Abstract
In yeast mitochondria the DEAD-box helicase Mss116p is essential for respiratory growth by acting as group I and group II intron splicing factor. Here we provide the first structure-based insights into how Mss116p assists RNA folding in vivo. Employing an in vivo chemical probing technique, we mapped the structure of the ai5γ group II intron in different genetic backgrounds to characterize its intracellular fold. While the intron adopts the native conformation in the wt yeast strain, we found that the intron is able to form most of its secondary structure, but lacks its tertiary fold in the absence of Mss116p. This suggests that ai5γ is largely unfolded in the mss116-knockout strain and requires the protein at an early step of folding. Notably, in this unfolded state misfolded substructures have not been observed. As most of the protein-induced conformational changes are located within domain D1, Mss116p appears to facilitate the formation of this largest domain, which is the scaffold for docking of other intron domains. These findings suggest that Mss116p assists the ordered assembly of the ai5γ intron in vivo.
Key words: RNA folding, group II intron, DEAD-box helicase, in vivo structural probing
Introduction
RNA folding is the most essential process underlying RNA function. Understanding RNA structure formation therefore provides deep insights into mechanisms of RNA-dependent processes. So far significant progress has been made in describing RNA structure and associated folding pathways in vitro.1–5 From these studies several interesting folding paradigms have emerged portraying the diversity of RNA biophysical behaviors. However, little is known about RNA structure formation in vivo and a comparison of RNA folding paradigms observed in vitro versus in vivo has also been the focus of only very few studies.6–12
A powerful model system for understanding the forces driving RNA folding in vitro has been a ribozyme derived from the yeast mitochondrial ai5γ group II intron.1 Like for other RNAs, an intricate network of tertiary contacts is essential for the assembly of individual domains into the splicing-competent structure.13–17 The ai5γ ribozyme D135 achieves its complex architecture by following a direct path, in which formation of the scaffolding domain (D1) constitutes a compact intermediate on the way from the unfolded to the native conformation.1,18 Although D135 folding is devoid of kinetic traps,18–20 this RNA collapses slowly to the compact intermediate, while subsequent docking of other domains is rapid.21 A small substructure within D1, the κ–ζ element, was found to control D135 compaction and to initiate a cascade of folding events.22 This folding control element serves as platform for active-site assembly during later stages of folding.23,24 Therefore, the group II intron appears to have evolved a sophisticated folding pathway, in which formation of an active-site docking region is rate-limiting.
In vivo efficient splicing of the ai5γ intron, like for all yeast mitochondrial group II and group I introns, depends on the protein Mss116p.25 MSS116 was identified in a screen for nuclear mutants that are unable to grow by respiration on nonfermentable carbon sources when their mtDNAs contain introns.26 In addition, Mss116p plays a role in mitochondrial translation and RNA processing.25 The splicing deficiency of mitochondrial introns is however not caused by the impaired mitochondrial translation, as splicing of mitochondrial introns continues in a ρ- strain encoding the wt MSS116 allele, but is inhibited in a ρ- strain with a mss116-knockout.25 Importantly, Mss116p also lowers the Mg2+ requirements for group II intron splicing in vitro,27,28 further implying a direct involvement of Mss116p in splicing of yeast mitochondrial introns. Mss116p belongs to the DEAD-box helicase family,29 which are known to be non-processive helicases and were recently suggested to be viewed as ATP-dependent RNA binding proteins.30 Indeed, Mss116p displays unwinding and annealing activity in vitro,27,28,31 whereby its ATPase activity is required to promote intron splicing.27 Two different models have been proposed to explain how Mss116p facilitates splicing of the ai5γ intron. One model suggests that the protein promotes folding of the intron by unwinding misfolded off-pathway intermediates (kinetic traps).28,32 The second model suggests that Mss116p acts more like a general RNA-binding protein and may in some cases stabilize folding intermediates. Multiple in vitro studies have provided experimental evidence in support of both mechanisms.25,27,28,32 Most recently, it has been shown that Mss116p stimulates ai5γ folding in vitro by accelerating the collapse to a near-native, compact intermediate state in an ATP-independent manner through stabilization of an early, unstable folding intermediate.33 Interestingly, the native state of the ai5γ ribozyme appears to be stabilized by binding of attached exons but not by Mss116p.33 At the same time, Mss116p influences the folding mechanism in another way: long exon sequences interfere with ai5γ splicing in vitro.34 Apparently, the unwinding activity of Mss116p is essential for exon unfolding, but not for intron folding in vitro. Despite these recent insights, so far direct studies of Mss116p-facilitated RNA folding in vivo have been lacking.
To elucidate how Mss116p facilitates folding of its natural target RNA in the cellular environment, we set out to first characterize the intracellular structure of the ai5γ group II intron in yeast mitochondria and compared it with the native conformation formed under optimal splicing conditions in vitro. Using chemical probing we found that the intron adopts a virtually identical conformation both in vivo and in vitro despite the very different folding environments, further implying that Mss116p promotes native state formation and in turn splicing. In order to gain the first structure-based insights into Mss116p's mechanism in vivo, we also monitored Mss116p-induced conformational changes within the intron in yeast. Unexpectedly, in the absence of Mss116p the ai5γ intron appears essentially unfolded, as most of the secondary but virtually no tertiary structure is formed. This suggests that the protein is required for folding of the RNA at an early step along the pathway.
Results
Establishing an in vivo assay for mapping RNA structure in yeast mitochondria.
In order to provide mechanistic insights into how Mss116p assists ai5γ splicing, we aimed at elucidating the Mss116p-induced structural changes within this RNA. The approach we chose was in vivo structural probing with dimethylsulfate (DMS).35,36 DMS readily penetrates cells, thereby modifying N1 of adenosines and N3 of cytidine if they are not engaged in H-bonding or protected by protein binding. Thus, this method provides information about RNA conformation both on the secondary and tertiary structure level as well as about RNA-protein interactions and associated structural alterations. Among others, DMS probing in vivo was previously used to study protein-induced conformational changes within the td intron in E. coli, thereby providing the first mechanistic insights into the action of an RNA chaperone on a misfolded intron.11 In the presence of the RNA chaperone StpA the td group I intron structure was significantly more open as indicated by an enhanced accessibility to DMS, revealing that StpA resolves tertiary contacts (e.g., the critical base-triples between the joining segment J3/4 and stem P6) enabling the RNA to refold. In this study we took the DMS probing in vivo method one step further and monitored RNA folding in vivo at true intracellular conditions. In other words, we were analyzing endogenous RNA levels (instead of overexpressing RNA and protein from a plasmid).
