Abstract
Sin Nombre hantavirus (SNV) is a New World hantavirus and causes hantavirus cardiopulmonary syndrome. The viral nucleocapsid protein (N) is an RNA chaperone and has multiple functions important in virus replication. The three negative sense RNA segments of hantaviruses form panhandle structures through imperfect hydrogen bonding of the 5′ and 3′ termini, and the chaperone activity of N can mediate correct panhandle formation. N also functions during transcription and translation initiation and the chaperone activity of N is likely to be involved in aspects of these processes. Using a series of mutations in the N gene we identified a region of N required for chaperone activity. The N-terminal 100 amino acids of N contain a domain that is both necessary and sufficient for RNA chaperone activity. We propose that this region of N may reside in one of two potential states. First, the region may be highly disordered and function in N-mediated RNA chaperone activity. Alternatively, in trimeric form, the region likely becomes ordered and serves in high affinity vRNA panhandle recognition.
Key words: hantavirus, bunyavirus, RNA chaperone, translation, transcription, cap-snatching
Introduction
RNA chaperones can mediate RNA refolding by actively accelerating correct base pairing. Alternatively, RNA chaperones can enable correct RNA structure by melting improperly formed base pairs thereby allowing the RNA to intrinsically fold correctly.1–3 RNA chaperones constitute a diverse set of proteins and lack an obvious consensus sequence or conserved RNA binding site for chaperone function. The ability of RNA chaperones to provide the energy to facilitate proper RNA folding, has led to a model positing that disordered regions of the protein are responsible for proper chaperone function.4,5 HIV NC and HCV core are two viral RNA chaperones that facilitate the proper folding of RNA.6–8 These molecules contain predicted disordered domains that are required for their chaperone function. These disordered domains can facilitate the destabilization and reformation of base pairs in the absence of ATP. The “entropy transfer” model is predicated on the idea that disordered regions of protein with high entropy can transfer their entropy to an RNA molecule simultaneously disordering the substrate RNA and ordering the protein. This allows the RNA to escape kinetic traps and to refold into the proper conformation. However, viable alternatives to the proposition are that RNA chaperones achieve RNA refolding through alternative RNA-protein association such as transient ionic interaction. Further, different specific RNA chaperones may well work through more than one mechanism.
The Bunyaviridae family of viruses is composed of negative stranded RNA viruses with three genome segments. The family includes select agents that can cause hemorrhagic fever or related pathogenesis when accidentally introduced into humans. Hantaviruses comprise a genus within this virus family. In contrast to the other genera of the family, which are arboviruses, hantaviruses are transmitted to humans directly from their natural rodent hosts. Such transmission can elicit either hemorrhagic fever with renal syndrome (HFRS) or hantavirus cardiopulmonary syndrome (HCPS). Sin nombre virus (SNV) is the prototype North American hantavirus and its natural host is the Sigmodontinae rodent Peromyscus maniculatus (the deer mouse).9–11
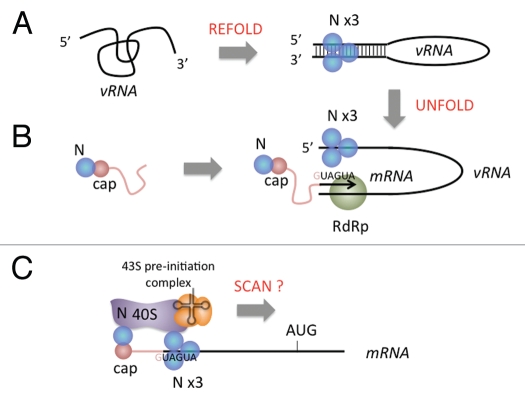
Hantavirus N is a RNA binding protein with an unexpected array of biological functions (Fig. 1). A primary role of N, which is shared by the proteins encoded by the homologous genes in the bunyavirus family and orthologs in other minus strand RNA viruses, is to recognize and protect the vRNA genome. The three genomic RNA segments of the bunyaviruses are encountered in higher order thermodynamically stable “panhandle” configuration arising through non-continuous base paring between the 5′ and 3′ end of individual segments12–15 and N can facilitate the formation of panhandles in misfolded vRNA.16,17 Genome recognition by SNV N is specific for the “viral panhandle” (Fig. 1A).18,19 N spontaneously forms trimers,20–24 and this form of N appears to bind with specificity to the panhandle. N also serves in the initiation of viral transcription (Fig. 1B). In this process N appears to function in two ways. Transcription in RNA viruses with segmented genomes, including the bunya-, orthomyxo- and arenaviruses, commences with the acquisition of capped cellular oligonucleotides through “cap-snatching”.25–32 In the hantaviruses, N plays a role in this process by protecting putative cellular caps from the cytoplasmic mRNA degradation apparatus of the cell.33 N also appears to play a role in transcription initiation by dissociating the panhandle of the viral RNA template.17 This activity may be necessary to render the 3′ terminus of the vRNA template accessible for viral mRNA synthesis. In addition to serving during genome recognition and the start of transcription SNV N can replace the cellular cap-binding complex (eIF4F) to facilitate preferential initiation and translational expression of viral mRNA (Fig. 1C).34 The three peptides of eIF4F (4E, 4G and 4A) mediate recognition of capped mRNA, recruitment of the 43S pre-initiation complex including the small ribosomal subunit and scanning from the 5′ cap to the start codon via RNA helicase activity, respectively. By extension, SNV N either replaces or more likely, bypasses the activities of the cap-binding complex.
Figure 1.
