]
- 18.Rein A, Henderson LE, Levin JG. Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: significance for viral replication. Trends Biochem Sci. 1998;23:297–301. doi: 10.1016/s0968-0004(98)01256-0. [DOI] [PubMed] [Google Scholar]
- 19.Cimarelli A, Darlix JL. Assembling the human immunodeficiency virus type 1. Cell Mol Life Sci. 2002;59:1166–1184. doi: 10.1007/s00018-002-8495-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muriaux D, Darlix JL, Cimarelli A. Targeting the assembly of the human immunodeficiency virus type 1. Curr Pharm Des. 2004;10:3725–3739. doi: 10.2174/1381612043382701. [DOI] [PubMed] [Google Scholar]
- 21.Morellet N, de Rocquigny H, Mely Y, Jullian N, Déméné H, Ottmann M, et al. Conformational behaviour of the active and inactive forms of the nucleocapsid NCp7 of HIV-1 studied by 1H NMR. J Mol Biol. 1994;235:287–301. doi: 10.1016/s0022-2836(05)80033-6. [DOI] [PubMed] [Google Scholar]
- 22.Morellet N, Jullian N, De Rocquigny H, Maigret B, Darlix JL, Roques BP. Determination of the structure of the nucleocapsid protein NCp7 from the human immunodeficiency virus type 1 by 1H NMR. EMBO J. 1992;11:3059–3065. doi: 10.1002/j.1460-2075.1992.tb05377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Summers MF, Henderson LE, Chance MR, Bess JWJ, South TL, Blake PR, et al. Nucleocapsid zinc fingers detected in retroviruses: EXAFS studies of intact viruses and the solution-state structure of the nucleocapsid protein from HIV-1. Protein Sci. 1992;1:563–574. doi: 10.1002/pro.5560010502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee BM, De Guzman RN, Turner BG, Tjandra N, Summers MF. Dynamical behavior of the HIV-1 nucleocapsid protein. J Mol Biol. 1998;279:633–649. doi: 10.1006/jmbi.1998.1766. [DOI] [PubMed] [Google Scholar]
- 25.Mely Y, Jullian N, Morellet N, et al. Spatial proximity of the HIV-1 nucleocapsid protein zinc fingers investigated by time-resolved fluorescence and fluorescence resonance energy transfer. Biochemistry. 1994;33:12085–12091. doi: 10.1021/bi00206a011. [DOI] [PubMed] [Google Scholar]
- 26.Ramboarina S, Srividya N, Atkinson RA, et al. Effects of temperature on the dynamic behaviour of the HIV-1 nucleocapsid NCp7 and its DNA complex. J Mol Biol. 2002;316:611–627. doi: 10.1006/jmbi.2001.5379. [DOI] [PubMed] [Google Scholar]
- 27.Bourbigot S, Ramalanjaona N, Boudier C, et al. How the HIV-1 nucleocapsid protein binds and destabilises the (-) primer binding site during reverse transcription. J Mol Biol. 2008;383:1112–1128. doi: 10.1016/j.jmb.2008.08.046. [DOI] [PubMed] [Google Scholar]
- 28.Spriggs S, Garyu L, Connor R, Summers MF. Potential intra- and intermolecular interactions involving the unique-5′ region of the HIV-1 5′-UTR. Biochemistry. 2008;47:13064–13073. doi: 10.1021/bi8014373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amarasinghe GK, De Guzman RN, Turner RB, et al. NMR structure of the HIV-1 nucleocapsid protein bound to stem-loop SL2 of the psi-RNA packaging signal. Implications for genome recognition. J Mol Biol. 2000;301:491–511. doi: 10.1006/jmbi.2000.3979. [DOI] [PubMed] [Google Scholar]
- 30.De Guzman RN, Wu ZR, Stalling CC, et al. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science. 1998;279:384–388. doi: 10.1126/science.279.5349.384. [DOI] [PubMed] [Google Scholar]
- 31.Morellet N, Demene H, Teilleux V, et al. Structure of the complex between the HIV-1 nucleocapsid protein NCp7 and the single-stranded pentanucleotide d(ACG CC) J Mol Biol. 1998;283:419–434. doi: 10.1006/jmbi.1998.2098. [DOI] [PubMed] [Google Scholar]
- 32.Bombarda E, Ababou A, Vuilleumier C, et al. Time-resolved fluorescence investigation of the human immunodeficiency virus type 1 nucleocapsid protein: influence of the binding of nucleic acids. Biophys J. 1999;76:1561–1570. doi: 10.1016/S0006-3495(99)77315-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aldovini A, Young RA. Mutations of RNA and protein sequences involved in human immunodeficiency virus type 1 packaging result in production of noninfectious virus. J Virol. 1990;64:1920–1926. doi: 10.1128/jvi.64.5.1920-1926.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Demene H, Dong CZ, Ottmann M, et al. 1H NMR structure and biological studies of the His23→Cys mutant nucleocapsid protein of HIV-1 indicate that the conformation of the first zinc finger is critical for virus infectivity. Biochemistry. 1994;33:11707–11716. doi: 10.1021/bi00205a006. [DOI] [PubMed] [Google Scholar]