To discern the influence of Mss116p on the intron fold, we compared the DMS pattern of the ai5γ intron in wild-type and mss116-knockout strains. As previously reported25 the ai5γ intron splices efficiently in the wt strain, while the knockout of MSS116 causes a severe splicing deficiency (Fig. 1). Notably, when treating cells with DMS, the splicing efficiency of ai5γ remained unchanged (data not shown). Two lines of evidence suggest that Mss116p acts directly on group II folding in yeast: the splicing deficiency of mt introns is not caused by an impaired mt translation25 and Mss116p is sufficient to promote efficient self-splicing at near-physiological conditions in vitro.27,28,32 Therefore, we assume that Mss116p directly contacts the intron RNA in vivo as well, thereby inducing the observed structural changes. In addition, Northern Blot analysis clearly revealed that comparable amounts of cox1 RNA are present in both wt and Δmss116 strains,25 indicating that the unspliced cox1 pre-RNA is not degraded in the knockout strain. In order to probe the ai5γ structure in vivo, yeast was grown in rich medium to log-phase and subsequently treated with DMS to modify the accessible atoms.35 After RNA preparation the modification sites are mapped by reverse transcription, as the bulky methyl-group at the Watson-Crick face of adenosines and cytidines causes the Reverse Transcriptase to stop the primer extension.35,37 The observed modifications were plotted onto the secondary structure map according to their relative intensity (Fig. 2B; Suppl. Figs. 2B and 3B). Upon comparing the modification pattern of the ai5γ intron for the different strains we gained significant insight into the role of Mss116p in group II intron folding.
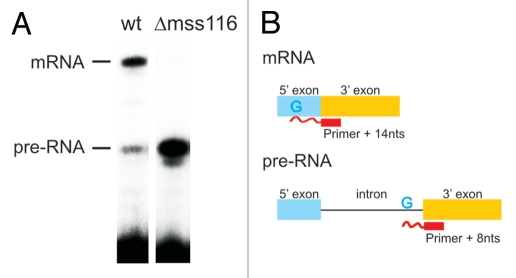
Figure 1.
Mss116p-promoted splicing of the ai5γ intron in yeast mitochondria. (A) Comparison of the in vivo splicing activity in a wt yeast strain and a mss116-knockout strain. In the presence of Mss116p 70–75% of the cox1 pre-RNA are spliced, while less than 1% of splicing occurs in the absence of Mss116p. (B) Scheme of the Poison Primer Assay used to assess in vivo splicing activities. A primer hybridizes to the 5′ end of the downstream exon and is extended by a Reverse Transcriptase in the presence of ddCTP (and dATP, dGTP, dTTP). The extension therefore stops at the first G in the template, which results in a Primer + 14 nts product for the ligated exons and Primer + 8 nts for the unspliced pre-RNA.
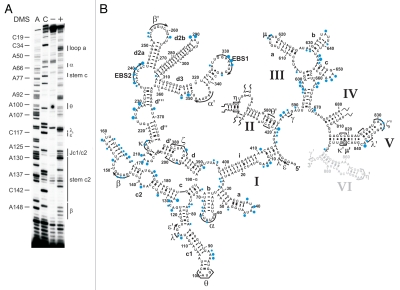
Figure 2.
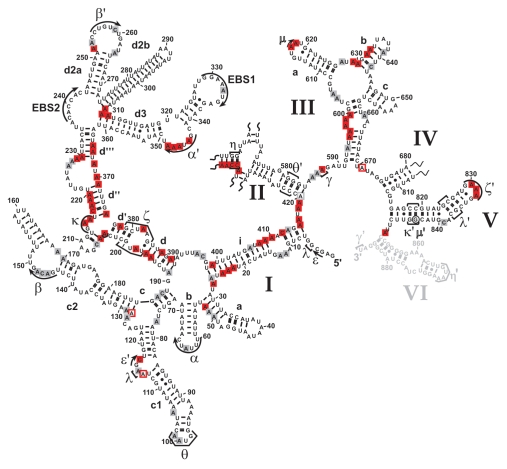
The intracellular structure of the ai5γ intron. (A) Representative primer extension gel showing the in vivo DMS modification pattern of the ai5γ intron - region at the c-c1-c2 three-way junction - in the wt yeast strain. (A and C) denote sequencing lanes. In the - lane natural stops of the Reverse Transcriptase are seen. In the + lane the in vivo DMS pattern is shown. Notably, comparing lanes 3 and 4 reveals the DMS-induced stops of the Reverse Transcriptase and thus accessible residues (N1-A, N3-C). Additional regions of the intron are shown in the subsequent figures (Figs. 3 and 4; Suppl. Figs. 2, 3 and 5). (B) Summary map in which residues methylated by DMS are indicated with blue filled circles. The size of the filled circles correlates with the relative modification intensity of individual bases. As the spliced intron form was analyzed, it was not possible to map the structure of domain 6 shown in gray (as it served as primer binding site to map the very 3′ part of the intron). D2 and D4 are shown in Supplementary Figures 2 and 3, respectively.
The intracellular structure of the ai5γ group II intron.
Group II introns have a conserved secondary structure organization consisting of six domains. The largest domain (D1) provides the scaffold for docking of other intron domains and for exon binding.21,38 Domain 5 is strictly required for catalysis, and together with constituents of D1 and D3, it forms the intron active-site, while Domain 6 harbors the branch point adenosine (reviewed in ref. 15, 39 and 40). A network of long-range tertiary interactions is essential for the assembly of the individual domains into the native, splicing-competent intron structure.14,15,41 In vitro the ai5γ intron requires non-physiological conditions, such as high-salt and elevated temperature, to adopt and maintain its native, splicing-competent conformation.20,42,43 While it is known that in vivo efficient splicing of yeast mitochondrial introns is dependent on both metal ion homeostasis44 and the cofactor Mss116p,25,26 the introns' intracellular fold has not been determined. We therefore mapped the structure of the ai5γ intron in a wt strain employing the in vivo DMS probing technique, thereby providing the first insights into the intracellular fold of a group II intron. The obtained in vivo modification pattern (Fig. 2; Suppl. Figs. 2B and 3B) was compared with the in vitro DMS pattern of the D123456 ribozyme folded in the presence of 100 mM Mg2+ (Suppl. Fig. 1). As the ribozyme was folded in vitro at reaction conditions that promote the formation of discrete tertiary interactions and in turn self-splicing,1 this gives rise to a distinct in vitro DMS modification pattern. As the in vivo and in vitro DMS patterns are almost identical, this suggests that the ai5γ intron forms the same array of interactions in vivo and in vitro. Thus, ai5γ adopts virtually the identical higher-order structure in the presence of Mss116p in vivo and at non-physiological conditions in vitro.
From the in vivo modification pattern it is evident that the secondary structure is properly formed (Fig. 2). Interestingly, some adenosines (e.g., A128 in stem c2, A420 in D2), which have been proposed to form terminal Watson-Crick base-pairs of their respective helices, were found to be intensely methylated. This points to a non-canonical configuration of these base-pairs, i.e., formation of a reverse Hoogsteen A.U pair, which is further supported by NAIM data for A128.45 Alternatively, the terminal base-pairs of duplexes may be “breathing”, thereby becoming modified in some intron molecules.
