Abstract
Aberrant expression of cyclin D1 protein is a common feature of breast cancer. However, the CCND1 gene encodes two gene products, cyclin D1a and cyclin D1b, which have discrete mechanisms of regulation and impact on cell behavior. A polymorphism at nucleotide 870 in the CCND1 gene, rs603965, influences the relative production of the encoded proteins and can impart increased risk for tumor development. Here, the impact of both the G/A870 polymorphism and cyclin D1b protein production on breast cancer risk, disease phenotype and patient outcome was analysed. In a large multiethnic case–control study, the G/A870 polymorphism conferred no significant risk for breast cancer overall or by stage or estrogen receptor (ER) status. However, the cyclin D1b protein was found to be upregulated in breast cancer, independent of cyclin D1a levels, and exhibited heterogeneous levels in breast cancer specimens. High cyclin D1a expression inversely correlated with the Ki67 proliferation marker and was not associated with clinical outcome. In contrast, elevated cyclin D1b expression was independently associated with adverse outcomes, including recurrence, distant metastasis and decreased survival. Interestingly, cyclin D1b was particularly associated with poor outcome in the context of ER-negative breast cancer. Thus, specific cyclin D1 isoforms are associated with discrete forms of breast cancer and high cyclin D1b protein levels hold prognostic potential.
Keywords: cyclin D1b, breast cancer, polymorphism, estrogen receptor, prognosis
Aberrant cellular proliferation is an inherent component of human cancer. Correspondingly, the deregulation of cell-cycle control pathways is an exceedingly common occurrence in cancer (Malumbres and Barbacid, 2001; Diehl, 2002; Cobrinik, 2005; Knudsen, 2006). The retinoblastoma tumor suppressor pathway is believed to function to restrain inappropriate proliferation and is inactivated at high-frequency in tumors (Cobrinik, 2005; Knudsen and Knudsen, 2006). The retinoblastoma protein limits proliferation by functioning as a transcriptional co-repressor of genes that are required for DNA replication and mitotic progression (Blais and Dynlacht, 2007; Iaquinta and Lees, 2007). After mitogenic signaling, this function of retinoblastoma is disrupted through CDK (cyclin-dependent kinase)-mediated phosphorylation. Particularly, the cyclin D1 protein is rate-limiting for the initiation of retinoblastoma phosphorylation and subsequent inactivation which facilitates cellular division (Cobrinik, 2005). Correspondingly, the levels of cyclin D1 protein are directly modulated by mitogenic and antiproliferative signaling pathways (Diehl, 2002; Arnold and Papanikolaou, 2005; Knudsen et al., 2006). Thus, cyclin D1 is a key regulator of cell-cycle progression, and functions as an oncogene in specific model systems.
Multiple studies indicate that cyclin D1 plays an important role in mammary biology and tumorigenesis. Mice deficient in cyclin D1 harbor surprisingly few defects; however, there is a pronounced deficit in mammary gland development (Sicinski et al., 1995). Correspondingly, cyclin D1-null mice are refractory to mammary tumor development driven by multiple oncogenes (Yu et al., 2001), whereas the transgenic expression of cyclin D1 is sufficient to induce mammary tumors in mice (Wang et al., 1994). Consistent with these functional studies, the cyclin D1 protein is overexpressed in >50% of breast tumors (Buckley et al., 1993; Gillett et al., 1994, 1996; Diehl, 2002; Sutherland and Musgrove, 2004; Arnold and Papanikolaou, 2005). The basis for the overexpression of cyclin D1 has not been fully determined, as the cyclin D1 locus is amplified in only 10–20% of breast cancer. Furthermore, the prognostic value of cyclin D1 overproduction in breast cancer patients remains somewhat controversial (Gillett et al., 1994, 1996; Arnold and Papanikolaou, 2005; Jirstrom et al., 2005; Roy and Thompson, 2006; Rudas et al., 2008).
It is now apparent that cyclin D1 exists in two isoforms: the conventional cyclin D1 (referred to as cyclin D1a) and cyclin D1b (Betticher et al., 1995; Knudsen, 2006; Knudsen et al., 2006). The cyclin D1b variant arises as a consequence of alternative splicing of the CCND1 transcript. This event leads to the loss of exon 5-encoded sequences and a unique C terminus that arises from translation of intron 4 (Betticher et al., 1995). Importantly, exon 5 harbors a number of regulatory motifs that are required for appropriate cyclin D1a regulation and protein turnover, including a putative PEST domain and the threonine 286 phosphorylation site, which is a critical effector of the subcellular localization and oncogenic potential of cyclin D1a (Diehl, 2002; Lu et al., 2003; Solomon et al., 2003; Knudsen et al., 2006). Consistent with these known structural alterations, cyclin D1b is constitutively nuclear and has enhanced activity for transforming fibroblastic cells (Lu et al., 2003; Solomon et al., 2003). Furthermore, cyclin D1b plays a potent role in modifying the requirement for anchorage dependence (Holley et al., 2005), and can effectively contribute to the bypass of estrogen antagonists in cell culture models of estrogen receptor (ER)-positive breast cancer (Wang et al., 2008). These findings, in particular, led us to evaluate the impact of cyclin D1b in breast cancer.