The multiple functions of Sin nombre hantavirus N and the known and possible roles of the RNA chaperone activity of N in these processes. (A) During substrate refolding, the low affinity RNA chaperone activity of N enables refolding of viral RNA (vRNA) to correct thermodynamically stable panhandle configuration.16 The RNA panhandle then can be bound at high affinity by trimeric N.19 (B) During transcription initiation N interacts with the cellular mRNA turnover machinery in cytoplasmic foci (processing bodies) to protect 5′ oligomeric RNA caps from degradation so that they can be uses in cap-snatching and transcription initiation.33 The RNA chaperone activity of N can dissociate the vRNA panhandle remaining bound to the 5′ end of the vRNA making the 3′ end of the vRNA accessible for use as a template by the viral RNA-dependent RNA polymerase (RdRp).17 (C) N is able to supplant the cellular cap-binding complex during translation initiation.34 It is not known whether N facilitates scanning of the small ribosomal subunit to the start codon in a manner analogous to the cap-binding complex. However, a possible additional role for the chaperone activity of N is to dissociate intramolecular interactions in the 5′ leader to facilitate access to the start codon. The sequence “GUAGUA,” which is present near the 5′ end of viral mRNA enables preferential recognition of viral mRNA by N.42
In this paper we demonstrate that an amino-terminal region of SNV N, which may be highly disordered, mediates RNA chaperone activity. In contrast to the requirement for N trimerization in vRNA panhandle recognition, N-mediated RNA chaperone activity does not require trimerization of N. We demonstrate that this amino-terminal region of N is both necessary and sufficient for RNA chaperone activity and that RNA refolding proceeds through destabilization of RNA/RNA duplexes and the inherent ability of the target RNA to refold into the correct configuration.
Results
The amino terminal 100 amino acids of N mediates RNA chaperone activity.
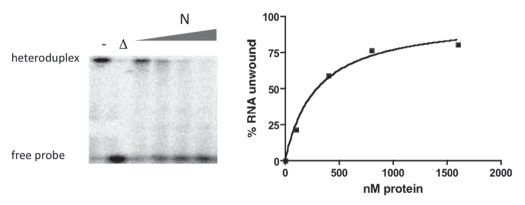
A useful general assay used to examine many RNA chaperones is based on the ability of chaperones to dissociate RNA heteroduplexes. Previously, we found that SNV N facilitates such RNA helix dissociation (RHD) in a concentration-dependent manner provided that there is a single-stranded region 3′ to the duplex.17 We used a heteroduplex RNA composed of a long, unlabeled RNA and a radioactively labeled 60 nucleotide long complementary probe in an attempt to identify regions of N required for RNA chaperone activity. In this assay, RHD can be assessed by electrophoretic detection of the liberated 60 nucleotide long RNA and subsequent quantification by phosphorimage analysis. Incubation of the heteroduplex with increasing amounts of full length N protein led to unwinding of the duplex (Fig. 2).
Figure 2.
Helix destabilization (HDS) assay used to examine the RNA chaperone activity of SNV N. The left part of the figures depicts helix dissociation as a function of increasing nM concentrations of N. Following incubation of 500 pM heteroduplex RNA with the indicated amounts of N, the products were analyzed by gel electrophoresis and quantified by phosporimaging as described in the Methods Section to produce the graph on the right. - is a control sample lacking N, Δ indicates heat denaturation of the heteroduplex.
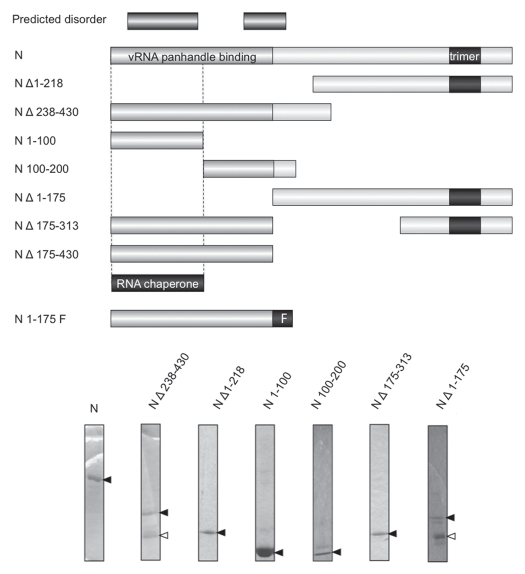
We first determined whether the RHD activity of N could be generally localized to the N- or C-terminal portion of the peptide. N Δ238–430 lacks approximately the C-terminal half of N and N Δ1–218 lacks the N-terminal half of N (Fig. 3). Both of these mutants have been characterized with respect to their interaction with the vRNA panhandle and both exhibit markedly diminished binding with the panhandle since an RNA binding domain near the amino terminus of N and a trimerization domain near the C-terminus of N are both required for panhandle recognition (Brown et al. submitted). In contrast, in the RHD assay, N Δ238–430 had near wild type levels of RHD activity while N Δ1–218 lacked detectible RNA chaperone function (Fig. 4A). This indicates that the C-terminus of N is dispensable for RHD activity.
Figure 3.
SNV mutants used to identify the region(s) of N required for chaperone activity. The segments of the N peptide present in the various N mutants are shown. The two longest putative disordered regions of N are shown the top and based on the PONDR plot of Figure 7 and Table 1. The regions of SNV N required for high affinity binding to the viral RNA panhandle are shown for reference. These are the aminoterminal 175 amino acids of N and the trimerization domain of N near the C-terminus (Brown and Panganiban, submitted). The N-terminal 175 amino acids of N also bind with high affinity to vRNA when the intrinsic trimerization domain is replaced with a foreign peptide that facilitates trimerization. Such a trimerization domain is labeled “F” at the bottom of the Figure. The major region of N required for RNA chaperone, activity based on the data of this manuscript, is also shown. Gel analysis of some of the peptides used in the analysis is shown at the bottom of the Figure. The dark arrows indicate N peptide as verified by western analysis (not shown). The white arrows indicate contaminating peptides as these species are not detected in western analysis.
Figure 4.