Apart from the secondary structure, tertiary interactions are in place as well. For example, the intra-domain long-range tertiary contact α-α', which is critical for exon-binding and for D135 compaction in vitro,45,46 is formed; this is exemplified by the lack of modification of residues A344, A346–A348 in the α' region and a weak modification of A63 and C65 in the α element (Figs. 2 and 4B; Suppl. Fig. 5A). Like α-α', the β-β' interaction is also formed by Watson-Crick base-pairing. In the β region we observed weak DMS modifications (Fig. 2). This might result from the fact that we probe an ensemble of intron molecules and β-β' might not be formed in all molecules (i.e., in the remaining pre-RNA). Alternatively, this interaction might be dynamic and not formed all the time. Notably, this interaction is not phylogenetically conserved occurring in only a subset of group II introns.13 Furthermore, the inter-domain long-range contacts are properly formed. For example, the θ-θ' interaction, which brings stem c1 harboring ε and λ' regions close to the D2 basal stem and the essential joining segment J2/3,47,48 is established as indicated by the lack of modification of bases A100–A101 in the θ loop (Fig. 2). Similarly, the in vivo modification pattern implies that the η-η' contact, which anchors D6 onto D2, thereby positioning the branch point adenosine A880 at the active-site,48–50 is likely to be in place, as the stem harboring the receptor η for the D6 tetraloop η' is formed (Suppl. Fig. 2A and B). Also, the coordination loop, which has been proposed to serve as binding site for the branch point A880 in Domain 6,49 adopts the same architecture both in vitro and in vivo (Fig. 2B; Suppl. Fig. 1B). Thus, it appears that the intricate network of long-range tertiary interactions is properly formed in vivo.
Figure 4.
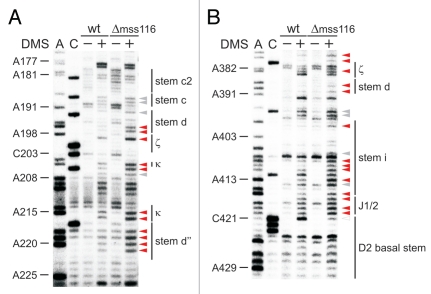
Tertiary structure elements are unfolded in the absence of Mss116p. (A) Representative gel showing the accessibility of nucleotides in the c-c1-c2 three-way junction, which harbors the θ, λ, ε′ and β elements forming long-range tertiary contacts with distinct regions of the intron, in the wt and mss116-knockout strain. Lanes are designated as in Figure 1; symbol code as in Figure 3. The normalized plot derived from this gel is shown (Suppl. Fig. 4C). (B) Representative gel showing the accessibility of residues in stem d3 including the exon binding site 1 (EBS1) and the α′ element, which forms an intra-domain contact with the closing loop of stem b. Lanes are designated as in Figure 1; symbol code as in Figure 3. The normalized plot derived from this gel is shown Supplementary Figure 4D.
The architecture of the active-site in vivo.
As the ai5γ intron splices efficiently in a wt strain (Fig. 1), this strongly implies that we monitored the structure of the spliced intron. Notably, it has previously been shown that most of the spliced ai5γ intron population is in the lariat form.51 Consistent with the fact that we probed the spliced intron, the exon-binding-sites (EBS1 and EBS2) were accessible to DMS (Figs. 2B and 4B). In contrast, from the DMS pattern of residues involved in active-site formation, we infer that the intron's catalytic center remains assembled.
For example, the distinct modification intensities of A382–A383 (Figs. 2B and 3B), which form an A·A platform, imply that the ζ receptor adopts a canonical conformation.52 Likewise, the weak accessibility of A198-N1, which is involved in a reverse-Hoogsteen-pair with U381, and of A830–A832 (Figs. 2B and 3A; Suppl. Fig. 5C) indicates that in the majority of the RNA population the tetraloop ζ' is interacting with its receptor ζ. Similarly, the inaccessibility of A204 and the weak modification of A214–A215 (Figs. 2B and 3A) indicate that the κ-κ' interaction is formed in the spliced intron in vivo. This suggests that the catalytic domain D5 is docked onto D1, which is a prerequisite for active-site formation. In line with this observation, the critical active-site contacts λ-λ' and ε-ε' are intact as well (Fig. 2): C117 is not methylated hinting to the correct formation of ε-ε', in which C117-N3 interacts with G3-N3,23,53,54 thereby rendering C117-N1 inaccessible. Along the same line, A115, which forms a base-triple with the base-pair C825–G836 in D5,55 is barely modified. Thus, in most molecules A115-N1 indeed contacts D5. In addition, residues involved in the ε-ε' and λ-λ' contacts are spatially close to the D5 dinucleotide bulge and J2/3, both of which are known key players in active-site organization.47,56 In the O. iheyensis group II intron crystal structure41 the invariant G5 (ε) interacts with A376 (A838 in ai5γ), which is extruded from the D5 helix. C377 (C839 in ai5γ), in contrast, stacks on top of G288 (G588 in ai5γ) and C289 (A589 in ai5γ) in J2/3, forming a triple-helix with the catalytic triad (C358–C360 in the Oi. intron and A816–C818 in ai5γ). Consistent with this architecture we observe a strong modification of A838, while A589 and C839 are barely methylated (Fig. 2B; Suppl. Fig. 5B and C), suggesting that these bases indeed form base-triples with A816–U847 and C818–G845, respectively. Also, the minor modification of A587 (Fig. 2B; Suppl. Fig. 5B) suggests that the γ-γ' contact between A587 in J2/3 and U887 is formed.
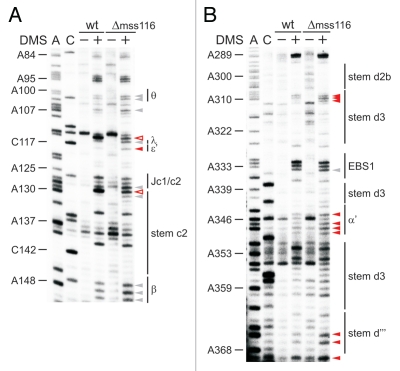
Figure 3.
The κ-ζ element depends on Mss116p for folding. Representative gel showing the modification intensity of nucleotides in the 5′ part (A) and 3′ part (B) of the D1 core structure composed of stems d, d′ and d″, in which the κ region, the ζ receptor and the coordination loop are embedded, in the wt and mss116-knockout strain. The arrow heads indicate residues, the accessibility of which changes due to the absence of Mss116p (filled arrow heads represent an increase in accessibility, while open ones highlight bases with reduced accessibility). Strong changes in accessibility are displayed in red (>2-fold); while smaller changes are shown in gray (1.5 to 2-fold). These values were derived from the normalized gel plots (see Suppl. Fig. 4A and B). Lanes are designated as in Figure 1 with the exception that − and + lanes are shown for both the wt and mss116-knockout strain. Comparing lanes 4 and 6 reveals the altered DMS modification pattern and thus conformational changes within the ai5γ intron due to the absence of Mss116p.