G/A870 polymorphism (rs603965) and breast cancer risk
The production of cyclin D1b can be modulated by a common G/A polymorphism at nucleotide 870 (rs603965) of the CCND1 gene (Betticher et al., 1995; Holley et al., 2001; Knudsen, 2006; Knudsen et al., 2006). This nucleotide is at a splice donor site, and although the G-allele represents an ideal consensus sequence for splicing, the A-allele is predicted to be less efficient for directing splicing(Knudsen et al., 2006). Findings from a number of laboratories have documented a role for the G/A870 polymorphism in cancer risk (Betticher et al., 1995; Matthias et al., 1998; Knudsen et al., 2006; Pabalan et al., 2008). Furthermore, specific studies have documented a trend toward allele-specific expression of cyclin D1b (Betticher et al., 1995; Holley et al., 2001); however, there are clearly modifiers of the splicing, as the cyclin D1b transcript can be observed in individuals or tumors harboring only the G-allele (Bala and Peltomaki, 2001; Carrere et al., 2005; Krieger et al., 2006; Gupta et al., 2008). To specifically investigate the impact of the G/A870 polymorphism, a large multiethnic, case–control study consisting of 1376 invasive breast cancer cases and 2583 controls was examined (Table 1). In this population, the rs603965 polymorphism was not associated with breast cancer risk, disease stage or ER status (Table 1), and was not modified by the age of diagnosis, body mass index, postmenopausal hormone use or family history of breast cancer (P-values for interaction >0.18, data not shown). These findings indicate that the G/A870 polymorphism is not a strong predictor of breast cancer risk or associated with the specific clinicopathological parameters evaluated in this cohort.
Table 1.
The association between rs603965 in cyclin D1 and breast cancer risk in the MEC
| Cyclin D1 genotype | Cases (n, %) | Controls (n, %) | ORa (95% CI) | ||||
|---|---|---|---|---|---|---|---|
| n=1376* | n=2583* | All cases | Localized cases n=956 | Advanced cases n=328 | ER+ cases n=849 | ER− cases n=252 | |
| All racial/ethnic groups | |||||||
| GG | 409 (32.9) | 869 (36.5) | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| AG | 609 (49.0) | 1111 (46.7) | 1.08 (0.92–1.27) | 1.11 (0.93–1.33) | 1.05 (0.80–1.37) | 1.18 (0.97–1.43) | 0.96 (0.70–1.31) |
| AA | 226 (18.2) | 401 (16.8) | 1.07 (0.87–1.33) | 1.09 (0.86–1.38) | 0.98 (0.68–1.41) | 1.17 (0.91–1.50) | 1.16 (0.78–1.74) |
| Per allele | 1.04 (0.94–1.16) | 1.05 (0.94–1.18) | 1.00 (0.84–1.19) | 1.09 (0.97–1.24) | 1.06 (0.87–1.30) | ||
| P=0.43 | P=0.39 | P=0.99 | P=0.16 | P=0.58 | |||
Abbreviations: CI, confidence interval; OR, odds ratio; MEC, Multiethnic Cohort.
Adjust for race/ethnicity and age (<55, 55–64 and 65+ years).
Numbers do not add up to total because of missing genotype data.
P-values for heterogeneity by race/ethnicity ≥0.07.
Relationship between cyclin D1b and cyclin D1a expression
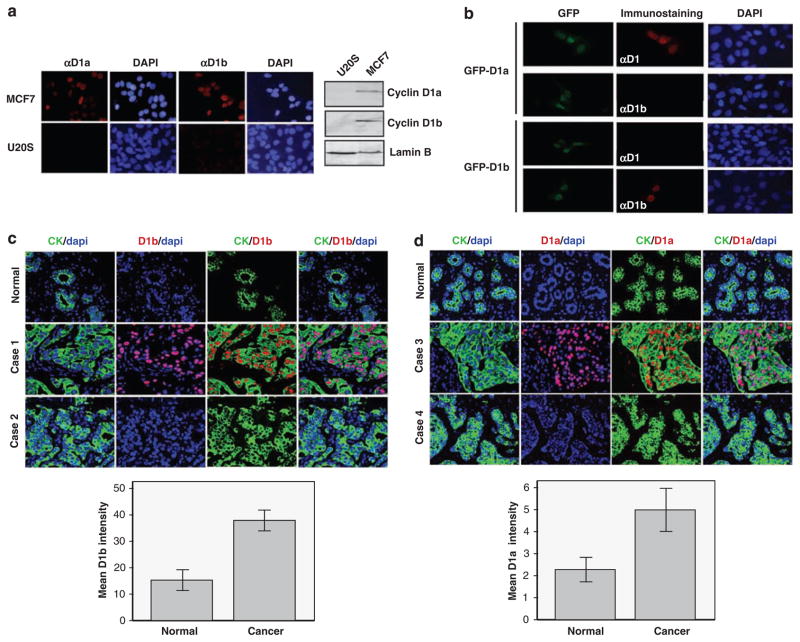
As there is an incomplete, and likely complex, link between the polymorphism and the production of cyclin D1b protein, direct analysis of this protein is pivotal to understanding its potential role in breast cancer. Here, we used a cyclin D1b-specific antibody, which has been utilized in a number of published studies, in which immunoblotting and immunostaining approaches have shown heterogeneity in protein expression in cell lines and tissue specimens (Burd et al., 2006; Marzec et al., 2006; Sanchez et al., 2008; Wang et al., 2008). However, to further confirm the specificity of the antibodies for immunostaining, antibody reactivity against MCF-7 and U2OS cells was compared. Earlier studies have shown that MCF-7 cells express robust levels of both cyclin D1 isoforms, whereas the levels of both proteins are barely detectable in U2OS cells (Wang et al., 2008). As shown in Figure 1a, both cyclin D1a and D1b nuclear reactivities were readily apparent in MCF-7 cells, but only minimal background staining was observed in U2OS cells. Thus, the amount of immunoreactivity observed in cytological analyses is consistent with the levels determined by immunoblotting (Figure 1a). To confirm that there was no cross-reactivity for the antibodies, U2OS cells were transfected with expression plasmids for green fluorescent protein (GFP)-cyclin D1a or GFP-cyclin D1b. Immunostaining with the cyclin D1a and cyclin D1b antibodies showed specific signals only in those cells transfected with appropriate GFP-cyclin D1a/D1b expression plasmids, indicating isoform specificity of the antibodies (Figure 1b).
Figure 1.