- 35.Dorfman T, Luban J, Goff SP, Haseltine WA, Gottlinger HG. Mapping of functionally important residues of a cysteine-histidine box in the human immunodeficiency virus type 1 nucleocapsid protein. J Virol. 1993;67:6159–6169. doi: 10.1128/jvi.67.10.6159-6169.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gorelick RJ, Nigida SM, Jr, Bess JW, Jr, et al. Noninfectious human immunodeficiency virus type 1 mutants deficient in genomic RNA. J Virol. 1990;64:3207–3211. doi: 10.1128/jvi.64.7.3207-3211.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ottmann M, Gabus C, Darlix JL. The central globular domain of the nucleocapsid protein of human immunodeficiency virus type 1 is critical for virion structure and infectivity. J Virol. 1995;69:1778–1784. doi: 10.1128/jvi.69.3.1778-1784.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gorelick RJ, Gagliardi TD, Bosche WJ, et al. Strict conservation of the retroviral nucleocapsid protein zinc finger is strongly influenced by its role in viral infection processes: characterization of HIV-1 particles containing mutant nucleocapsid zinc-coordinating sequences. Virology. 1999;256:92–104. doi: 10.1006/viro.1999.9629. [DOI] [PubMed] [Google Scholar]
- 39.Tanchou V, Decimo D, Pechoux C, et al. Role of the N-terminal zinc finger of human immunodeficiency virus type 1 nucleocapsid protein in virus structure and replication. J Virol. 1998;72:4442–4447. doi: 10.1128/jvi.72.5.4442-4447.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thomas JA, Gorelick RJ. Nucleocapsid protein function in early infection processes. Virus Res. 2008;134:39–63. doi: 10.1016/j.virusres.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gabus C, Auxilien S, Péchoux C, Dormont D, Swietnicki W, Morillas M, et al. The prion protein has DNA strand transfer properties similar to retroviral nucleocapsid protein. J Mol Biol. 2001;307:1011–1021. doi: 10.1006/jmbi.2001.4544. [DOI] [PubMed] [Google Scholar]
- 42.Krishnamoorthy G, Roques B, Darlix JL, Mely Y. DNA condensation by the nucleocapsid protein of HIV-1: a mechanism ensuring DNA protection. Nucleic Acids Res. 2003;31:5425–5432. doi: 10.1093/nar/gkg738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lapadat-Tapolsky M, De Rocquigny H, Van Gent D, et al. Interactions between HIV-1 nucleocapsid protein and viral DNA may have important functions in the viral life cycle. Nucleic Acids Res. 1993;21:831–839. doi: 10.1093/nar/21.4.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muriaux D, Mirro J, Harvin D, Rein A. RNA is a structural element in retrovirus particles. Proc Natl Acad Sci USA. 2001;98:5246–5251. doi: 10.1073/pnas.091000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mark-Danieli M, Laham N, Kenan-Eichler M, Castiel A, Melamed D, Landau M, et al. Single point mutations in the zinc finger motifs of the human immunodeficiency virus type 1 nucleocapsid alter RNA binding specificities of the gag protein and enhance packaging and infectivity. J Virol. 2005;79:7756–7767. doi: 10.1128/JVI.79.12.7756-7767.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jouvenet N, Simon SM, Bieniasz PD. Imaging the interaction of HIV-1 genomes and Gag during assembly of individual viral particles. Proc Natl Acad Sci USA. 2009;106:19114–19119. doi: 10.1073/pnas.0907364106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ivanyi-Nagy R, Davidovic L, Khandjian EW, Darlix JL. Disordered RNA chaperone proteins: from functions to diseases. Cell Mol Life Sci. 2005;62:1409–1417. doi: 10.1007/s00018-005-5100-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tompa P, Csermely P. The role of structural disorder in the function of RNA and protein chaperones. FASEB J. 2004;18:1169–1175. doi: 10.1096/fj.04-1584rev. [DOI] [PubMed] [Google Scholar]
- 49.Levin JG, Guo J, Rouzina I, Musier-Forsyth K. Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: critical role in reverse transcription and molecular mechanism. Prog Nucleic Acid Res Mol Biol. 2005;80:217–286. doi: 10.1016/S0079-6603(05)80006-6. [DOI] [PubMed] [Google Scholar]