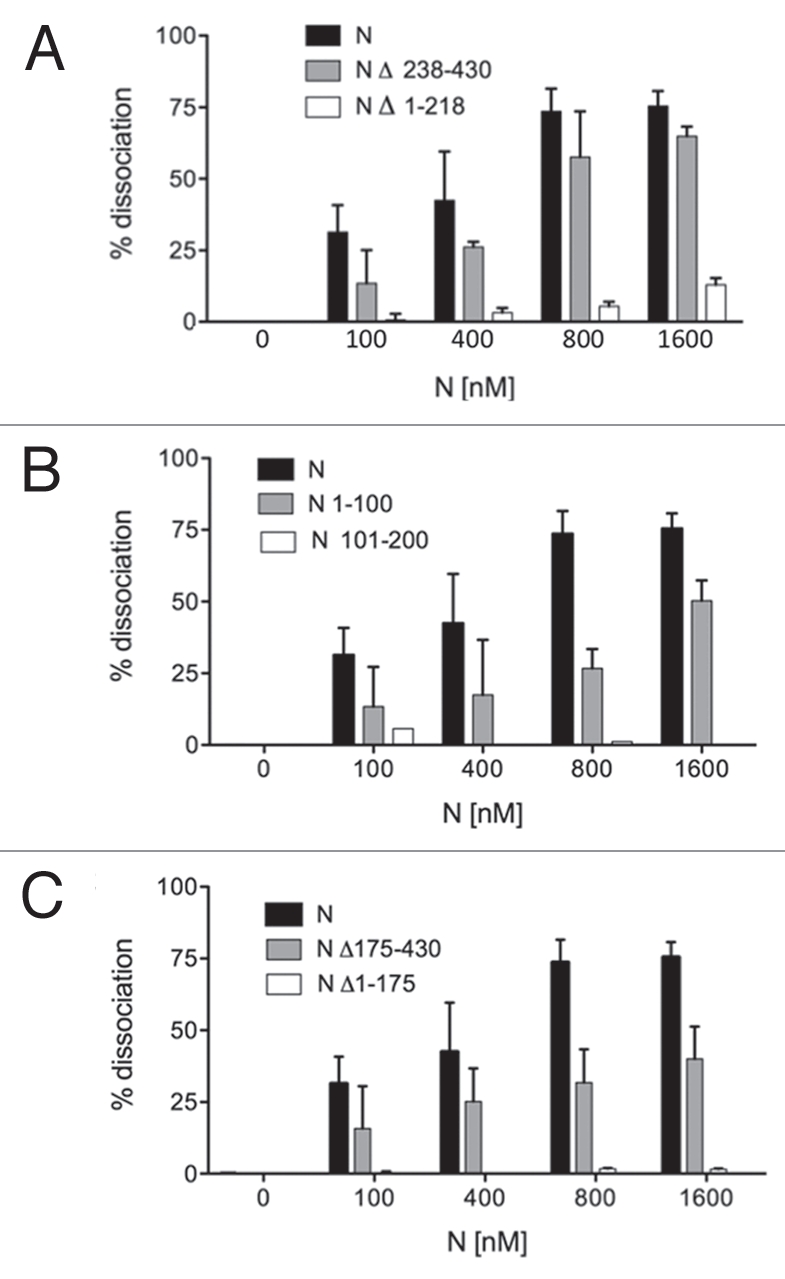
The amino-terminal 100 amino acids of N are required for RNA chaperone activity. (A) depicts the effect of N- and C-terminal deletions on the RHD activity of SNV N. Heteroduplex RNA was incubated with increasing amounts of full length N, N Δ236–430 or N Δ1–218. (B) depicts RHD activity as a function of increasing concentrations of N 1–100 or N 100–200. In (C) N Δ175–430 and N Δ1–175 were evaluated to establish the possible contribution of a central RNA binding region of N to RNA chaperone activity. All assays were performed a minimum of three times.
As will be described below, there are two regions in the aminoterminus of SNV N, amino acids 18–94 and 144–175, that may be disordered. Assignment of the RHD domain to the aminoterminal half of N raises the question of whether either of these two hypothetical disordered regions near the N-terminus may be required for helix disassociation. We constructed two peptides containing either amino acids 1–100 (N 1–100) or amino acids 101–200 (N 101–200) (Fig. 3) and examined the ability of both peptides to mediate RNA helix destabilization. We found that N 1–100 was functional in helix dissociation but somewhat less active than full length N (Fig. 4B). In contrast, residues 101–200 of N displayed little or no capacity to dissociate the RNA duplex. Thus, the first 100 amino acids of N is sufficient for RNA substantial helix dissociation activity.
Hantaan virus (HTNV) contains an RNA binding domain that has been mapped to amino acids 175 to 218.35 This region is surprisingly dispensable for high affinity binding and recognition of the vRNA panhandle by trimeric SNV N (Brown and Panganiban, submitted). However, as this central portion of N binds RNA with low affinity it is plausible that this region might function in the RNA chaperone activity of N. N Δ175–430 contains the first 175 amino-terminal amino acids of N and its complement, N Δ1–175, contains residues 175 to 430 including the region corresponding to the HTNV RNA binding domain. As might be expected in light of the results with N 1–100, N Δ1–175 was able to mediate significant RHD activity. However, as with N 1–100, this activity was less efficient than that of wild type N (Fig. 4C). N Δ1–175, exhibited no detectable RHD activity (Fig. 4C). These data are again consistent with assignment of the RNA chaperone activity of N principally to the amino-terminal 100 amino acids of N. However, the data are also consistent with an ancillary role of amino acids 175–218 in RHD activity since N Δ238–430, a peptide containing the amino terminal 100 amino acids as well as the central region, exhibited RHD activity similar to full-length N.
Addition of a heterologous trimerization domain inhibits the RNA chaperone activity of N.
The first 175 amino acids of SNV N are sufficient for high affinity vRNA panhandle recognition and binding provided that this amino-terminal portion of N is maintained in trimeric conformation (Brown BA and Panganiban AT, submitted). Trimerization could be successfully mediated either by the presence of the native trimerization domain of N located near the C-terminus or by a foreign trimerization domain in the form of the 27 amino acid long phage T4 fibritin trimerization domain (the “foldon” peptide) (Fig. 3). It is noteworthy that the RHD activity of N appeared to occur efficiently without trimerization of N, as both N Δ238–430 and N Δ175–430 display significant RHD activity even though they are deficient in N trimerization (Fig. 4). We wanted to further investigate the relationship between N trimerization and the RNA chaperone activity. Thus, we examined the activities of N Δ175–313 and a counterpart containing the foldon peptide (N 1–175F) (Fig. 3). N Δ175–313 contains the first 175 amino acids of N and along with the bone fide SNV trimerization domain. N 175F is a chimeric N peptide composed of the first 175 amino acids of N appended with the foldon trimerization peptide. As might be expected, N Δ175–313 exhibited RHD activity, although its activity was somewhat less than that of full length N (Fig. 5). Surprisingly, the presence of the foldon trimerization domain profoundly inhibited RHD activity. On the surface, these data appear to be contradictory. The presence of the native trimerization domain in N Δ175–313 does not ablate RNA chaperone activity of the amino-terminus of N, whereas a heterologous trimerization domain markedly interferes with RHD activity. These observations are considered further in the Discussion.