Cyclin D1a and cyclin D1b expression is elevated in breast cancer. (a) U2OS and MCF-7 cells were stained with cyclin D1a or cyclin D1b antibodies as indicated (left panel). Representative images taken at equal exposures for cyclin D1/cyclin D1b are shown. U2OS and MCF-7 cell lysates were subjected to immunoblotting with the indicated antibodies (right panel). (b) U2OS cells were transfected with the GFP-cyclin D1a or GFP-cyclin D1b expression plasmids as indicated. These cell populations were then stained with cyclin D1a or cyclin D1b antibodies. Representative images taken at equal exposure are shown. (c) Slides were co-stained for cyclin D1b and cytokeratin (CK) to detect epithelial cells. DAPI staining was used to detect all nuclei in the section. Representative images of specific breast cancer cases and normal controls are shown (top panel). Quantitation of the D1b signal in the CK-positive compartment was carried out and data were obtained from 39 normal specimens and 150 invasive ductal carcinomas (bottom panel; error bars indicate 95% CI, P<0.001). Student’s t-test was carried out. (d) Slides were co-stained for cyclin D1a and cytokeratin (CK) to detect epithelial cells. DAPI staining was utilized to detect all nuclei in the section. Representative images of specific breast cancer cases and normal controls are shown (top panel). Quantitation of the D1a signal in the CK-positive compartment was carried out from 44 normal specimens and 148 invasive ductal carcinomas (bottom panel; error bars indicate 95% CI, P<0.001). Student’s t-test was carried out.
These antibodies were first used to evaluate a cohort containing44 normal breast tissue specimens and 150 cancer cases, as summarized in Supplementary Table 1. The staining for cyclin D1b was quantified using the AQUA fluorescence-based immunohistochemistry platform (Dolled-Filhart et al., 2006). Cyclin D1b reactivity is located in the cytokeratin-positive carcinoma cells and is restricted from the stroma (Figure 1c). Moreover, cyclin D1b exhibits a nuclear staining pattern in all positive-staining sections analysed (Figure 1c). Overall, the levels of cyclin D1b expression were heterogeneous between tumors, suggesting that alterations in the levels of cyclin D1b expression could be associated with specific features of breast cancer (Figure 1c). Importantly, these analyses show that cyclin D1b protein levels are upregulated in breast cancer relative to normal breast tissue (P<0.001) and are consistent with recent findings (Wang et al., 2008). As cyclin D1b is transcribed from the same gene as cyclin D1a, the association with breast cancer may be merely reflective of the overall overexpression of cyclin D1 in the same tumor. Therefore, the expression of cyclin D1a was determined in parallel on the same quantitative platform. In this context, cyclin D1a also showed elevated expression among breast cancer specimens (Figure 1d, P<0.001), showing an overall distribution profile similar to cyclin D1b.
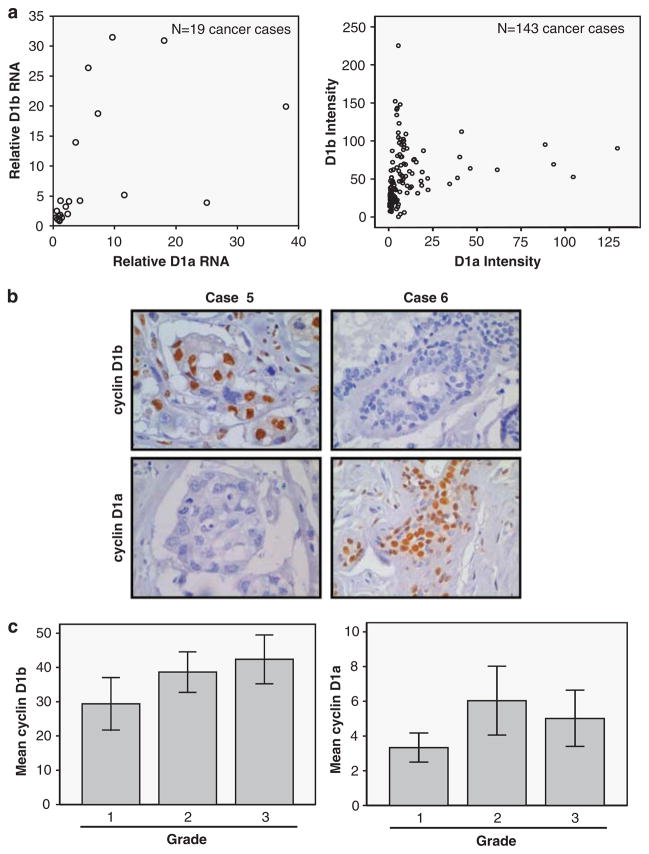
These findings suggested that the expression of cyclin D1b could be simply a by-product of elevated levels of cyclin D1. Such a concern has also been noted with respect to the low-molecular-weight forms of cyclin E observed in breast cancer specimens (Spruck et al., 2006). Using quantitative reverse transcription PCR analyses of the cyclin D1 transcripts in a limited number of breast tumor specimens (n=19), we observed that there was not a strong direct relationship between the levels of cyclin D1a and cyclin D1b RNA (Figure 2a). Furthermore, quantitative assessment of staining for cyclin D1a vs cyclin D1b in each tumor indicated that protein levels are not directly correlated (Figure 2a). Although many cases expressed both cyclin D1a and cyclin D1b proteins, in specific cases, the expression of a single cyclin D1 species appeared to predominate, as shown in representative stained chromagen sections (Figure 2b). Thus, although both cyclin D1a and cyclin D1b levels are elevated in breast cancer, the relative abundance of each species could reflect distinct subtypes of the disease.
Figure 2.
Cyclin D1b defines a population of breast cancer distinct from cyclin D1a. (a) Quantitative reverse transcription PCR was performed to determine the relative levels of cyclin D1a- and cyclin D1b-specific RNA, wherein signal from non-neoplastic was set to ‘1’ (left panel). The absolute intensity of cyclin D1b vs cyclin D1a in a total of 143 breast cancer specimens is shown (right panel). (b) Specific cases show preferentially the expressions of cyclin D1b (case 5) and cyclin D1a (case 6). (c) Cyclin D1b or cyclin D1a intensity was determined as a function of tumor grade (error bars indicate 95% CI). Cyclin D1b levels are associated with increased tumor grade (P=0.014) and cyclin D1a levels are not associated with increased tumor grade (P>0.5). ANOVA statistical test was carried out.