- 50.Cristofari G, Darlix JL. The ubiquitous nature of RNA chaperone proteins. Prog Nucleic Acid Res Mol Biol. 2002;72:223–268. doi: 10.1016/s0079-6603(02)72071-0. [DOI] [PubMed] [Google Scholar]
- 51.Cruceanu M, Urbaneja MA, Hixson CV, Johnson DG, Datta SA, Fivash MJ, et al. Nucleic acid binding and chaperone properties of HIV-1 Gag and nucleocapsid proteins. Nucleic Acids Res. 2006;34:593–605. doi: 10.1093/nar/gkj458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Williams MC, Rouzina I, Wenner JR, Gorelick RJ, Musier-Forsyth K, Bloomfield VA. Mechanism for nucleic acid chaperone activity of HIV-1 nucleocapsid protein revealed by single molecule stretching. Proc Natl Acad Sci USA. 2001;98:6121–6126. doi: 10.1073/pnas.101033198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cruceanu M, Stephen AG, Beuning PJ, Gorelick RJ, Fisher RJ, Williams MC. Single DNA molecule stretching measures the activity of chemicals that target the HIV-1 nucleocapsid protein. Anal Biochem. 2006;358:159–170. doi: 10.1016/j.ab.2006.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Godet J, de Rocquigny H, Raja C, Glasser N, Ficheux D, Darlix JL, Mély Y. During the early phase of HIV-1 DNA synthesis, nucleocapsid protein directs hybridization of the TAR complementary sequences via the ends of their double-stranded stem. J Mol Biol. 2006;356:1180–1192. doi: 10.1016/j.jmb.2005.12.038. [DOI] [PubMed] [Google Scholar]
- 55.Ramalanjaona N, de Rocquigny H, Millet A, Ficheux D, Darlix JL, Mély Y. Investigating the mechanism of the nucleocapsid protein chaperoning of the second strand transfer during HIV-1 DNA synthesis. J Mol Biol. 2007;374:1041–1053. doi: 10.1016/j.jmb.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 56.Beltz H, Azoulay J, Bernacchi S, Clamme JP, Ficheux D, Roques B, et al. Impact of the terminal bulges of HIV-1 cTAR DNA on its stability and the destabilizing activity of the nucleocapsid protein NCp7. J Mol Biol. 2003;328:95–108. doi: 10.1016/s0022-2836(03)00244-4. [DOI] [PubMed] [Google Scholar]
- 57.de Rocquigny H, Shvadchak V, Avilov S, Dong CZ, Dietrich U, Darlix JL, Mély Y. Targeting the viral nucleocapsid protein in anti-HIV-1 therapy. Mini Rev Med Chem. 2008;8:24–35. doi: 10.2174/138955708783331603. [DOI] [PubMed] [Google Scholar]
- 58.Ivanyi-Nagy R, Darlix JL. Intrinsic Disorder In The Core Proteins Of Flaviviruses. Protein Pept Lett. 2010 May 10; doi: 10.2174/092986610791498911. [DOI] [PubMed] [Google Scholar]
- 59.Gabus C, Derrington E, Leblanc P, Chnaiderman J, Dormont D, Swietnicki W, et al. The prion protein has RNA binding and chaperoning properties characteristic of nucleocapsid protein NCP7 of HIV-1. J Biol Chem. 2001;276:19301–19309. doi: 10.1074/jbc.M009754200. [DOI] [PubMed] [Google Scholar]
- 60.Marylène Mougel, Andrea Cimarelli, Jean-Luc Darlix. Implications of the Nucleocapsid and the Microenvironment in Retroviral Reverse Transcription. Viruses. 2010;2:939–960. doi: 10.3390/v2040939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Demirov DG, Freed EO. Retrovirus budding. Virus Res. 2004;106:87–102. doi: 10.1016/j.virusres.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 62.Klein KC, Reed JC, Lingappa JR. Intracellular destinies: degradation, targeting, assembly and endocytosis of HIV Gag. AIDS Rev. 2007;9:150–161. [PubMed] [Google Scholar]
- 63.Chu H, Wang JJ, Spearman P. Human immunodeficiency virus type-1 Gag and host vesicular trafficking pathways. Curr Top Microbiol Immunol. 2009;339:67–84. doi: 10.1007/978-3-642-02175-6_4. [DOI] [PubMed] [Google Scholar]
- 64.Bieniasz PD. The cell biology of HIV-1 virion genesis. Cell Host Microbe. 2009;5:550–558. doi: 10.1016/j.chom.2009.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ono A. HIV-1 Assembly at the Plasma Membrane: Gag Trafficking and Localization. Future Virol. 2009;4:241–257. doi: 10.2217/fvl.09.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ganser-Pornillos BK, Yeager M, Sundquist WI. The structural biology of HIV assembly. Curr Opin Struct Biol. 2008;18:203–217. doi: 10.1016/j.sbi.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Briggs JA, Riches JD, Glass B, Bartonova V, Zanetti G, Kräusslich HG. Structure and assembly of immature HIV. Proc Natl Acad Sci USA. 2009;106:11090–11095. doi: 10.1073/pnas.0903535106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Morita E, Sundquist WI. Retrovirus budding. Annu Rev Cell Dev Biol. 2004;20:395–425. doi: 10.1146/annurev.cellbio.20.010403.102350. [DOI] [PubMed] [Google Scholar]
