Abstract
Numerous physiological and pathological stimuli promote the rearrangement of the actin cytoskeleton, thereby modulating cellular motile functions. Although it seems intuitively obvious that cell motility requires coordinated protein biosynthesis, until recently the linkage between cytoskeletal actin dynamics and correlated gene activities remained unknown. This knowledge gap was filled in part by the discovery that globular actin polymerization liberates myocardin-related transcription factor (MRTF) cofactors, thereby inducing the nuclear transcription factor serum response factor (SRF) to modulate the expression of genes encoding structural and regulatory effectors of actin dynamics. This insight stimulated research to better understand the actin–MRTF–SRF circuit and to identify alternative mechanisms that link cytoskeletal dynamics and genome activity.
Both during embryonic development and as functional components of mature multicellular organisms, individual cells continuously undergo physical changes in appearance, shape, position and contact with extracellular structures including other cells (BOX 1). These physical changes require cellular motile functions, which are the driving force of many dynamic cell behaviours such as cell migration, guided movement, engulfment, adhesion and contraction. The motile functions of cells are regulated by both physiological and pathological stimuli. The physical basis for cellular motile functions is provided by macromolecular assemblies, including cytoskeletal scaffold structures of actin micro filaments. These scaffolds undergo dynamic changes in both polymerization and their interaction with specific binding proteins. Regarding the actin microfilament, poly merization of monomeric globular actin (G-actin) into a filamentous actin (F-actin) fibre is influenced by local intracellular concentrations of ATP-bound G-actin and by the activity of many actin-binding proteins (ABPs). This determines the rate and direction of polymerization, as well as the shape of the newly generated filament1. Such dynamic rearrangements of actin filaments generate the physical force needed for cells to protrude filopodial or lamellipodial membrane extensions and to readjust adhesive contacts, known as focal adhesions, to the cellular environment, as required for motile cell functions.
Box 1 | Motile functions of cells.
The living eukaryotic cell must be viewed as a dynamic structure experiencing frequent changes in macromolecular contacts, signal inputs and metabolic flux. As a consequence, cells display continued physical rearrangement and adjustment. Such motility functions require supporting intracellular counter-forces, as generated by dynamic rearrangements of the actin microfilament. Polymerization of monomeric globular actin (G-actin) into filamentous actin (F-actin) polymers is reversible and both reactions are facilitated by numerous actin binding proteins (ABPs).
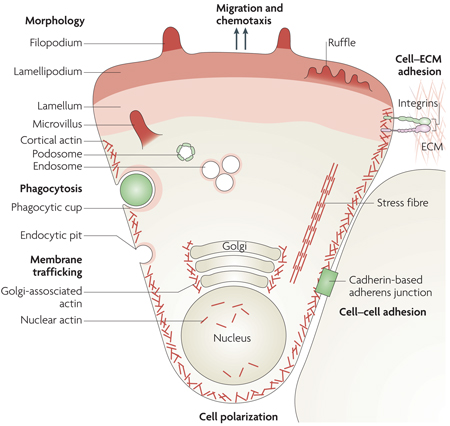
Cellular motile functions involving dynamic actin microfilament rearrangements include cell migration, spreading, adhesion, contraction and polarization, cell–cell contact and cell–extracellular matrix (ECM) interactions (see the figure). These microfilament rearrangements result in the processes of chemotaxis, mechanotaxis (cell stretching and overload), maintenance of cellular tone, shape changes (branching), engulfment (by phagocytosis), neurite or vascular tip cell extension, axon guidance, morphological rearrangements (such as those required for epithelial–mesenchymal transitions) and intracellular transport (such as RNA localization, protein delivery, membrane trafficking, endocytosis, internalization, secretion and organelle positioning).
Dynamic changes of the actin microfilament contribute to each of these motile functions, either as structural components or as platforms for signal transduction. Microfilament polymerization and depolymerization, accompanied by the association and dissociation of interacting molecular structures, provide the physical scaffold for dynamic changes underlying motility. As discussed in this review, the multitude of cell behaviours involving actin dynamics requires gene expression and de novo protein biosynthesis. Selected ABPs function as courier proteins to communicate to the cellular genome the state of actin rearrangements. Such transcriptional relay systems modulate the expression of genes that encode products which contribute to either the execution or regulation of cellular motility functions. Actin structures are indicated in the figure in different shades of red. Image is reproduced, with permission, from Nature Cell Biology REF. 142 (2007) Macmillan Publishers Ltd. All rights reserved.
Although actin microfilament turnover involves the regeneration of depleted ATP–actin levels from ADP–actin pools, enabled by the ABP profilin, cell motility also requires the de novo biosynthesis of G-actin. Similarly, activation of protein synthesis is required for the timely generation of other structural components of the actin microfilament network (for example, myosins, actin assembly and disassembly factors, capping proteins and F-actin cross-linking proteins) as well as regulatory components (for example, kinases, phosphatases and myosin light chains (MLCs)). Therefore, cell motility requires the tight temporal coupling of actin dynamics and transcriptional activity.
The requirement for the temporal linkage of actin turnover and transcription necessitates signal transduction mechanisms that communicate the cyto plasmic actin polymerization status to the nuclear genome. How does the transcriptional machinery sense the need for the biosynthesis of components of both the dynamic actin network and the motility apparatus? To approach this question we first review the various cellular receptors that are activated by motility-inducing signals and subsequently lead to activation of the central intracellular regulatory proteins of the Rho family of GTP-binding proteins (FIG. 1). We then focus on relay systems that enable dynamic rearrangements of the cytoskeletal actin microfilament to be communicated to the nucleus by release and nuclear translocation of specific ABPs, thereby eliciting defined changes in the expression of specific gene profiles (FIG. 2).
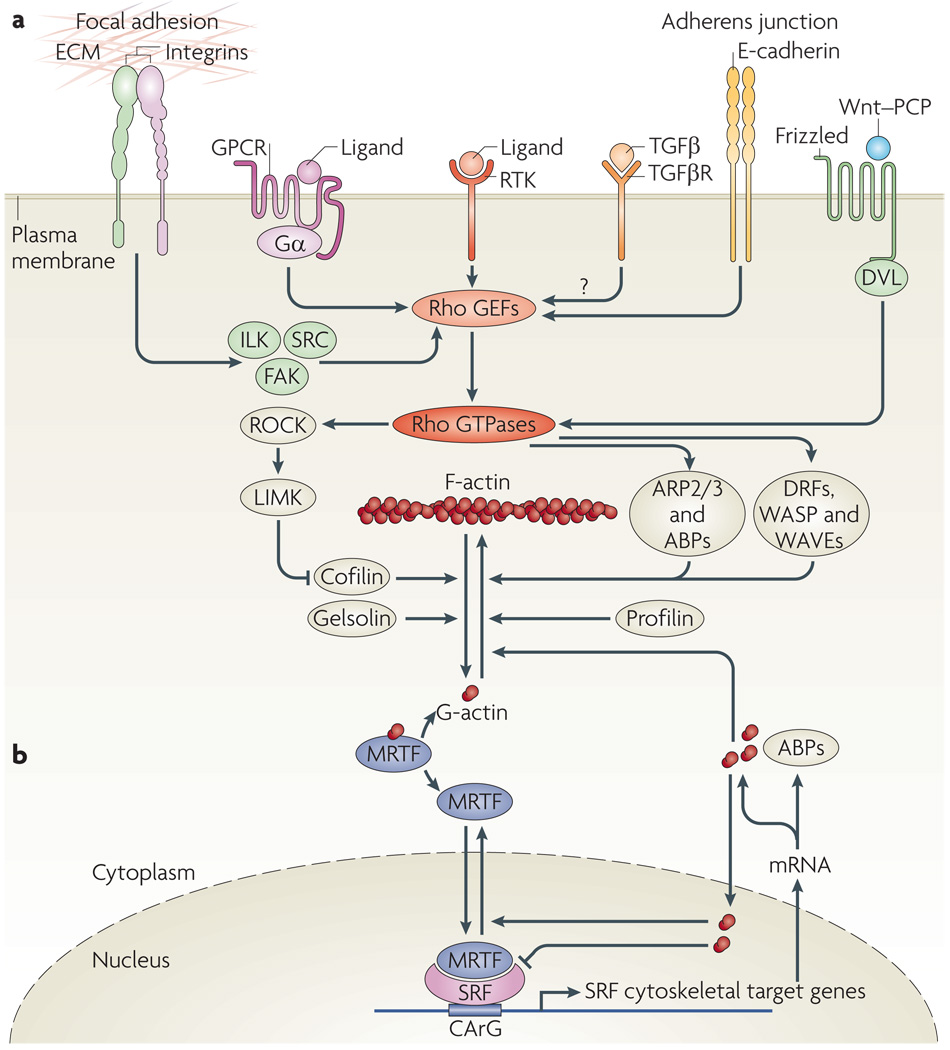
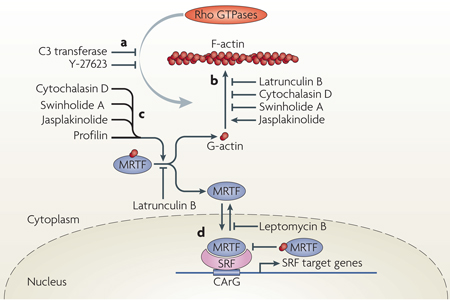
Figure 1. receptors affecting actin dynamics and MrTF-mediated regulation of SrF target genes.
a | Cytoskeletal actin microfilament dynamics are affected by the activation of six classes of plasma membrane receptor: receptor Tyr kinases (RTKs), G protein-coupled receptors (GPCRs; with α-subunits of Gα12/13, Gαq/11 or Gαi/0), integrins as structural mediators of focal adhesions, transforming growth factor-β receptors (TGFβRs), E-cadherins at adherens junctions and Frizzled, which mediates the non-canonical Wnt–planar cell polarity (PCP) pathway involving Dishevelled (DVL). These receptors modulate the activity of Rho GTPases2 through Rho guanine nucleotide exchange factors (GEFs). Effectors of Rho GTPases, including Rho-associated kinases (ROCKs), formins (such as Diaphanous-related formins (DRFs)), Wiskott–Aldrich syndrome protein (WASP), WASP-family verprolin homologues (WAVEs) and the actin-related protein 2/3 (ARP2/3) complex and other actin-binding proteins (ABPs), orchestrate actin polymerization by incorporating globular actin (G-actin) into the filamentous actin (F-actin) polymer3. High levels of cytoplasmic G-actin retain serum response factor (SRF) cofactor proteins, myocardin-related transcription factors (MRTFs), in the cytoplasm. Incorporation of G-actin into the F-actin filament liberates MRTFs to enter the nucleus and interact with the transcription factor SRF58. This triggers expression of a subset of SRF target genes, namely cytoskeletal genes. b | Activation of SRF class II target genes. Nuclear MRTF can be complexed by nuclear G-actin, which inhibits MRTF-mediated stimulation of SRF-dependent transcription and facilitates MRTF nuclear export76. SRF class II target genes that are transcribed as a result of MRTF–SRF activation include actin itself and many genes that modulate actin dynamics, such as gelsolin and vinculin. These newly made proteins, with increasing time and concentration, might stimulate cytoplasmic actin polymerization, complex cytoplasmic MRTF or elevate levels of nuclear G-actin to downregulate MRTF-mediated transcription and stimulate nuclear export of MRTF. FAK, focal adhesion kinase; ILK, integrin-linked protein kinase; LIMK, LIM domain kinase.
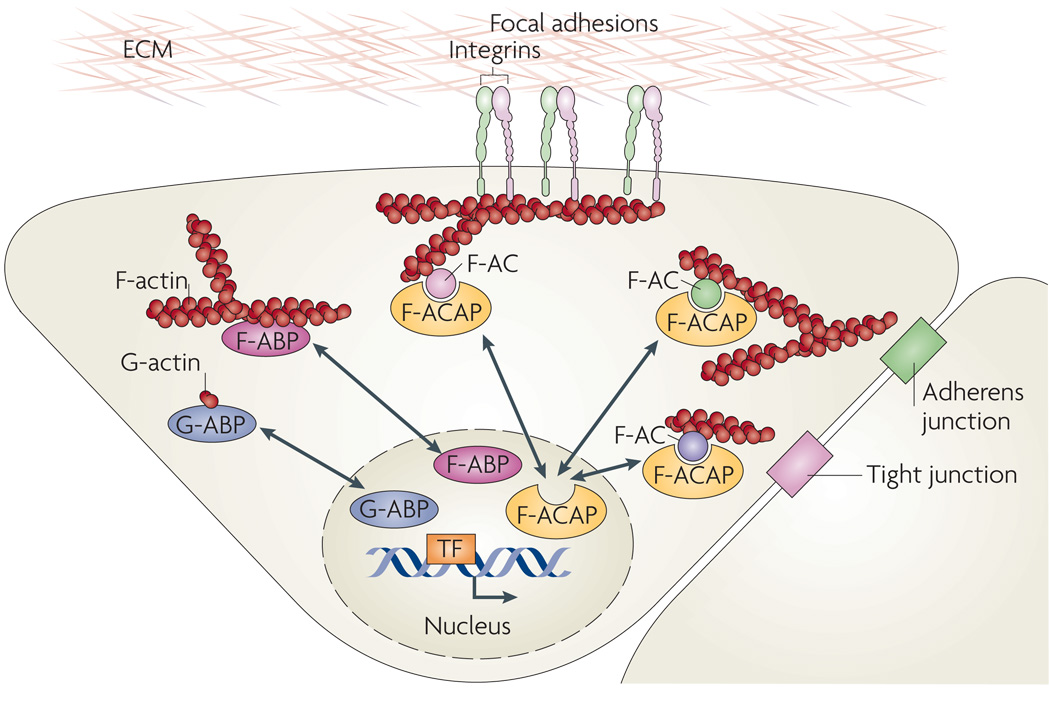
Figure 2. Actin-binding proteins as microfilament messengers.