To initially investigate the relationships between the different cyclin D1 proteins and selected properties of breast cancer, these tumors were evaluated for other molecular markers associated with disease. These analyses revealed no relationship of cyclin D1b with ER status, progesterone receptor (PR) expression, or Her2/Neu (data not shown). However, there was a modest quantitative relationship of increasing cyclin D1b levels with increased grade (r=0.258, P=0.017), and the difference in cyclin D1b levels between grade 1 and grade 3 invasive ductal carcinoma was statistically significant (P=0.014, Figure 2c). In contrast, cyclin D1a levels were not statistically different between breast cancer cases grouped by grade (P=0.108, Figure 2c). Thus, on the basis of the differential expression patterns of cyclins D1a and D1b across cancer cases, cyclin D1b may harbor prognostic power in breast cancer that is distinct from cyclin D1a.
Cyclin D1b expression and breast cancer outcome
To specifically define the impact of cyclin D1b vs D1a expression on clinical outcome, expression levels were analysed in an independent cohort of 175 primary breast cancers with a median follow-up of 75 months (Supplementary Table 2). The levels of cyclin D1 protein reactivity were quantified as described in the Supplementary Materials and Methods, and the slides were scored by an expert breast cancer pathologist (EKAM). On this platform, high nuclear cyclin D1b expression (that is, average intensity >2+) was not associated with clinicopathological features (assessed by the Mann–Whitney non-parametric test), including tumor size (P=0.21), age (P=0.67), lymph node status (P=0.08), ER (P=0.59), PR (P=0.57) or HER2 amplification (χ2-test, P=0.79). With this scoring criterion, tumors exhibiting high cyclin D1b levels were not significantly associated with grade III disease (P=0.34). Furthermore, the expression of cyclin D1b was not associated with the level of other cell-cycle regulatory factors (for example, cyclin E and p27Kip1), including cyclin D1a (P=0.46), in agreement with the data in Figure 2.
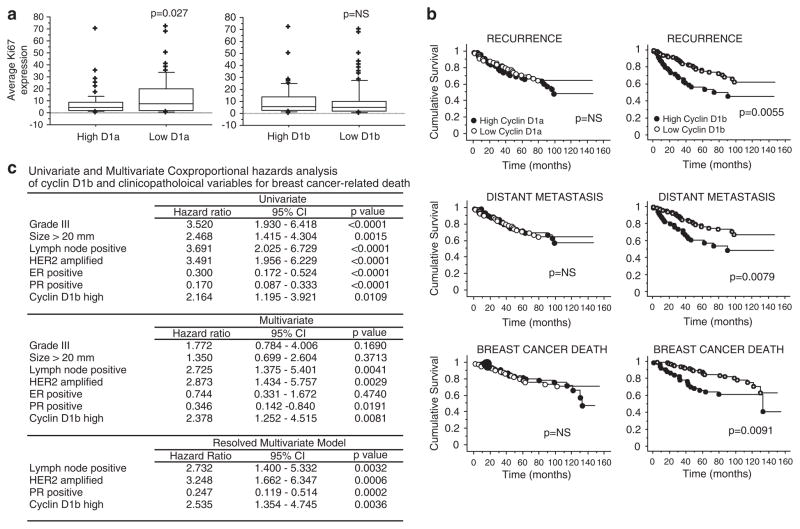
As both cyclin D1a and cyclin D1b have been associated with stimulating proliferative responses in cell culture models (Lu et al., 2003; Solomon et al., 2003; Wang et al., 2008), the association of either cyclin D1 variant was evaluated against the proliferation marker Ki67 (Figure 3a). These analyses showed that levels of cyclin D1a staining were inversely correlated with Ki67 (assessed by the Mann–Whitney non-parametric test), consistent with the earlier reported relationship with the ER-positive, low-proliferation phenotype (Barnes and Gillett, 1998; Roy and Thompson, 2006). In contrast, cyclin D1b levels were not associated with Ki67 staining. These findings further indicated that cyclin D1a and D1b may be disparately associated with breast cancer subtypes and resultant clinical outcomes.
Figure 3.