- 69.Usami Y, Popov S, Popova E, Inoue M, Weissenhorn W, Göttlinger HG. The ESCRT pathway and HIV-1 budding. Biochem Soc Trans. 2009;37:181–184. doi: 10.1042/BST0370181. [DOI] [PubMed] [Google Scholar]
- 70.Yuan B, Campbell S, Bacharach E, Rein A, Goff SP. Infectivity of Moloney murine leukemia virus defective in late assembly events is restored by late assembly domains of other retroviruses. J Virol. 2000;74:7250–7260. doi: 10.1128/jvi.74.16.7250-7260.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yuan B, Fassati A, Yueh A, Goff SP. Characterization of Moloney murine leukemia virus p12 mutants blocked during early events of infection. J Virol. 2002;76:10801–10810. doi: 10.1128/JVI.76.21.10801-10810.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Scheifele LZ, Kenney SP, Cairns TM, Craven RC, Parent LJ. Overlapping roles of the Rous sarcoma virus Gag p10 domain in nuclear export and virion core morphology. J Virol. 2007;81:10718–10728. doi: 10.1128/JVI.01061-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Phillips JM, Murray PS, Murray D, Vogt VM. A molecular switch required for retrovirus assembly participates in the hexagonal immature lattice. EMBO J. 2008;27:1411–1420. doi: 10.1038/emboj.2008.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Burniston MT, Cimarelli A, Colgan J, Curtis SP, Luban J. Human immunodeficiency virus type 1 Gag polyprotein multimerization requires the nucleocapsid domain and RNA and is promoted by the capsid-dimer interface and the basic region of matrix protein. J Virol. 1999;73:8527–8540. doi: 10.1128/jvi.73.10.8527-8540.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cimarelli A, Sandin S, Höglund S, Luban J. Basic residues in human immunodeficiency virus type 1 nucleocapsid promote virion assembly via interaction with RNA. J Virol. 2000;74:3046–3057. doi: 10.1128/jvi.74.7.3046-3057.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Muriaux D, Costes KS, Nagashima J, Mirro E, Cho S, Lockett SJ, Rein A. Role of murine leukemia virus nucleocapsid protein in virus assembly. J Virol. 2004;78:12378–12385. doi: 10.1128/JVI.78.22.12378-12385.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lee EG, Linial ML. Basic residues of the retroviral nucleocapsid play different roles in gag-gag and Gag-Psi RNA interactions. J Virol. 2004;78:8486–8495. doi: 10.1128/JVI.78.16.8486-8495.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bohmovα K, Hadravovα R, Stokrovα J, Tuma R, Ruml T, Pichovα I, Rumlovα M. Effect of dimerizing domains and basic residues on in vitro and in vivo assembly of Mason-Pfizer monkey virus and human immunodeficiency virus. J Virol. 2010;84:1977–1988. doi: 10.1128/JVI.02022-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang SW, Aldovini A. RNA incorporation is critical for retroviral particle integrity after cell membrane assembly of Gag complexes. J Virol. 2002;76:11853–11865. doi: 10.1128/JVI.76.23.11853-11865.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang SW, Noonan K, Aldovini A. Nucleocapsid-RNA interactions are essential to structural stability but not to assembly of retroviruses. J Virol. 2004;78:716–723. doi: 10.1128/JVI.78.2.716-723.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yovandich JL, Chertova EN, Kane BP, Gagliardi TD, Bess JW, Jr, Sowder RC, 2nd, et al. Alteration of zinc-binding residues of simian immunodeficiency virus p8(NC) results in subtle differences in gag processing and virion maturation associated with degradative loss of mutant NC. J Virol. 2001;75:115–124. doi: 10.1128/JVI.75.1.115-124.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lee EG, Linial ML. Deletion of a Cys-His motif from the Alpharetrovirus nucleocapsid domain reveals late domain mutant-like budding defects. Virology. 2006;347:226–233. doi: 10.1016/j.virol.2005.11.048. [DOI] [PubMed] [Google Scholar]