Figure 5.
Addition of a heterologous trimerization domain to the RNA chaperone region of N abrogates RHD activity. N Δ175–313 contains the intrinsic trimerization domain of N and N 175F contains the foldon trimerization domain. RNA chaperone (RHD) activity was determined as in Figure 4. Data for the RHD activity of N-His175F are also included in the graph but not visible as RHD activity was undetectible. All assays were performed a minimum of three times.
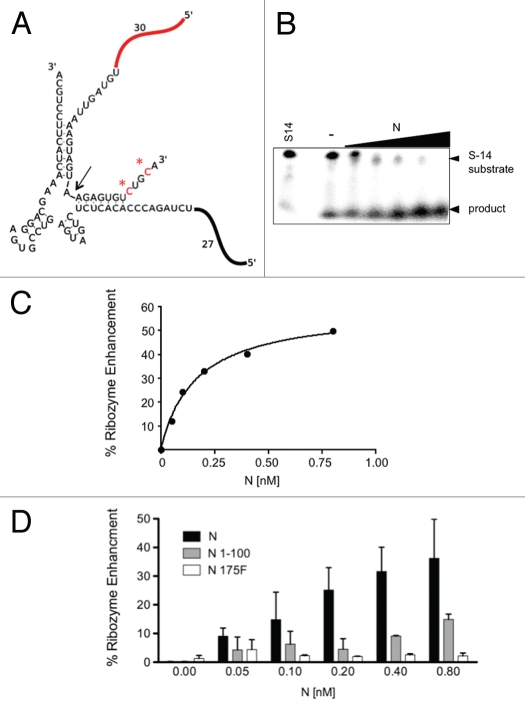
To further verify the RNA chaperone activity of N we used a second assay that has been used previously to monitor the activity of RNA chaperones. Hammerhead ribozymes are self-catalytic RNAs that can be constructed to mediate trans-cleavage of RNA substrates following correct hydrogen bonding with the ribozyme core (Fig. 6A). RNA chaperones can accelerate the activity of ribozymes by increasing the rate of RNA substrate association and with dissociation of the cleaved RNA products. In essence, RNA chaperones increase ribozyme formation and turnover thereby enhancing ribozyme activity. N was able to augment ribozyme activity by up to 50% over the ribozyme alone (Fig. 6B and C). Moreover, a significant increase in ribozyme activity was conferred by the N-terminal 100 amino acids of N albeit at a lower efficiency that that of wild type N (Fig. 6D). Further, N 175F did not significantly enhance RNA chaperone activity. Thus, the results of this ribozyme enhancement assay for these key N derivatives are in accord with the results from the RHD assay.
Figure 6.
Enhancement of hammerhead ribozyme activity by N. We used an RNA chaperone assay developed by Betrand and Rossi (Bertrand and Rossi 1994), to further examine the RNA chaperone activity of N. (A) In this assay, a functional ribozyme is formed through the hydrogen bonding of a 58 nucleotide long radioactively labeled substrate RNA (designated substrate 14, “S-14”) with the unlabeled remainder of the ribozyme. The radioactively labeled C residues in S-14 are highlighted with asterisks. Cleavage of S-14 takes place 13 nucleotides from the 3′ end (indicated by arrow) and the labeled uncleaved and 45 nucleotide long cleavage product can be detected and quantified by electrophoresis and phosphorimaging as described in the Methods. (B) Denaturing gel showing the effect of increasing concentrations of N on ribozyme activity. The first lane contains S-14 RNA alone without the remainder of the ribozyme. - indicates ribozyme activity in the absence of N. The remaining lanes depict ribozyme activity with increasing amounts of N. The quantified data were used to generate the graph shown in (C). In the graph ribozyme activity in the absence of N is set to zero and the percentage of activity attributable to N is then plotted as a function of increasing N concentration. (D) Bar graph comparing the effect of two key N mutants (N Δ1–100 and N 175F) on ribozyme activity. The data are based on a minimum of three experiments.
Defined structure and potential disorder in SNV N.
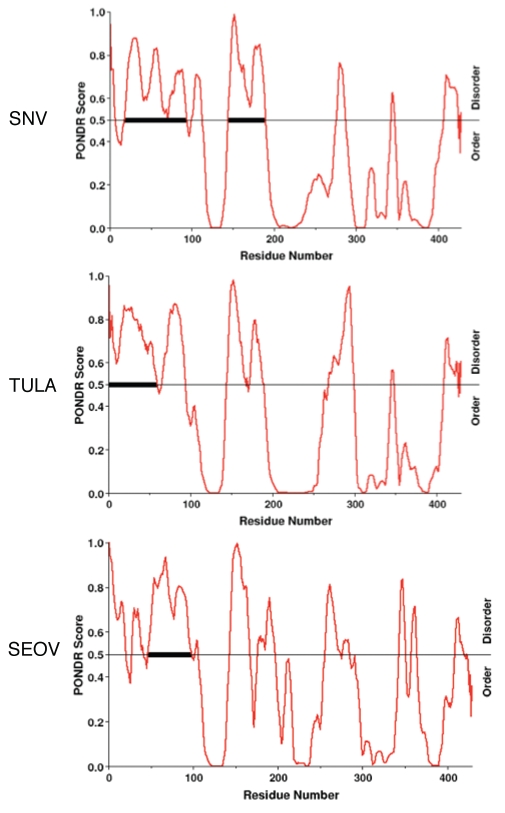
The amino-terminal seventy five amino acids of SNV N can form an intramolecular coiled-coil and be crystallized.22 However, a predictor for naturally disordered regions along polypeptide sequences, PONDR® VL-XT,36,37 indicates that the region may, under some circumstances, assume a disordered configuration (Fig. 7). In addition, smaller regions of potentially disordered amino acid sequence are situated in the middle and C-terminus of N (a.a. 276 to 287 and a.a. 407 to 424). Members of the hanta-virus genus are typically subdivided into three diverse groups based on the rodent hosts that they infect in nature. SNV and its relatives infect Sigmodontinae rodents in a species-specific manner, while other hantaviruses infect Arvocolinae and Murinae rodents. We generated similar PONDR-plots for the N proteins of more than fifteen additional viruses from all three host groups and these yielded profiles that also indicated potential disorder within the amino-terminal region of N (data not shown). The profiles for the two hantaviruses that exhibit the lowest apparent disorder in the amino-terminus of N are presented in Figure 7 along with SNV. These are for Tula and Seoul (SEOV) viruses that infect members of the Arvocolinae and Murinae, respectively.
