Abstract
The ribosome binding site of Escherichia coli rpoS mRNA, encoding the stationary sigma-factor RpoS, is sequestered by an inhibitory stem-loop structure (iss). Translational activation of rpoS mRNA at low temperature and during exponential growth includes Hfq-facilitated duplex formation between rpoS and the small regulatory RNA DsrA as well as a concomitant re-direction of RNase III cleavage in the 5′-untranslated region of rpoS upon DsrA·rpoS annealing. In this way, DsrA-mediated regulation does not only activate rpoS translation by disrupting the inhibitory secondary structure but also stabilizes the rpoS transcript. Although minor structural changes by Hfq have been observed in rpoS mRNA, a prevailing question concerns unfolding of the iss in rpoS at low growth temperature. Here, we have identified the DEAD-box helicase CsdA as an ancillary factor required for low temperature activation of RpoS synthesis by DsrA. The lack of RpoS synthesis observed in the csdA mutant strain at low growth temperature could be attributed to a lack of duplex formation between rpoS and DsrA, showing that at low temperature the sole action of Hfq is not sufficient to permit DsrA·rpoS annealing. An interactome study has previously indicated an association between Hfq and CsdA. However, immunological assays did not reveal a physical interaction between Hfq and CsdA. These findings add to a model, wherein Hfq binds upstream of the rpoS iss and presents DsrA in a conformation receptive to annealing. Melting of the iss by CsdA may then permit DsrA·rpoS duplex formation, and consequently rpoS translation.
Key words: riboregulation, Hfq, translational activation, rpoS, DEAD box helicase CsdA
Introduction
Bacterial trans-acting small regulatory RNAs (sRNA) are generally synthesized in response to certain growth or stress conditions.1 For instance, E. coli sRNAs are known to be synthesized during iron depletion, oxidative stress, outer membrane stress, elevated glycine levels, changes in glucose concentration and elevated glucose-phosphate levels.2–6
The majority of the hitherto characterized sRNAs in E. coli and several other Bacteria act as negative regulators by preventing ribosome loading onto the mRNA through base-pairing with, or in the vicinity of the ribosome binding site (rbs). As a result, the respective mRNA is prone to rapid decay.7–9 Variations of negative regulation by sRNAs exerted distantly from the rbs can further include binding to and obstructing of an upstream ribosome loading site,10 obstruction of a C/A rich element11 as well as binding to the coding region.12 In addition, indirect translational silencing by preventing translation of an upstream reading frame to which the target gene is translationally coupled has also been reported.13
Positive regulation by sRNAs appears to be less frequent. In some cases the sRNAs act by an “anti-antisense” mechanism and open up intramolecular inhibitory stem-loop structures (iss), which block ribosome binding.14–16 Examples include translation activation of (1) hla mRNA by RNAIII in S. aureus,16 (2) vca0939 mRNA by Qrr1 in V. cholerae,17 (3) glmS mRNA by GlmZ,18 (4) shiA mRNA by RyhB19 and (5) rpoS mRNA by DsrA,20 RprA21 or ArcZ,22 with the three latter regulatory events occurring in E. coli.
A well studied model system is the translational activation of rpoS mRNA, encoding the stationary phase sigma-factor, RpoS, by the sRNA DsrA. DsrA is predominantly transcribed at low growth temperature (25°C) and its stability decreases with increasing temperature.23 At low growth temperature, the DsrA·rpoS interaction was shown to relieve an iss that impedes ribosome access to the rpoS rbs.20 Later on it became apparent that the hexameric RNA chaperone Hfq stimulates DsrA-mediated activation of rpoS translation.24 More recently, the double strand-specific nuclease RNase III was shown to cleave rpoS mRNA upstream of the rbs, which in turn leads to a rapid inactivation of the mRNA. In contrast, base-pairing of DsrA with the rpoS leader creates an alternative RNase III cleavage site within the DsrA·rpoS duplex that prevents reuse of DsrA.25
The sRNA DsrA binds to Hfq on the positively charged proximal site,26 whereas poly(A) oligonucleotides bind with high affinity to the distal site.26 A structural study by Link et al.27 suggested that the distal binding site can accommodate (A-R-N)i or (A-R-N-N)i tracts, with R being a purine nucleotide and N being any nucleotide. Binding of different RNA sequences to the distal site could thus explain why Hfq can interact with many different RNA ligands.27 On the other hand, the dedicated RNA binding surfaces on either site of the Hfq-hexamer could allow to bring two RNAs together in a pair-wise fashion, which can be reconciled with several studies wherein Hfq was shown to stimulate annealing between two RNA ligands.28–33 It was shown that high affinity binding of Hfq to the rpoS leader depends on an A6 segment and on AAN(4) repeats situated upstream of the DsrA·rpoS base-pairing site.30 RpoS leaders containing these sequences formed stable ternary complexes with Hfq and DsrA. In a recent study Soper et al.22 provided evidence that Hfq in fact increases the stability of the DsrA-rpoS complex by binding to this upstream A-rich regions in the leader rather than the kinetics of DsrA·rpoS duplex formation. Even though Hfq was shown to induce structural changes in mRNAs targeted by sRNAs,34 footprinting studies30,35 did not strongly support the idea that Hfq is sufficient to open up the iss in rpoS. In addition, the requirement for Hfq in rpoS translation could be bypassed when DsrA was over-produced.22 Although the spatial positioning of the rpoS-Hfq-DsrA complex upstream of the annealing site could bring the interaction sites in close proximity and thereby accelerate DsrA·rpoS annealing,30 it remained puzzling how the rpoS iss opens at low temperature to allow anti-antisense annealing between DsrA and rpoS.
The E. coli CsdA DEAD-box helicase is induced upon a temperature downshift.36 The CsdA protein has been implicated in ribosome biogenesis,37–40 in mRNA turnover upon cold-adaptation41,42 as well as in translation initiation.43 Most likely, these functions are rooted in the ATP-dependent helicase activity of CsdA, which has been shown to resolve secondary structures in RNA.36,44,45 Interestingly, an interactome study46 identified CsdA as one of the ∼70 proteins associated with Hfq.