A model summarizing the nucleus–cytoplasm shuttling of three different types of actin-binding proteins (ABPs): globular actin (G-actin)-binding proteins (G-ABPs), filamentous actin (F-actin) binding proteins (F-ABPs) and F-actin complex-associated proteins (F-ACAPs). Examples of G-ABPs include myocardin-related transcription factors (MRTFs), striated muscle activator of Rho-dependent signalling (STARS, also known as ABRA), junction-mediating and regulatory protein (JMY), β-thymosin, profilin, neural Wiskott–Aldrich syndrome protein (N-WASP), the actin-related protein 2/3 (ARP2/3) complex and spire31. Examples of F-ABPs include the actin-binding LIM proteins (ABLIMs) cofilin, gelsolin, filamin, α-actinin, supervillin and LIM and SH3 domain protein 1 (LASP1)31,32, and examples of F-ACAPs include ABL1, integrin cytoplasmic domain-associated protein 1 (ICAP1α), LIM domain proteins and p120-catenin44. The shuttling LIM domain proteins zyxin, lipoma-preferred partner (LPP), Cys-rich proteins (CRPs), Hic-5 (also known as TGFB1I1), antileukoproteinase (ALP), paxilin and LIM and SH3 domain protein 1 (LASP1) were shown to directly bind F-actin, whereas LIM kinase, particularly interesting new Cys-His protein 1 (PINCH; also known as LIMS1) and four and a half LIM domains protein 2 (FHL2) are probably indirectly linked to F-ACs44,45. F-ACs assemble at the cytoplasmic sides of focal adhesions, cadherin-mediated cell–cell adherens junctions and cell–cell tight junctions. TF, transcription factor.
General influences of nuclear actin and nuclear ABPs on overall gene expression are not discussed in detail. Instead, this review emphasizes recent insights into the specific function of G-actin to control both nucleus–cytoplasm shuttling and the nuclear activity of transcriptional cofactors of the myocardin protein family (myocardin itself, plus the myocardin-related transcription factors (MRTFs)) (FIG. 3). These proteins, in turn, control the activity of serum response factor (SRF), a nuclear transcription factor. Thereby, the cytoplasmic concentration of G-actin is reflected by the concentration of MRTF retained in the cytoplasm and is a direct measure of actin dynamics. The release and nuclear translocation of cytoplasmic MRTF on actin polymerization elicits SRF-directed target gene activation. Thereby, the actin–MRTF–SRF circuit allows for the precise modulation of gene expression in concert with cytoskeletal assembly and disassembly. Since SRF target genes encode structural components of the microfilament (for example, actin itself) and regulators of actin dynamics (for example, gelsolin and vinculin), as well as microRnAs (miRnAs) that provide feedback regu lation in these pathways, the MRTF–SRF circuit assumes central importance in directing essential gene activities required for the execution of motile cell functions.
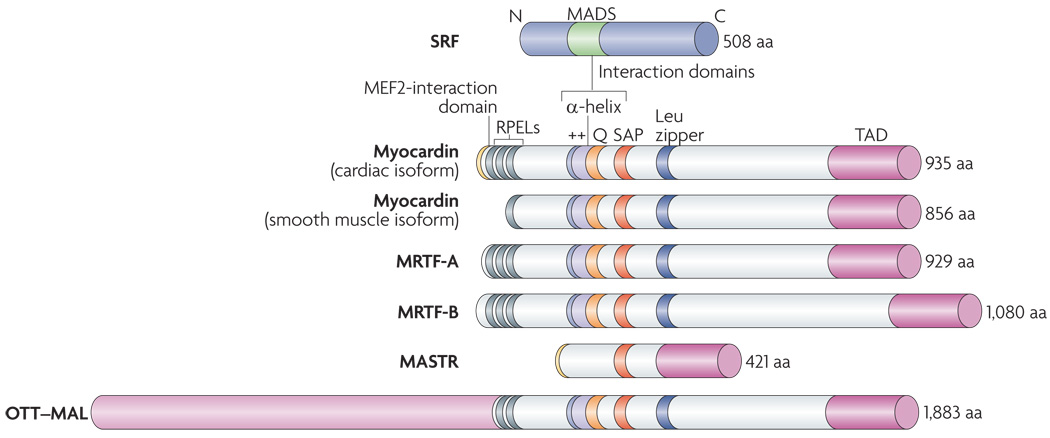
Figure 3. Structure of myocardin family members.
Functional domains of homology among the myocardin family proteins are shown and the numbers of amino acids are indicated. Myocardin-related transcription factors (MRTFs) are potent transcriptional coactivators that associate with serum response factor (SRF) through a basic region (++) and an adjacent Glu-rich domain (Q). Between these domains is a short α-helical region with similar secondary structure to a domain in the ternary complex factor protein ELK1, known as a B box, which mediates their interaction with SRF52,68,71. Myocard in family proteins contain Arg-Pro-X-X-X-Glu-Leu (RPEL) motifs, which mediate their interact ion with globular actin (G-actin). Members of the myocardin family have a homologous SAP domain, named after SAFA or SAFB, acinus and PIAS, which participates in different kinds of chromosomal DNA metabolism. Deletion of this region disrupts the ability of myocardin to activate a subset of SRF-dependent genes68. Myocardin and MRTFs contain powerful transcriptional activation domains (TADs) required for the stimulation of SRF activity. A dimerization motif resembling a Leu zipper mediates homo- and heterodimerization of myocardin and MRTFs. Alternative usage of 5′ exons in the myocardin gene gives rise to proteins with different amino termini. A cardiac-specific splice variant of myocard in contains a unique amino-terminal sequence that confers the ability to interact with the myocyte-specific enhancer factor 2 (MEF2) transcription factor, a MADS-boxtranscription factor related to SRF. This MEF2-interaction domain is also contained in a divergent member of the myocard in family called MEF2-activating SAP transcriptional regulator (MASTR). MASTR lacks the SRF-interaction domain.The OTT (also known as RBM1B)-MAL (also known as MRTF-A) fusion protein of AMKL leukaemia cells contains MRTF-A sequences.
Signal regulation of actin dynamics
The diverse cellular motile functions (BOX 1) are elicited by a host of extracellular stimuli. These stimuli are perceived by cognate receptor proteins that activate different members of the Rho GTPase family through selective Rho guanine nucleotide exchange factors (GeFs) (FIG. 1). The Rho GTPases encompass members of the Rho, Rac and Cdc42 subfamilies2, which regulate effector proteins that modulate the polymerization equilibrium of G-actin and F-actin in the cytoplasm. G-actin forms complexes with different ABPs, including the nucleating factors profilin, formins and the actin-related protein 2/3 (ARP2/3) complex 1,3. Activation of Rho GTPases promotes actin polymerization by two downstream signalling modules, one involving the Rho-associated kinase (RoCK)–LiM kinase–cofilin pathway (cofilin is an ABP that can stimulate actin depolymerization and thereby enhance actin poly merization elsewhere), and the other mediated by formin (such as Diaphanous-related formin (DRF)) ABPs. Experimentally, actin dynamics can be effectively modulated using clostridial toxins and other actin-targeting natural compounds4 (BOX 2).
Box 2 | In vitro modulators of actin dynamics and MRTF–SRF activity.
Elucidation of the myocardin-related transcription factor (MRTF)-mediated communication between actin dynamics and transcriptional activation has been helped greatly by the use of compounds that modulate actin activity51,52,75,76. The effect of Rho GTPases on actin polymerization can be inhibited by clostridial toxins such as the RHOA inhibitor C3 transferase (ADP-ribosytransferase) or by the Rho-associated kinase (ROCK)-inhibiting substance Y-27,623 (see the figure, part a). Actin-targeting natural compounds impair actin polymerization (latrunculin B, cytochalasin D and swinholide A) or stabilize the filamentous actin (F-actin) polymer (jasplakinolide)4 (see the figure, part b). Interestingly, these compounds also affect the cytoplasmic MRTF–actin complex (see the figure, part c). Cytochalasin D, swinholide A and jasplakinolide disrupt the MRTF–globular actin (G-actin) complex, thereby liberating MRTF, whereas latrunculin B inhibits the dissociation of MRTF from G-actin52,75,76. Nuclear G-actin also complexes with MRTF, thereby inhibiting the activation potential of serum response factor (SRF) and stimulating nuclear export of MRTF (see the figure, part d). Nuclear MRTF–G-actin complexes76 are affected by actin-targeting compounds in a smilar manner to cytoplasmic complexes. Importantly, these studies reveal that many compounds that were previously thought to primarily modulate actin polymerization equilibrium, also influence the stabilities of actin–actin binding protein (ABP) complexes.
Mutant variants of actin have also been useful in studying the MRTF-dependent regulation of SRF. Actin variants Glu13Arg and Arg62Asp are impaired in polymerization and therefore inhibit MRTF activity, whereas Val159Asn and Ser14Cys stimulate F-actin formation and activate MRTF function50. Glu15Ser also stabilizes F-actin while also apparently interacting more tightly with MRTF and promoting its nuclear entry75. MRTF variants defective in interacting with actin display constitutive nuclear localization74. Structural information on MRTF–G-actin was provided recently by co-crystallization of MRTF-derived RPEL peptides and G-actin in the presence of latrunculin B77.
As outlined in FIG. 1, several types of plasma membrane receptors modulate actin polymerization and cell ular motility by modulating Rho GTPase activity. in the examples discussed below, the link between these receptors and the MRTF–SRF module is emphasized.
Signalling by G protein-coupled receptors
G protein-coupled receptors (GPCRs) are a large family of membrane receptors displaying seven transmembrane helical domains that can couple to heterotrimeric G proteins (guanine nucleotide-binding proteins; consisting of α-, β-, and γ-subunits). Heterotrimeric G proteins with Gα subunits of the types Gα12/13, Gαq/11 and Gαi/0 contribute to actin turnover by engaging Rho GTPases5 (FIG. 1). GPCRs interact with many ligands, including hormones, chemokines, bioactive lipids, gastrointestinal peptides, platelet activators and orphan ligands of the developing brain6. Activation of the Rho–actin–MRTF–SRF pathway has been clearly demonstrated for the GPCR ligands lysophosphatidic acid (LPA)7, sphinghosine-1-phosphate (S1P)7,8 and a neuronal orphan ligand6.
Receptor Tyr kinase signalling
Numerous Tyr kinase receptors link to Rho GTPases, including those for platelet-derived growth factor (PDGF), insulin, ephrin A (EphA) and ephB proteins, TRKA, TRKB and TRKC, epidermal growth factor (EGF), fibroblast growth factor (FGF), vascular endothelial growth factor (VEGF) and MET9. Many of these receptors have essential influences on motile cell functions. Activation of the actin–MRTF–SRF pathway has been shown by VEGF receptor signalling in endothelial cells10, EphA signalling in neurons11,12 and neurotrophin signalling in dorsal root ganglia (probably through the TRKA, TRKB and TRKC receptors)13. The central importance of SRF signalling in these types of signalling events is exemplified by the abnormalities in vasculogenesis14,15, neuronal axon guidance and synaptic targeting11,13,16 in Srf knock-out mice.
Integrin signalling
Integrin receptors link cells to the extracellular matrix (ECM) and are the main structural organizing components of focal adhesions. Integrin signalling has a big impact on the reorganization of the actin cytoskeleton. Integrin-mediated actin dynamics is pri marily achieved by activating the RHOA and RAC1 GTPases, which involves the regulation of Rho GEFs by the kinases integrin-linked kinase (ILK), focal adhesion kinase (FAK) and SRC17 (FIG. 1). Integrins also communicate external and internal mechanical stress to the actin cyto skeleton. Such mechano-sensing was shown to activate the actin–MRTF–SRF pathway18.