Figure 7.
Regions of likely disorder in three diverse members of the hantavirus genus. Regions of hypothetically disordered regions in the N proteins of three hantaviruses that infect Sigmodontinae, Arvocolinae, Murinae hosts. Plots used PONDR® VL-XT as described in the text. In these plots increasing contiguous length of amino acids in disordered array correspondingly increases the likelihood that a region can assume a disordered state.
PONDR VL-XT predictor integrates feedforward neural networks composed of non-redundant sets of order and disordered sequences to identify long regions of potential disorder. The accuracy of the prediction increases as the length of the hypothetically disordered region increases. The contiguous relevant regions of Tula and SEOV are about forty amino acids in length and PONDR VL-XT accuracy for predicting disorder is about 99% for regions of this length. The contiguous putatively disordered region in the first 100 amino acids of SNV N is 76 residues in length (Table 1). We used three additional PONDR algorithms (XL1-XT, CAN-XT and VL3-BA) to examine the set of hantavirus N proteins and these produced plots similar to those using VL-XT (data not shown). In sum, it is unequivocal that the amino terminus of N is able to form an ordered structure. However, N molecules may assume a disordered configuration under some circumstances.
Table 1.
PONDR® VL-XT disorder prediction
| Predicted disorder segment | Avg. strength |
| 18–94 | 0.6874 |
| 100–112 | 0.635 |
| 144–190 | 0.7466 |
| 276–287 | 0.5783 |
| 407–424 | 0.6436 |
| Longest Disordered Region | 77 |
| Overall percent disordered | 42.06 |
Discussion
RNA chaperones play an important role in rescuing misfolded RNA from thermodynamically stable kinetic traps. The entropy transfer model proposed by Tompa and Csermely proposes that intrinsically disordered proteins are capable of unwinding stable RNA/RNA duplexes through entropy transfer.4 During entropy transfer disordered regions of the protein would transiently interact with misfolded ordered RNA. After interaction the chaperone would decrease in entropy while concurrently increasing the entropy of the RNA/RNA duplex. This transfer of entropy enables unfolding of the RNA allowing the RNA to refold into its active secondary or tertiary structure. SNV N is an RNA chaperone that binds misfolded vRNA and unwinds the misfolded RNA an allowing the panhandles to form.16 Using RHD and ribozyme enhancement assays we identified a restricted region of SNV N responsible for its RNA chaperone activity. Full length N at a concentration of 400 nM is able to efficiently disassociate a majority of the RNA/RNA duplex (protein:nucleotide ratio of 1:0.5). This protein to nucleotide ratio is similar to that reported for the RNA chaperones, Hepatitis C virus (HCV) and West Nile virus (WNV) core peptides, for similar strand displacement activity.6,8 SNV N, HCV core, WNV core, HIV NC and a host of other RNA chaperones are predicted to be a largely disordered.38–40 This is in keeping with the general model proposed by Tompa and Csermely. PONDR® identified several domains of potentially significant disorder in SNV N (Fig. 7 and Table 1). The N-terminal 100 amino acids of N contains the longest predicted disordered region of N and the principle RNA chaperone activity of N appears to also be conferred by this region (Fig. 3A).
It is worth comparing and contrasting the high affinity binding of N to the vRNA panhandle with the lower affinity, nonspecific binding to RNA that takes place during N-mediated RNA chaperone activity. Binding to the panhandle is mediated by a region restricted to the 175 amino terminal residues of N and trimerization is required for high affinity binding. RNA chaperone activity, which is not substrate-specific, also requires a limited N-terminal portion of N but trimerization is not required and trimerization may even abrogate RNA chaperone activity as evidenced by the inability of N 175F to serve as a chaperone. In striking contrast, this same chimeric N peptide (N175F) binds with high affinty to the vRNA panhandle (Brown BA and Panganiban AT, submitted).
Significantly, a crystal structure of the first 73 amino acids of for SNV N indicates that the region features a coiled coil.22 Moreover, this structure would encompass a large portion of the putative N-terminal disordered domain, which includes residues 18–94. Protein crystallization requires the presence of a specific structure. Disordered domains cannot be crystallized because multiple conformations (disorder) are present in the protein population. We think the paradoxical concept of N order and disorder may be explained by the ability of the amino terminal portion of N to reside in two alternative states. First, the region may assume a highly ordered three-dimensional conformation in trimeric form. This would effectively constitute the high affinity vRNA panhandle binding domain, as specific binding to the panhandle would likely mandate that there are unique contacts between the vRNA and N. Alternatively, the amino terminal region of N may be disordered in monomeric form and in this state possess RNA chaperone activity. To enable crystallization of N, Boudko, et al. attached the amino-terminal 94 residues of N to the T4 fibritin trimerization domain (foldon peptide).22 We used a foldon derivative of the T4 fibritin peptide similar to that used for crystallization of N to generate N trimers. The presence of the foldon domain may directly induce order within the amino terminus of N. This would be analogous to the conversion of HCV core from a highly disordered peptide to a more structured peptide following addition of n-dodecyl β-D-maltoside or interaction with lipid droplets.6 Potentially the foldon domain may also limit the ability of the amino-terminus of N to move freely in solution and to function as a chaperone. This would lower the number of conformational states of the peptide decreasing the entropy that could be transferred to an RNA/RNA duplex.