Moreover, the catalytic effector D3 contributes to active-site formation.54,57 Of particular importance are the internal bulge of the basal stem and the pentaloop (N3-A617; N3-A618), which forms the ε-ε' interaction with G844-2′OH in D5.58 The minor modification of A617–A618 (Fig. 2B; Suppl. Fig. 5B) is in good agreement with an intact ε-ε' interaction in vivo. As for the internal D3 bulge, two consecutive sheared A·A base-pairs are essential for enhancing the rate of catalysis.58 Their formation involves the Hoogsteen face of A599 and A662 H-bonding with the minor groove edge of A661 and A598, respectively.58 Also, this internal loop (A599-N1; A662-N1) appears to contact the tandem G-C base-pairs of the ε-ε′ motif.54 Interestingly, bases within the internal bulge were equally methylated (Fig. 2B, Suppl. Fig. 5B) in contrast to the in vitro DMS pattern, in which only A599 is modified (Suppl. Fig. 1). As the sheared A.A base-pairs do not involve the N1 of respective adenosines, we cannot evaluate their conformation; however the observed modification pattern suggests that the D1–D3 contact either does not involve A599-N1 and A662-N1 or it does not form in vivo (at least in the spliced form). Thus, in vivo D3 adopts a conformation slightly different than the one observed in vitro. Aside from the notable change in the D3 bulge, the in vivo structural probing data strongly suggest that the spliced intron maintains a structure expected for a functional molecule.
Mapping the intracellular structure of the ai5γ intron we also aimed at determining whether Mss116p is bound to the spliced intron, thereby protecting distinct residues from modification and resulting in a specific footprint. Upon comparing the in vitro and in vivo modification patterns of the most essential domains (D1, D3 and D5) we did not observe an Mss116p-specific footprint. Therefore, we were particularly interested in D2 and D4, since other group II intron splicing cofactors have binding sites in these domains.59 Surprisingly, we did not detect an Mss116p-footprint either in D4 or D2 (Suppl. Figs. 2 and 3), suggesting that the protein does not remain bound to the intron in the native state. Possibly, Mss116p binding has a high koff. Alternatively, the interaction between Mss116p and the intron might not involve functional groups which we can assess with DMS.
The DEAD-box protein is essential for the formation of the κ-ζ folding control element.
That the κ-ζ element is of utmost importance for ai5γ folding in vitro specifically sparked our interest into how Mss116p influences the formation of this critical substructure at the core of the D1 scaffold. Therefore, we compared the in vivo DMS pattern of the wt and mss116-knockout strains (Figs. 3 and 4; Suppl. Figs. 2A, 3A and 5A–C). In the absence of Mss116p the modification intensity of many residues was altered (e.g., A195–A196, A214–A215, C217-A220; indicated with arrow heads in Fig. 3A), while others remained unaffected (e.g., A176–A177, A208–A210 or A225 in Fig. 3A). The observed conformational changes were summarized in a differential map (Fig. 5, Suppl. Figs. 2C and 3C), which is based on normalized plots (Suppl. Figs. 2D, 3D, 4 and 6) derived from the modification gels (representative ones are shown in Figs. 3, 4, Suppl. Figs. 2A, 3A and 5A–C). In this differential map an increase or decrease in modification intensity of residues in the Δmss116 strain is indicated with a closed and open square, respectively, while residues which are equally modified (e.g., A92–A95 or A331–A332) or protected (A121–A122 or A300–A305) in both strains are not highlighted (Fig. 5, Suppl. Figs. 2C and 3C).
Figure 5.
Mss116p-induced conformational changes within the ai5γ intron in vivo. Differential summary map: residues, the modification intensity of which changes in the absence of Mss116p (Δmss116 strain), are highlighted. This map is based on normalized plots (see Suppl. Figs. 4 and 6), which had been derived from the modification gels. The closed squares indicate an increase in accessibility; while open squares represent bases with reduced accessibility in the absence of Mss116p. The (A and C) residues whose modification remains unaltered (i.e., equally modified or protected in both strains) are not highlighted. Color code as in Figure 3. D2 and D4 are shown in Supplementary Figures 2 and 3, respectively.
First of all, it is important to mention that in the Δmss116 strain extensive formation of secondary structure elements is observed throughout the molecule except for the κ-ζ element and some other regions (Suppl. Fig. 7). This conformation is highly reminiscent of the state, which the ai5γ ribozyme adopts in vitro in the presence of KCl only.22 Of the observed conformational changes, most residues, which show the strongest increase in accessibility to DMS in the absence of Mss116p, indeed clustered within the κ-ζ element (red squares; Fig. 5). Adenosines both in the κ and ζ regions (A198, A204, A214, A215 and A382) are significantly more accessible in the Δmss116 strain (Figs. 3 and 5), implying that D5 is not docked onto D1. The increased accessibility of adenosines in the D5 tetraloop (A830–A832) further supports this observation (Suppl. Fig. 5C). Consistent with this the active-site constituents ε-ε′ and λ-λ′ are not formed either, as exemplified by the increased accessibility of A115 and C117 (Fig. 4A). Due to the lack of the λ-λ′ contact A114-N1 is not pushed out to the solvent resulting in a reduced A114 modification. The reduced accessibility of A671 in J3/4 could be explained in a manner comparable to that of A114. In case of A128 and A420, these adenosines are likely to form Watson-Crick base pairs with U185 and U583, respectively, in the unfolded state (i.e., in the Δmss116 strain), while these rearrange to non-canonical base pairs upon native state formation (as previously observed for A875 of the td group I intron12 and as seen for A128 in Suppl. Fig. 1A, comparing lane 0 and lane 100). Aside from the lack of critical tertiary interactions, it is also evident that even the secondary structure, into which the κ-ζ element is embedded, does not form without Mss116p (enhanced modification of e.g., A195–A196, C217-A220; Figs. 3 and 5). Strikingly, in vitro these stems are dependent on Mg2+ ions for their formation and play an essential role in the D135 collapse.22,45 These data indicate that Mss116p is essential for the formation of the folding control element. In light of the in vitro folding pathway it is likely that the element itself may be of importance for ai5γ folding in vivo as well.
Mss116p facilitates formation of tertiary and some secondary structure.
As the κ-ζ element initiates a cascade of folding events in the D135 ribozyme in vitro,21,22,45 we aimed at understanding whether Mss116p depletion broadly affects the intron structure. Assessing the modification pattern of known tertiary contacts revealed that these interactions do not form without Mss116p as seen in the differential map in Figure 5. The α-α′ and θ-θ′ interactions are open; residues within these interaction partners (e.g., A146–A148, A346–A348; Figs. 4 and 5; Suppl. Fig. 5A) are strongly modified in the Δmss116 strain. As A100–A101 in the θ region (Figs. 4A and 5) as well as C464–C465 of the η region (Suppl. Fig. 2A and C) are significantly more accessibility to DMS in the Δmss116 strain, this suggests that the tetraloopreceptor interactions θ-θ′ and η-η′ do not form without Mss116p. Also, adenosines in the D6 tetraloop (A868–A869) are strongly modified in the absence of Mss116p (Suppl. Fig. 5D). Along the same line, the coordination loop (A227–A229, A368, A370) is also more exposed in the Δmss116 strain (Fig. 5). Furthermore, the strong increase in accessibility of A617-N1 and A618-N1 (Suppl. Fig. 5B) suggests that the D3–D5 contact ε-ε′ does not form in the mss116-knockout strain. These findings imply that the active-site is not assembled in the absence of Mss116p. Consistent with this both EBS sites are modified in the Δmss116 strain (Fig. 4B and data not shown).