TGFβ signalling
The transforming growth factor-β (TGFβ) superfamily of cytokines includes TGFβ-family polypeptides, bone morphogenetic proteins (BMPs) and activins. These ligands act through heterotetrameric complexes of Ser/Thr kinase receptors, which activate different signalling pathways and, in addition, modulate Rho GTPase-mediated actin dynamics19. TGFβ-induced alterations in cellular architecture and motility contribute to cellular epithelial–mesenchymal transitions (EMT), which are biologically linked to embryonic tissue movements and to tumour cell invasiveness and metastasis. Activation of the MRTF–SRF module was observed on TGFβ20–22 and BMP signalling23.
E-cadherin signalling
Epithelial cell–cell junctions (adherens junctions) are mediated by homophilic inter actions of epithelial cadherins (E-cadherins). Cadherins are functionally linked to the actomyosin cytoskeletal scaffold through catenins1. Assembly and disassembly of cell–cell contacts both requires and elicits extensive actin dynamics and activates the MRTF–SRF pathway through the GTPase RAC1 (REFS 20, 24, 25). The dis assembly of epithelial cell contacts is an initiating event in EMT. The release of catenins (α-catenin, β-catenin and p120-catenin) activates actin dynamics and can directly modulate nuclear protein function26.
Non-canonical Wnt signalling
The non-canonical, β-catenin-independent Wnt–planar cell polarity (PCP) signalling pathway uses the Wnt signalling component Dishevelled to activate regulators of actin dynamics, including the Rho, Rac and Cdc42 GTPases, ROCK27 and the formin-related protein Dishevelled-associated activator of morphogenesis 1 (DAAM1)28. This pathway was originally identified in Drosophila melanogaster in the establishment of spatial tissue organization, specifically the planar cell polarity of pupal fly wing cells29. In vertebrates, Wnt signalling modulates actin dynamics for polarized cell movements during gastrulation and organogenesis30. The link of Wnt–PCP signalling to the MRTF–SRF relay is still hypothetical.
Microfilament-to-genome relay systems
In this section, we review the range of currently characterized mechanisms by which cells communicate the dynamic status of their actin microfilaments to the genome.
Cytoplasmic actin dynamics cause changes in both G-actin concentration and F-actin structure; the latter can, in turn, lead to an altered structure or composition of F-actin complexes (F-ACs), such as focal adhesion complexes. Actin is recognized by ABPs, which, for clarity, we suggest are classified here as G-actin binding proteins (G-ABPs), F-actin binding proteins (F-ABPs) and F-actin complex-associated proteins (F-ACAPs) (FIG. 2).
Conceptually, changes in actin dynamics may be communicated to the genome in three ways. First, changes in cytoplasmic monomeric G-actin concentrations can elicit corresponding changes in nuclear G-actin levels. Second, changes in cytoplasmic monomeric G-actin levels can be sensed by G-ABPs, whereby polymerization of G-actin into F-actin can result in the release of G-ABPs, their nuclear translocation and the subsequent modulation of nuclear transcription factors (FIG. 2). This mechanism is best characterized by the MRTF–SRF circuit (FIG. 1; FIG. 3). Last, the status of cytoplasmic actin dynamics can be scored directly by cellular sensors of F-actin filament structure and density. Thus, architectural rearrangements of cytoplasmic F-actin fibres can lead to the release of F-ABPs or F-ACAPs, followed by their nuclear translocation (FIG. 2). In the following section, we provide a brief overview of different relay systems linking actin dynamics to transcriptional genome activity, according to the mechanisms conceptualized above.
Nuclear actin and ABPs
Nuclear actin, in both monomeric G-actin and polymeric F-actin forms, profoundly affects transcriptional responses in general terms31,32. it influences gene expression by at least three mechanisms: actin is a component of all three types of RNA polymerases (classes I, II, and III), it contributes to ATP-dependent chromatin remodelling and it binds different ribonucleoprotein (RNP) complexes in eukaryotic cell nuclei, including small nuclear RNAs (snRNAs) and heterogeneous nuclear RNAs (hnRNAs). In these activities, actin functions jointly with both G-ABPs and F-ABPs. G-actin can enter and exit nuclei by diffusion; however, use of its nuclear export sequence, partly in conjunction with members of the exportin protein family or with profilin, regulates its export. This controls the nuclear exit of other G-ABPs, such as MRTFs, which are exported bound to G-actin (see below).
JNK signalling
Signals that elicit dynamic actin rearrangements on activation of Rho GTPases, like the above-mentioned non-canonical Wnt–PCP pathway, were shown to also result in the activation of the Jun N-terminal kinases (JNKs)33–35, which regulate gene transcription. In different experimental settings, GTPase-mediated JNK activation involved mediation by the kinases mixed lineage kinase 3 (MLK3; also known as MAP3K11)36 and ROCKs37, or the adaptor protein CRK38. It remains to be shown unequivocally that such mechanisms of JNK activation strictly correlate with the stimulation of actin dynamics.
NF-κB signalling
Cytoplasm-to-nucleus translocation of the transcription factor nuclear factor-κB (NF-κB) can be elicited by regulators of actin dynamics. Expression in endothelial cells of intercellular adhesion molecule 1 (ICAM1), on stimulation by thrombin, involves RHOA–ROCK-mediated activation of nuclear NF-κB39. Also, disruption of epithelial cell–cell contacts and associated actin reorganization leads to RHOA–ROCK-mediated activation of protein kinase D (PKD1), resulting in the activation of NF-κB40. This scenario may reflect loss of cell–cell contacts during pathological stimulation of endothelial cell motility.
Nuclear hormone receptors
Nuclear hormone receptors, specifically the androgen receptor (AR), are modulated in their transcriptional activity by actin and ABPs, in addition to being under the control of their cognate hormone ligand. Regulation of AR by F-ABPs and G- ABPs can be exerted at the level of cytoplasm-to-nuclear translocation, or by influencing the transcriptional activity of a promoter-bound receptor. ABPs shown to interact with the AR include gelsolin, α-actinin 2, supervilllin, filamin and transgelin32. G-actin is part of the AR transcription complex.
The DNA damage response, Nck and JMY
Genotoxic insults affect actin dynamics and elicit transcriptional responses41,42. DNA damaging treatment with ultraviolet (UV) light causes two cytoplasmic ARP2/3 effector proteins, junction-mediating and regulatory protein (JMY) and the NCK1–suppressor of cytokine signalling 7 (SOCS7) complex, to translocate to the nucleus. JMY is a G-ABP that can stimulate cell motility. On nuclear translocation, JMY acts as a cofactor of the p53–p300 complex to stimulate the transcription of pro-apoptotic genes41. Nuclear translocation of NCK1–SOCS7 follows UV-induced separation from septin proteins43, which act as F-ABPs. Activation of the DNA damage check-point response requires an as yet undefined nuclear function of NCK1. This highlights new facets of actin microfilamant-to-genome communication that specifically contribute to the cellular genotoxic stress response.
LIM domain-containing proteins
Integrin-mediated engagement of cell–ECM contacts at focal adhesions causes the nuclear translocation of cytoskeletal F-ACAPs44,45. The best studied examples are LIM domain-containing proteins and include zyxin, lipoma-preferred partner (LPP), thyroid receptor-interacting protein 6 (TRIP6), paxilin, Hic-5 (also known as TGFB1I1), Cys-rich protein 1 (CRP1), CRP2, antileukoproteinase (ALP), four and a half LIM domains protein 2 (FHL2), LIM and SH3 domain protein 1 (LASP1) and particularly interesting new Cys–His protein 1 (PINCH; also known as LIMS1). These proteins engage in the formation of protein–protein assemblies at focal adhesion-associated actin fibres and, alternatively, can act as nuclear cofactors of transcription or as regulators of nuclear mRNA export. Interestingly, some of these LIM proteins are either cofactors of the transcription factor SRF (for example FHL and CRP proteins)46,47 or are encoded by SRF-regulated genes (for example zyxin and FHL2)47,48. The precise mechanisms of liberation from the focal adhesion are not clear for each of these F-ACBPs, nor are the affected target genes. It seems that LiM protein release from focal adhesions is a mechanism by which changes in the structure and/or density of the F-actin filament are relayed to the nucleus by certain F-ACAP courier proteins for the purpose of modulating gene expression.
The MRTF–SRF circuit
The appreciation that the transcription factor SRF is subject to regulation by Rho GTPases49 and actin dynamics50,51 provided the first clue to a hitherto undetected link between the actin cyto-architecture and gene activity. Subsequent identification of G-actin-regulated nucleus–cytoplasm shuttling of the SRF cofactors, MRTFs, revealed for the first time a direct mechanism enabling cytoplasmic G-actin to control nuclear transcriptional activity52. MRTFs are G-ABPs, and this regulatory system seems to be of prime importance for the fuelling of actin-based cellular motile function. Thus, the remainder of this Review concentrates on the MRTF–SRF circuit.
Molecular and biological functions of SRF
SRF is a versatile transcription factor, encoded by a single gene that is abundantly expressed in many cell types53,54. SRF homologues are found in all species investigated, ranging from yeast to humans. Homodimeric SRF binds with high affinity and specificity to the palindromic CC(A/T)6GG DnA sequence, called the CArG-box55,56, and directs the transcription of numerous target genes on stimulation by various signalling cascades47,57–61. SRF associates with target gene promoters to build a regulatory platform for the recruitment of various cofactors58,62. Differential cofactor recruitment is best exemplified by the ternary complex factor (TCF) subclass of Ets-type cofactors63–65 and the myocardin family of co-activators (myocardin and MRTFs)66–69. These two types of SRF cofactors respond to distinct signalling inputs (including mitogen-activated protein kinase (MAPK) signalling and actin signalling) and, accordingly, enable SRF to direct the expression of different sets of target genes70. MRTFs and TCFs display mutually exclusive interactions with SRF71,72.
SRF target genes fulfil essential functions (see below), explaining, in part, the dramatic phenotypes seen on Srf deletion in mice73. A broad range of biological processes is supported by SRF, including gastrulation, development and function of the heart and the cardiovascular system, functioning of all three types of muscle cells, endothelial cell biology and vascularization, development and regeneration of the liver, T cell and B cell activities of the immune system and neuronal functions of the developing and adult brain. At the cellular level, many of these phenotypes can be attributed at least in part to impaired functioning of the actin microfilament48. Given the importance of SRF in regulating actin cytoskeletal dynamics, there are nevertheless functions of SRF that go beyond the transcriptional control of cytoskeletal target genes, including contributions to cell survival and apoptosis.
Molecular and biological functions of MRTF proteins
Myocardin, the founding member of the MRTF family (FIG. 3), is expressed specifically in the cardiovascular system66–68, whereas other MRTF family members display more widespread expression patterns. Myocardin shares homology with MRTF-A (also known as MAL, MKL1 and BSAC) and MRTF-B (also known as MKL2 and MAL16) in a series of conserved domains; to harmonize the nomenclature we suggest using the MRTF designation, which is based on homology criteria of the gene family69. These potent transcriptional co-activators associate with SRF through a basic region and an adjacent Glu-rich domain (FIG. 3).
The amino-termini of MRTFs contain three RPEL domains, which form a stable complex with monomeric G-actin52,74–77, resulting in the sequestration of MRTFs in the cytoplasm. Myocardin contains divergent RPeL domains that do not bind actin efficiently; consequently, homodimeric myocardin is constitutively nuclear and insensitive to cellular stimuli that modulate actin polymerization68,78. However, heterodimerization between myocardin and other MRTFs has been proposed to facilitate cooperativity between CArG boxes in SRF-regulated muscle genes79. Thereby, myocardin might be subjected indirectly to the regulatory mechanisms operating on MRTFs, specifically G-actin binding.
Knockout studies of myocardin (Myocd) and MRTF genes in mice have shown that these SRF co-activators are uniquely required in vivo in different cell types and at different developmental stages. Nearly all of the Myocd mutant phenotypes reflect abnormalities in aspects of motile cell functions and regulation of SRF-dependent contractile and cytoskeletal genes80–82. Mice lacking MRTF-A are viable, but females are unable to nurse their offspring owing to impairments in mammary myoepithelial cells83,84. The global deletion of MRTF-B results in embryonic lethality owing to cardiovascular defects, including abnormal patterning of the branchial arch arteries, a double-outlet right ventricle, ventricular septal defects and a thin-walled myocardium85.
Actin-mediated regulation of MRTF nuclear shuttling
At low actin polymerization states, MRTFs are held in an inactive state in the cytoplasm by reversible complex formation with G-actin50,52,75. Thus, MRTFs are bona fide G-ABPs. Stimulation of Rho GTPases feeds G-actin into the F-actin filament, thereby liberating MRTFs from G-actin and allowing the nuclear import of MRTF and subsequent activation of SRF-dependent transcription (FIG. 1).