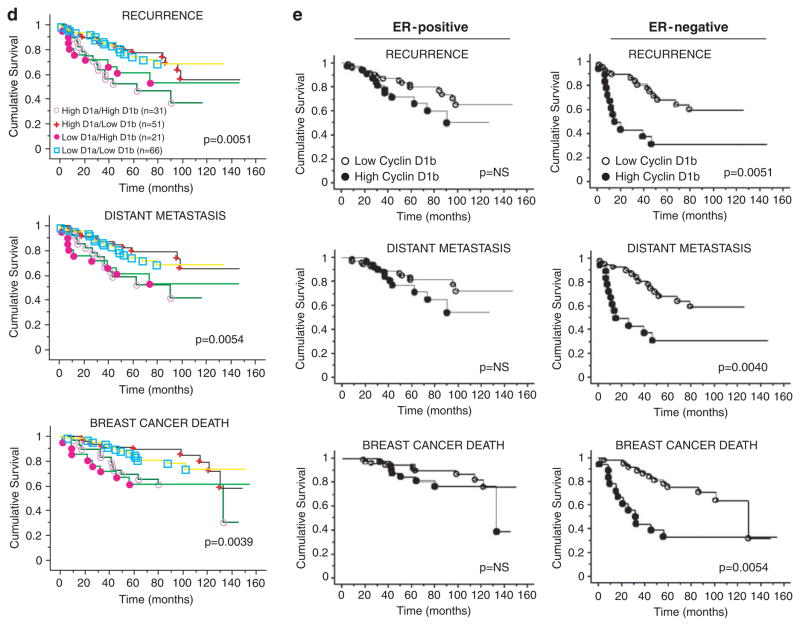
Cyclin D1b is associated with poor outcome in breast cancer. (a) Relationship between cyclin D1a and cyclin D1b levels and the proliferation marker Ki67 were monitored. Box plots depict data for cyclin D1a low (n=73), cyclin D1a high (n=74), dichotomized on the median H-score, and cyclin D1b low (n=98) and cyclin D1b high (n=50) dichotomized at an intensity >2+, as related to the percentage of Ki67-positive cells. A significant inverse correlation between cyclin D1a levels and Ki67 was detected (P=0.0256) using the Mann–Whitney non-parametric test. (b) The Kaplan–Meier analysis for cyclin D1a and cyclin D1b expression was carried out in the entire cohort. Graphs represent analyses carried out for recurrence, distant metastasis and breast cancer-related death. Left panel: high cyclin D1a expressors (n=82) are represented by ● and low cyclin D1a expressors (n=87) are represented by ○;. Right panel: high cyclin D1b expressors (n=52) are represented by ● and low cyclin D1b expressors (n=123) are represented by ○. Statistical analyses were carried out using the log-rank test. (c) Summary of statistical analyses of cyclin D1b by univariate and multivariate analyses. Details of the analyses are provided in the Supplementary Materials and methods. (d) The Kaplan–Meier analysis was carried out for distinct cyclin D1a/D1b-isoform subsets in the entire cohort. High cyclin D1a/high cyclin D1b expressors (n=31) are represented by ○, high cyclin D1a/low cyclin D1b expressors (n=51) are represented by +, low cyclin D1a/high cyclin D1b expressors (n=21) by ●, and low cyclin D1a/low cyclin D1b expressors (n=66) are represented by □. Statistical analyses were carried out using the log-rank test. (e) Kaplan–Meier graphs for cyclin D1b expression stratified by ER status. Graphs represent analyses carried out for recurrence, distant metastasis and breast cancer-related death among ER-positive or ER-negative cases. High cyclin D1b expressors are represented by ● and low cyclin D1b expressors by ○. Statistical analyses were carried out using the log-rank test.
In univariate analysis, high cyclin D1a levels were not associated with recurrence, metastases or cancer-specific death (P>0.05) (Figure 3b). In contrast, high levels of cyclin D1b were associated with adverse patient outcome for recurrence (P=0.0055), distant metastases (P=0.0079) and breast cancer-specific death (P=0.0091) (Figure 3b). Furthermore, in univariate analysis incorporating traditional prognostic variables, including tumor size, grade, lymph node status, ER and PR status and HER2 amplification, high expression of cyclin D1b was a statistically significant predictor of poor outcome from breast cancer (HR (hazard ratio): 2.164, 95% CI (confidence interval): 1.195–3.921, P=0.011) (Figure 3c). To assess if cyclin D1b was an independent prognostic factor in this cohort, and not the result of other confounding variables, Cox proportional hazard models were constructed with stepwise removal of redundant variables until resolution (Figure 3c). The initial model incorporating all the significant variables on univariate analysis showed that high expression of cyclin D1b remained significant on multivariate analysis (HR 2.378, 95% CI 1.252–4.515, P=0.0081; Figure 3c). The resolved model showed that high levels of cyclin D1b remain associated with poor survival together with the well-validated prognostic markers, such as lymph node status, HER2 amplification and PR status. Thus, cyclin D1b protein levels are independently associated with poor survival in breast cancer.
To determine whether the effects of cyclin D1b on survival were modified by other factors associated with breast cancer pathogenesis, specific subset analyses were carried out. Initially, as cyclin D1b is related in structure and function to cyclin D1a, the four subsets of relative levels of the isoforms were compared (Figure 3d). These data showed that cyclin D1b was predictive for disease outcomes irrespective of the relative cyclin D1a levels. Subsequently, as high levels of cyclin D1b protein are observed in both ER-positive and ER-negative tumors, the impact of cyclin D1b on both forms of disease was analysed. As shown in Figure 3e, ER-positive tumors with high levels of cyclin D1b tended to have a poor prognosis, but this result did not reach statistical significance. In contrast, in ER-negative tumors, high-level expression of cyclin D1b was associated with poor outcome for recurrence (P=0.0051), distant metastases (P=0.004) and breast cancer death (P=0.0054) (Figure 3e). Thus, cyclin D1b is associated with poor disease outcome in breast cancer in a manner that is not modified by the relative level of cyclin D1a; however, the association of high cyclin D1b levels with disease is most apparent in the context of ER-negative disease.
Summary
The relevance of G/A870 polymorphism to breast cancer has been the subject of several earlier studies (Krippl et al., 2003; Ceschi et al., 2005; Shu et al., 2005; Naidu et al., 2008; Yu et al., 2008). Although the overall conclusions of these studies have been disparate, a recent meta-analysis of published cases suggested that the polymorphism is weakly related to increased breast cancer risk (Pabalan et al., 2008). Our findings are in agreement, in revealing a negligible impact of the polymorphism on breast cancer risk. Owing to the size and ethnic heterogeneity of our cohort, we believe that a consensus is arising related to the modest impact of the G/A870 polymorphism in breast cancer, which contrasts with the stronger influence observed in other tumor types (Pabalan et al., 2008).