Using the DsrA/rpoS model system, we addressed the question, whether CsdA might represent an auxiliary factor involved in translational activation. We show that at low growth temperature synthesis of RpoS requires CsdA and that DsrA·rpoS duplex formation does not occur in a csdA mutant strain. As no evidence was obtained for a physical interaction between Hfq and CsdA we infer that the CsdA-mediated destabilization of the iss in rpoS precedes DsrA·rpoS annealing.
Results
CsdA is required for low temperature synthesis of rpoS in vivo and stimulates rpoS synthesis in vitro.
Given that Hfq was found to be associated with the dead box helicase CsdA46 and that Hfq did not significantly perturb the iss in rpoS mRNA,30,35 we asked whether CsdA might represent an ancillary factor involved in Hfq-mediated translational activation of rpoS mRNA by DsrA at low temperature. As shown previously,23 during exponential growth RpoS synthesis was increased at 24°C when compared to 37°C (Fig. 1, lanes 4 and 5). Whereas no significant differences in the RpoS levels were observed at 37°C in both, the csdA mutant strain and the wild-type strain (Fig. 1, lanes 3 and 4), hardly any RpoS synthesis was observed in the csdA- strain at 24°C (Fig. 1, lane 6). Ectopic expression of csdA in the csdA- background did not affect RpoS synthesis at 37°C but restored it at 24°C (Fig. 1, lanes 2 and 7).
Figure 1.

Steady-state levels of RpoS determined by quantitative western-blotting in the wt strain and the isogenic csdA mutant. The E. coli strains WJW45 (wt), the isogenic csdA deficient strain WJW45ΔcsdA (csdA−) as well as WJW45ΔcsdA harbouring plasmid pUCdeaD38 (csdA−/pUCdeaD) were grown to early log phase (OD600 of 0.4) either at 37°C (lane 2–4) or at 24°C (lane 5–7), respectively. Cell extracts of the rpoS mutant strain RH90,53 served as a negative control (lane 1). Equal amounts of total cellular proteins were loaded in each lane of the SDS-polyacrylamide gel. The RpoS protein was detected by immunological means as described in Materials and Methods. Only the relevant section of the immunoblot is shown.
Next, we tested whether the stimulatory effect of CsdA on DsrA-mediated rpoS synthesis at low temperature could be recapitulated in an in vitro translation system (PURESYSTEM) at 24°C. However, at this temperature the in vitro translation system was hardly active and effects of CsdA on rpoS mRNA translation were difficult to discern (not shown) despite the presence of Hfq in the PURESYSTEM (Suppl. Fig. S1). As CsdA is active at 37°C,45 we tested whether addition of CsdA would stimulate rpoS synthesis in the presence of DsrA at the elevated temperature. As shown in Figure 2, the addition of CsdA at a molecular ratio of 1:1 to rpoS mRNA led to an ∼2-fold increase of RpoS synthesis (lane 6), whereas the same amount of heat-inactivated CsdA (lane 7) resulted in RpoS levels comparable to that obtained in the absence of CsdA (lane 4).
Figure 2.
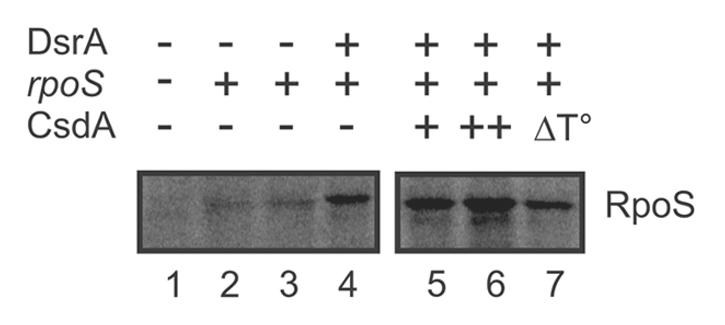
CsdA stimulates rpoS synthesis in vitro. 10 pmol of in vitro synthesized full length rpoS mRNA were translated using the PURESYSTEM. Translation of rpoS mRNA was performed in the absence (lanes 2–3) or in the presence of equimolar amounts of DsrA (lanes 4–7). Lane 1, no mRNA was added to the reaction mixture. Lane 2, equimolar amounts of the sRNA RyhB, which does not stimulate rpoS translation, was added. Lane 5 and 6, translation in the presence of 5 (+) and 10 pmol (++) CsdA, respectively. Lane 7, 10 pmol of heat-inactivated CsdA protein (ΔT°) was added to the reaction mixture. The protein samples were resolved on a 12% SDS-polyacrylamide gel. The signals were visualized by a PhosporImager (Molecular Dynamics). The experiment was performed in triplicate. Only the relevant section of one representative autoradiogram is shown. The position of the RpoS is indicated.
CsdA is required for DsrA·rpoS duplex formation.
We next asked whether the absence of RpoS synthesis in the CsdA-strain at low temperature is attributable to a lack of annealing between rpoS mRNA and DsrA. We addressed this question by utilizing RNase III clevage upon rpoS·DsrA duplex formation25 as a diagnostic marker. The E. coli strains WJW45 and WJW45ΔcsdA were grown to an OD600 of 0.4 at 24°C, as this temperature leads to a strong induction of DsrA synthesis.23 Total RNA obtained from either strain was purified and RNase III-dependent cleavage of rpoS was assessed by primer extension using a [32P] 5′end-labeled rpoS-specific primer. Compared with the wild-type strain, the primer extension signal for the 5′-end of rpoS mRNA (i.e., the steady state levels of rpoS mRNA) was approximately 40–50% reduced in the csdA- strain. Nevertheless, RNase III dependent cleavage in rpoS mRNA was not observed in the csdA- strain (Fig. 3, lane 6) but, as previously shown, occurred in the wild-type strain at G−11225 (Fig. 3, lane 5). As the levels of both, DsrA and Hfq, were not grossly altered at 24°C in either strain (Fig. 3), we conclude that CsdA is required for DsrA·rpoS duplex formation.
Figure 3.