Nuclear G-actin also modulates MRTF functions in multiple ways76. First, nuclear export of MRTFs is facilitated by nuclear G-actin. Second, nuclear G-actin prevents nuclear MRTF from activating SRF target genes, so that liberation of the nuclear actin–MRTF complex is required to stimulate SRF. Thus, cellular G-actin regulates MRTFs at three levels: nuclear import, nuclear export and nuclear activation or inactivation of MRTF-SRF-dependent transcription76 (FIG. 1b). It has also been suggested that MAPK phosphorylation of MRTF stimulates its nuclear export by enhancing the inter action of MRTF with nuclear actin86. Therefore, in light of the actin genes themselves being under MRTF-SRF control, a G-actin-dependent, kinetically tuned auto-regulatory feedback loop emerges for Rho-directed control of transcription mediated by MRTF–SRF. Rho activates the initial synthesis of G-actin and ABPs to stimulate F-actin polymerization in the cytoplasm as stimulus-elicited cell biological requirements dictate. With ongoing actin synthesis, G-actin levels rise in both the cytoplasm and the nucleus, thereby downregulating the SRF response by simultaneously impairing the nuclear function and facilitating the export of nuclear MRTF, and by en suring efficient retention of cytoplasmic MRTF (FIG. 1). Accordingly, nucleus–cytoplasm shuttling of MRTF is an essential component of this actin-directed feedback circuit.
The above regulatory mechanisms have been described most thoroughly in muscle cells and cultured fibroblasts. By contrast, in primary neurons a constitutively high nuclear level of MRTF is seen in the absence of stimuli. It will be interesting to identify modulators influencing nuclear MRTF activity in neurons and to explore the potential contributions of nuclear actin12,76 and nuclear MRTF phosphorylation86,87 to the neuronal functions of MRTFs.
Positive and negative regulators of MRTF–SRF activity
Numerous cofactors in addition to TCFs and MRTFs exert positive and negative effects on SRF activity. In the cytoplasm of muscle cells, striated muscle activator of Rho-dependent signalling (STARS; also known as ABRA) is a G-ABP that is upregulated during cardiomyopathy88. STARS is localized to the sarcomere and, by interacting with monomeric G-actin, promotes nuclear translocation of MRTF-A. STARS thereby syner gizes with MRTF-A to stimulate SRF-mediated transcription. Positive nuclear cofactors of SRF include the Nkx2–5 family of homeodomain proteins89, members of the GATA family of zinc-finger proteins90,91 and the CRP family of Cys-rich LIM-only proteins46. Negative co factors include the LIM-only protein FHL2 (REFS 47,92), histone deacetylase 4 (HDAC4)93, the homeo domain protein MSX1 (REF. 94) and the zinc-finger protein krueppel-like factor 4 (KLF4)95. Homeodomain-only protein (HOP) dampens SRF activity in cardiac muscle cells by competing with it for myocardin and MRTF-A interaction, and also by recruiting HDAC2 to SRF target genes96–98. Members of the myocardin family also engage various partners in addition to or in conjunction with SRF. These include the histone acetyltransferase p300 (REFS 99,100), class II HDACs99, SMAD4 (REF. 101) (a transcription factor regulated by TGFβ signalling), forkhead box protein O4 (FOXO4)95 and GATA transcription factors102. Such combinatorial associations of SRF with positive and negative partners expand the regulatory potential of SRF by providing cell type specificity, temporal control and signal responsiveness to SRF target genes.
MRTF–SRF target gene functions
SRF target genes, containing functional CArG boxes for SRF binding, have been identified by candidate gene characterization, genome-wide expression profiling of SRF-deficient cells and in silico genome queries47,48,59,60 (for review see REF. 57). Based on these studies, we estimate the number of SRF target genes to be around 300. Chromatin immunoprecipitation (ChIP) with anti-SRF antisera, which provides stringent support for direct gene regulation by SRF, has identified more than 200 SRF target genes103. Selective cofactor recruitment (TCF versus myocardin and MRTFs) directs signal-specifi c regulation of different classes of SRF target genes. Class I target genes, which primarily encode proteins with ‘immediate–early’ cellular functions, including the rapid transcriptional activation of genes encoding proteins involved in the G0–G1 transition, are aided by TCF cofactors62,63. Class ii target genes, which are regulated by SRF in conjunction with myocardin or other MRTF cofactors57, encode at least three types of proteins, namely those involved in muscle-specific and contractile functions, actin microfilament dynamics and cell motility, and miRNA activities. In the following, we restrict our discussion to class II SRF target genes to emphasize their regulation by the myocardin-family transcriptional co-activators.
Muscle-specific and contractile genes
Nearly all smooth muscle-specific genes and many cardiac and skeletal muscle genes are controlled by CArG boxes. Myocardin is a particularly potent activator of SRF-dependent smooth muscle gene expression79,84,104–106. Forced overexpression of MRTFs also activates smooth muscle genes in transfected cells, which raises the question of why the endogenous levels of MRTF in many non-muscle cells are not sufficient to drive the expression of muscle genes59,69,107,108. It is likely that endogenous MRTF levels are regulated by actin signalling, whereas overexpressed MRTFs bypass this regulatory mechanism to directly activate downstream muscle genes. MRTFs are also required for skeletal muscle differentiation, as shown by the inhibition of skeletal muscle gene expression on expression of dominant-negative MRTF mutants in vivo and in vitro59,108. Many of the contractile functions of muscle cells are enabled by myocardin–SRF- and MRTF–SRF-regulated genes57.
Genes affecting actin dynamics and cell motility
SRF target genes, which are regulated on recruitment of MRTF cofactors, encode proteins related to actin cytoskeletal activities. SRF-regulated gene products can be grouped into three categories according to their contributions to actin microfilament function: structural (for example, actin), effectors of actin turnover (for example, cofilin 1) and regulators of actin dynamics (for example, talin 1). TABLE 1 lists some well-characterized examples of cytoskeletal SRF target genes. In biological terms, the encoded proteins contribute to development (mesodermal patterning), functioning of the contractile apparatus (muscle cell contraction, vascular tone) and ECM-mediated adhesion and tissue remodelling.
Table 1.
SRF–MRTF-regulated genes encode actin microfilament effectors
| Gene symbol* | Gene name | Function |
|---|---|---|
| Structural components of microfilament function | ||
| ACTA1 | Actin, alpha 1 | Microfilament building |
| ACTA2 | Actin, alpha 2 | Microfilament building |
| ACTB | Actin, beta | Microfilament building |
| ACTG1 | Actin, gamma 1 | Microfilament building |
| ACTG2 | Actin, gamma 2 | Microfilament building |
| CALD1 | Caldesmon 1 | Muscle cell contraction |
| DMD | Dystrophin | Membrane anchorage |
| ITGB1 | Integrin, beta 1 | Focal adhesion component |
| MYH11 | Myosin heavy chain 11 | Actomyosin structure |
| MYH9 | Myosin heavy chain 9 | Actomyosin structure |
| MYL9 | Myosin light chain 9 | Actomyosin structure |
| TAGLN (SM22A) | Transgelin | Muscle cell contraction |
| SMTN | Smoothelin | Muscle cell contraction |
| TPM1 | Tropomyosin 1 | Muscle cell contraction |
| VCL | Vinculin | Linking integrin to microfilaments |
| Effectors of actin turnover | ||
| ACTR3 (ARP3) | Actin-related protein 3 | Microfilament turnover and branching |
| CFL1 | Cofilin 1 | Microfilament turnover |
| GSN | Gelsolin | Microfilament turnover |
| VIL1 | Villin 1 | Microfilament reorganization |
| SVIL | Supervillin | Microfilament reorganization |
| Regulators of actin dynamics | ||
| CTGF | Connective tissue growth factor |
An ECM protein that is an integrin ligand and modulates cellular migratory activity |
| CYR61 (CCN1) | Cys-rich protein 61 | An ECM protein that is an integrin ligand and modulates cellular migratory activity |
| FHL1 and FHL2 | Four-and-half LIM domains 1 and 2 |
Adherens junction signalling and transcriptional regulation (through SRF binding) |
| FLNA | Filamin A | Integrin signalling |
| MMP9 | Matrix metalloprotease 9 | ECM remodelling and cell migration |
| MYLK | Myosin light chain kinase | Actomyosin regulation |
| SRF | Serum response factor | Transcriptional regulation |
| TLN1 | Talin 1 | Integrin signalling |
ECM, extracellular matrix.
All the genes listed have promoters with functional serum response factor (SRF)-binding sites, as confirmed by chromatin immunoprecipitation (ChIP). Myocardin-related transcription factor (MRTF) ChIP is currently available only for a subset of these genes, see also REF. 57. Alternative gene symbols are provided in brackets.
Expression profiling using modulators of MRTF activity (dominant-negative MRTF and treatment with cytocalasin D in the absence or presence of latrunculin B; see BOX 2) identified a subset of ∼ 30 MRTF-dependent SRF target genes7,59,109. We expect this number to grow with ongoing investigations110. ChIP-based evidence for the association of MRTF with SRF at CArG boxes is available but is limited103. Despite this limitation, identified MRTF-dependent SRF target genes contribute to cell–ECM adhesion and actomyosin activity. Many structural components of focal adhesions are encoded by SRF–MRTF-regulated genes, including α1, α5, α9 and β1 integrins, as well as talin 1, vinculin, and the syndecan proteins 2 and 4 (REF. 57), such that SRF deficiency leads to impaired formation of focal adhesions48.
miRNAs
Recent studies have revealed central roles for miRNAs in the cellular circuitry through which SRF and MRTFs control muscle gene expression, cytoskeletal dynamics and stress responsiveness111. SRF directly activates the expression of two bicistronic miRNA genes that encode pairs of homologous miRNAs (miR-1-1 and miR133a-2, and miR-1–2 and miR133a-1)112 (FIG. 4a). These miRNAs, in turn, regulate a large collection of mRNAs, many of which encode proteins that impinge on the SRF–MRTF signalling pathway. miR-133, for example, represses the expression of SRF, thereby establishing a negative feedback loop to precisely titrate SRF expression in muscle cells113. A particularly intriguing finding is that miR-1 can substitute for SRF in the induction of mesoderm from mouse embryonic stem cells114.
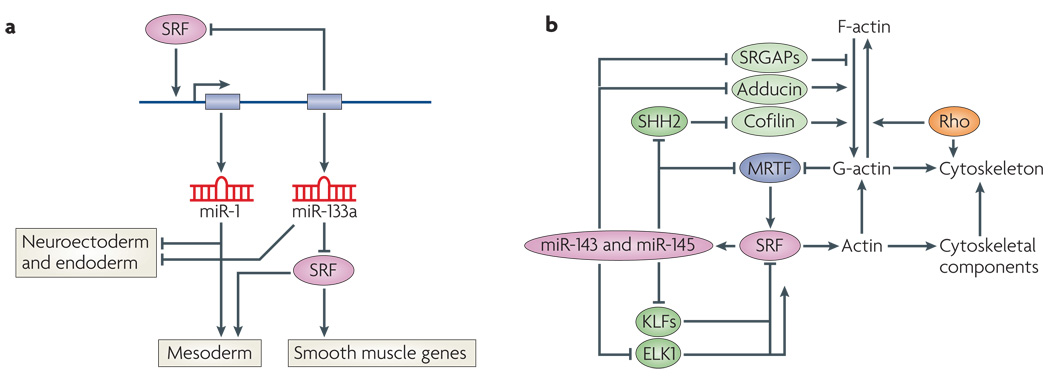
Figure 4. SRF-mediated regulation of miRNAs.
a | Regulation of microRNA-1 (miR-1) and miR-133 by sereum response factor (SRF). SRF activates transcription of a bicistronic miRNA cluster encoding miR-1 and miR-133 (REFS 112,113). miR-133a inhibits SRF expression, establishing a precisely titrated feedback loop to modulate SRF activity. There are two clusters of miR-1 and miR-133a genes in mammalian genomes, which are expressed specifically in cardiac and skeletal muscle cells. A third homologous pair of miRNAs, miR-206 and miR-133b, is expressed specifically in skeletal muscle independently of SRF. Genetic deletion of miR-1 results in phentoypes that suggest it has a role in mesoderm formation. Genetic deletion of the two miR-133a genes results in perinatal lethality owing to cardiac defects. miR-1 and miR-133a also repress neuroectoderm and endoderm genes and promote mesoderm gene expression. miR-1 can substitute for SRF to regulate downstream genes involved in mesoderm development by an undefined mechanism. b | Regulation of miR-143 and miR-145 by SRF. SRF activates transcription of a bicistronic miRNA cluster encoding miR-143 and miR-145, which are expressed specifically in cardiac and smooth muscle cells115,117. These miRNAs, regulate the expression of numerous mRNAs encoding regulators of actin signalling and myocardin-related transcription factor (MRTF)–SRF activity, thereby establishing an elaborate series of feedback loops to modulate the actin–MRTF–SRF signalling pathway. Targets of miR-143 or miR-145 include the zinc-finger proteins krueppel-like factor 4 (KLF4) and KLF5 (which inhibit SRF), MRTF-B, slingshot 2 phosphatase (SHH2; which controls actin polymerization by cofilin phosphorylation), adducin 3 (which promotes actin polymerization) and Slit-Robo Rho GTPase-activating protein 1 (SRGAP1) and SRGAP2 (which inhibit Rho signalling)115. This pathway has been shown to be essential for vascular remodelling in response to injury. In the absence of miR-143 and miR-145, actin stress fibre formation is disrupted, rendering smooth muscle cells insensitive to mechanical stimuli that typically cause vascular stenosis.