- 83.Grigorov B, Décimo D, Smagulova F, Mougel MD, Muriaux JL, Darlix JL. Intracellular localization of Gag is impaired by mutations in the nucleocapsid zinc fingers. Retrovirology. 2007;4:54. doi: 10.1186/1742-4690-4-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hogue IB, Hoppe A, Ono A. Quantitative fluorescence resonance energy transfer microscopy analysis of the human immunodeficiency virus type 1 Gag-Gag interaction: relative contributions of the CA and NC domains and membrane binding. J Virol. 2009;83:7322–7336. doi: 10.1128/JVI.02545-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ott DE, Coren LV, Shatzer T. The nucleocapsid region of human immunodeficiency virus type 1 Gag assists in the coordination of assembly and Gag processing: role for RNA-Gag binding in the early stages of assembly. J Virol. 2009;83:7718–7727. doi: 10.1128/JVI.00099-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ono A, Waheed AA, Joshi A, Freed EO. Association of human immunodeficiency virus type 1 gag with membrane does not require highly basic sequences in the nucleocapsid: use of a novel Gag multimerization assay. J Virol. 2005;79:14131–14140. doi: 10.1128/JVI.79.22.14131-14140.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Popov S, Popova E, Inoue M, Göttlinger HG. Divergent Bro1 domains share the capacity to bind human immunodeficiency virus type 1 nucleocapsid and to enhance virus-like particle production. J Virol. 2009;83:7185–7193. doi: 10.1128/JVI.00198-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dussupt V, Javid MP, Abou-Jaoudé G, Jadwin JA, de La Cruz J, Nagashima K, Bouamr F. The nucleocapsid region of HIV-1 Gag cooperates with the PTAP and LYPXnL late domains to recruit the cellular machinery necessary for viral budding. PLoS Pathog. 2009;5:1000339. doi: 10.1371/journal.ppat.1000339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Popova E, Popov S, Göttlinger HG. Human immunodeficiency virus type 1 nucleocapsid p1 confers ESCRT pathway dependence. J Virol. 2010;84:6590–6597. doi: 10.1128/JVI.00035-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Resh MD. Intracellular trafficking of HIV-1 Gag: how Gag interacts with cell membranes and makes viral particles. AIDS Rev. 2005;7:84–91. [PubMed] [Google Scholar]
- 91.Saad JS, Miller J, Tai J, Kim A, Ghanam RH, Summers MF. Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly. Proc Natl Acad Sci USA. 2006;103:11364–11369. doi: 10.1073/pnas.0602818103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Alfadhli A, Dhenub TC, Still A, Barklis E. Analysis of human immunodeficiency virus type 1 Gag dimerization-induced assembly. J Virol. 2005;79:14498–14506. doi: 10.1128/JVI.79.23.14498-14506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ott DE, Coren LV, Gagliardi TD. Redundant roles for nucleocapsid and matrix RNA-binding sequences in human immunodeficiency virus type 1 assembly. J Virol. 2005;79:13839–13847. doi: 10.1128/JVI.79.22.13839-13847.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sherer NM, Swanson CM, Papaioannou S, Malim MH. Matrix mediates the functional link between human immunodeficiency virus type 1 RNA nuclear export elements and the assembly competency of Gag in murine cells. J Virol. 2009;83:8525–8535. doi: 10.1128/JVI.00699-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jin J, Sturgeon T, Weisz OA, Mothes W, Montelaro RC. HIV-1 matrix dependent membrane targeting is regulated by Gag mRNA trafficking. PLoS One. 2009;4:655196. doi: 10.1371/journal.pone.0006551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Muriaux D, Mirro J, Nagashima K, Harvin D, Rein A. Murine leukemia virus nucleocapsid mutant particles lacking viral RNA encapsidate ribosomes. J Virol. 2002;76:11405–11413. doi: 10.1128/JVI.76.22.11405-11413.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Poon DT, Chertova EN, Ott DE. Human immunodeficiency virus type 1 preferentially encapsidates genomic RNAs that encode Pr55(Gag): functional linkage between translation and RNA packaging. Virology. 2002;293:368–378. doi: 10.1006/viro.2001.1283. [DOI] [PubMed] [Google Scholar]