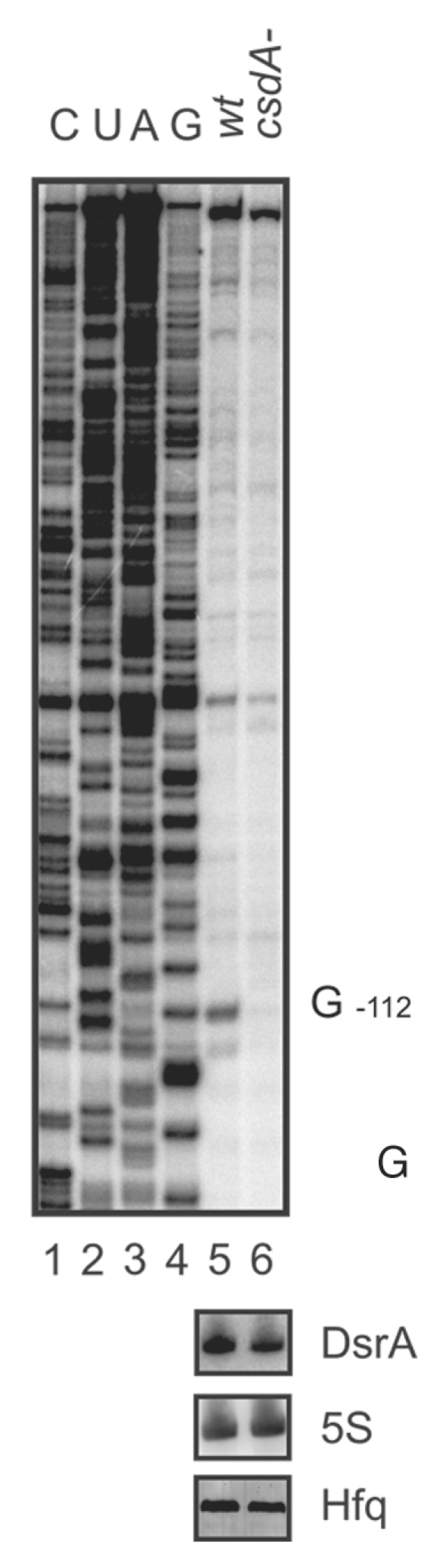
Primer extension analysis of total RNA isolated from the E. coli strain WJW45 (wt) and the csdA deficient strain WJW45ΔcsdA (csdA-) grown to an OD600 of 0.4 at 24°C. Top: The product of the RNase III cleavage within the rpoS leader (position G−112) accumulated only in the wild-type strain (lane 5) and was hardly detectable in the csdA deficient strain (lane 6). Lanes 1–4, sequencing ladder. The experiment was performed in triplicate. One representative autoradiograph is shown. Bottom: The DsrA RNA levels and 5S rRNA levels (loading control) of the wt and csdA-strain grown at 24°C were determined by northern-blot analysis as described in Materials and Methods. The Hfq protein levels in both strains were determined by western-blotting using equal amounts of total protein of the two strains at the time the cells were harvested for preparation of total RNA.
Hfq and CsdA do not physically interact.
Next, we asked whether Hfq and CsdA interact directly with each other or whether the observed association between both proteins is RNA mediated. To distinguish between these possibilities we used Hfq and CsdA-His6 proteins purified to homogeneity with and without micrococcal nuclease (MN) treatment and performed Far-western-blot analyses and co-immunoprecipitation studies. For Far-western analysis, the CsdA-His6 protein treated or left untreated with MN was loaded on a 10% SDS polyacrylamide gel and then transferred onto a nitrocellulose membrane. After denaturation and renaturation, membrane bound CsdA-His6 was incubated with Hfq, which was treated with MN. Then anti-Hfq-antibodies were used to detect Hfq protein eventually bound to CsdA-His6. Hfq binding to CsdA-His6 was only observed when CsdA-His6 was not treated with MN (Fig. 4A, lane 1). However, Hfq binding to CsdA-His6 was not detected after treatment of both proteins with nuclease (Fig. 4A, lane 2). As the anti-Hfq antibody did not recognize CsdA-His6 (not shown), the Far-western studies suggested that the observed CsdA-His6-Hfq association is RNA-mediated.
Figure 4.
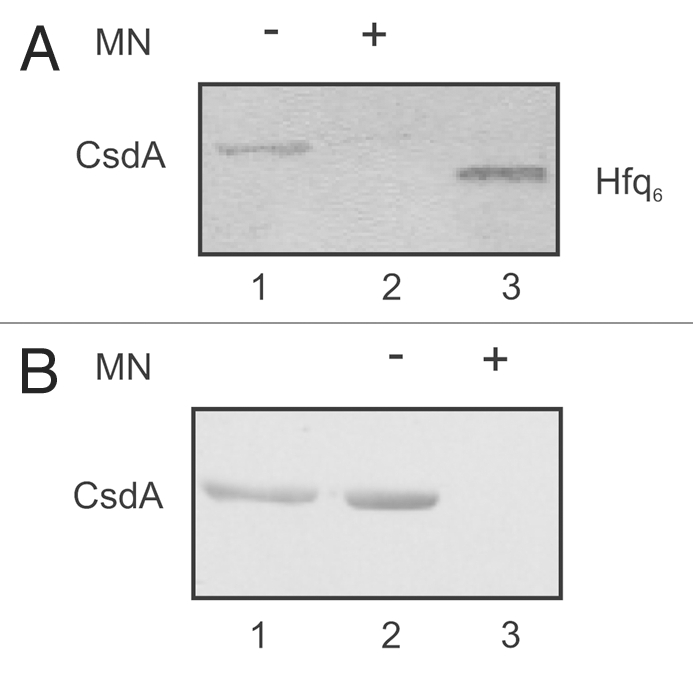
The interaction between CsdA and Hfq is RNA mediated. (A) Far western-blotting does not reveal an interaction between CsdA and Hfq. 25 pmol of CsdA-His6 (lane 1) or 25 pmol of CsdA-His6 (lanes 2) treated with micrococcal nuclease (MN) and 25 pmol of Hfq (lane 3, control) were separated by SDS-polyacrylamide gel electrophoresis and then electroblotted onto nitrocellulose membranes. After re-naturation of the proteins the membranes were incubated with Hfq (150 pmol Hfq as hexamer) as described in Materials and Methods. The CsdAHis6-Hfq complex was visualized by immuno-detection using anti-Hfq antibodies (lane 1), whereas no CsdA-Hfq complex could be detected with anti-Hfq antibodies after treatment with MN (lane 2). Lane 3, Hfq protein (control) was detected with anti-Hfq antibodies. (B) Co-immunoprecipitation assay with protein CsdA-His6 and Hfq using anti-Hfq antibodies. Lane 1, CsdA-His6 (10 pmol) was loaded as a positive control for the anti-His antibodies. 25 pmol of purified hexameric Hfq were incubated together with 50 pmol CsdA-His6 (lane 2) or 50 pmol CsdA-His6 treated with MN (lane 3) followed by immunoprecipitation using anti-Hfq antibodies bound to the Dynabeads (see Materials and Methods). The co-immunoprecipitated CsdA-His6 was detected on the western-blot with anti-His antibodies. Only the relevant sections of the immunoblots are shown.