A screen for miRNAs regulated by MRTF-A in cardiac myocytes identified several miRNAs with key roles in muscle differentiation, proliferation and phenotypic switching115. Many of the same miRNAs were down-regulated in SRF-null compared to wild-type mouse embryonic stem cells. Bioinformatics also showed a disproportionate number of predicted SRF-binding sites associated with miRNA genes116.
miR-143 and miR-145 are encoded by a bicistronic miRNA gene, controlled by a distal upstream CArG box (FIG. 4b). These two miRNAs are expressed in early cardiac progenitors and throughout the embryonic heart before becoming restricted to all vascular and visceral smooth muscle cells115,117. miR-145 knockdown in cultured cells was reported to block smooth muscle gene activation by myocardin, pointing to this miRNA as an essential cofactor of myocardin117,118. By contrast, knockout mice lacking miR-143, miR-145 or both are viable and express smooth muscle SRF target genes normally115. However, these mice are resistant to vessel remodelling in response to vascular injury. The insensitivity of these mutant mice to vascular injury seems to reflect abnormal regulation of a collection of proteins involved in the modulation of actin cytoskeletal dynamics and stress fibre formation, rendering the mutant mice insensitive to deformation of the vessel wall. For example, these miRNAs target adducin 3, which caps barbed ends of actin filaments and acts as a bridge between the membrane and actin cytoskeleton115. Slingshot 2, another target of miR-143 and miR-145, promotes cell motility and enhances F-actin reorganization by dephosphorylating and activating cofilin, an actin severing factor. miR-145 targets KLF4 and KLF5, which repress SRF activity, as well as Slit-Robo GTPase-activating proteins, which modulate cell migration by inactivating the small GTPase Cdc42 and inhibiting actin polymerization115. Finally, these miRNAs target MRTF-B, the upstream regulator of their expression, providing a negative feedback loop to precisely titrate miR-143 and miR-145 expression and actin dynamics115 (FIG. 4b).
Mice lacking miR-143 and miR-145 display a reduction in blood pressure owing to a decreased vascular tone. These findings point to a role for miR-145, and to a lesser extent miR-143, in the control of a feedback loop that modulates a cytoskeletal–transcriptional circuit regulated by SRF.
Another miRNA regulated by SRF together with MRTF is miR-486, a cardiac and skeletal muscle-enriched miRNA expressed from an alternative promoter in an intron of ankyrin 1 (REF. 119). miR-486 acts as a downstream effector for the SRF–MRTF pathway and promotes phosphoinositide 3-kinase (PI3K)–AKT signalling by inhibiting the expression of phosphatase and tensin homologue (PTEN) and FOXO1A, which act as negative regulators of PI3K signalling.
By regulating the expression of miRNAs that affect the signalling system which controls MRTF–SRF activity, the MRTF–SRF partnership generates a balanced regulatory network to govern cytoskeletal function and signal responsiveness. It is curious that SRF, more so than other transcription factors, is so tightly intertwined with miRNA regulatory circuits. Perhaps this reflects the importance of maintaining tight control over SRF and its downstream targets.
The actin–MRTF–SRF circuit in pathology
MRTF-mediated communication between the actin cytoskeleton and the genome has broad implications for many motile functions of cells. Cell type-specific mouse knock-out studies of Srf and MRTF genes suggest that the MRTF–SRF circuit contributes to human pathology. Available evidence supporting this notion is discussed below in the context of two major threats to human health: cardiovascular diseases and cancer.
Cardiovascular diseases
Given the plethora of genes encoding contractile proteins that are regulated by SRF and members of the myocardin family, it is not surprising that this regulatory pathway has a central role in force- and mechano-sensing of muscle cells, such that dysregulation of SRF-dependent gene expression contributes to numerous disease models of the cardiovascular system. During pathological remodel ling of the heart in the settings of hypertrophy and heart failure, SRF-dependent genes are dysregulated. STARS and myocardin, two positive modulators of SRF function, are upregulated during cardiomyopathy and hypertrophy, respectively88,120,121. Caspase 3 cleavage of SRF in a failing heart generates a dominant-negative form of SRF that may suppress expression of sarcomeric genes122. Thus, SRF is not only required for the appropriate expression of cardiac contractile proteins, but excessive SRF is pathogenic, indicating the requirement of MRTF–SRF activity for cardiac homeostasis. This may explain why miRnAs, which provide robustness and stability to gene regulatory programmes, are so intimately integrated into the MRTF–SRF signalling pathway.
Recently, SRF and myocardin were shown to be overexpressed in small cerebral arteries during Alzheimer’s disease, and to have a key role in the progression of the disease by enhancing arterial hypercontractility through the activation of SRF-dependent smooth muscle contractile genes, thereby diminishing cerebral blood flow123. in addition, SRF and myocardin control amyloid-β peptide clearance from cerebral vascular smooth muscle cells124.
SRF also functions as a central regulator of smooth muscle cell phenotypes in response to injury and contributes to pathological smooth muscle cell proliferation in the vessel wall. Signals that diminish myocardin and MRTF activity promote excessive proliferation of smooth muscle cells, as occurs during atherosclerosis and restenosis125. The forkhead transcription factor FOXO4 and the zinc-finger protein KLF4 are upregulated during de differentiation of vascular smooth muscle cells in vivo 95, resulting in suppression of SRF-dependent transcription. Thus, the MRTF–SRF circuit functions as a nodal sensor of mechanical stress and growth factor signalling to modulate phenotypic switching of smooth muscle cells.
Cancer
Associations of the actin–MRTF–SRF circuit with human cancer biology have been identified, suggesting an MRTF–SRF involvement in the neoplastic process. However, definitive evidence for causative links to clinically relevant carcinogenesis is still lacking.
Infant acute megakaryoblastic leukaemia (AMKL), which correlates with poor prognosis, is associated with the balanced chromosomal translocation t(1;22) (p13;q13), in which the MRTF-A gene (MKL1; also known as MAL) is fused to the genomic fusion partner OTT (also known as RBM15)126–128 (FIG. 3). OTT–MAL is a constitutively nuclear, deregulated activator of SRF, which is not subject to control by G-actin129–131. OTT–MAL activates the transcription factor Jκ recombination signal-binding protein (RBP-Jκ), which is normally controlled by canonical notch signalling and is overridden by OTT–MAL to cause abnormal megakaryopoiesis132.
Myocardin was argued to be a differentiation-inducing tumour suppressor protein that is downregulated in mesenchyme-derived sarcomas133. By contrast, leiomyosarcoma (LMS) smooth muscle tumours display overexpression of myocardin134. In human prostate cancer, androgen receptor activity was suggested to regulate SRF-dependent expression of FHL2 (REF. 135), which itself is a regulator of SRF activity47,92 and integrin signalling; the expression of either SRF or FHL2 correlates with poor prognosis.
The general association of MRTF–SRF activity with actin-regulated cellular motile functions links actin–MRTF activity to cancer metastasis. Accordingly, depletion of either MRTF or SRF caused reduced adhesion, spreading, motility, invasion and colonization of metastatic tumour cells in an experimental in vivo metastasis assay109. In support of MRTF–SRF activity contributing to tumour metastasis, suppressor of cancer cell invasion (SCAI) was found to repress MRTF activity. SCAI was postulated to downregulate MRTF–SRF-mediated expression of β1 integrin, thereby acting as a suppressor of tumour cell invasion136.
Conclusions and future directions
Although previously unappreciated, it now seems obvious that actin-driven cell motility requires genomic support. Several independent mechanisms can now be outlined that communicate the dynamic status of the actin cytoskeleton to the genome. Microfilament-to-genome communication is mediated by different courier proteins, representing G-ABPs, F-ABPs or F-ACAPs, the cytoplasmic release of which for nuclear translocation is dependent on changes in G-actin levels or F-actin structure and composition. Such courier proteins can function inside nuclei as transcriptional cofactors, as clearly shown for the CRP, FHL2, JMy and MRTF proteins. The control of SRF activity by multiple cofactor interactions and signalling inputs serves as a paradigm for understanding the logic of connecting genome activity with actin cytoskeletal dynamics. The intimate functional link between actin dynamics, MRTF–SRF-regulated gene expression and cellular motile behaviour has been confirmed by genetic means in several cell and organ systems, including embryonic stem cells48, the developing mouse embryo73,116,137, muscle tissues82,138 and the brain11,139,140. However, numerous molecular details of the regulatory interactions involved remain to be worked out in the MRTF–SRF circuit and in all the other above-mentioned systems.
We anticipate that new actin-dependent courier proteins will be identified. For each relay system it will be important to characterize comprehensively the associated differential gene expression profiles. The example of MRTF–SRF-directed expression of miRnA-encoding genes indicates that surprising mechanistic insight is to be derived from future studies in this area of cell biology.
Microbial pathogens, both bacteria (for example, Listeria Spp., Samonella Spp. and enteropathogenic Escherichia coli) and viruses (for example, vaccinia virus), subvert actin cytoskeletal functions of the infected host cell by modulating its actin dynamics (for review see REF. 141). it is likely that these influences on micro filament remodelling are accompanied by changes in gene activity and we postulate that this involves the MRTF–SRF circuit. It is equally possible that other, hitherto undetected, G-ABPs might be involved in such events of host cell infection. In more general terms, actin dynamics might communicate to the genome and thereby combat other cellular insults, as already seen in the context of UV-induced DNA damage.
Vesicle trafficking such as endocytosis, exocytosis and Golgi-to-endoplasmic reticulum transport, which are dependent on dynamic actin rearrangements, might also elicit and even require changes in gene expression. This poorly studied aspect of membrane trafficking warrants closer investigation and may offer surprising new insights.
Embryonic development requires cell movement and motile functions at many essential steps. The precise contributions of microfilament-to-genome communication to these activities are still inadequately understood. Although participation of SRF in mouse gastrulation was noticed early on73, the MRTF–SRF circuit is expected to contribute to development in many additional ways, including enabling functions during EMT22 and stem cell programming and reprogramming117. Deeper insight into these issues will prove rewarding.
The remarkable progress in identifying actin-dependent SRF cofactors and determining their mechanisms of action has provided new insights into the myriad aspects of cell physiology, development and disease. Given the likely involvement of the actin–MRTF–SRF pathway in tissue remodelling during disease, it will be important to determine how this and similar pathways of cytoskeleton-to-nucleus communication can be therapeutically modulated.
Acknowledgements
We apologize to all colleagues whose work could not be cited in full owing to space restrictions. We thank G. Posern and B. Knöll for comments on the manuscript. E.N.O. was supported by grants from the National Institutes of Health, the American Heart Association, the Robert A. Welch Foundation and the Leducq Foundation. A.N. acknowledges financial support by the DFG (No120/12-3 and SFB 773/A3).