- 98.Balvay L, Lopez Lastra M, Sargueil B, Darlix JL, Ohlmann T. Translational control of retroviruses. Nat Rev Microbiol. 2007;5:128–140. doi: 10.1038/nrmicro1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ono A, Ablan SD, Lockett SJ, Nagashima K, Freed EO. Phosphatidylinositol(4,5)bisphosphate regulates HIV-1 Gag targeting to the plasma membrane. Proc Natl Acad Sci USA. 2004;101:14889–14894. doi: 10.1073/pnas.0405596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Freed EO. HIV-1 Gag: flipped out for PI(4,5)P. Proc Natl Acad Sci USA. 2006;103:11101–11102. doi: 10.1073/pnas.0604715103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hamard-Péron E, Juillard F, Saad JS, Roy C, Roingeard P, Summers MF, et al. Targeting of MuLV Gag to the plasma membrane is mediated by PIP2/PS and a poly-basic region in the Matrix. J Virol. 2010;84:503–515. doi: 10.1128/JVI.01134-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chukkapalli V, Oh SJ, Ono A. Opposing mechanisms involving RNA and lipids regulate HIV-1 Gag membrane binding through the highly basic region of the matrix domain. Proc Natl Acad Sci USA. 2010;107:1600–1605. doi: 10.1073/pnas.0908661107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Camus G, Segura-Morales C, Molle D, Lopez-Vergès S, Begon-Pescia C, Cazevieille C, et al. The clathrin adaptor complex AP-1 binds HIV-1 and MLV Gag and facilitates their budding. Mol Biol Cell. 2007;18:3193–3203. doi: 10.1091/mbc.E06-12-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Batonick M, Favre M, Boge M, Spearman P, Höning S, Thali M. Interaction of HIV-1 Gag with the clathrin-associated adaptor AP-2. Virology. 2005;342:190–200. doi: 10.1016/j.virol.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 105.Dong X, Li H, Derdowski A, Ding L, Burnett A, Chen X, et al. AP-3 directs the intracellular trafficking of HIV-1 Gag and plays a key role in particle assembly. Cell. 2005;120:663–674. doi: 10.1016/j.cell.2004.12.023. [DOI] [PubMed] [Google Scholar]
- 106.Joshi A, Garg H, Nagashima K, Bonifacino JS, Freed EO. GGA and Arf proteins modulate retrovirus assembly and release. Mol Cell. 2008;30:227–238. doi: 10.1016/j.molcel.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Grigorov B, Arcanger F, Roingeard P, Darlix JL, Muriaux D. Assembly of infectious HIV-1 in human epithelial and lymphoblastic cell lines. J Mol Biol. 2006;359:848–862. doi: 10.1016/j.jmb.2006.04.017. [DOI] [PubMed] [Google Scholar]
- 108.Molle D, Segura-Morales C, Camus G, Berlioz-Torrent C, Kjems J, Basyuk E, Bertrand E. Endosomal trafficking of HIV-1 gag and genomic RNAs regulates viral egress. J Biol Chem. 2009;284:19727–19743. doi: 10.1074/jbc.M109.019844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lehmann M, Milev MP, Abrahamyan L, Yao XJ, Pante N, Mouland AJ. Intracellular transport of human immunodeficiency virus type 1 genomic RNA and viral production are dependent on dynein motor function and late endosome positioning. J Biol Chem. 2009;284:14572–14585. doi: 10.1074/jbc.M808531200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tritel M, Resh M. Kinetic analysis of HIV-1 assembly reveals the presence of sequential intermediates. J Virol. 2000;74:5845–5855. doi: 10.1128/jvi.74.13.5845-5855.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lingappa JR, Thielen BK. Assembly of immature HIV-1 capsids using a cell-free system. Methods Mol Biol. 2009;485:185–195. doi: 10.1007/978-1-59745-170-3_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cochrane AW, McNally MT, Mouland AJ. The retrovirus RNA trafficking granule: from birth to maturity. Retrovirology. 2006;3:18. doi: 10.1186/1742-4690-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mouland AJ, Mercier J, Luo M, Bernier L, DesGroseillers L, Cohen EA. The double-stranded RNA-binding protein Staufen is incorporated in human immunodeficiency virus type 1: evidence for a role in genomic RNA encapsidation. J Virol. 2000;74:5441–5451. doi: 10.1128/jvi.74.12.5441-5451.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chatel-Chaix L, Clément JF, Martel C, Bériault V, Gatignol A, DesGroseillers L, Mouland AJ. Identification of Staufen in the human immunodeficiency virus type 1 Gag ribonucleoprotein complex and a role in generating infectious viral particles. Mol Cell Biol. 2004;24:2637–2648. doi: 10.1128/MCB.24.7.2637-2648.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chatel-Chaix L, Boulay K, Mouland AJ, Desgroseillers L. The host protein Staufen1 interacts with the Pr55Gag zinc fingers and regulates HIV-1 assembly via its N-terminus. Retrovirology. 2008;5:41. doi: 10.1186/1742-4690-5-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ueno T, Tokunaga K, Sawa H, Maeda M, Chiba J, Kojima A, et al. Nucleolin and the packaging signal, psi, promote the budding of human immunodeficiency virus type-1 (HIV-1) Microbiol Immunol. 2004;48:111–118. doi: 10.1111/j.1348-0421.2004.tb03496.x. [DOI] [PubMed] [Google Scholar]