It is now clear from a number of studies that modifiers external to the polymorphism are associated with the accumulation of cyclin D1b. For example, factors associated with chromatin remodeling and translation potently modulate the abundance of the cyclin D1b isoform (Batsche et al., 2006; Sanchez et al., 2008). Specifically, loss of the Brm chromatin remodeling protein results in a substantial increase in the cyclin D1b transcript as a result of less efficient splicing. Through an independent mechanism, the Ewing’s sarcoma oncogene and the ets family transcription factor (FLI1) results in the upregulation of cyclin D1b through the alteration of transcript elongation. In addition, a dissociation between the G/A870 genotype and the relative expression of cyclin D1b has been observed in multiple tumor types, including colorectal, mantle cell lymphoma and non-small-cell lung cancer (Bala and Peltomaki, 2001; Carrere et al., 2005; Krieger et al., 2006; Gupta et al., 2008). In this context, it is important to appreciate that both genotypic and histochemical based analyses have merit in dissecting an impact on cancer risk or disease outcome. However, owing to the lack of direct correlation between genotype and protein production, direct analyses of the cyclin D1b protein product is perhaps the only means to define the impact of the protein on disease.
As cyclin D1a and cyclin D1b proteins are closely related (they derive from the same primary transcript), it is critical to discern whether the expressions of cyclin D1a and D1b are coupled or independent factors. A clear advantage in the analyses of cyclin D1b is the possible use of cyclin D1a- and D1b-specific antibodies. As described earlier, and further validated herein, the available reagents can clearly differentiate between cyclin D1a and D1b (Burd et al., 2006; Marzec et al., 2006; Sanchez et al., 2008; Wang et al., 2008). Using such reagents, we quantitatively determined the levels of each protein unambiguously, and these results were further supported by quantitative analyses on RNA transcripts from primary breast cancer. These analyses show that the expressions of cyclin D1a and cyclin D1b are not related by means of a simple linear relationship. Such a finding is supported by our analyses of cyclin D1b levels in breast cancer, and similar analyses in mantle cell lymphoma (Carrere et al., 2005; Krieger et al., 2006) and prostate cancer (Comstock et al., 2009). Consistent with these results, in cell culture models, the levels of cyclin D1 isoforms are differentially regulated by ER antagonists and DNA damage signals (Wang et al., 2008). Thus, the relative production of cyclin D1a and D1b may reflect differences in the tumor-specific signaling pathways or tissue context of the tumor. As such, outcomes associated with cyclin D1b are not merely a reflection of an overabundance of cyclin D1 protein.
The importance of cyclin D1 as a marker for outcome in breast cancer is somewhat controversial. Although >50% of breast cancers express cyclin D1 at elevated levels, a significant majority of these tumors are of the ER-positive luminal subtype, which is generally associated with better prognosis (Gillett et al., 1994, 1996; Barnes and Gillett, 1998; Roy and Thompson, 2006). From our analyses, we found that cyclin D1a levels were elevated predominantly in ER-positive tumors, were inversely correlated with Ki67 and had little impact on disease outcome in the cohort analysed when dichotomized about the median into high and low expressors. In contrast, elevated levels of cyclin D1b protein were independent of a variety of clinicopathological features, including Ki67 and ER. However, in marked contrast to cyclin D1a, elevated cyclin D1b was associated with poor overall survival, recurrence and metastasis independent of other commonly analysed prognostic variables. Thus, cyclin D1b identifies a unique subset of tumors associated with increased disease progression. In keeping with cyclin D1b acting independently of cyclin D1a, the association of cyclin D1b with disease outcome was not modulated by the status of cyclin D1a. In addition, cyclin D1b levels were independent of Her2. However, owing to the limited number of Her2-positive tumors in our cohorts, we were unable to address whether cyclin D1b status influenced disease outcome in this subset. In contrast, stratification of patients based on ER status provides a compelling rationale for investigating cyclin D1b function in discrete subtypes of breast cancer. Among ER-positive breast cancers, high levels of cyclin D1b, although trending toward poor outcome, were not significant. On the basis of the preclinical studies (Wang et al., 2008), it will be crucial to specifically interrogate the association of cyclin D1b levels to the response to endocrine therapies (for example, tamoxifen) in a setting free of cytotoxic agents. Clearly, larger studies will be required to specifically define the influence of cyclin D1b levels in ER-positive breast cancer and address the critical question of response to hormonal therapy. In contrast with ER-positive cancers, high levels of cyclin D1b were strongly associated with poor disease outcome in ER-negative breast cancers. As this form of the disease is characterized by relatively low cyclin D1a expression, this finding suggests that cyclin D1b may be particularly relevant in the etiology, progression and the ultimate poor prognosis associated with ER-negative breast cancer. Presumably, the differing survival could reflect an important impact of cyclin D1b on therapeutic interventions involved in the treatment of ER-negative breast cancer. However, this possibility remains to be explored in preclinical models.
Combined, these studies provide critical insight into the association of the G/A870 polymorphism and cyclin D1 isoform expression with risk and progression of breast cancer, and in so doing show that cyclin D1b levels are independently associated with adverse disease outcome.
Supplementary Material
Acknowledgments
The authors thank all of the laboratory and administrative staff that contributed to the editing and preparation of the manuscript. This work was supported by the following grants: the National Health and Medical Research Council of Australia (Grant no. 276408), the Cancer Institute NSW, the Petre Foundation and the RT Hall Trust, the National Institutes of Health Grant CA104213 (ESK), the National Institutes of Health Grant CA099996 (KEK). We thank Drs Paul Crea and Davendra Segara of St Vincent’s Clinic for access to patient material and data.