Aside from tertiary interactions several helices cannot form without Mss116p (Figs. 3–5; Suppl. Fig. 5). In addition to the afore mentioned helices (d, d′, d″), stem i, which protrudes from the five-way junction in D1, is largely unformed, as indicated by methylation of e.g., residues A22–A24 and A408–A413 in the Δmss116 strain (Figs. 3B and 5; Suppl. Fig. 5A). Also, residues in stems a and c (A50–A52 and A187–C188) are somewhat more accessible to DMS, suggesting that they are to some extent less stable and might be “breathing” in the absence of Mss116p (Figs. 3A and 5; Suppl. Fig. 5A). Interestingly, Mg2+ ions are strictly required for the formation of these helices (except for stem i) in vitro.22 In other words the structural conformation, which the ai5γ intron is capable of adopting in the absence of Mss116p in vivo (Suppl. Fig. 7), strongly correlates with the unfolded state of the D135 ribozyme obtained in vitro in the presence of monovalent ions only.22
Besides D1, D3—its four-way junction together with helix b and the internal loop—appears partially unfolded (e.g., increased modification of A596–A600; A627 through A633; Fig. 5; Suppl. Fig. 5B). Interestingly, in D2 the structural alteration is limited to the stem containing η and some other subtle changes (e.g., A441–A450; Suppl. Figs. 2A and C). Similarly, within D4 only A801–A803 is more intensely modified in the Δmss116 strain (Suppl. Fig. 3C). Hence, Mss116p seems to be less important for the structural organization of D2 and D4. Also, formation of the D5 and D6 secondary structure is not dependent on Mss116p. Thus, the data suggest that Mss116p actively promotes the formation of the D1 scaffold followed by assembly of the ai5γ intron tertiary structure.
It is noteworthy that in the absence of Mss116p the intron is substantially unfolded, but in this unfolded state (Δmss116 strain) we did not detect any misfolded substructures; i.e., a region, which should be single-stranded in the native intron molecule, forming base-pairs with some other element (like an altP3,60 equivalent) or that only one part of a helix is protected from modification while its counterpart is accessible (implying that they do not interact with each other). In contrast, most residues are methylated to a comparable extent in the Δmss116 strain. In brief, in the absence of Mss116p the ai5γ intron appears largely unfolded, suggesting that the protein is required for intron folding at an early step.
Discussion
RNA folding, which is most integral to RNA function, has so far mostly been studied in vitro.1–5 Of the studies that monitored intracellular RNA structure formation, only few have attempted a comprehensive correlation of in vitro versus in vivo folding paradigms.6,12 As the folding pathway of the ai5γ ribozyme has been thoroughly studied in vitro at non-physiological conditions,1 we aimed at elucidating how this large, multidomain RNA, which depends on the helicase Mss116p for efficient splicing in vivo, folds in yeast. Within this initial study we have already observed some interesting parallels between the in vitro and in vivo folding pathways. Thus, at least in case of the ai5γ intron, some aspects of the in vitro folding paradigm apply to folding in the living cell. Additional studies are required to determine the extent of correlation between in vivo and in vitro folding pathways.
Here we initially describe the first intracellular structure of a group II intron, namely that of the ai5γ intron in yeast using in vivo chemical probing. Notably, we worked with endogenous RNA levels to obtain a biologically relevant picture of intracellular RNA folding. Monitoring the ai5γ structure in a wt genetic background revealed that the intron adopts a conformation expected for a catalytically active intron. Indeed, comparing the in vivo modification pattern of the ai5γ intron (Fig. 2) with that of the D123456 ribozyme folded in vitro at conditions optimal for self-splicing (Suppl. Fig. 1) implied that in both molecules tertiary contacts as well as the active-site are properly assembled despite the different folding environments. This suggests that Mss116p, which is encoded in the wt strain, substitutes for the high [Mg2+] required for folding of ai5γ ribozymes in vitro.20,42,43 Consistent with this, Mss116p is capable of promoting ai5γ splicing at near-physiological conditions in vitro.27,28,32
As the ai5γ intron splices efficiently in the wt strain, our results indicate that the spliced intron retains its native conformation. This finding is in excellent agreement with ai5γ being capable of reverse splicing.61 This process and associated intron homing is a long known characteristic of group II introns, for which most introns depend on a maturase for assistance.39 Although ai5γ is not mobile, our results might still have implications for homing, as we provide the first insights into the architecture of a reverse splicing-competent intron in vivo. It appears that group II introns might not have to undergo large-scale rearrangements to act as mobile genetic elements after splicing.
Efficient splicing of yeast mitochondrial introns depends on the nuclear-encoded DEAD-box protein Mss116p. Helicases are an ubiquitous protein family involved in virtually all cellular processes by promoting RNA structural rearrangements and RNP remodeling.62 Therefore, Mss116p-promoted ai5γ splicing represents an interesting model system to assess general principles of intracellular folding and the novel role of helicases in this fundamental aspect of RNA function. By mapping the ai5γ structure in a mss116-knockout strain, we were able to present the first structural basis for helicase-facilitated folding of a complex RNA in vivo. In the absence of Mss116p the intron is largely unfolded (except for most of the secondary structure), suggesting that the protein is required at an early step in the folding pathway. As outlined before, we have not found any evidence for misfolding of the ai5γ intron. Instead our observations are more in line with the intron being inherently unstable. This is in excellent agreement with the loss of splicing activity. Also, as a non-functional ai5γ intron interferes with cytochrome oxidase I assembly in yeast mitochondria, depending on an intron, of which >90% of the population misfolds, might not be favorable for cell viability. Consistent with this, it has been shown that misfolded RNA is rapidly degraded in yeast.63 Thus, our data suggest that Mss116p assists intron folding and in turn splicing by promoting the ordered assembly of the RNA. This could be achieved through stabilizing the intron RNA during folding or Mss116p might act as a RNA chaperone. Interestingly, in a previous study the RNA chaperone StpA-induced conformational changes in the td group I intron were assessed by in vivo chemical probing.11 In contrast to Mss116p's action on ai5γ, the RNA chaperone StpA was observed to loosen discrete tertiary interactions in the td intron, thereby giving the RNA a chance to refold and to reach the native conformation.11 In case of Mss116p, however we only observed that residues are becoming significantly less accessible to DMS modification in the presence of the protein. Also, StpA did resolve the structure of the spliced form of the td intron,11 while the spliced ai5γ intron maintains its native structure in the presence of Mss116p (Fig. 2). Together with the fact that the ai5γ intron adopts a highly comparable conformation both in vitro in the absence of Mg2+ and in vivo in the absence of Mss116p, this points to a rather stabilizing role of Mss116p in ai5γ folding. However, in light of the fact that Mss116p's unwinding activity is necessary to promote splicing of pre-RNA with long exons in vitro,34 it is possible that inhibitory structures are present in the exons flanking the ai5γ intron in the Δmss116 strain, which are otherwise unwound by Mss116p in the wt strain, thereby further assisting intron splicing.