- 117.Bacharach E, Gonsky J, Alin K, Orlova M, Goff SP. The carboxy-terminal fragment of nucleolin interacts with the nucleocapsid domain of retroviral gag proteins and inhibits virion assembly. J Virol. 2000;74:11027–11039. doi: 10.1128/jvi.74.23.11027-11039.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zimmerman C, Klein KC, Kiser PK, Singh AR, Firestein BL, Riba SC, Lingappa JR. Identification of a host protein essential for assembly of immature HIV-1 capsids. Nature. 2002;415:88–92. doi: 10.1038/415088a. [DOI] [PubMed] [Google Scholar]
- 119.Lingappa J, Dooher J, Newman M, Kiser P, Klein K. Basic residues in the nucleocapsid domain of Gag are required for interaction of HIV-1 gag with ABCE1 (HP68), a cellular protein important for HIV-1 capsid assembly. J Biol Chem. 2006;281:3773–3784. doi: 10.1074/jbc.M507255200. [DOI] [PubMed] [Google Scholar]
- 120.Dooher JE, Schneider BL, Reed JC, Lingappa JR. Host ABCE1 is at plasma membrane HIV assembly sites and its dissociation from Gag is linked to subsequent events of virus production. Traffic. 2007;8:195–211. doi: 10.1111/j.1600-0854.2006.00524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ott DE. Cellular proteins detected in HIV-1. Rev Med Virol. 2008;18:159–175. doi: 10.1002/rmv.570. [DOI] [PubMed] [Google Scholar]
- 122.Lehmann MJ, Sherer NM, Marks CB, Pypaert M, Mothes W. Actin- and myosin-driven movement of viruses along filopodia precedes their entry into cells. J Cell Biol. 2005;170:317–325. doi: 10.1083/jcb.200503059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chen C, Weisz OA, Stolz DB, Watkins SC, Montelaro RC. Differential effects of actin cytoskeleton dynamics on equine infectious anemia virus particle production. J Virol. 2004;78:882–891. doi: 10.1128/JVI.78.2.882-891.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chen C, Jin J, Rubin M, Huang L, Sturgeon T, Weixel KM, et al. Association of gag multimers with filamentous actin during equine infectious anemia virus assembly. Curr HIV Res. 2007;5:315–323. doi: 10.2174/157016207780636542. [DOI] [PubMed] [Google Scholar]
- 125.Gladnikoff M, Shimoni E, Gov NS, Rousso I. Retroviral assembly and budding occur through an actin-driven mechanism. Biophys J. 2009;97:2419–2428. doi: 10.1016/j.bpj.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Leblanc P, Baas D, Darlix JL. Analysis of the interactions between HIV-1 and the cellular prion protein in a human cell line. J Mol Biol. 2004;337:1035–1051. doi: 10.1016/j.jmb.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 127.Lötscher M, Recher M, Lang KS, Navarini A, Hunziker L, Santimaria R, et al. Induced prion protein controls immune-activated retroviruses in the mouse spleen. PLoS One. 2007;2:1158. doi: 10.1371/journal.pone.0001158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Perugi F, Muriaux D, Ramirez BC, Chabani S, Decroly E, Darlix JL, et al. Human Discs Large is a new negative regulator of HIV-1 infectivity. Mol Biol Cell. 2009;20:498–508. doi: 10.1091/mbc.E08-02-0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Leung K, Kim JO, Ganesh L, Kabat J, Schwartz O, Nabel GJ. HIV-1 assembly: viral glycoproteins segregate quantally to lipid rafts that associate individually with HIV-1 capsids and virions. Cell Host Microbe. 2008;3:285–292. doi: 10.1016/j.chom.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Henriet S, Mercenne G, Bernacchi S, Paillart JC, Marquet R. Tumultuous relationship between the human immunodeficiency virus type 1 viral infectivity factor (Vif) and the human APOBEC-3G and APOBEC-3F restriction factors. Microbiol Mol Biol Rev. 2009;73:211–232. doi: 10.1128/MMBR.00040-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zennou V, Perez-Caballero D, Gottlinger H, Bieniasz PD. APOBEC3G incorporation into human immunodeficiency virus type 1 particles. J Virol. 2004;78:12058–12061. doi: 10.1128/JVI.78.21.12058-12061.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Burnett A, Spearman P. APOBEC3G multimers are recruited to the plasma membrane for packaging into human immunodeficiency virus type 1 virus-like particles in an RNA-dependent process requiring the NC basic linker. J Virol. 