The Far-western studies were verified by co-immunoprecipitation experiments using purified Hfq and CsdA-His6 proteins either treated or untreated with MN. The proteins were mixed together (see Materials and Methods) and anti-Hfq antibodies bound to “Dynabeads protein G” were used for immunoprecipitation. The protein immunoprecipitates were electrophoresed on a 10% SDS-polyacrylamide gel. Then, a western-blot was performed with anti-His antibodies with the aim to detect CsdA-His6 eventually co-immunoprecipitated with Hfq. As shown in Figure 4B, lane 2, when both, Hfq and CsdA-His6 were not treated with MN, CsdA-His6 co-immunoprecipitated with Hfq. As CsdA-His6 was not immunoprecipitated with the anti-Hfq antibodies bound “Dynabeads protein G” alone (not shown), Hfq was apparently in complex with CsdA-His6. However, when both proteins were treated with MN, no co-immunoprecipitation of CsdA with Hfq was observed (Fig. 4B, lane 3). A reciprocal co-immunoprecipitation experiment (not shown) where anti-His-antibodies bound to Dynabeads protein G were used for immunoprecpitation and anti-Hfq antibodies were used to detect on a western-blot Hfq possibly co-immunoprecipitated with CsdA-His6 resulted in the same outcome. These studies corroborated the Far-western studies in that the association between Hfq and CsdA does not result from direct protein-protein contacts but that is most likely brought about by RNA(s) associated with the proteins after purification.
Discussion
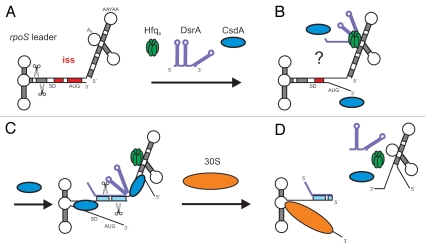
This study implicates the dead box helicase CsdA in Hfq-mediated low temperature activation of rpoS mRNA by DsrA. Soper et al.22 have recently put forward a model, in which the Hfq-DsrA complex binds to A-rich segments in the rpoS leader, i.e., upstream of the DsrA·rpoS annealing site (Fig. 5). Although structural probing of rpoS mRNA upon incubation with Hfq suggested a less tightly folded iss,30 it remained uncertain whether these perturbations are sufficient for relieving the iss, which is in turn required for DsrA·rpoS annealing. Based on our observations, we hypothesize that CsdA is involved in opening the iss at low temperature and that DsrA bound by Hfq at the A-rich segment(s) in the rpoS leader can then base-pair with rpoS opposite of the rbs (Fig. 5). Clearly, as opening of stem-loop 1 of DsrA appears to be required for DsrA·rpoS duplex formation,35 it cannot be excluded that CsdA also acts by unfolding DsrA. Nevertheless, Hfq was shown to induce conformational changes in DsrA31 and Hfq at least partially destabilized stem-loop 1 in DsrA.35 In addition, NMR studies of a complex between a C-terminally truncated Hfq protein (aa 1–65) and a 34 nucleotide DsrA RNA revealed no imino-shifts arising from nucleotide base-pairing, also indicating that stem-loop 2 is unfolded upon Hfq binding (Beich-Frandsen M, et al. unpublished). Thus, it is conceivable the rpoS-Hfq-DsrA complex situated in the leader presents DsrA in a manner that allows partial duplex formation opposite of the rbs of rpoS. It is interesting to note that the minimal base-pairing region required for the interaction between the sRNA SgrS with its target mRNA ptgS was recently determined with 14 nucleotides.47 Thus, the proximal base-pairing nucleotides upstream of the Hfq binding site in DsrA35 could suffice to initiate duplex formation opposite of the rpoS rbs. This event may dislodge DsrA from Hfq (Fig. 5). In addition, it seems plausible that the action of CsdA contributes to recycling of Hfq from the leader region upon DsrA is released (Fig. 5). As CsdA is known to function as a bidirectional helicase44 this might occur through unwinding of the 5′leader. Alternatively, it is also possible that CsdA acts by displacement of Hfq without unwinding the leader, a function that has been attributed to other helicases.48 In any case, the involvement of CsdA in low temperature activation of rpoS adds a new actor to riboregulation. This observation could even have implications in understanding the mode of action of sRNAs in bacteria that lack apparent Hfq homologues but posses helicases.
Figure 5.
Model for translational activation of rpoS mRNA by DsrA, Hfq and CsdA. (A) At low temperature and during exponential growth RNase III cleavage occurs in the rpoS leader, which primes rapid decay of the mRNA.25 As DsrA·rpoS annealing (Fig. 3) and consequently rpoS translation (Fig. 1) did not occur in the csdA- strain, intramolecular cleavage in the rpoS leader most likely accounts for the reduced steady state levels of rpoS mRNA in the latter when compared to the wild-type strain (see Fig. 3). (B) Hfq-DsrA binding to A-rich segments (A6 and/or AAN(4)30) in the rpoS leader occurs at low temperature when DsrA is expressed30 (for clarity only one possible Hfq binding site is occupied by the Hfq-DsrA complex). CsdA aids in melting the iss and perhaps in unfolding of DsrA. (C) Annealing between DsrA and the rpoS mRNA occurs opposite of the rbs. (D) Translation of rpoS ensues and intermolecular RNase III cleavage of the DsrA·rpoS duplex (at G−112 in rpoS mRNA) prevents reuse of DsrA. Hfq is eventually recycled as a consequence of DsrA·rpoS base-pairing and/or by CsdA action. The segment of the iss is in red. Scissors signify RNase III cleavage. All other components are denoted.