In vitro Mss116p assists the formation of an early, unstable folding intermediate, which is followed by a cascade of folding events resulting in the formation of the compact near-native state.33 The near-native state formed in the presence of Mss116p is identical to that formed more slowly without Mss116p,19,33 suggesting that the initial steps in the ai5γ folding pathway are highly similar at near-physiological conditions in the absence and presence of Mss116p in vitro. Importantly, high [Mg2+] is able to accelerate the ai5γ collapse in vitro as well (rate constant for compaction are 1.9 min−1 and 0.004 min−1 at non- and near-physiological conditions, respectively19,21), further implying that Mss116p replaces the function of high metal ion concentration. In vivo the RNA is largely unfolded in the absence of Mss116p, suggesting that the cofactor protein is required at an early step in ai5γ folding in yeast mitochondria as well. As the strongest protein-induced structural changes occur in the D1 scaffold (Fig. 5), this indicates that Mss116p assists in the assembly of D1, which is a prerequisite for docking of other intron domains onto the D1 scaffold during later stages of folding. As intracellular RNA folding occurs co-transcriptionally, D1 is likely to play a central role in intron folding, since it is transcribed first. Accordingly, D1 has to fold early. That most of the strongest modifications within D1 are clustered at the κ-ζ element in the Δmss116 strain points to a central role in the formation of the D1 scaffold in vivo. However, the precise site of action of Mss116p during the early folding steps remains enigmatic both in vitro and in vivo. In any case folding of the scaffolding domain appears to be a critical step at non-physiological conditions,21 near-physiological conditions with3 and without Mss116p19 as well as in yeast mitochondria. Aside from accelerating the Mg2+-induced collapse of ai5γ in an ATP-independent manner, it has been shown that ATP hydrolysis is necessary for the protein's turnover and that Mss116p does not stabilize the native state of the ribozyme, but that such stabilization results from binding of flanking exons.33 In light of these findings, it is tempting to speculate that we do not observe an Mss116p-specific footprint in the native ai5γ intron, because the protein might not remain bound to the spliced intron. Indeed, ATP hydrolysis was found to play a role in promoting splicing in vivo as well.25 To this end, although we have yet not identified an intermediate state in vivo, Mss116p appears to be important early along both the in vitro33 and intracellular folding pathways and is perhaps also less important during later folding stages in yeast mitochondria. The details of the in vivo folding pathway of the ai5γ intron have yet to be explored.
Materials and Methods
Strains and plasmids.
The yeast strains used in this study were a generous gift from Phil Perlman.25 The wt strain (MATa ade1 lys1) had previously been used to create the mss116-knockout strain (MATa ade1 lys1 mss116Δ::kanr). The plasmid pEL85 containing the D123456 ribozyme was kindly provided by Anna M. Pyle. The BamHI-linearized plasmids were used for in vitro transcription.
In vivo DMS structural probing.
A detailed description of this method is found in reference 35 and 36. In brief, yeast was grown to OD600 nm of 1 in YP medium with 2% Raffinose and then incubated with DMS at a final concentration of 50 mM (30°C, 2 minutes). After quenching the reaction with β-mercaptoethanol, the cell pellet was frozen at −80°C for 15–30 minutes and then total RNA was isolated using the ‘RiboPure™-Yeast’ Kit (Ambion). Importantly, an untreated control, in which RNA was isolated from cells that were not treated with DMS, was also prepared.
Chemical probing in vitro.
10 pmol D123456 ribozyme were folded by first denaturing the RNA in the presence of 0.5 M KCl and 50 mM Cacodylate buffer pH 7.0 at 95°C for one minute followed by 2 minutes at room temperature. MgCl2 or EDTA pH 8.0 were then added to 100 mM or 0.1 mM (f.c.), respectively, in order to yield native or unfolded ribozymes (100 and 0 lanes in Suppl. Fig. 1). The samples were incubated at 42°C for 20 minutes. After folding the DMS modification was performed by adding DMS (f.c. 29 mM) and incubating the samples at room temperature for 20 minutes. The reaction was stopped using β-mercaptoethanol (f.c. 280 mM). After precipitation the RNA was used for Reverse Transcription.
Reverse transcription.
Either 40 εg of total RNA or 1 pmol of D123456 RNA was used for reverse transcription, which was essentially performed as described in reference 35. RT stop controls were obtained from unmodified RNA to detect natural stops of the AMV Reverse Transcriptase (Promega) or Transcriptor Reverse Transcriptase (Roche) along the ai5γ RNA template. Primers used for reverse transcription are summarized in Supplementary Table 1. Analysis of polyacrylamide gels was done with ImageQuant software (GE Healthcare).
In vivo splicing assay.
This assay was performed as described in reference 11 with the notable exception that ddCTP was used to stop extension of the primer at the first G in the template RNA (Fig. 1).
Acknowledgements
Phil Perlman and Anna Pyle are acknowledged for their generous gifts of yeast strains and plasmids, respectively. We are grateful to Olga Fedorova for critically reading the manuscript. This work was supported by the Austrian Science Foundation FWF [grant Y401 to C.W.].