Glossary
- F-actin
A flexible, helical polymer of G-actin monomers that is 5–9 nm in diameter. An F-actin polymer is polar, displaying a plus (+) and a minus (−) end
- Focal adhesion
A cellular structure that links the ECM on the outside of the cell to the actin cytoskeleton inside the cell through integrin receptors
- Guanine nucleotide exchange factor
A protein that activates a specific small GTPase by catalysing the exchange of bound GDP for GTP
- Heterotrimeric G protein
One of a family of heterotrimeric cytoplasmic signal mediators composed of an α-subunit containing a GTP-binding site and intrinsic GTPase activity together with a hydrophobic and an often acylated β- and γ-protein subcomplex. The β- and γ-protein subcomplex may be activated by the dissociation of α–GTP, which is initiated by GTP exchange on the α-subunit
- Epithelial–mesenchymal transition
The transformation of an epithelial cell into a mesenchymal cell with migratory and invasive properties
- Adherens junction
A cell–cell adhesion complex that contains classical cadherins and catenins that are attached to cytoplasmic actin filaments
- SAP domain
(SAFA or SAFB, acinus and PIAS domain). A peptide motif found in several proteins known to contact DNA
- Leu zipper
A Leu-rich domain in a protein that binds to other proteins with a similar domain
- LIM domain
(Lin11, Isl1 and Mec3 domain) A zinc-binding protein domain that ligates two zinc ions. LIM domains mediate interactions with other proteins and have diverse functions as regulators of gene expression, cell adhesion and motility, and signal transduction
- RPEL domain
A protein domain containing the amino acid sequence Arg-Pro-X-X-X-Glu-Leu that is found triplicated in the myocardin family of transcriptional coactivators. RPEL domains form stable complexes with G-actin
- Barbed end
The plus (+) end of the polar F-actin polymer that, in contrast to the minus (−) end, is more active with regard to incorporation of G-actin into the polymer (polymer elongation)
- Leiomyosarcoma
A rare type of cancer (a soft tissue sarcoma) that is a malignant neoplasm of smooth muscle origin.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
DATABASES
UniProtKB: http://www.uniprot.org CRK | DAAM1 | FAK | FHL2 | ILK | JMY | KLF4 | MRTF-A | MRTF-B | NCK1 | SRC | SRF | STARS
FURTHER INFORMATION
Eric N. Olson’s homepage: www4.utsouthwestern.edu/olsonlab/index.htm
Alfred Nordheim’s homepage: www.molbio.uni-tuebingen.de
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
References
- 1.Le Clainche C, Carlier MF. Regulation of actin assembly associated with protrusion and adhesion in cell migration. Physiol. Rev. 2008;88:489–513. doi: 10.1152/physrev.00021.2007. [DOI] [PubMed] [Google Scholar]
- 2.Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annu. Rev. Cell Dev. Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- 3.Pollard TD. Regulation of actin filament assembly by Arp2/3 complex and formins. Annu. Rev. Biophys. Biomol. Struct. 2007;36:451–477. doi: 10.1146/annurev.biophys.35.040405.101936. [DOI] [PubMed] [Google Scholar]
- 4.Allingham JS, Klenchin VA, Rayment I. Actin-targeting natural products: structures, properties and mechanisms of action. Cell. Mol. Life Sci. 2006;63:2119–2134. doi: 10.1007/s00018-006-6157-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cotton M, Claing A. G protein-coupled receptors stimulation and the control of cell migration. Cell. Signal. 2009;21:1045–1053. doi: 10.1016/j.cellsig.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 6.Iguchi T, et al. Orphan G protein-coupled receptor GPR56 regulates neural progenitor cell migration via a Gα12/13 and Rho pathway. J. Biol. Chem. 2008;283:14469–14478. doi: 10.1074/jbc.M708919200. [DOI] [PubMed] [Google Scholar]
- 7.Descot A, et al. Negative regulation of the EGFR-MAPK cascade by actin-MAL-mediated Mig6/Errfi-1 induction. Mol. Cell. 2009;35:291–304. doi: 10.1016/j.molcel.2009.07.015. [DOI] [PubMed] [Google Scholar]
- 8.Lockman K, et al. Sphingosine 1-phosphate stimulates smooth muscle cell differentiation and proliferation by activating separate serum response factor co-factors. J. Biol. Chem. 2004;279:42422–42430. doi: 10.1074/jbc.M405432200. [DOI] [PubMed] [Google Scholar]
- 9.Schiller MR. Coupling receptor tyrosine kinases to Rho GTPases — GEFs what’s the link. Cell. Signal. 2006;18:1834–1843. doi: 10.1016/j.cellsig.2006.01.022. [DOI] [PubMed] [Google Scholar]
- 10.Chai J, Jones MK, Tarnawski AS. Serum response factor is a critical requirement for VEGF signaling in endothelial cells and VEGF-induced angiogenesis. Faseb J. 2004;18:1264–1266. doi: 10.1096/fj.03-1232fje. [DOI] [PubMed] [Google Scholar]
- 11. Knoll B, et al. Serum response factor controls neuronal circuit assembly in the hippocampus. Nature Neurosci. 2006;9:195–204. doi: 10.1038/nn1627. This paper shows for the first time the essential role of SRF-controlled actin dynamics for the regulation of mouse neuronal development, neurite outgrowth and axonal guidance. See also references 12 and 13.
- 12.Stern S, et al. A nuclear actin function regulates neuronal motility by serum response factor-dependent gene transcription. J. Neurosci. 2009;29:4512–4518. doi: 10.1523/JNEUROSCI.0333-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wickramasinghe SR, et al. Serum response factor mediates NGF-dependent target innervation by embryonic DRG sensory neurons. Neuron. 2008;58:532–545. doi: 10.1016/j.neuron.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franco CA, et al. Serum response factor is required for sprouting angiogenesis and vascular integrity. Dev. Cell. 2008;15:448–461. doi: 10.1016/j.devcel.2008.07.019. [DOI] [PubMed] [Google Scholar]
- 15.Holtz ML, Misra RP. Endothelial-specific ablation of serum response factor causes hemorrhaging, yolk sac vascular failure, and embryonic lethality. BMC Dev. Biol. 2008;8:65–78. doi: 10.1186/1471-213X-8-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knoll B, Nordheim A. Functional versatility of transcription factors in the nervous system: the SRF paradigm. Trends Neurosci. 2009;32:432–442. doi: 10.1016/j.tins.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 17.Huveneers S, Danen EH. Adhesion signaling — crosstalk between integrins, Src and Rho. J. Cell Sci. 2009;122:1059–1069. doi: 10.1242/jcs.039446. [DOI] [PubMed] [Google Scholar]
- 18.Zhao XH, et al. Force activates smooth muscle α-actin promoter activity through the Rho signaling pathway. J. Cell Sci. 2007;120:1801–1809. doi: 10.1242/jcs.001586. [DOI] [PubMed] [Google Scholar]
- 19.Moustakas A, Heldin CH. Dynamic control of TGF-β signaling and its links to the cytoskeleton. FEBS Lett. 2008;582:2051–2065. doi: 10.1016/j.febslet.2008.03.027. [DOI] [PubMed] [Google Scholar]
- 20.Fan L, et al. Cell contact-dependent regulation of epithelial-myofibroblast transition via the Rho-Rho kinase-phospho-myosin pathway. Mol. Biol. Cell. 2007;18:1083–1097. doi: 10.1091/mbc.E06-07-0602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hinson JS, Medlin MD, Lockman K, Taylor JM, Mack CP. Smooth muscle cell-specific transcription is regulated by nuclear localization of the myocardin-related transcription factors. Am. J. Physiol. Heart Circ. Physiol. 2007;292:H1170–H1180. doi: 10.1152/ajpheart.00864.2006. [DOI] [PubMed] [Google Scholar]
- 22.Morita T, Mayanagi T, Sobue K. Dual roles of myocardin-related transcription factors in epithelial mesenchymal transition via slug induction and actin remodeling. J. Cell Biol. 2007;179:1027–1042. doi: 10.1083/jcb.200708174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lagna G, et al. Control of phenotypic plasticity of smooth muscle cells by bone morphogenetic protein signaling through the myocardin-related transcription factors. J. Biol. Chem. 2007;282:37244–37255. doi: 10.1074/jbc.M708137200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Busche S, Descot A, Julien S, Genth H, Posern G. Epithelial cell-cell contacts regulate SRF-mediated transcription via Rac-actin-MAL signalling. J. Cell Sci. 2008;121:1025–1035. doi: 10.1242/jcs.014456. [DOI] [PubMed] [Google Scholar]
- 25.Sebe A, et al. Rac, PAK and p38 regulate cell contact-dependent nuclear translocation of myocardin-related transcription factor. FEBS Lett. 2008;582:291–298. doi: 10.1016/j.febslet.2007.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perez-Moreno M, Fuchs E. Catenins: keeping cells from getting their signals crossed. Dev. Cell. 2006;11:601–612. doi: 10.1016/j.devcel.2006.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Winter CG, et al. Drosophila Rho-associated kinase (Drok) links Frizzled-mediated planar cell polarity signaling to the actin cytoskeleton. Cell. 2001;105:81–91. doi: 10.1016/s0092-8674(01)00298-7. [DOI] [PubMed] [Google Scholar]
- 28.Lai SL, Chien AJ, Moon RT. Wnt/Fz signaling and the cytoskeleton: potential roles in tumorigenesis. Cell Res. 2009;19:532–545. doi: 10.1038/cr.2009.41. [DOI] [PubMed] [Google Scholar]
- 29.Simons M, Mlodzik M. Planar cell polarity signaling: from fly development to human disease. Annu. Rev. Genet. 2008;42:517–540. doi: 10.1146/annurev.genet.42.110807.091432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roszko I, Sawada A, Solnica-Krezel L. Regulation of convergence and extension movements during vertebrate gastrulation by the Wnt/PCP pathway. Semin. Cell Dev. Biol. 2009;20:986–997. doi: 10.1016/j.semcdb.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gieni RS, Hendzel MJ. Actin dynamics and functions in the interphase nucleus: moving toward an understanding of nuclear polymeric actin. Biochem. Cell Biol. 2009;87:283–306. doi: 10.1139/O08-133. [DOI] [PubMed] [Google Scholar]
- 32.Zheng B, Han M, Bernier M, Wen JK. Nuclear actin and actin-binding proteins in the regulation of transcription and gene expression. FEBS J. 2009;276:2669–26685. doi: 10.1111/j.1742-4658.2009.06986.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coso OA, et al. The small GTP-binding proteins Rac1 and Cdc42 regulate the activity of the JNK/ SAPK signaling pathway. Cell. 1995;81:1137–1146. doi: 10.1016/s0092-8674(05)80018-2. [DOI] [PubMed] [Google Scholar]
- 34.Minden A, Lin A, Claret FX, Abo A, Karin M. Selective activation of the JNK signaling cascade and c-Jun transcriptional activity by the small GTPases Rac and Cdc42Hs. Cell. 1995;81:1147–1157. doi: 10.1016/s0092-8674(05)80019-4. [DOI] [PubMed] [Google Scholar]
- 35.Rosso SB, Sussman D, Wynshaw-Boris A, Salinas PC. Wnt signaling through Dishevelled, Rac and JNK regulates dendritic development. Nature Neurosci. 2005;8:34–42. doi: 10.1038/nn1374. [DOI] [PubMed] [Google Scholar]
- 36.Teramoto H, et al. Signaling from the small GTP-binding proteins Rac1 and Cdc42 to the c-Jun N-terminal kinase/stress-activated protein kinase pathway. A role for mixed lineage kinase 3/proteintyrosine kinase 1, a novel member of the mixed lineage kinase family. J. Biol. Chem. 1996;271:27225–27228. doi: 10.1074/jbc.271.44.27225. [DOI] [PubMed] [Google Scholar]
- 37.Marinissen MJ, et al. The small GTP-binding protein RhoA regulates c-Jun by a ROCK-JNK signaling axis. Mol. Cell. 2004;14:29–41. doi: 10.1016/s1097-2765(04)00153-4. [DOI] [PubMed] [Google Scholar]
- 38.Girardin SE, Yaniv M. A direct interaction between JNK1 and CrkII is critical for Rac1-induced JNK activation. EMBO J. 2001;20:3437–3446. doi: 10.1093/emboj/20.13.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fazal F, et al. Essential role of cofilin-1 in regulating thrombin-induced RelA/p65 nuclear translocation and intercellular adhesion molecule 1 (ICAM-1) expression in endothelial cells. J. Biol. Chem. 2009;284:21047–21056. doi: 10.1074/jbc.M109.016444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cowell CF, et al. Loss of cell-cell contacts induces NF-κB via RhoA-mediated activation of protein kinase D1. J. Cell Biochem. 2009;106:714–728. doi: 10.1002/jcb.22067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coutts AS, Weston L, La Thangue NB. A transcription co-factor integrates cell adhesion and motility with the p53 response. Proc. Natl Acad. Sci. USA. 2009;106:19872–19877. doi: 10.1073/pnas.0906785106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zuchero JB, Coutts AS, Quinlan ME, Thangue NB, Mullins RD. p53-cofactor JMY is a multifunctional actin nucleation factor. Nature Cell Biol. 2009;11:451–459. doi: 10.1038/ncb1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kremer BE, Adang LA, Macara IG. Septins regulate actin organization and cell-cycle arrest through nuclear accumulation of NCK mediated by SOCS7. Cell. 2007;130:837–850. doi: 10.1016/j.cell.2007.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hervy M, Hoffman L, Beckerle MC. From the membrane to the nucleus and back again: bifunctional focal adhesion proteins. Curr. Opin. Cell Biol. 2006;18:524–532. doi: 10.1016/j.ceb.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 45.Kadrmas JL, Beckerle MC. The LIM domain: from the cytoskeleton to the nucleus. Nature Rev. Mol. Cell Biol. 2004;5:920–931. doi: 10.1038/nrm1499. [DOI] [PubMed] [Google Scholar]
- 46.Chang DF, et al. Cysteine-rich LIM-only proteins CRP1 and CRP2 are potent smooth muscle differentiation cofactors. Dev. Cell. 2003;4:107–118. doi: 10.1016/s1534-5807(02)00396-9. [DOI] [PubMed] [Google Scholar]
- 47.Philippar U, et al. The SRF target gene Fhl2 antagonizes RhoA/MAL-dependent activation of SRF. Mol. Cell. 2004;16:867–880. doi: 10.1016/j.molcel.2004.11.039. [DOI] [PubMed] [Google Scholar]
- 48. Schratt G, et al. Serum response factor is crucial for actin cytoskeletal organization and focal adhesion assembly in embryonic stem cells. J. Cell Biol. 2002;156:737–750. doi: 10.1083/jcb.200106008. Using SRF-deficient embryonic stem cells, this paper uncovers the essential function of SRF in the control of actin cytoskeletal assembly, cell migration and cell adhesion.