Several proteins including RNA polymerase, ribosomal protein S1,49 RNase E,50 polyA-polymerase (PAP I) and polynucleotide-phosphorylase (PNPase)51 have been found in complex with Hfq. However, follow up studies did neither reveal a physical interaction between Hfq and ribosomal protein S1,52 nor with RNase E.53 Most likely, these complexes are as infered for the CsdA-Hfq association (Fig. 4) RNA-mediated and result from the spatial association of the transcriptional, translational and RNA decay machineries.
Materials and Methods
Bacterial strains and growth conditions.
The E. coli strains WJW45, the isogenic csdA deletion strain WJW45ΔcsdA37 the rpoS mutant strain RH 90,54 as well as plasmid pUCdeaD38 have been previously described. The bacterial strains were grown in Luria-Bertani (LB) medium supplemented with ampicillin (100 µg/ml) or tetracycline (20 µg/ml) where appropriate.
Western-blot analysis.
Samples of the bacterial cultures grown in LB medium at 24°C or at 37°C were harvested in early logarithmic growth phase (OD600 of 0.4), pelleted and boiled in protein sample buffer. Equal amounts of total protein were separated on 12% SDS-polyacrylamide gels, blotted onto a nitrocellulose membrane and probed with anti-RpoS antibodies (provided by F. Norel, Pasteur Institute, Paris), or specific antibodies against Hfq (Pineda, Berlin). The antibody-antigen complexes were visualized with alkaline-phosphatase conjugated secondary antibodies using the chromogenic substrates NBT and BCIP as described.31
RNA preparation for in vitro studies.
For full length rpoS mRNA synthesis, the plasmid pUrpos16,31 cleaved with EcoRI was used as template for in vitro transcription with T7 RNA polymerase (Fermentas). Synthesis of DsrA and RhyB RNA was performed as described.25,31 The run-off transcripts were further gel-purified following standard procedures and the RNA concentration was determined by measuring the A260.
In vitro translation assay.
The PURESYSTEM classic II (PURE2030C; Wako Chemicals GmbH, Germany) was used for the in vitro translation assays according to the manufacturer's instructions. The 25 µl reaction contained 10 pmol of in vitro transcribed full length rpoS mRNA template, 10 pmol DsrA or RyhB (internal control; Fig. 2, lane 2), 10 pmol L-[35S]-Methionine (Hartmann Analytic, Germany) and two different concentrations of CsdA as indicated in the legend to Figure 2. As an additional mock control CsdA was heat-inactivated (Fig. 2, lane 7) by boiling for 10 min at 100°C and subsequent cooling on ice before use. The components were mixed together on ice and incubated for 2 hour at 37°C. The in vitro translation reactions were terminated with four volumes of acetone, kept on ice and the proteins were precipitated by centrifugation (16,000 g, 15 min, 4°C), and then separated on 12% SDS-polyacrylamide gel. The signals were visualized by a PhosporImager (Molecular Dynamics) and quantified by ImageQuant software.
RNA isolation and primer extension analysis.
The E. coli strains WJW45 and WJW45ΔcsdA were grown to an OD600 of 0.4 at 24°C to induce low temperature DsrA synthesis.23 Isolation of total RNA and primer extension analysis to detect the RNase III specific cleavage signals at position G−112 in rpoS mRNA were performed as recently described.25,52 In brief, total RNA was purified from culture aliquots using the hot phenol method.55 Primer extension analysis was performed using AMV reverse transcriptase (Promega) together with 15 µg of purified total RNA primed with the rpoS-specific [32P] 5′-end labeled oligonucleotide (5′-TCC GTT CTC ATC AAA TTC CGC ATC-3′). The extension products along with a sequencing ladder, which was prepared using the 5′segment (nt −564 to +188) of rpoS mRNA as a template, were resolved on a 6% sequencing gel. The resulting signals were visualized by a PhosphorImager (Molecular Dynamics).
Northern-blot analysis.
The steady-state levels of DsrA in strains WJW45 and WJW45ΔcsdA were determined by northern-blot analysis using 15 µg RNA of the same RNA preparations used for the primer extension analysis (see above). The RNA samples were denatured for 5 min at 82°C in RNA-loading buffer containing 50% formamide, separated on 8% polyacrylamide/8 M urea gels, and then transferred to nylon membranes by electroblotting. The RNA was crosslinked to the membrane by exposure to UV light. The membrane was hybridized with DsrA-specific [32P] 5′-end labeled oligonucleotide (5′-TCG TTA CAC CAG GAA ATC TGA TGT-3′), and as a loading control with 5S rRNA-specific oligonucleotide R25 (5′-GGT GGG ACC ACC GCG CTA CGG CCG CCA GGC-3′). The hybridization signals were visualized using a PhosphorImager (Molecular Dynamics).
Purification of CsdA and Hfq.
The purification of CsdA was performed as described by Bizzebard et al.44 with some modifications. The plasmid pCsdA44 was transformed into E. coli hfq-strain JW4130 (Keio gene knockout collection, NBRP, Japan) and incubated at 28°C. At an OD600 of 0.6, the synthesis of CsdA was induced by addition of IPTG to a final concentration of 1 mM. After an additional hour of incubation at 28°C, the cells were harvested by centrifugation at 4,000x g for 10 minutes at 4°C. All subsequent procedures were performed at 4°C. Approximately 7 g of cells were resuspended in 20 ml lysis buffer (500 mM NaCl, 50 mM NaH2PO4, pH 8.0, 200 µg/ml PMSF, 5 µg/ml lysozyme, 20 µg/ml DNaseI, 20 mM imidazole) and then lysed using a French press (SimAminco) at 10,000 psi. The lysate was centrifuged for 30 minutes at 10,000, 15,000 and 30,000x g respectively, to remove cellular debris. The supernatant (16 ml) was incubated over-night at 4°C with 2 ml Ni-NTA agarose (QIAGEN). Then, the lysate-Ni-NTA mixture was loaded on a column according to the protocol of the manufacturer and washed with washing buffer (500 mM NaCl, 50 mM NaH2PO4, pH 8.0) containing 20 mM imidazole. The proteins were eluted with elution buffer, consisting of washing buffer with increasing concentrations (60–500 mM) of imidazole. The fractions obtained from step-wise elution were analysed by SDS-PAGE. The purest fractions were pooled, dialysed against CsdA storage buffer (1 M NaCl, 20 mM Tris-HCl, pH 7.5, 0.1 mM EDTA, 1 mM DTT), and subsequently stored in the same buffer. The activity of CsdA was tested as specified by Turner et al.45 Hfq was purified as described by Vecerek et al.51
Micrococcal nuclease treatment of Hfq and CsdA.