2007;81:5000–5013. doi: 10.1128/JVI.02237-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Simon JH, Carpenter EA, Fouchier RA, Malim MH. Vif and the p55Gag polyprotein of human immunodeficiency virus type 1 are present in colocalizing membrane-free cytoplasmic complexes. J Virol. 1999;73:2667–2674. doi: 10.1128/jvi.73.4.2667-2674.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zhang H, Pomerantz RJ, Dornadula G, Sun Y. Human immunodeficiency virus type 1 Vif protein is an integral component of an mRNP complex of viral RNA and could be involved in the viral RNA folding and packaging process. J Virol. 2000;74:8252–8826. doi: 10.1128/jvi.74.18.8252-8261.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Henriet S, Richer D, Bernacchi S, Decroly E, Vigne R, Ehresmann B, et al. Cooperative and specific binding of Vif to the 5′ region of HIV-1 genomic RNA. J Mol Biol. 2005;354:55–72. doi: 10.1016/j.jmb.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 136.Neil SJ, Zang T, Bieniasz PD. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 2008;451:425–430. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- 137.Neil SJ, Eastman SW, Jouvenet N, Bieniasz PD. HIV-1 Vpu promotes release and prevents endocytosis of nascent retrovirus particles from the plasma membrane. PLoS Pathog. 2006;2:39. doi: 10.1371/journal.ppat.0020039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Perez-Caballero D, Zang T, Ebrahimi A, McNatt MW, Gregory DA, Johnson MC, Bieniasz PD. Tetherin inhibits HIV-1 release by directly tethering virions to cells. Cell. 2009;139:499–511. doi: 10.1016/j.cell.2009.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Perlman M, Resh MD. Identification of an intracellular trafficking and assembly pathway for HIV-1 Gag. Traffic. 2006;7:731–745. doi: 10.1111/j.1398-9219.2006.00428.x. [DOI] [PubMed] [Google Scholar]
- 140.Ehrlich LS, Medina GN, Khan MB, Powell MD, Mikoshiba K, Carter CA. Activation of the inositol (1,4,5)-triphosphate calcium gate receptor is required for HIV-1 Gag release. J Virol. 2010;84:6438–6451. doi: 10.1128/JVI.01588-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Abbink TE, Ooms M, Haasnoot PC, Berkhout B. The HIV-1 leader RNA conformational switch regulates RNA dimerization but does not regulate mRNA translation. Biochemistry. 2005;44:9058–9066. doi: 10.1021/bi0502588. [DOI] [PubMed] [Google Scholar]
- 142.Brasey A, Lopez-Lastra M, Ohlmann T, Beerens N, Berkhout B, Darlix JL, Sonenberg N. The leader of human immunodeficiency virus type 1 genomic RNA harbors an internal ribosome entry segment that is active during the G2/M phase of the cell cycle. J Virol. 2003;77:3939–3949. doi: 10.1128/JVI.77.7.3939-3949.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Larson DR, Johnson MC, Webb WW, Vogt VM. Visualization of retrovirus budding with correlated light and electron microscopy. Proc Natl Acad Sci USA. 2005;102:15453–15458. doi: 10.1073/pnas.0504812102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Jouvenet N, Bieniasz PD, Simon SM. Imaging the biogenesis of individual HIV-1 virions in live cells. Nature. 2008;454:236–240. doi: 10.1038/nature06998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ivanchenko S, Godinez WJ, Lampe M, Kräusslich HG, Eils R, Rohr K, et al. Dynamics of HIV-1 assembly and release. PLoS Pathog. 2009;5:1000652. doi: 10.1371/journal.ppat.1000652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Gousset K, Ablan SD, Coren LV, Ono A, Soheilian F, Nagashima K, et al. Real-time visualization of HIV-1 Gag trafficking in infected macrophages. PLoS Pathog. 2008;4:1000015. doi: 10.1371/journal.ppat.1000015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Milev MP, Brown CM, Mouland AJ. Live cell visualization of the interactions between HIV-1 Gag and the cellular RNA-binding protein Staufen1. Retrovirology. 2010;7:41. doi: 10.1186/1742-4690-7-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Jin J, Sherer NM, Heidecker G, Derse D, Mothes W. Assembly of the murine leukemia virus is directed towards sites of cell-cell contact. PLoS Biol. 2009;7:1000163. doi: 10.1371/journal.pbio.1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]