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/13484
Supplementary Material
References
- 1.Pyle AM, Fedorova O, Waldsich C. Folding of group II introns: a model system for large, multidomain RNAs? Trends Biochem Sci. 2007;32:138–145. doi: 10.1016/j.tibs.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 2.Shcherbakova I, Mitra S, Laederach A, Brenowitz M. Energy barriers, pathways and dynamics during folding of large, multidomain RNAs. Curr Opin Chem Biol. 2008;12:655–666. doi: 10.1016/j.cbpa.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sosnick TR. Kinetic barriers and the role of topology in protein and RNA folding. Protein Sci. 2008;17:1308–1318. doi: 10.1110/ps.036319.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Treiber DK, Williamson JR. Beyond kinetic traps in RNA folding. Curr Opin Struct Biol. 2001;11:309–314. doi: 10.1016/s0959-440x(00)00206-2. [DOI] [PubMed] [Google Scholar]
- 5.Woodson SA. Metal ions and RNA folding: a highly charged topic with a dynamic future. Curr Opin Chem Biol. 2005;9:104–109. doi: 10.1016/j.cbpa.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 6.Donahue CP, Fedor MJ. Kinetics of hairpin ribozyme cleavage in yeast. RNA. 1997;3:961–973. [PMC free article] [PubMed] [Google Scholar]
- 7.Donahue CP, Yadava RS, Nesbitt SM, Fedor MJ. The kinetic mechanism of the hairpin ribozyme in vivo: influence of RNA helix stability on intracellular cleavage kinetics. J Mol Biol. 2000;295:693–707. doi: 10.1006/jmbi.1999.3380. [DOI] [PubMed] [Google Scholar]
- 8.Mahen EM, Harger JW, Calderon EM, Fedor MJ. Kinetics and thermodynamics make different contributions to RNA folding in vitro and in yeast. Mol Cell. 2005;19:27–37. doi: 10.1016/j.molcel.2005.05.025. [DOI] [PubMed] [Google Scholar]
- 9.Yadava RS, Choi AJ, Lebruska LL, Fedor MJ. Hairpin ribozymes with four-way helical junctions mediate intracellular RNA ligation. J Mol Biol. 2001;309:893–902. doi: 10.1006/jmbi.2001.4713. [DOI] [PubMed] [Google Scholar]
- 10.Adilakshmi T, Lease RA, Woodson SA. Hydroxyl radical footprinting in vivo: mapping macromolecular structures with synchrotron radiation. Nucleic Acids Res. 2006;34:64. doi: 10.1093/nar/gkl291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Waldsich C, Grossberger R, Schroeder R. RNA chaperone StpA loosens interactions of the tertiary structure in the td group I intron in vivo. Genes Dev. 2002;16:2300–2312. doi: 10.1101/gad.231302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Waldsich C, Masquida B, Westhof E, Schroeder R. Monitoring intermediate folding states of the td group I intron in vivo. EMBO J. 2002;21:5281–5291. doi: 10.1093/emboj/cdf504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Michel F, Ferat JL. Structure and activities of group II introns. Annu Rev Biochem. 1995;64:435–461. doi: 10.1146/annurev.bi.64.070195.002251. [DOI] [PubMed] [Google Scholar]
- 14.Qin PZ, Pyle AM. The architectural organization and mechanistic function of group II intron structural elements. Curr Opin Struct Biol. 1998;8:301–308. doi: 10.1016/s0959-440x(98)80062-6. [DOI] [PubMed] [Google Scholar]
- 15.Lehmann K, Schmidt U. Group II introns: structure and catalytic versatility of large natural ribozymes. Crit Rev Biochem Mol Biol. 2003;38:249–303. doi: 10.1080/713609236. [DOI] [PubMed] [Google Scholar]
- 16.Toor N, Rajashankar K, Keating KS, Pyle AM. Structural basis for exon recognition by a group II intron. Nat Struct Mol Biol. 2008;15:1221–1222. doi: 10.1038/nsmb.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Michel F, Costa M, Westhof E. The ribozyme core of group II introns: a structure in want of partners. Trends Biochem Sci. 2009;34:189–199. doi: 10.1016/j.tibs.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 18.Steiner M, Karunatilaka KS, Sigel RK, Rueda D. Single-molecule studies of group II intron ribozymes. Proc Natl Acad Sci USA. 2008;105:13853–13858. doi: 10.1073/pnas.0804034105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fedorova O, Waldsich C, Pyle AM. Group II intron folding under near-physiological conditions: collapsing to the near-native state. J Mol Biol. 2007;366:1099–1114. doi: 10.1016/j.jmb.2006.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Swisher J, Su L, Brenowitz M, Anderson V, Pyle A. productive folding to the native state by a group II intron ribozyme. J Mol Biol. 2002;315:297–310. doi: 10.1006/jmbi.2001.5233. [DOI] [PubMed] [Google Scholar]
- 21.Su LJ, Waldsich C, Pyle AM. An obligate intermediate along the slow folding pathway of a group II intron ribozyme. Nucleic Acids Res. 2005;33:6674–6687. doi: 10.1093/nar/gki973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Waldsich C, Pyle AM. A kinetic intermediate that regulates proper folding of a group II intron RNA. J Mol Biol. 2008;375:572–580. doi: 10.1016/j.jmb.2007.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boudvillain M, Pyle AM. Defining functional groups, core structural features and inter-domain tertiary contacts essential for group II intron self-splicing: a NAIM analysis. EMBO J. 1998;17:7091–7104. doi: 10.1093/emboj/17.23.7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Costa M, Michel F. Frequent use of the same tertiary motif by self-folding RNAs. EMBO J. 1995;14:1276–1285. doi: 10.1002/j.1460-2075.1995.tb07111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang HR, Rowe CE, Mohr S, Jiang Y, Lambowitz AM, Perlman PS. The splicing of yeast mitochondrial group I and group II introns requires a DEAD-box protein with RNA chaperone function. Proc Natl Acad Sci USA. 2005;102:163–168. doi: 10.1073/pnas.0407896101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seraphin B, Boulet A, Simon M, Faye G. Construction of a yeast strain devoid of mitochondrial introns and its use to screen nuclear genes involved in mitochondrial splicing. Proc Natl Acad Sci USA. 1987;84:6810–6814. doi: 10.1073/pnas.84.19.6810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Solem A, Zingler N, Pyle AM. A DEAD protein that activates intron self-splicing without unwinding RNA. Mol Cell. 2006;24:611–617. doi: 10.1016/j.molcel.2006.10.032. [DOI] [PubMed] [Google Scholar]
- 28.Halls C, Mohr S, Del Campo M, Yang Q, Jankowsky E, Lambowitz AM. Involvement of DEAD-box proteins in group I and group II intron splicing. Biochemical characterization of Mss116p, ATP hydrolysis-dependent and -independent mechanisms and general RNA chaperone activity. J Mol Biol. 2007;365:835–855. doi: 10.1016/j.jmb.2006.09.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seraphin B, Simon M, Boulet A, Faye G. Mitochondrial splicing requires a protein from a novel helicase family. Nature. 1989;337:84–87. doi: 10.1038/337084a0. [DOI] [PubMed] [Google Scholar]