To remove remaining RNA, the purified CsdA (CsdA-His6) and Hfq proteins were treated with microccocal nuclease (Fermentas) and re-purified on a Ni-NTA agarose column. Briefly, 700 pmol of protein in storage buffer were treated in the presence of 10 mM CaCl2 with 100 U microccocal nuclease for one hour at 37°C. The protein fractions were then incubated with 200 µl Ni-NTA overnight at four 4°C or for two hours on ice. The protein-Ni-NTA mixture was then loaded on a Spin-X column (Costar), washed with lysis buffer (500 mM NaCl, 50 mM NaH2PO4, pH 8.0, 20 mM imidazole) and eluted with lysis buffer containing 100 mM imidazol. The eluted proteins were further dialysed against the corresponding storage buffers and used for the experiments.
Far-western analysis.
Far-western analysis was performed according to the protocol of Wu et al.56 with some modifications described by Vecerek et al.51 Briefly, CsdA (25 pmol) treated or left untreated with micrococcal nuclease was separated on a 10% SDS-polyacrylamide gel and then electroblotted onto a nitrocellulose membrane at 15V for 30 minutes. De-naturation and re-naturation of the proteins bound to the membrane was performed three times in AC buffer (100 mM NaCl, 20 mM Tris-HCl pH 7.6, 0.5 mM EDTA, 1 mM DTT, 10% glycerol, 0.1% Tween-20 and 2% milk powder) with decreasing concentrations of Guanidine-HCl.56 After the final wash with AC buffer the nitrocellulose membrane was incubated for three hours at RT with Hfq (150 pmol as hexamer) in 10 ml AC buffer, followed by several washing steps. Bound Hfq protein was detected following a standard western-blot protocol with anti-Hfq antibodies and visualized as described above.
Co-immunoprecipitation.
25 pmol of purified hexameric Hfq protein was incubated in 100 µl AC buffer (10% glycerol, 100 mM NaCl, 20 mM Tris-HCl pH 7.6, 0.5 mM EDTA, 0.1% Tween-20, 1 mM DTT) at room temperature together with 50 pmol CsdA protein for one hour. Anti-Hfq antibodies were bound to Dynabeads according to the manufacture's instructions. The Dynabeads-anti-Hfq antibody complex was added to the protein mixture and incubated for an additional hour at room temperature. Non-specifically bound proteins were removed from the beads by magnetic separation and by washing the pellets three times with AC buffer. The washed Dynabeads-Ab-Ag complex and the co-immunoprecipitated proteins were resuspended in Laemmli buffer and analysed by immunoblotting with anti-His antibodies directed against CsdA-His6.
Acknowledgements
We are grateful to Dr. T. Bizebard and M. Dreyfus for providing materials. This study was supported by grant F 1720 from the Austrian Science Fund to U.B.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/13768
Supplementary Material
References
- 1.Repoila F, Darfeuille F. Small regulatory non-coding RNAs in bacteria: physiology and mechanistic aspects. Biol Cell. 2009;101:117–131. doi: 10.1042/BC20070137. [DOI] [PubMed] [Google Scholar]
- 2.De Lay N, Gottesman S. The Crp-activated small non-coding regulatory RNA CyaR (RyeE) links nutritional status to group behavior. J Bacteriol. 2009;191:461–476. doi: 10.1128/JB.01157-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gottesman S. Micros for microbes: non-coding regulatory RNAs in bacteria. Trends Genet. 2005;21:399–404. doi: 10.1016/j.tig.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 4.Johansen J, Rasmussen AA, Overgaard M, Valentin-Hansen P. Conserved small non-coding RNAs that belong to the sigmaE regulon: role in downregulation of outer membrane proteins. J Mol Biol. 2006;364:1–8. doi: 10.1016/j.jmb.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 5.Urbanowski ML, Stauffer LT, Stauffer GV. The gcvB gene encodes a small untranslated RNA involved in expression of the dipeptide and oligopeptide transport systems in Escherichia coli. Mol Microbiol. 2000;37:856–868. doi: 10.1046/j.1365-2958.2000.02051.x. [DOI] [PubMed] [Google Scholar]
- 6.Görke B, Vogel J. Noncoding RNA control of the making and breaking of sugars. Genes Dev. 2008;22:2914–2925. doi: 10.1101/gad.1717808. [DOI] [PubMed] [Google Scholar]
- 7.Gottesman S. The small RNA regulators of Escherichia coli: roles and mechanisms. Annu Rev Microbiol. 2004;58:303–328. doi: 10.1146/annurev.micro.58.030603.123841. [DOI] [PubMed] [Google Scholar]
- 8.Kaberdin VR, Bläsi U. Translation initiation and the fate of bacterial mRNAs. FEMS Microbiol Rev. 2006;30:967–979. doi: 10.1111/j.1574-6976.2006.00043.x. [DOI] [PubMed] [Google Scholar]
- 9.Papenfort K, Vogel J. Multiple target regulation by small noncoding RNAs rewires gene expression at the post-transcriptional level. Res Microbiol. 2009;160:278–287. doi: 10.1016/j.resmic.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 10.Darfeuille F, Unoson C, Vogel J, Wagner EG. An antisense RNA inhibits translation by competing with standby ribosomes. Mol Cell. 2007;26:381–392. doi: 10.1016/j.molcel.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 11.Sharma CM, Darfeuille F, Plantinga TH, Vogel J. A small RNA regulates multiple ABC transporter mRNAs by targeting C/A-rich elements inside and upstream of ribosome-binding sites. Genes Dev. 2007;21:2804–2817. doi: 10.1101/gad.447207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bouvier M, Sharma CM, Mika F, Nierhaus KH, Vogel J. Small RNA binding to 5′ mRNA coding region inhibits translational initiation. Mol Cell. 2008;32:827–837. doi: 10.1016/j.molcel.2008.10.027. [DOI] [PubMed] [Google Scholar]