- 49. Hill CS, Wynne J, Treisman R. The Rho family GTPases RhoA, Rac1, and CDC42Hs regulate transcriptional activation by SRF. Cell. 1995;81:1159–1170. doi: 10.1016/s0092-8674(05)80020-0. This paper describes for the first time that SRF is a nuclear target of Rho-mediated signalling.
- 50.Posern G, Sotiropoulos A, Treisman R. Mutant actins demonstrate a role for unpolymerized actin in control of transcription by serum response factor. Mol. Biol. Cell. 2002;13:4167–4178. doi: 10.1091/mbc.02-05-0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sotiropoulos A, Gineitis D, Copeland J, Treisman R. Signal-regulated activation of serum response factor is mediated by changes in actin dynamics. Cell. 1999;98:159–169. doi: 10.1016/s0092-8674(00)81011-9. This paper reveals the regulation of SRF activity by G-actin and actin dynamics, consistent with the subsequent discovery that MRTFs are a key intermediary between actin dynamics and SRF activation. See also references 50, 74 and 75.
- 52. Miralles F, Posern G, Zaromytidou AI, Treisman R. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell. 2003;113:329–342. doi: 10.1016/s0092-8674(03)00278-2. This important paper shows that MRTF-A binds monomeric G-actin in the cytoplasm through the N-terminal RPEL motifs and translocates to the nucleus to interact with SRF in response to actin polymerization. See also reference 128.
- 53. Norman C, Runswick M, Pollock R, Treisman R. Isolation and properties of cDNA clones encoding SRF, a transcription factor that binds to the c-Fos serum response element. Cell. 1988;55:989–1003. doi: 10.1016/0092-8674(88)90244-9. This paper describes the initial cloning of SRF and its mechanism of action as a dimeric transcriptional activator through binding to the CArG box DNA motif.
- 54.Shore P, Sharrocks AD. The MADS-box family of transcription factors. Eur. J. Biochem. 1995;229:1–13. doi: 10.1111/j.1432-1033.1995.tb20430.x. [DOI] [PubMed] [Google Scholar]
- 55.Pellegrini L, Tan S, Richmond TJ. Structure of serum response factor core bound to DNA. Nature. 1995;376:490–498. doi: 10.1038/376490a0. [DOI] [PubMed] [Google Scholar]
- 56. Treisman R. Identification of a protein-binding site that mediates transcriptional response of the c-fos gene to serum factors. Cell. 1986;46:567–574. doi: 10.1016/0092-8674(86)90882-2. This report describes for the first time the binding of a transcription factor, later identified as SRF (see reference 53), to the c-fos transcriptional regulatory element termed SRE
- 57.Miano JM, Long X, Fujiwara K. Serum response factor: master regulator of the actin cytoskeleton and contractile apparatus. Am. J. Physiol. Cell Physiol. 2007;292:C70–C81. doi: 10.1152/ajpcell.00386.2006. [DOI] [PubMed] [Google Scholar]
- 58.Posern G, Treisman R. Actin’ together: serum response factor, its cofactors and the link to signal transduction. Trends Cell Biol. 2006;16:588–596. doi: 10.1016/j.tcb.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 59.Selvaraj A, Prywes R. Expression profiling of serum inducible genes identifies a subset of SRF target genes that are MKL dependent. BMC Mol. Biol. 2004;5:13–28. doi: 10.1186/1471-2199-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sun Q, et al. Defining the mammalian CArGome. Genome Res. 2006;16:197–207. doi: 10.1101/gr.4108706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stritt C, et al. Paracrine control of oligodendrocyte differentiation by SRF-directed neuronal gene expression. Nature Neurosci. 2009;12:418–427. doi: 10.1038/nn.2280. [DOI] [PubMed] [Google Scholar]
- 62. Shaw PE, Schroter H, Nordheim A. The ability of a ternary complex to form over the serum response element correlates with serum inducibility of the human c-Fos promoter. Cell. 1989;56:563–572. doi: 10.1016/0092-8674(89)90579-5. This is the first report of SRF recruiting a cofactor (named TCF) to regulate target gene activity
- 63.Buchwalter G, Gross C, Wasylyk B. Ets ternary complex transcription factors. Gene. 2004;324:1–14. doi: 10.1016/j.gene.2003.09.028. [DOI] [PubMed] [Google Scholar]
- 64.Dalton S, Treisman R. Characterization of SAP-1, a protein recruited by serum response factor to the c-Fos serum response element. Cell. 1992;68:597–612. doi: 10.1016/0092-8674(92)90194-h. [DOI] [PubMed] [Google Scholar]
- 65.Hipskind RA, Rao VN, Mueller CG, Reddy ES, Nordheim A. Ets-related protein Elk-1 is homologous to the c-Fos regulatory factor p62TCF. Nature. 1991;354:531–534. doi: 10.1038/354531a0. [DOI] [PubMed] [Google Scholar]
- 66.Parmacek MS. Myocardin-related transcription factors: critical coactivators regulating cardiovascular development and adaptation. Circ. Res. 2007;100:633–644. doi: 10.1161/01.RES.0000259563.61091.e8. [DOI] [PubMed] [Google Scholar]
- 67.Pipes GC, Creemers EE, Olson EN. The myocardin family of transcriptional coactivators: versatile regulators of cell growth, migration, and myogenesis. Genes Dev. 2006;20:1545–1556. doi: 10.1101/gad.1428006. [DOI] [PubMed] [Google Scholar]
- 68. Wang D, et al. Activation of cardiac gene expression by myocardin, a transcriptional cofactor for serum response factor. Cell. 2001;105:851–862. doi: 10.1016/s0092-8674(01)00404-4. This important paper reports the discovery of myocardin, the founding member of the myocardin family of co-activators and a powerful activator of SRF-dependent transcription in cardiac muscle cells.
- 69. Wang DZ, et al. Potentiation of serum response factor activity by a family of myocardin-related transcription factors. Proc. Natl Acad. Sci. USA. 2002;99:14855–14860. doi: 10.1073/pnas.222561499. This paper reports the identification, structure, function and expression of MRTF-A and MRTF-B, showing their ability to act as potent co-activators of SRF.
- 70.Gineitis D, Treisman R. Differential usage of signal transduction pathways defines two types of serum response factor target gene. J. Biol. Chem. 2001;276:24531–24539. doi: 10.1074/jbc.M102678200. [DOI] [PubMed] [Google Scholar]
- 71.Wang Z, et al. Myocardin and ternary complex factors compete for SRF to control smooth muscle gene expression. Nature. 2004;428:185–189. doi: 10.1038/nature02382. [DOI] [PubMed] [Google Scholar]
- 72.Zaromytidou AI, Miralles F, Treisman R. MAL and ternary complex factor use different mechanisms to contact a common surface on the serum response factor DNA-binding domain. Mol. Cell. Biol. 2006;26:4134–4148. doi: 10.1128/MCB.01902-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Arsenian S, Weinhold B, Oelgeschlager M, Ruther U, Nordheim A. Serum response factor is essential for mesoderm formation during mouse embryogenesis. EMBO J. 1998;17:6289–6299. doi: 10.1093/emboj/17.21.6289. Through the creation of SRF-deficient embryos, this paper shows for the first time that SRF is essential for mesoderm formation during embryogenesis, probably reflecting in part its requisite role for the activation of cytoskeletal gene expression and cell migration
- 74.Guettler S, Vartiainen MK, Miralles F, Larijani B, Treisman R. RPEL motifs link the serum response factor cofactor MAL but not myocardin to Rho signaling via actin binding. Mol. Cell. Biol. 2008;28:732–742. doi: 10.1128/MCB.01623-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Posern G, Miralles F, Guettler S, Treisman R. Mutant actins that stabilise F-actin use distinct mechanisms to activate the SRF coactivator MAL. EMBO J. 2004;23:3973–3983. doi: 10.1038/sj.emboj.7600404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Vartiainen MK, Guettler S, Larijani B, Treisman R. Nuclear actin regulates dynamic subcellular localization and activity of the SRF cofactor MAL. Science. 2007;316:1749–1752. doi: 10.1126/science.1141084. This manuscript describes a role for nuclear G-actin in regulating the activity of nuclear MRTF.
- 77.Mouilleron S, Guettler S, Langer CA, Treisman R, McDonald NQ. Molecular basis for G-actin binding to RPEL motifs from the serum response factor coactivator MAL. EMBO J. 2008;27:3198–3208. doi: 10.1038/emboj.2008.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kuwahara K, Barrientos T, Pipes GC, Li S, Olson EN. Muscle-specific signaling mechanism that links actin dynamics to serum response factor. Mol. Cell. Biol. 2005;25:3173–3181. doi: 10.1128/MCB.25.8.3173-3181.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Wang Z, Wang DZ, Pipes GC, Olson EN. Myocardin is a master regulator of smooth muscle gene expression. Proc. Natl Acad. Sci. USA. 2003;100:7129–7134. doi: 10.1073/pnas.1232341100. This paper shows that myocardin acts as a master regulator of smooth muscle gene expression and can reprogramme fibroblasts to adopt a smooth muscle fate. See also references 104–106.