While the entropy transfer model and protein disorder in RNA chaperone activity is attractive, it has yet to be established that such disorder actually functions during the reconfiguration of misfolded RNA for any cellular or viral RNA chaperone. Thus, it is possible that intrinsic disorder is not required for RNA chaperone function. In such a scenario, the first 73 amino acids of N, in structured form could function in RNA chaperone activity. However, such a model does not easily account for the strongly negative effect of the foldon domain on chaperone activity and, conversely the strong positive effect on panhandle binding.
HTNV N contains a central RNA binding region within amino acids 175–218.35 However, the corresponding region of SNV N is not sufficient for chaperone activity as evidenced by the inability of N Δ1–175 to unwind RNA/RNA duplexes (Fig. 4C). On the other hand, this central region may work in conjunction with the amino-terminal 100 amino acids of N to enable full RNA chaperone activity. Since this central region binds to RNA it may augment the chaperone activity resident in the N-terminal 100 amino acids of the protein by increasing the overall affinity of N for RNA.
The PONDR® scores for N from Tula and SEOV, which are both old world hantaviruses, demonstrate the same general pattern of disorder in the N-terminus as SNV N (Fig. 7). However, N-terminal disordered domains in N are not a characteristic of the other genera of Bunyaviridae, such as bunyamwera or tomato spot wilt virus (TSWV) (data not shown). It will be interesting to see whether RNA chaperone activity is conserved among other members of the virus family.
A full understanding of the role of chaperone activity in hantavirus replication requires an understanding of mechanism of action helix dissociation. We have mapped the activity to the first 100 amino acids and to the first predicted disorder domain for SNV N. This activity could play an important function in genome replication, packaging and transcription of viral genomes and requires in vivo characterization to determine the scope of its biological function.
Materials and Methods
Wild-type N and N mutants.
N, N Δ1–175, N Δ175–430, N Δ1–218, N Δ238–430, N Δ175–313, N Δ175–430 and N 175 Foldon (F) were all created by PCR based cloning as previously described in (Brown BA and Panganiban AT, submitted), and all contained a C-terminal his8 tag. N 1–100 and N 101–200 were generated by PCR amplification of the n gene from N. For N 1–100 was amplified using primers 2.13 R (GAC GAA TTC TTA TTT CCA TAT CTC AAT GAT GAC) and 2.13 F (AAG GAG ATA TCC CAT GGG CAC CCT CAA AGA AGT GCA A) then digested with Nco I and Eco RI. N 101–200 was amplified with 2.14R (TTT GAA TTC TTT GTA ACG GAA CCT TCC GGG A) and 2.14 F (AAA CCA TGG AAG TCC TTG ATG TAA ATT CCA) after which it was digested with Nco I and Eco RI. The PCR products were then ligated into pTri-Ex 1.1 (Novagen).
N mutants were purified by metal affinity chromatography as described previously.17 In brief, the N derivatives were transformed into Rosetta pLac I cells (Novagen). Single colonies were picked and grown overnight in 5 mL of LB with ampicillin and chloramphenicol. The 5 mL cultures were then used to inoculate 1 liter of LB with the appropriate antibiotics. The culture was grown to an OD600 = 0.6 and induced with 1 mM IPTG. The cells were then lysed in 8 M urea overnight. The lysed cells were centrifuged at 10,000x g for 30 minutes and the supernatant was passed through a 0.2 µM filter. The lysate was then passed through a His-Trap column (GE Healthcare) and eluted using a 10 mM to 500 mM imidazole gradient. Fractions containing N were identified using SDS gels and Coomassie staining and the protein-containing fractions were combined and renatured by step-wise dialysis.
RNA.
The larger unlabeled RNA used in the helix destabilization assay was transcribed from pT-GFP, which had been cut with Pvu II. The cut plasmid was purified by agarose gel electrophoresis and transcribed using T7 express RNA transcription kits (Promega). The transcription reaction was then treated with DNase I for 30 minutes at 37°C and the RNA purified by Trizol (Invitrogen). The smaller labeled “probe” RNA was transcribed from a PCR product arising from amplification with primers pT-GFP-F (ACT ACT TAA TAC GAC TCA TAT AGG G) and pT-GFP-R (CCT TGG GGC GGA CTG GGT GCT CTG GT) and pT-GFP template DNA. The RNA was transcribed by a T7 transcription kit (Fermentas) and then DNase I treated for 30 minutes at 37°C according to the manufacture's protocols. The RNA was then purified by Trizol.
RNA/RNA duplexes were generated by combining equal molar amounts of the large GFP RNA and 60 nt probe RNA, heating at 95°C for 5 minutes and allowing annealing of the heteroduplex by incubation at 42°C for 3 hours. Finally the duplex was purified from free 60 nt probe using RNeasy columns (Qiagen). The duplex was then stored at −80 for up to 2 weeks.
RNA helix destabilization assay. Standard RNA helix destabilization (RHD) assays were carried out in 20 µl reactions. RNA/RNA duplexes were incubated with varying amounts of protein and 1x RNA chaperone buffer (40 mM HEPES at pH 7.4, 80 mM NaCl, 20 mM KCl, 10 mM MgCl and 1 mM DTT). The reaction mixture was incubated at 37°C for 30 minutes and 10 µl of 2x Lameli buffer was added to stop the reaction. The RNA was then loaded onto a 12% acrylamide gel and electrophoresed at 200 V for 2 hours. The gel was exposed to a phosphor storage screen and RNA unwinding was quantified based on the appearance of dissociated probe using Image Quant software (v.5.4).
Prediction of protein disorder.
The SNV N protein disorder was calculated using the predictor for natural disorder (PONDR®) using the VL-XT network.36,37,41
R3 ribozyme and S14 substrate.