- 13.Vecerek B, Moll I, Blasi U. Control of Fur synthesis by the non-coding RNA RyhB and iron-responsive decoding. EMBO J. 2007;26:965–975. doi: 10.1038/sj.emboj.7601553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Repoila F, Majdalani N, Gottesman S. Small non-coding RNAs, co-ordinators of adaptation processes in Escherichia coli: the RpoS paradigm. Mol Microbiol. 2003;48:855–861. doi: 10.1046/j.1365-2958.2003.03454.x. [DOI] [PubMed] [Google Scholar]
- 15.Urban JH, Vogel J. Two seemingly homologous non-coding RNAs act hierarchically to activate glmS mRNA translation. PLoS Biol. 2008;6:64. doi: 10.1371/journal.pbio.0060064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morfeldt E, Taylor D, von Gabain A, Arvidson S. Activation of alpha-toxin translation in Staphylococcus aureus by the trans-encoded antisense RNA, RNAIII. EMBO J. 1995;14:4569–4577. doi: 10.1002/j.1460-2075.1995.tb00136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lenz DH, Mok KC, Lilley BN, Kulkarni RV, Wingreen NS, Bassler BL. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell. 2004;118:69–82. doi: 10.1016/j.cell.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 18.Kalamorz F, Reichenbach B, Marz W, Rak B, Görke B. Feedback control of glucosamine-6-phosphate synthase GlmS expression depends on the small RNA GlmZ and involves the novel protein YhbJ in Escherichia coli. Mol Microbiol. 2007;65:1518–1533. doi: 10.1111/j.1365-2958.2007.05888.x. [DOI] [PubMed] [Google Scholar]
- 19.Prevost K, Salvail H, Desnoyers G, Jacques JF, Phaneuf E, Masse E. The small RNA RyhB activates the translation of shiA mRNA encoding a permease of shikimate, a compound involved in siderophore synthesis. Mol Microbiol. 2007;64:1260–1273. doi: 10.1111/j.1365-2958.2007.05733.x. [DOI] [PubMed] [Google Scholar]
- 20.Lease RA, Cusick ME, Belfort M. Riboregulation in Escherichia coli: DsrA RNA acts by RNA:RNA interactions at multiple loci. Proc Natl Acad Sci USA. 1998;95:12456–12461. doi: 10.1073/pnas.95.21.12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Majdalani N, Chen S, Murrow J, St John K, Gottesman S. Regulation of rpoS by a novel small RNA: the characterization of RprA. Mol Microbiol. 2001;39:1382–1394. doi: 10.1111/j.1365-2958.2001.02329.x. [DOI] [PubMed] [Google Scholar]
- 22.Soper T, Mandin P, Majdalani N, Gottesman S, Woodson SA. Positive regulation by small RNAs and the role of Hfq. Proc Natl Acad Sci USA. 2010;107:9602–9607. doi: 10.1073/pnas.1004435107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sledjeski DD, Gupta A, Gottesman S. The small RNA, DsrA, is essential for the low temperature expression of RpoS during exponential growth in Escherichia coli. EMBO J. 1996;15:3993–4000. [PMC free article] [PubMed] [Google Scholar]
- 24.Sledjeski DD, Whitman C, Zhang A. Hfq is necessary for regulation by the untranslated RNA DsrA. J Bacteriol. 2001;183:1997–2005. doi: 10.1128/JB.183.6.1997-2005.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Resch A, Afonyushkin T, Lombo TB, McDowall KJ, Blasi U, Kaberdin VR. Translational activation by the noncoding RNA DsrA involves alternative RNase III processing in the rpoS 5′-leader. RNA. 2008;14:454–459. doi: 10.1261/rna.603108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mikulecky PJ, Kaw MK, Brescia CC, Takach JC, Sledjeski DD, Feig AL. Escherichia coli Hfq has distinct interaction surfaces for DsrA, rpoS and poly(A) RNAs. Nat Struct Mol Biol. 2004;11:1206–1214. doi: 10.1038/nsmb858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Link TM, Valentin-Hansen P, Brennan RG. Structure of Escherichia coli Hfq bound to polyriboadenylate RNA. Proc Natl Acad Sci USA. 2009;106:19292–19297. doi: 10.1073/pnas.0908744106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arluison V, Hohng S, Roy R, Pellegrini O, Regnier P, Ha T. Spectroscopic observation of RNA chaperone activities of Hfq in post-transcriptional regulation by a small non-coding RNA. Nucleic Acids Res. 2007;35:999–1006. doi: 10.1093/nar/gkl1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moller T, Franch T, Hojrup P, Keene DR, Bachinger HP, Brennan RG, et al. Hfq: a bacterial Sm-like protein that mediates RNA-RNA interaction. Mol Cell. 2002;9:23–30. doi: 10.1016/s1097-2765(01)00436-1. [DOI] [PubMed] [Google Scholar]
- 30.Soper TJ, Woodson SA. The rpoS mRNA leader recruits Hfq to facilitate annealing with DsrA sRNA. RNA. 2008;14:1907–1917. doi: 10.1261/rna.1110608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vecerek B, Rajkowitsch L, Sonnleitner E, Schroeder R, Bläsi U. The C-terminal domain of Escherichia coli Hfq is required for regulation. Nucleic Acids Res. 2008;36:133–143. doi: 10.1093/nar/gkm985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang A, Wassarman KM, Ortega J, Steven AC, Storz G. The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Mol Cell. 2002;9:11–22. doi: 10.1016/s1097-2765(01)00437-3. [DOI] [PubMed] [Google Scholar]