- 80.Huang J, et al. Myocardin regulates expression of contractile genes in smooth muscle cells and is required for closure of the ductus arteriosus in mice. J. Clin. Invest. 2008;118:515–525. doi: 10.1172/JCI33304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Huang J, et al. Myocardin is required for cardiomyocyte survival and maintenance of heart function. Proc. Natl Acad. Sci. USA. 2009;106:18734–18739. doi: 10.1073/pnas.0910749106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li S, Wang DZ, Wang Z, Richardson JA, Olson EN. The serum response factor coactivator myocardin is required for vascular smooth muscle development. Proc. Natl Acad. Sci. USA. 2003;100:9366–9370. doi: 10.1073/pnas.1233635100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sun Y, et al. Acute myeloid leukemia-associated Mkl1 (Mrtf-a) is a key regulator of mammary gland function. Mol. Cell. Biol. 2006;26:5809–5826. doi: 10.1128/MCB.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li S, Chang S, Qi X, Richardson JA, Olson EN. Requirement of a myocardin-related transcription factor for development of mammary myoepithelial cells. Mol. Cell. Biol. 2006;26:5797–5808. doi: 10.1128/MCB.00211-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Oh J, Richardson JA, Olson EN. Requirement of myocardin-related transcription factor-B for remodeling of branchial arch arteries and smooth muscle differentiation. Proc. Natl Acad. Sci. USA. 2005;102:15122–15127. doi: 10.1073/pnas.0507346102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Muehlich S, et al. Serum-induced phosphorylation of the serum response factor coactivator MKL1 by the extracellular signal-regulated kinase 1/2 pathway inhibits its nuclear localization. Mol. Cell. Biol. 2008;28:6302–6313. doi: 10.1128/MCB.00427-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kalita K, Kharebava G, Zheng JJ, Hetman M. Role of megakaryoblastic acute leukemia-1 in ERK1/2-dependent stimulation of serum response factor-driven transcription by BDNF or increased synaptic activity. J. Neurosci. 2006;26:10020–10032. doi: 10.1523/JNEUROSCI.2644-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kuwahara K, et al. Modulation of adverse cardiac remodeling by STARS, a mediator of MEF2 signaling and SRF activity. J. Clin. Invest. 2007;117:1324–1334. doi: 10.1172/JCI31240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chen CY, Schwartz RJ. Recruitment of the tinman homolog Nkx-2.5 by serum response factor activates cardiac α-actin gene transcription. Mol. Cell. Biol. 1996;16:6372–6384. doi: 10.1128/mcb.16.11.6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Belaguli NS, et al. Cardiac tissue enriched factors serum response factor and GATA-4 are mutual coregulators. Mol. Cell. Biol. 2000;20:7550–7558. doi: 10.1128/mcb.20.20.7550-7558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sepulveda JL, Vlahopoulos S, Iyer D, Belaguli N, Schwartz RJ. Combinatorial expression of GATA4, Nkx2-5, and serum response factor directs early cardiac gene activity. J. Biol. Chem. 2002;277:25775–25782. doi: 10.1074/jbc.M203122200. [DOI] [PubMed] [Google Scholar]
- 92.Neuman NA, et al. The four-and-a-half LIM domain protein 2 regulates vascular smooth muscle phenotype and vascular tone. J. Biol. Chem. 2009;284:13202–13212. doi: 10.1074/jbc.M900282200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Davis FJ, Gupta M, Camoretti-Mercado B, Schwartz RJ, Gupta MP. Calcium/calmodulin-dependent protein kinase activates serum response factor transcription activity by its dissociation from histone deacetylase, HDAC4. Implications in cardiac muscle gene regulation during hypertrophy. J. Biol. Chem. 2003;278:20047–20058. doi: 10.1074/jbc.M209998200. [DOI] [PubMed] [Google Scholar]
- 94.Hayashi K, Nakamura S, Nishida W, Sobue K. Bone morphogenetic protein-induced MSX1 and MSX2 inhibit myocardin-dependent smooth muscle gene transcription. Mol. Cell. Biol. 2006;26:9456–9470. doi: 10.1128/MCB.00759-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu Y, et al. Kruppel-like factor 4 abrogates myocardin-induced activation of smooth muscle gene expression. J. Biol. Chem. 2005;280:9719–9727. doi: 10.1074/jbc.M412862200. [DOI] [PubMed] [Google Scholar]
- 96.Chen F, et al. Hop is an unusual homeobox gene that modulates cardiac development. Cell. 2002;110:713–723. doi: 10.1016/s0092-8674(02)00932-7. [DOI] [PubMed] [Google Scholar]
- 97.Kook H, et al. Cardiac hypertrophy and histone deacetylase-dependent transcriptional repression mediated by the atypical homeodomain protein HOP. J. Clin. Invest. 2003;112:863–871. doi: 10.1172/JCI19137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shin CH, et al. Modulation of cardiac growth and development by HOP, an unusual homeodomain protein. Cell. 2002;110:725–735. doi: 10.1016/s0092-8674(02)00933-9. [DOI] [PubMed] [Google Scholar]
- 99.Cao D, et al. Modulation of smooth muscle gene expression by association of histone acetyltransferases and deacetylases with myocardin. Mol. Cell. Biol. 2005;25:364–376. doi: 10.1128/MCB.25.1.364-376.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hanna M, et al. Mechanical regulation of the proangiogenic factor CCN1/CYR61 gene requires the combined activities of MRTF-A and CREB-binding protein histone acetyltransferase. J. Biol. Chem. 2009;284:23125–23136. doi: 10.1074/jbc.M109.019059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Qiu P, et al. Myocardin enhances Smad3-mediated transforming growth factor-β1 signaling in a CArG box-independent manner: Smad-binding element is an important cis element for SM22α transcription in vivo. Circ. Res. 2005;97:983–991. doi: 10.1161/01.RES.0000190604.90049.71. [DOI] [PubMed] [Google Scholar]
- 102.Oh J, et al. Target gene-specific modulation of myocardin activity by GATA transcription factors. Mol. Cell. Biol. 2004;24:8519–8528. doi: 10.1128/MCB.24.19.8519-8528.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cooper SJ, Trinklein ND, Nguyen L, Myers RM. Serum response factor binding sites differ in three human cell types. Genome Res. 2007;17:136–144. doi: 10.1101/gr.5875007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chen J, Kitchen CM, Streb JW, Miano JM. Myocardin: a component of a molecular switch for smooth muscle differentiation. J. Mol. Cell. Cardiol. 2002;34:1345–1356. doi: 10.1006/jmcc.2002.2086. [DOI] [PubMed] [Google Scholar]
- 105.Du KL, et al. Myocardin is a critical serum response factor cofactor in the transcriptional program regulating smooth muscle cell differentiation. Mol. Cell. Biol. 2003;23:2425–2437. doi: 10.1128/MCB.23.7.2425-2437.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yoshida T, et al. Myocardin is a key regulator of CArG-dependent transcription of multiple smooth muscle marker genes. Circ. Res. 2003;92:856–864. doi: 10.1161/01.RES.0000068405.49081.09. [DOI] [PubMed] [Google Scholar]
- 107.Du KL, et al. Megakaryoblastic leukemia factor-1 transduces cytoskeletal signals and induces smooth muscle cell differentiation from undifferentiated embryonic stem cells. J. Biol. Chem. 2004;279:17578–17586. doi: 10.1074/jbc.M400961200. [DOI] [PubMed] [Google Scholar]
- 108.Li S, et al. Requirement for serum response factor for skeletal muscle growth and maturation revealed by tissue-specific gene deletion in mice. Proc. Natl Acad. Sci. USA. 2005;102:1082–1087. doi: 10.1073/pnas.0409103102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Medjkane S, Perez-Sanchez C, Gaggioli C, Sahai E, Treisman R. Myocardin-related transcription factors and SRF are required for cytoskeletal dynamics and experimental metastasis. Nature Cell Biol. 2009;11:257–268. doi: 10.1038/ncb1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Verdoni AM, Aoyama N, Ikeda A, Ikeda S. Effect of destrin mutations on the gene expression profile in vivo. Physiol. Genomics. 2008;34:9–21. doi: 10.1152/physiolgenomics.00285.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Niu Z, Li A, Zhang SX, Schwartz RJ. Serum response factor micromanaging cardiogenesis. Curr. Opin. Cell Biol. 2007;19:618–627. doi: 10.1016/j.ceb.2007.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–220. doi: 10.1038/nature03817. This paper shows for the first time that SRF regulates the expression of miRNAs. The identified miRNA, miR-1, controls cardiomyocyte proliferation.
- 113. Liu N, et al. MicroRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev. 2008;22:3242–3254. doi: 10.1101/gad.1738708. This paper shows that the miRNA miR-133a is required for cardiac development and negatively regulates SRF. See also reference 116.
- 114.Ivey KN, et al. MicroRNA regulation of cell lineages in mouse and human embryonic stem cells. Cell Stem Cell. 2008;2 doi: 10.1016/j.stem.2008.01.016. 2192-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Xin M, et al. MicroRNAs miR-143 and miR-145 modulate cytoskeletal dynamics and responsiveness of smooth muscle cells to injury. Genes Dev. 2009;23:2166–2178. doi: 10.1101/gad.1842409. This paper shows that SRF, together with members of the myocardin family, directly activates a bicistronic cluster of miRNAs, encoding miR-143 and miR-145, which in turn control smooth muscle phenotype switching through the regulation of numerous regulators of actin dynamics. See also references 117 and 118.
- 116.Niu Z, et al. Serum response factor orchestrates nascent sarcomerogenesis and silences the biomineralization gene program in the heart. Proc. Natl Acad. Sci. USA. 2008;105:17824–17829. doi: 10.1073/pnas.0805491105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Cordes KR, et al. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460:705–710. doi: 10.1038/nature08195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Boettger T, et al. Acquisition of the contractile phenotype by murine arterial smooth muscle cells depends on the Mir143/145 gene cluster. J. Clin. Invest. 2009;119:2634–2647. doi: 10.1172/JCI38864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Small EM, et al. Regulation of PI3-kinase/Akt signaling by muscle-enriched microRNA-486. Proc. Natl Acad. Sci. USA. 2010;107:4218–4223. doi: 10.1073/pnas.1000300107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Torrado M, et al. Myocardin mRNA is augmented in the failing myocardium: expression profiling in the porcine model and human dilated cardiomyopathy. J. Mol. Med. 2003;81:566–577. doi: 10.1007/s00109-003-0470-7. [DOI] [PubMed] [Google Scholar]
- 121.Xing W, et al. Myocardin induces cardiomyocyte hypertrophy. Circ. Res. 2006;98:1089–1097. doi: 10.1161/01.RES.0000218781.23144.3e. [DOI] [PubMed] [Google Scholar]
- 122.Chang J, et al. Inhibitory cardiac transcription factor, SRF-N, is generated by caspase 3 cleavage in human heart failure and attenuated by ventricular unloading. Circulation. 2003;108:407–413. doi: 10.1161/01.CIR.0000084502.02147.83. [DOI] [PubMed] [Google Scholar]
- 123.Chow N, et al. Serum response factor and myocardin mediate arterial hypercontractility and cerebral blood flow dysregulation in Alzheimer’s phenotype. Proc. Natl Acad. Sci. USA. 2007;104:823–828. doi: 10.1073/pnas.0608251104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bell RD, et al. SRF and myocardin regulate LRP-mediated amyloid-β clearance in brain vascular cells. Nature Cell Biol. 2009;11:143–153. doi: 10.1038/ncb1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Owens GK, Kumar MS, Wamhoff BR. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol. Rev. 2004;84:767–801. doi: 10.1152/physrev.00041.2003. [DOI] [PubMed] [Google Scholar]
- 126.Carroll A, et al. The t(1;22) (p13;q13) is nonrandom and restricted to infants with acute megakaryoblastic leukemia: a pediatric oncology group study. Blood. 1991;78:748–752. [PubMed] [Google Scholar]
- 127.Ma Z, et al. Fusion of two novel genes, RBM15 and MKL1, in the t(1;22)(p13;q13) of acute megakaryoblastic leukemia. Nature Genet. 2001;28:220–221. doi: 10.1038/90054. [DOI] [PubMed] [Google Scholar]
- 128.Mercher T, et al. Recurrence of OTT-MAL fusion in t(1;22) of infant AML-M7. Genes Chromosomes Cancer. 2002;33:22–28. doi: 10.1002/gcc.1208. [DOI] [PubMed] [Google Scholar]
- 129.Cen B, et al. Megakaryoblastic leukemia 1, a potent transcriptional coactivator for serum response factor (SRF), is required for serum induction of SRF target genes. Mol. Cell. Biol. 2003;23:6597–6608. doi: 10.1128/MCB.23.18.6597-6608.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Descot A, et al. OTT-MAL is a deregulated activator of serum response factor-dependent gene expression. Mol. Cell. Biol. 2008;28:6171–6181. doi: 10.1128/MCB.00303-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sawada T, et al. Fusion of OTT to BSAC results in aberrant up-regulation of transcriptional activity. J. Biol. Chem. 2008;283:26820–26828. doi: 10.1074/jbc.M802315200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Mercher T, et al. The OTT-MAL fusion oncogene activates RBPJ-mediated transcription and induces acute megakaryoblastic leukemia in a knockin mouse model. J. Clin. Invest. 2009;119:852–864. doi: 10.1172/JCI35901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Milyavsky M, et al. Inactivation of myocardin and p 16 during malignant transformation contributes to a differentiation defect. Cancer Cell. 2007;11:133–146. doi: 10.1016/j.ccr.2006.11.022. [DOI] [PubMed] [Google Scholar]
- 134.Perot G, et al. Strong smooth muscle differentiation is dependent on myocardin gene amplification in most human retroperitoneal leiomyosarcomas. Cancer Res. 2009;69:2269–2278. doi: 10.1158/0008-5472.CAN-08-1443. [DOI] [PubMed] [Google Scholar]
- 135.Heemers HV, Regan KM, Dehm SM, Tindall DJ. Androgen induction of the androgen receptor coactivator four and a half LIM domain protein-2: evidence for a role for serum response factor in prostate cancer. Cancer Res. 2007;67:10592–10599. doi: 10.1158/0008-5472.CAN-07-1917. [DOI] [PubMed] [Google Scholar]
- 136. Brandt DT, et al. SCAI acts as a suppressor of cancer cell invasion through the transcriptional control of β-integrin. Nature Cell Biol. 2009;11:557–568. doi: 10.1038/ncb1862. This paper uncovers a role for MRTF in the control of cancer cell metastasis as a consequence of its regulation by SCAI, a transcriptional repressor that competes with MRTF for interaction with SRF. See also reference 109
- 137.Parlakian A, et al. Targeted inactivation of serum response factor in the developing heart results in myocardial defects and embryonic lethality. Mol. Cell. Biol. 2004;24:5281–5289. doi: 10.1128/MCB.24.12.5281-5289.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Miano JM, et al. Restricted inactivation of serum response factor to the cardiovascular system. Proc. Natl Acad. Sci. USA. 2004;101:17132–17137. doi: 10.1073/pnas.0406041101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Alberti S, et al. Neuronal migration in the murine rostral migratory stream requires serum response factor. Proc. Natl Acad. Sci. USA. 2005;102:6148–6153. doi: 10.1073/pnas.0501191102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ramanan N, et al. SRF mediates activity-induced gene expression and synaptic plasticity but not neuronal viability. Nature Neurosci. 2005;8:759–767. doi: 10.1038/nn1462. [DOI] [PubMed] [Google Scholar]
- 141.Bhavsar AP, Guttman JA, Finlay BB. Manipulation of host-cell pathways by bacterial pathogens. Nature. 2007;449:827–834. doi: 10.1038/nature06247. [DOI] [PubMed] [Google Scholar]
- 142.Chhabra ES, Higgs HN. The many faces of actin: matching assembly factors with cellular structures. Nature Cell Biol. 2007;9:1110–1121. doi: 10.1038/ncb1007-1110. [DOI] [PubMed] [Google Scholar]