The ribozyme R3 template was generated by PCR amplification of HH template as described in reference 7. PrimerF-HH-S14 (ACA CTA ATA CGA CTC ACT ATA GGG) and PrimerR-HH (TGC AGG AAG TAG TTT CGT CCT). The S14 ribozyme substrate template was amplified by amplification of the S14 template with PrimerF-HH-S14 (ACA CTA ATA CGA CTC ACT ATA GGG) and Primer R-S14 (TGC AGA CAC TCT TAC TAC TTT AA). The PCR products were directly transcribed by T7 polymerase (Promega), DNAse treated and purified by removal of nucleotides with Rneasy columns (Qiagen). For ribozyme enhancement assays 10 nM of R3 in RNA chaperone buffer was added to 30 nM S14 substrate with varying N concentrations in a volume of 5 µl. The reaction was incubated at 37°C for 30 minutes and stopped by the addition of 5 µl EDTA and 5 µl Lamelli buffer and incubated on ice until loading onto gels. The samples were electrophoresed in a 9% acrylamide, 7 M urea TBE gel for 2 hours, exposed to a storage phosphor screen overnight and quantitated using Image Quant (v. 5.4).
Acknowledgements
We thank Dave Bear, Dave Peabody and Jesse Summers for discussion. This work was supported by research grant R01AI074011 from the NIH.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/13862
References
- 1.Rajkowitsch L, Chen D, Stampfl S, Semrad K, Waldsich C, Mayer O, et al. RNA chaperones, RNA annealers and RNA helicases. RNA Biol. 2007;4:118–130. doi: 10.4161/rna.4.3.5445. [DOI] [PubMed] [Google Scholar]
- 2.Rajkowitsch L, Schroeder R. Dissecting RNA chaperone activity. RNA. 2007;13:2053–2060. doi: 10.1261/rna.671807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rajkowitsch L, Semrad K, Mayer O, Schroeder R. Assays for the RNA chaperone activity of proteins. Biochem Soc Trans. 2005;33:450–456. doi: 10.1042/BST0330450. [DOI] [PubMed] [Google Scholar]
- 4.Tompa P, Csermely P. The role of structural disorder in the function of RNA and protein chaperones. Faseb J. 2004;18:1169–1175. doi: 10.1096/fj.04-1584rev. [DOI] [PubMed] [Google Scholar]
- 5.Cristofari G, Darlix JL. The ubiquitous nature of RNA chaperone proteins. Prog Nucleic Acid Res Mol Biol. 2002;72:223–268. doi: 10.1016/s0079-6603(02)72071-0. [DOI] [PubMed] [Google Scholar]
- 6.Ivanyi-Nagy R, Lavergne JP, Gabus C, Ficheux D, Darlix JL. RNA chaperoning and intrinsic disorder in the core proteins of Flaviviridae. Nucleic Acids Res. 2008;36:712–725. doi: 10.1093/nar/gkm1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bertrand EL, Rossi JJ. Facilitation of hammerhead ribozyme catalysis by the nucleocapsid protein of HIV-1 and the heterogeneous nuclear ribonucleoprotein A1. EMBO J. 1994;13:2904–2912. doi: 10.1002/j.1460-2075.1994.tb06585.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cristofari G, Ivanyi-Nagy R, Gabus C, Boulant S, Lavergne JP, Penin F, et al. The hepatitis C virus Core protein is a potent nucleic acid chaperone that directs dimerization of the viral (+) strand RNA in vitro. Nucleic Acids Res. 2004;32:2623–2631. doi: 10.1093/nar/gkh579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Botten J, Mirowsky K, Kusewitt D, Bharadwaj M, Yee J, Ricci R, et al. Experimental infection model for Sin Nombre hantavirus in the deer mouse (Peromyscus maniculatus) Proc Natl Acad Sci USA. 2000;97:10578–10583. doi: 10.1073/pnas.180197197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmaljohn C, Hjelle B. Hantaviruses: a global disease problem. Emerg Infect Dis. 1997;3:95–104. doi: 10.3201/eid0302.970202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamada T, Hjelle B, Lanzi R, Morris C, Anderson B, Jenison S. Antibody responses to Four Corners hantavirus infections in the deer mouse (Peromyscus maniculatus): identification of an immunodominant region of the viral nucleocapsid protein. J Virol. 1995;69:1939–1943. doi: 10.1128/jvi.69.3.1939-1943.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hewlett MJ, Pettersson RF, Baltimore D. Circular forms of Uukuniemi virion RNA: an electron microscopic study. J Virol. 1977;21:1085–1093. doi: 10.1128/jvi.21.3.1085-1093.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Obijeski JF, Bishop DH, Murphy FA, Palmer EL. Structural proteins of La Crosse virus. J Virol. 1976;19:985–997. doi: 10.1128/jvi.19.3.985-997.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pettersson RF, von Bonsdorff CH. Ribonucleoproteins of Uukuniemi virus are circular. J Virol. 1975;15:386–392. doi: 10.1128/jvi.15.2.386-392.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raju R, Kolakofsky D. The ends of La Crosse virus genome and antigenome RNAs within nucleocapsids are base paired. J Virol. 1989;63:122–128. doi: 10.1128/jvi.63.1.122-128.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mir MA, Panganiban AT. The bunyavirus nucleocapsid protein is an RNA chaperone: possible roles in viral RNA panhandle formation and genome replication. Rna. 2006;12:272–282. doi: 10.1261/rna.2101906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mir MA, Panganiban AT. Characterization of the RNA chaperone activity of hantavirus nucleocapsid protein. J Virol. 2006;80:6276–6285. doi: 10.1128/JVI.00147-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mir MA, Panganiban AT. Trimeric hantavirus nucleocapsid protein binds specifically to the viral RNA panhandle. J Virol. 2004;78:8281–8288. doi: 10.1128/JVI.78.15.8281-8288.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mir MA, Panganiban AT. The hantavirus nucleocapsid protein recognizes specific features of the viral RNA panhandle and is altered in conformation upon RNA binding. J Virol. 2005;79:1824–1835. doi: 10.1128/JVI.79.3.1824-1835.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alfadhli A, Love Z, Arvidson B, Seeds J, Willey J, Barklis E. Hantavirus nucleocapsid protein oligomerization. J Virol. 2001;75:2019–2023. doi: 10.1128/JVI.75.4.2019-2023.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alfadhli A, Steel E, Finlay L, Bachinger HP, Barklis E. Hantavirus nucleocapsid protein coiled-coil domains. J Biol Chem. 2002;277:27103–27108. doi: 10.1074/jbc.M203395200. [DOI] [PubMed] [Google Scholar]