- 33.Rajkowitsch L, Schroeder R. Dissecting RNA chaperone activity. RNA. 2007;13:2053–2060. doi: 10.1261/rna.671807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moll I, Leitsch D, Steinhauser T, Blasi U. RNA chaperone activity of the Sm-like Hfq protein. EMBO Rep. 2003;4:284–289. doi: 10.1038/sj.embor.embor772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lease RA, Woodson SA. Cycling of the Sm-like protein Hfq on the DsrA small regulatory RNA. J Mol Biol. 2004;344:1211–1223. doi: 10.1016/j.jmb.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 36.Jones PG, Mitta M, Kim Y, Jiang W, Inouye M. Cold shock induces a major ribosomal-associated protein that unwinds double-stranded RNA in Escherichia coli. Proc Natl Acad Sci USA. 1996;93:76–80. doi: 10.1073/pnas.93.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Charollais J, Dreyfus M, Iost I. CsdA, a cold-shock RNA helicase from Escherichia coli, is involved in the biogenesis of 50S ribosomal subunit. Nucleic Acids Res. 2004;32:2751–2759. doi: 10.1093/nar/gkh603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moll I, Grill S, Grundling A, Bläsi U. Effects of ribosomal proteins S1, S2 and the DeaD/CsdA DEAD-box helicase on translation of leaderless and canonical mRNAs in Escherichia coli. Mol Microbiol. 2002;44:1387–1396. doi: 10.1046/j.1365-2958.2002.02971.x. [DOI] [PubMed] [Google Scholar]
- 39.Peil L, Virumae K, Remme J. Ribosome assembly in Escherichia coli strains lacking the RNA helicase DeaD/CsdA or DbpA. The FEBS J. 2008;275:3772–3782. doi: 10.1111/j.1742-4658.2008.06523.x. [DOI] [PubMed] [Google Scholar]
- 40.Toone WM, Rudd KE, Friesen JD. DeaD, a new Escherichia coli gene encoding a presumed ATP-dependent RNA helicase, can suppress a mutation in rpsB, the gene encoding ribosomal protein S2. J Bacteriol. 1991;173:3291–3302. doi: 10.1128/jb.173.11.3291-3302.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Awano N, Xu C, Ke H, Inoue K, Inouye M, Phadtare S. Complementation analysis of the cold-sensitive phenotype of the Escherichia coli csdA deletion strain. J Bacteriol. 2007;189:5808–5815. doi: 10.1128/JB.00655-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prud'homme-Genereux A, Beran RK, Iost I, Ramey CS, Mackie GA, Simons RW. Physical and functional interactions among RNase E, polynucleotide phosphorylase and the cold-shock protein, CsdA: evidence for a ‘cold shock degradosome’. Mol Microbiol. 2004;54:1409–1421. doi: 10.1111/j.1365-2958.2004.04360.x. [DOI] [PubMed] [Google Scholar]
- 43.Butland G, Krogan NJ, Xu J, Yang WH, Aoki H, Li JS, et al. Investigating the in vivo activity of the DeaD protein using protein-protein interactions and the translational activity of structured chloramphenicol acetyltransferase mRNAs. J Cell Biochem. 2007;100:642–652. doi: 10.1002/jcb.21016. [DOI] [PubMed] [Google Scholar]
- 44.Bizebard T, Ferlenghi I, Iost I, Dreyfus M. Studies on three E. coli DEAD-box helicases point to an unwinding mechanism different from that of model DNA helicases. Biochemistry. 2004;43:7857–7866. doi: 10.1021/bi049852s. [DOI] [PubMed] [Google Scholar]
- 45.Turner AM, Love CF, Alexander RW, Jones PG. Mutational analysis of the Escherichia coli DEAD box protein CsdA. J Bacteriol. 2007;189:2769–2776. doi: 10.1128/JB.01509-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Butland G, Peregrin-Alvarez JM, Li J, Yang W, Yang X, Canadien V, et al. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature. 2005;433:531–537. doi: 10.1038/nature03239. [DOI] [PubMed] [Google Scholar]
- 47.Maki K, Murita T, Otaka A, Aiba H. A minimal base-pairing region of a bacterial small RNA SgrS required for translational repression of ptsG mRNA. Mol Microbiol. 2010;76:782–792. doi: 10.1111/j.1365-2958.2010.07141.x. [DOI] [PubMed] [Google Scholar]
- 48.Fairman ME, Maroney PA, Wang W, Bowers HA, Gollnick P, Nilsen TW, et al. Protein displacement by DExH/D “RNA helicases” without duplex unwinding. Science. 2004;304:730–734. doi: 10.1126/science.1095596. [DOI] [PubMed] [Google Scholar]
- 49.Sukhodolets MV, Garges S. Interaction of Escherichia coli RNA polymerase with the ribosomal protein S1 and the Sm-like ATPase Hfq. Biochemistry. 2003;42:8022–8034. doi: 10.1021/bi020638i. [DOI] [PubMed] [Google Scholar]
- 50.Morita T, Maki K, Aiba H. RNase E-based ribonucleoprotein complexes: mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes Dev. 2005;19:2176–2186. doi: 10.1101/gad.1330405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mohanty BK, Maples VF, Kushner SR. The Sm-like protein Hfq regulates polyadenylation dependent mRNA decay in Escherichia coli. Mol Microbiol. 2004;54:905–920. doi: 10.1111/j.1365-2958.2004.04337.x. [DOI] [PubMed] [Google Scholar]
- 52.Vecerek B, Beich-Frandsen M, Resch A, Bläsi U. Translational activation of rpoS mRNA by the non-coding RNA DsrA and Hfq does not require ribosome binding. Nucleic Acids Res. 2010;38:1284–1293. doi: 10.1093/nar/gkp1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Worrall JA, Gorna M, Crump NT, Phillips LG, Tuck AC, Price AJ, et al. Reconstitution and analysis of the multienzyme Escherichia coli RNA degradosome. J Mol Biol. 2008;382:870–883. doi: 10.1016/j.jmb.2008.07.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lange R, Hengge-Aronis R. Identification of a central regulator of stationary-phase gene expression in Escherichia coli. Mol Microbiol. 1991;5:49–59. doi: 10.1111/j.1365-2958.1991.tb01825.x. [DOI] [PubMed] [Google Scholar]
- 55.Lin-Chao S, Bremer H. Effect of the bacterial growth rate on replication control of plasmid pBR322 in Escherichia coli. Mol Gen Genet. 1986;203:143–149. doi: 10.1007/BF00330395. [DOI] [PubMed] [Google Scholar]
- 56.Wu Y, Li Q, Chen XZ. Detecting protein-protein interactions by far western blotting. Nat Protoc. 2007;2:3278–3284. doi: 10.1038/nprot.2007.459. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.