References
- Arnold A, Papanikolaou A. Cyclin D1 in breast cancer pathogenesis. J Clin Oncol. 2005;23:4215–4224. doi: 10.1200/JCO.2005.05.064. [DOI] [PubMed] [Google Scholar]
- Bala S, Peltomaki P. Cyclin D1 as a genetic modifier in hereditary nonpolyposis colorectal cancer. Cancer Res. 2001;61:6042–6045. [PubMed] [Google Scholar]
- Barnes DM, Gillett CE. Cyclin D1 in breast cancer. Breast Cancer Res Treat. 1998;52:1–15. doi: 10.1023/a:1006103831990. [DOI] [PubMed] [Google Scholar]
- Batsche E, Yaniv M, Muchardt C. The human SWI/SNF subunit Brm is a regulator of alternative splicing. Nat Struct Mol Biol. 2006;13:22–29. doi: 10.1038/nsmb1030. [DOI] [PubMed] [Google Scholar]
- Betticher DC, Thatcher N, Altermatt HJ, Hoban P, Ryder WD, Heighway J. Alternate splicing produces a novel cyclin D1 transcript. Oncogene. 1995;11:1005–1011. [PubMed] [Google Scholar]
- Blais A, Dynlacht BD. E2F-associated chromatin modifiers and cell cycle control. Curr Opin Cell Biol. 2007;19:658–662. doi: 10.1016/j.ceb.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley MF, Sweeney KJ, Hamilton JA, Sini RL, Manning DL, Nicholson RI, et al. Expression and amplification of cyclin genes in human breast cancer. Oncogene. 1993;8:2127–2133. [PubMed] [Google Scholar]
- Burd CJ, Petre CE, Morey LM, Wang Y, Revelo MP, Haiman CA, et al. Cyclin D1b variant influences prostate cancer growth through aberrant androgen receptor regulation. Proc Natl Acad Sci USA. 2006;103:2190–2195. doi: 10.1073/pnas.0506281103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrere N, Belaud-Rotureau MA, Dubus P, Parrens M, de Mascarel A, Merlio JP. The relative levels of cyclin D1a and D1b alternative transcripts in mantle cell lymphoma may depend more on sample origin than on CCND1 polymorphism. Haematologica. 2005;90:854–856. [PubMed] [Google Scholar]
- Ceschi M, Sun CL, Van Den Berg D, Koh WP, Yu MC, Probst-Hensch N. The effect of cyclin D1 (CCND1) G870A-polymorphism on breast cancer risk is modified by oxidative stress among Chinese women in Singapore. Carcinogenesis. 2005;26:1457–1464. doi: 10.1093/carcin/bgi093. [DOI] [PubMed] [Google Scholar]
- Cobrinik D. Pocket proteins and cell cycle control. Oncogene. 2005;24:2796–2809. doi: 10.1038/sj.onc.1208619. [DOI] [PubMed] [Google Scholar]
- Comstock CES, Augello MA, Pe Benito R, Karch J, Tran TH, Utama FE, et al. Cyclin D1 splice variants: polymorphism, risk, and isoform specific regulation in prostate cancer. Clin Cancer Res. 2009 doi: 10.1158/1078-0432.CCR-08-2865. (submitted) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl JA. Cycling to cancer with cyclin D1. Cancer Biol Ther. 2002;1:226–231. doi: 10.4161/cbt.72. [DOI] [PubMed] [Google Scholar]
- Dolled-Filhart M, McCabe A, Giltnane J, Cregger M, Camp RL, Rimm DL. Quantitative in situ analysis of beta-catenin expression in breast cancer shows decreased expression is associated with poor outcome. Cancer Res. 2006;66:5487–5494. doi: 10.1158/0008-5472.CAN-06-0100. [DOI] [PubMed] [Google Scholar]
- Gillett C, Fantl V, Smith R, Fisher C, Bartek J, Dickson C, et al. Amplification and overexpression of cyclin D1 in breast cancer detected by immunohistochemical staining. Cancer Res. 1994;54:1812–1817. [PubMed] [Google Scholar]
- Gillett C, Smith P, Gregory W, Richards M, Millis R, Peters G, et al. Cyclin D1 and prognosis in human breast cancer. Int J Cancer. 1996;69:92–99. doi: 10.1002/(SICI)1097-0215(19960422)69:2<92::AID-IJC4>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Gupta VK, Feber A, Xi L, Pennathur A, Wu M, Luketich JD, et al. Association between CCND1 G/A870 polymorphism, allele-specific amplification, cyclin D1 expression, and survival in esophageal and lung carcinoma. Clin Cancer Res. 2008;14:7804–7812. doi: 10.1158/1078-0432.CCR-08-0744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holley SL, Heighway J, Hoban PR. Induced expression of human CCND1 alternative transcripts in mouse Cyl-1 knockout fibroblasts highlights functional differences. Int J Cancer. 2005;114:364–370. doi: 10.1002/ijc.20750. [DOI] [PubMed] [Google Scholar]
- Holley SL, Parkes G, Matthias C, Bockmuhl U, Jahnke V, Leder K, et al. Cyclin D1 polymorphism and expression in patients with squamous cell carcinoma of the head and neck. Am J Pathol. 2001;159:1917–1924. doi: 10.1016/S0002-9440(10)63038-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iaquinta PJ, Lees JA. Life and death decisions by the E2F transcription factors. Curr Opin Cell Biol. 2007;19:649–657. doi: 10.1016/j.ceb.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirstrom K, Stendahl M, Ryden L, Kronblad A, Bendahl PO, Stal O, et al. Adverse effect of adjuvant tamoxifen in premenopausal breast cancer with cyclin D1 gene amplification. Cancer Res. 2005;65:8009–8016. doi: 10.1158/0008-5472.CAN-05-0746. [DOI] [PubMed] [Google Scholar]
- Knudsen ES, Knudsen KE. Retinoblastoma tumor suppressor: where cancer meets the cell cycle. Exp Biol Med (Maywood) 2006;231:1271–1281. doi: 10.1177/153537020623100713. [DOI] [PubMed] [Google Scholar]