- 30.Liu F, Putnam A, Jankowsky E. ATP hydrolysis is required for DEAD-box protein recycling but not for duplex unwinding. Proc Natl Acad Sci USA. 2008;105:20209–20214. doi: 10.1073/pnas.0811115106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang Q, Del Campo M, Lambowitz AM, Jankowsky E. DEAD-box proteins unwind duplexes by local strand separation. Mol Cell. 2007;28:253–263. doi: 10.1016/j.molcel.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 32.Del Campo M, Tijerina P, Bhaskaran H, Mohr S, Yang Q, Jankowsky E, et al. Do DEAD-box proteins promote group II intron splicing without unwinding RNA? Mol Cell. 2007;28:159–166. doi: 10.1016/j.molcel.2007.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fedorova O, Solem A, Pyle AM. Protein-facilitated folding of group II intron ribozymes. J Mol Biol. 2010;397:799–813. doi: 10.1016/j.jmb.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zingler N, Solem A, Pyle AM. Dual roles for the Mss116 cofactor during splicing of the ai5gamma group II intron. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liebeg A, Waldsich C. Probing RNA structure within living cells. Methods Enzymol. 2009;468:219–238. doi: 10.1016/S0076-6879(09)68011-3. [DOI] [PubMed] [Google Scholar]
- 36.Wells SE, Hughes JM, Igel AH, Ares M., Jr Use of dimethyl sulfate to probe RNA structure in vivo. Methods Enzymol. 2000;318:479–493. doi: 10.1016/s0076-6879(00)18071-1. [DOI] [PubMed] [Google Scholar]
- 37.Brunel C, Romby P. Probing RNA structure and RNA-ligand complexes with chemical probes. Methods Enzymol. 2000;318:3–21. doi: 10.1016/s0076-6879(00)18040-1. [DOI] [PubMed] [Google Scholar]
- 38.Qin PZ, Pyle AM. Stopped-flow fluorescence spectroscopy of a group II intron ribozyme reveals that domain 1 is an independent folding unit with a requirement for specific Mg2+ ions in the tertiary structure. Biochemistry. 1997;36:4718–4730. doi: 10.1021/bi962665c. [DOI] [PubMed] [Google Scholar]
- 39.Pyle AM, Lambowitz AM. Group II introns: ribozymes that splice RNA and invade DNA. In: Gesteland RF, Cech TR, Atkins JF, editors. The RNA World. Cold Spring Harbor, New York: Cold Spring Harbor Laborartory Press; 2006. pp. 469–534. [Google Scholar]
- 40.Michel F, Umesono K, Ozeki H. Comparative and functional anatomy of group II catalytic introns—a review. Gene. 1989;82:5–30. doi: 10.1016/0378-1119(89)90026-7. [DOI] [PubMed] [Google Scholar]
- 41.Toor N, Keating KS, Taylor SD, Pyle AM. Crystal structure of a self-spliced group II intron. Science. 2008;320:77–82. doi: 10.1126/science.1153803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Su LJ, Brenowitz M, Pyle AM. An alternative route for the folding of large RNAs: apparent two-state folding by a group II intron ribozyme. J Mol Biol. 2003;334:639–652. doi: 10.1016/j.jmb.2003.09.071. [DOI] [PubMed] [Google Scholar]
- 43.Swisher J, Duarte C, Su L, Pyle A. Visualizing the solvent-inaccessible core of a group II intron ribozyme. EMBO J. 2001;20:2051–2061. doi: 10.1093/emboj/20.8.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gregan J, Kolisek M, Schweyen RJ. Mitochondrial Mg(2+) homeostasis is critical for group II intron splicing in vivo. Genes Dev. 2001;15:2229–2237. doi: 10.1101/gad.201301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Waldsich C, Pyle AM. A folding control element for tertiary collapse of a group II intron ribozyme. Nat Struct Mol Biol. 2007;14:37–44. doi: 10.1038/nsmb1181. [DOI] [PubMed] [Google Scholar]
- 46.Su L. Biochemistry and Molecular Biophysics. New York: Columbia University; 2002. The folding pathway and core structure assembly of a group II intron ribozyme; p. 187. [Google Scholar]
- 47.Fedorova O, Mitros T, Pyle AM. Domains 2 and 3 interact to form critical elements of the group II intron active site. J Mol Biol. 2003;330:197–209. doi: 10.1016/s0022-2836(03)00594-1. [DOI] [PubMed] [Google Scholar]
- 48.Costa M, Deme E, Jacquier A, Michel F. Multiple tertiary interactions involving domain II of group II self-splicing introns. J Mol Biol. 1997;267:520–536. doi: 10.1006/jmbi.1996.0882. [DOI] [PubMed] [Google Scholar]
- 49.Hamill S, Pyle AM. The receptor for branch-site docking within a group II intron active site. Mol Cell. 2006;23:831–840. doi: 10.1016/j.molcel.2006.07.017. [DOI] [PubMed] [Google Scholar]
- 50.Chanfreau G, Jacquier A. An RNA conformational change between the two chemical steps of group II selfsplicing. EMBO J. 1996;15:3466–3476. [PMC free article] [PubMed] [Google Scholar]
- 51.Podar M, Chu VT, Pyle AM, Perlman PS. Group II intron splicing in vivo by first step hydrolysis. Nature. 1998;391:915–918. doi: 10.1038/36142. [DOI] [PubMed] [Google Scholar]
- 52.Cate JH, Gooding AR, Podell E, Zhou K, Golden BL, Kundrot CE, et al. Crystal structure of a group I ribozyme domain: principles of RNA packing. Science. 1996;273:1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- 53.Jacquier A, Michel F. Base-pairing interactions involving the 5′ and 3′-terminal nucleotides of group II selfsplicing introns. J Mol Biol. 1990;213:437–447. doi: 10.1016/S0022-2836(05)80206-2. [DOI] [PubMed] [Google Scholar]
- 54.Fedorova O, Pyle AM. A conserved element that stabilizes the group II intron active site. RNA. 2008;14:1048–1056. doi: 10.1261/rna.942308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boudvillain M, Delencastre A, Pyle AM. A new RNA tertiary interaction that links active-site domains of a group II intron and anchors them at the site of catalysis. Nature. 2000;406:315–318. doi: 10.1038/35018589. [DOI] [PubMed] [Google Scholar]
- 56.de Lencastre A, Pyle AM. Three essential and conserved regions of the group II intron are proximal to the 5′-splice site. RNA. 2008;14:11–24. doi: 10.1261/rna.774008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Koch JL, Boulanger SC, Dib-Hajj SD, Hebbar SK, Perlman PS. Group II Introns deleted for multiple substructures retain self-splicing activity. Mol Cell Biol. 1992;12:1950–1958. doi: 10.1128/mcb.12.5.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fedorova O, Pyle AM. Linking the group II intron catalytic domains: tertiary contacts and structural features of domain 3. EMBO J. 2005;24:3906–3916. doi: 10.1038/sj.emboj.7600852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Solem A, Zingler N, Pyle AM J. L-P-T. Group II introns and their protein collaborators. In: Walter NG, Woodson SA, Batey RT, editors. Non-protein coding RNAs. Berlin Heidelberg: Springer-Verlag; 2009. pp. 167–182. [Google Scholar]
- 60.Pan J, Woodson SA. Folding intermediates of a self-splicing RNA: mispairing of the catalytic core. J Mol Biol. 1998;280:597–609. doi: 10.1006/jmbi.1998.1901. [DOI] [PubMed] [Google Scholar]
- 61.Roitzsch M, Pyle AM. The linear form of a group II intron catalyzes efficient autocatalytic reverse splicing, establishing a potential for mobility. RNA. 2009;15:473–482. doi: 10.1261/rna.1392009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 63.Jackson SA, Koduvayur S, Woodson SA. Self-splicing of a group I intron reveals partitioning of native and misfolded RNA populations in yeast. RNA. 2006;12:2149–2159. doi: 10.1261/rna.184206. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.