- 22.Boudko SP, Kuhn RJ, Rossmann MG. The coiled-coil domain structure of the Sin Nombre virus nucleocapsid protein. J Mol Biol. 2007;366:1538–1544. doi: 10.1016/j.jmb.2006.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaukinen P, Koistinen V, Vapalahti O, Vaheri A, Plyusnin A. Interaction between molecules of hantavirus nucleocapsid protein. J Gen Virol. 2001;82:1845–1853. doi: 10.1099/0022-1317-82-8-1845. [DOI] [PubMed] [Google Scholar]
- 24.Kaukinen P, Vaheri A, Plyusnin A. Mapping of the regions involved in homotypic interactions of Tula hantavirus N protein. J Virol. 2003;77:10910–10916. doi: 10.1128/JVI.77.20.10910-10916.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duijsings D, Kormelink R, Goldbach R. In vivo analysis of the TSWV cap-snatching mechanism: single base complementarity and primer length requirements. EMBO J. 2001;20:2545–2552. doi: 10.1093/emboj/20.10.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Estabrook EM, Tsai J, Falk BW. In vivo transfer of barley stripe mosaic hordeivirus ribonucleotides to the 5′ terminus of maize stripe tenuivirus RNAs. Proc Natl Acad Sci USA. 1998;95:8304–8309. doi: 10.1073/pnas.95.14.8304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garcin D, Lezzi M, Dobbs M, Elliott RM, Schmaljohn C, Kang CY, et al. The 5′ ends of Hantaan virus (Bunyaviridae) RNAs suggest a prime-and-realign mechanism for the initiation of RNA synthesis. J Virol. 1995;69:5754–5762. doi: 10.1128/jvi.69.9.5754-5762.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hagen M, Chung TD, Butcher JA, Krystal M. Recombinant influenza virus polymerase: requirement of both 5′ and 3′ viral ends for endonuclease activity. J Virol. 1994;68:1509–1515. doi: 10.1128/jvi.68.3.1509-1515.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jin H, Elliott RM. Characterization of Bunyamwera virus S RNA that is transcribed and replicated by the L protein expressed from recombinant vaccinia virus. J Virol. 1993;67:1396–1404. doi: 10.1128/jvi.67.3.1396-1404.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li ML, Ramirez BC, Krug RM. RNA-dependent activation of primer RNA production by influenza virus polymerase: different regions of the same protein subunit constitute the two required RNA-binding sites. EMBO J. 1998;17:5844–5852. doi: 10.1093/emboj/17.19.5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramirez BC, Garcin D, Calvert LA, Kolakofsky D, Haenni AL. Capped nonviral sequences at the 5′ end of the mRNAs of rice hoja blanca virus RNA4. J Virol. 1995;69:1951–1954. doi: 10.1128/jvi.69.3.1951-1954.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vialat P, Bouloy M. Germiston virus transcriptase requires active 40S ribosomal subunits and utilizes capped cellular RNAs. J Virol. 1992;66:685–693. doi: 10.1128/jvi.66.2.685-693.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mir MA, Duran WA, Hjelle BL, Ye C, Panganiban AT. Storage of cellular 5′ mRNA caps in P bodies for viral cap-snatching. Proc Natl Acad Sci USA. 2008;105:19294–19299. doi: 10.1073/pnas.0807211105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mir MA, Panganiban AT. A protein that replaces the entire cellular eIF4F complex. EMBO J. 2008;27:3129–3139. doi: 10.1038/emboj.2008.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu X, Severson W, Villegas N, Schmaljohn CS, Jonsson CB. The RNA binding domain of the hantaan virus N protein maps to a central, conserved region. J Virol. 2002;76:3301–3308. doi: 10.1128/JVI.76.7.3301-3308.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Romero P, Obradovic Z, Li X, Garner EC, Brown CJ, Dunker AK. Sequence complexity of disordered protein. Proteins. 2001;42:38–48. doi: 10.1002/1097-0134(20010101)42:1<38::aid-prot50>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 37.Li X, Romero P, Rani M, Dunker AK, Obradovic Z. Predicting Protein Disorder for N-, C- and Internal Regions. Genome Inform Ser Workshop Genome Inform. 1999;10:30–40. [PubMed] [Google Scholar]
- 38.Dokland T, Walsh M, Mackenzie JM, Khromykh AA, Ee KH, Wang S. West Nile virus core protein; tetramer structure and ribbon formation. Structure. 2004;12:1157–1163. doi: 10.1016/j.str.2004.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma L, Jones CT, Groesch TD, Kuhn RJ, Post CB. Solution structure of dengue virus capsid protein reveals another fold. Proc Natl Acad Sci USA. 2004;101:3414–3419. doi: 10.1073/pnas.0305892101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Herschlag D, Khosla M, Tsuchihashi Z, Karpel RL. An RNA chaperone activity of non-specific RNA binding proteins in hammerhead ribozyme catalysis. EMBO J. 1994;13:2913–2924. doi: 10.1002/j.1460-2075.1994.tb06586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Romero P, Obradovic Z, Dunker K. Sequence Data Analysis for Long Disordered Regions Prediction in the Calcineurin Family. Genome Inform Ser Workshop Genome Inform. 1997;8:110–124. [PubMed] [Google Scholar]
- 42.Mir AM, Panganiban AT. The Triplet Repeats of the Sin Nombre Hantavirus 5 Untranslated Region Are Sufficient in cis for Nucleocapsid-Mediated Translation Initiation. J Virol. 2010 doi: 10.1128/JVI.02720-09. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]