- Knudsen KE. The cyclin D1b splice variant: an old oncogene learns new tricks. Cell Div. 2006;1:15. doi: 10.1186/1747-1028-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen KE, Diehl JA, Haiman CA, Knudsen ES. Cyclin D1: polymorphism, aberrant splicing and cancer risk. Oncogene. 2006;25:1620–1628. doi: 10.1038/sj.onc.1209371. [DOI] [PubMed] [Google Scholar]
- Krieger S, Gauduchon J, Roussel M, Troussard X, Sola B. Relevance of cyclin D1b expression and CCND1 polymorphism in the pathogenesis of multiple myeloma and mantle cell lymphoma. BMC Cancer. 2006;6:238. doi: 10.1186/1471-2407-6-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krippl P, Langsenlehner U, Renner W, Yazdani-Biuki B, Wolf G, Wascher TC, et al. The 870G>A polymorphism of the cyclin D1 gene is not associated with breast cancer. Breast Cancer Res Treat. 2003;82:165–168. doi: 10.1023/B:BREA.0000004372.20461.33. [DOI] [PubMed] [Google Scholar]
- Lu F, Gladden AB, Diehl JA. An alternatively spliced cyclin D1 isoform, cyclin D1b, is a nuclear oncogene. Cancer Res. 2003;63:7056–7061. [PubMed] [Google Scholar]
- Malumbres M, Barbacid M. To cycle or not to cycle: a critical decision in cancer. Nat Rev Cancer. 2001;1:222–231. doi: 10.1038/35106065. [DOI] [PubMed] [Google Scholar]
- Marzec M, Kasprzycka M, Lai R, Gladden AB, Wlodarski P, Tomczak E, et al. Mantle cell lymphoma cells express predominantly cyclin D1a isoform and are highly sensitive to selective inhibition of CDK4 kinase activity. Blood. 2006;108:1744–1750. doi: 10.1182/blood-2006-04-016634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthias C, Branigan K, Jahnke V, Leder K, Haas J, Heighway J, et al. Polymorphism within the cyclin D1 gene is associated with prognosis in patients with squamous cell carcinoma of the head and neck. Clin Cancer Res. 1998;4:2411–2418. [PubMed] [Google Scholar]
- Naidu R, Yip CH, Taib NA. Polymorphisms of HER2 Ile655Val and cyclin D1 (CCND1) G870A are not associated with breast cancer risk but polymorphic allele of HER2 is associated with nodal metastases. Neoplasma. 2008;55:87–95. [PubMed] [Google Scholar]
- Pabalan N, Bapat B, Sung L, Jarjanazi H, Francisco-Pabalan O, Ozcelik H. Cyclin D1 Pro241Pro (CCND1-G870A) polymorphism is associated with increased cancer risk in human populations: a meta-analysis. Cancer Epidemiol Biomarkers Prev. 2008;17:2773–2781. doi: 10.1158/1055-9965.EPI-08-0169. [DOI] [PubMed] [Google Scholar]
- Roy PG, Thompson AM. Cyclin D1 and breast cancer. Breast. 2006;15:718–727. doi: 10.1016/j.breast.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Rudas M, Lehnert M, Huynh A, Jakesz R, Singer C, Lax S, et al. Cyclin D1 expression in breast cancer patients receiving adjuvant tamoxifen-based therapy. Clin Cancer Res. 2008;14:1767–1774. doi: 10.1158/1078-0432.CCR-07-4122. [DOI] [PubMed] [Google Scholar]
- Sanchez G, Bittencourt D, Laud K, Barbier J, Delattre O, Auboeuf D, et al. Alteration of cyclin D1 transcript elongation by a mutated transcription factor up-regulates the oncogenic D1b splice isoform in cancer. Proc Natl Acad Sci USA. 2008;105:6004–6009. doi: 10.1073/pnas.0710748105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu XO, Moore DB, Cai Q, Cheng J, Wen W, Pierce L, et al. Association of cyclin D1 genotype with breast cancer risk and survival. Cancer Epidemiol Biomarkers Prev. 2005;14:91–97. [PubMed] [Google Scholar]
- Sicinski P, Donaher JL, Parker SB, Li T, Fazeli A, Gardner H, et al. Cyclin D1 provides a link between development and oncogenesis in the retina and breast. Cell. 1995;82:621–630. doi: 10.1016/0092-8674(95)90034-9. [DOI] [PubMed] [Google Scholar]
- Solomon DA, Wang Y, Fox SR, Lambeck TC, Giesting S, Lan Z, et al. Cyclin D1 splice variants. Differential effects on localization, RB phosphorylation, and cellular transformation. J Biol Chem. 2003;278:30339–30347. doi: 10.1074/jbc.M303969200. [DOI] [PubMed] [Google Scholar]
- Spruck C, Sun D, Fiegl H, Marth C, Mueller-Holzner E, Goebel G, et al. Detection of low molecular weight derivatives of cyclin E1 is a function of cyclin E1 protein levels in breast cancer. Cancer Res. 2006;66:7355–7360. doi: 10.1158/0008-5472.CAN-05-3240. [DOI] [PubMed] [Google Scholar]
- Sutherland RL, Musgrove EA. Cyclins and breast cancer. J Mammary Gland Biol Neoplasia. 2004;9:95–104. doi: 10.1023/B:JOMG.0000023591.45568.77. [DOI] [PubMed] [Google Scholar]
- Wang TC, Cardiff RD, Zukerberg L, Lees E, Arnold A, Schmidt EV. Mammary hyperplasia and carcinoma in MMTV-cyclin D1 transgenic mice. Nature. 1994;369:669–671. doi: 10.1038/369669a0. [DOI] [PubMed] [Google Scholar]
- Wang Y, Dean JL, Millar EK, Tran TH, McNeil CM, Burd CJ, et al. Cyclin D1b is aberrantly regulated in response to therapeutic challenge and promotes resistance to estrogen antagonists. Cancer Res. 2008;68:5628–5638. doi: 10.1158/0008-5472.CAN-07-3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu CP, Yu JC, Sun CA, Tzao C, Ho JY, Yen AM. Tumor susceptibility and prognosis of breast cancer associated with the G870A polymorphism of CCND1. Breast Cancer Res Treat. 2008;107:95–102. doi: 10.1007/s10549-007-9522-y. [DOI] [PubMed] [Google Scholar]
- Yu Q, Geng Y, Sicinski P. Specific protection against breast cancers by cyclin D1 ablation. Nature. 2001;411:1017–1021. doi: 10.1038/35082500. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.